Abstract
The genome of hepatitis C virus (HCV) contains cis-acting replication elements (CREs) comprised of RNA stem-loop structures located in both the 5′ and 3′ noncoding regions (5′ and 3′ NCRs) and in the NS5B coding sequence. Through the application of several algorithmically independent bioinformatic methods to detect phylogenetically conserved, thermodynamically favored RNA secondary structures, we demonstrate a long-range interaction between sequences in the previously described CRE (5BSL3.2, now SL9266) with a previously predicted unpaired sequence located 3′ to SL9033, approximately 200 nucleotides upstream. Extensive reverse genetic analysis both supports this prediction and demonstrates a functional requirement in genome replication. By mutagenesis of the Con-1 replicon, we show that disruption of this alternative pairing inhibited replication, a phenotype that could be restored to wild-type levels through the introduction of compensating mutations in the upstream region. Substitution of the CRE with the analogous region of different genotypes of HCV produced replicons with phenotypes consistent with the hypothesis that both local and long-range interactions are critical for a fundamental aspect of genome replication. This report further extends the known interactions of the SL9266 CRE, which has also been shown to form a “kissing loop” interaction with the 3′ NCR (P. Friebe, J. Boudet, J. P. Simorre, and R. Bartenschlager, J. Virol. 79:380-392, 2005), and suggests that cooperative long-range binding with both 5′ and 3′ sequences stabilizes the CRE at the core of a complex pseudoknot. Alternatively, if the long-range interactions were mutually exclusive, the SL9266 CRE may function as a molecular switch controlling a critical aspect of HCV genome replication.
Hepatitis C virus (HCV), a flavivirus in the genus Hepacivirus, possesses a positive (mRNA)-sense genome of approximately 9.6 kb encoding a single polyprotein. This polyprotein is cleaved co- and posttranslationally to generate proteins that form the enveloped virus particle and those that replicate the genome. Polyprotein translation is initiated within a highly structured internal ribosome entry site (IRES) occupying much of the 5′ noncoding region (5′ NCR). The 5′ NCR also contains sequences required for genome replication (9, 19, 26), and like functionally analogous regions in the 3′ NCR, these form defined stem-loop structures that operate in cis and are known or suspected to recruit cellular or viral proteins (5, 10, 30). In addition to these cis-acting replication elements (CREs) in the noncoding extremes of the genome, there is evidence that additional RNA structures exist within the coding regions. The latter structure is of two types, phylogenetically conserved well-defined structures occupying the 5′ and 3′ regions of the sense strand of the coding region of HCV (23, 31, 32, 36) and a less well-characterized but much more extensive set of RNA secondary structures, collectively designated genome-scale ordered RNA structure (GORS), that spans the entire coding region of HCV (29).
The potential functional role(s) of phylogenetically conserved RNA secondary structures in coding regions has been analyzed extensively by reverse genetic analysis, predominantly using antibiotic resistance or a luciferase-encoding subgenomic replicon system (18), and more recently, in analysis of structures in the core-encoding region using the HCV replication system (17, 23, 25, 34). Several groups have reported that a short stem-loop structure in the NS5B coding region, variously designated 5BSL3.2 or SL-V (16, 36), has a clearly defined function in genome replication. This structure, henceforth designated SL9266 (see Materials and Methods for details of a unified numbering scheme), forms a stem-loop with two short base-paired helices, separated by a 8-nucleotide (nt) bulge loop on the 3′ side, and capped with a 12-nt terminal loop (16, 36). Extensive mutagenesis has demonstrated that the structural integrity of the element must be retained for replication. In addition, substitutions within the two unpaired loop or bulge regions can also be deleterious, which implies that these regions also contribute important functions during replication. SL9266 therefore forms a cis-acting replication element, though its precise function during genome replication has yet to be determined. SL9266 is the penultimate of five phylogenetically conserved RNA structures in the region encoding NS5B. Limited mutagenesis of the upstream adjacent structure (SL9217), which has also been designated SL-VII (16) or 5BSL3.1 (36), have produced contradictory results, and further studies are required to unequivocally demonstrate a role in genome replication.
Functional analysis of the SL9266 CRE and related RNA structures in the NS5B coding region necessitates the introduction of mutations that leave the underlying coding sequence intact. The restriction of mutagenesis to synonymous substitutions naturally places some limits on the substitutions that can be tested. However, Friebe and colleagues (8) have demonstrated that SL9266 can be functionally moved to the 3′ NCR, albeit with a reduction in replication efficiency. This suggests that the function of this structure is at least partially position dependent but does allow more extensive mutagenic studies. The position dependence could be due to a requirement for a spatially dependent interaction with another region of the virus genome; indeed, they have demonstrated a functionally required kissing loop (tertiary RNA structure) interaction between the terminal unpaired loop of SL9266 and SL2 in the X tail of the 3′ NCR (8).
We have developed novel bioinformatic strategies to detect phylogenetically conserved long-range RNA-RNA interactions. These approaches are based upon well-established and accepted thermodynamic methodologies but extend them to take advantage of the wealth of sequence data available for HCV. Using this information, we have investigated the structure and function of SL9266. We demonstrate that the relatively weak prediction of SL9266 using standard bioinformatic methods can be explained by the structure adopting an additional alternative and potentially metastable pairing with sequences situated approximately 200 nt upstream. Mutagenesis of the two interacting sequences provides genetic support for the interaction and also demonstrated some sequence specificity within SL9266. Duplex formation with the upstream sequences and the 3′ X tail involves distinct regions of SL9266, and the revised model presented here does not preclude the existence of a combined kissing loop interaction with SL2 in the 3′ untranslated region and a pseudoknot interaction of the CRE bulge sequence upstream to form a complex long-range pseudoknot.
MATERIALS AND METHODS
Sequence alignments.
Sequence data sets were initially compiled from all available epidemiologically unlinked variants of all six genotypes (those showing >2% sequence divergence from each other) that were >95% complete between nucleotide positions 9001 and 9377 and a second set between nucleotide positions 8204 and 9377. Nucleotides were numbered with reference to the H77 complete genome sequence, GenBank accession no. AF011753 (15). Representative subsets of sequences within each alignment were used for RNA structure determination. GenBank accession numbers of analyzed sequences are provided in the supplemental material.
Stem-loop nomenclature.
Several methods have been used to describe stem-loops in NS5B and elsewhere in the HCV genome (16, 32, 36). Following the adoption of a standardized system for numbering HCV sequences (15), it had been proposed that stem-loops are numbered based on the position of the first 5′ paired base in the structure (16a). Accordingly, stem-loops previously referred to as 5BSL1, 5BSL2, 5BSL3.1 to 5BSL3.3 (36), SLIV to SLVII (16) or SL8828, SL8926, SL9011, SL9061, and SL9118 (16, 31, 32) are redesignated as SL9033, SL9132, SL9217, SL9266, and SL9324, respectively, in the current study. Likewise, SL2 in the 3′ X tail is renumbered SL9571.
RNA structure prediction.
RNA structures were predicted using MFOLD through the web interface at http://frontend.bioinfo.rpi.edu/applications/mfold/. Automated analysis of most energetically stable RNA structures was performed using the program StructureDist v. 1.3 (available at http://www.picornavirus.org/). SFOLD analysis was conducted using the program Srna on the server at http://sfold.wadsworth.org/srna.pl. PFOLD analysis used the web interface at http://www.daimi.au.dk/∼compbio/rnafold/. All programs were run with default settings.
Cell culture, plasmids, and mutagenesis.
Monolayers of the human hepatoma cell line Huh7 (kindly provided by R. Bartenschlager) were maintained in Dulbecco's modified minimal essential medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum, 1% nonessential amino acids, 100 U pencillin/100 μg streptomycin, and 2 mM l-glutamine (Invitrogen) (DMEM P/S). Cells were passaged after treatment with trypsin-EDTA and seeded at a dilution of 1:3 to 1:5.
The parental, genotype 1b, neomycin-encoding replicon, designated pFK-I389neo/NS3-3′/wt was generously provided by R. Bartenschlager and has been fully described by Lohmann et al. (18). The cDNA was modified by the introduction of a previously described cell culture adaptive change of serine for isoleucine as residue 2204 of the polyprotein (1). A derivative replicon, designated pFKnt341-sp-PI-lucEI3420-9605/5.1, expressing a firefly luciferase reporter gene (kindly provided by GlaxoSmithKline, United Kingdom) consisted (5′ to 3′) of the HCV 5′ NCR, a 63-nt spacer, the poliovirus IRES, and luciferase gene, followed by an encephalomyocarditis virus IRES, the NS3-NS5B coding region, and 3′ NCR of HCV. Derivatives of both replicons carrying substitutions (GDD to GND) of the active site of the NS5B RNA-dependent RNA polymerase were used as controls where appropriate.
All site-directed mutagenesis was conducted on a unique SpeI-XhoI fragment (nt 5582 to 8005), subcloned in pBluescript II SK(+), using Stratagene QuikChange site-directed mutagenesis. All mutations were detected and confirmed by sequencing, rebuilt into the appropriate subgenomic replicon, and sequenced again.
Substitution of SL9266 with the analogous sequence of other HCV genotypes was achieved using a cassette system. Briefly, a 528-nt KpnI-SpeI fragment spanning SL9266 was subcloned into pBluescript II SK(+) (Invitrogen) and used as a template for PCR with primers BsmBI-1F (GCGTCTCTGTTCATGTGGTGCCTACTCC) and BsmBI-2R (GCGTCTCTTAACCAGCAACGAACCAGCT). The blunt ends of the reaction product were ligated to create a plasmid in which SL9266 was precisely replaced with a stuffer fragment containing two BsmBI restriction sites. This cassette vector was cleaved with BsmBI and ligated with complementary oligonucleotides for the stem-loop sequences from other genotypes. The sequences are illustrated below in Fig. 6A. After sequencing, the KpnI-SpeI fragment was rebuilt into pFK-I389neo/NS3-3′/wt.
FIG. 6.
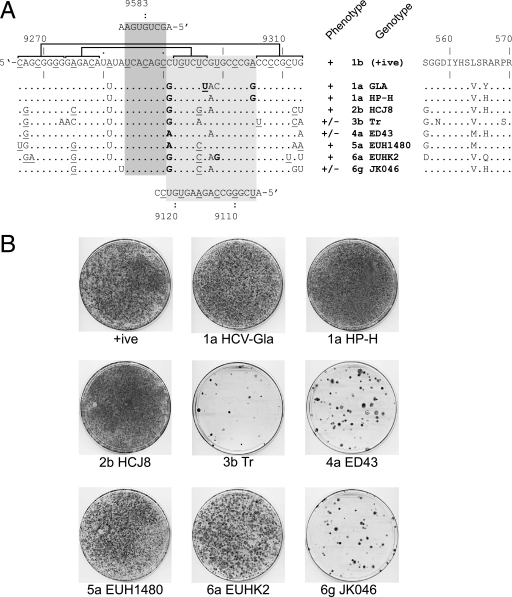
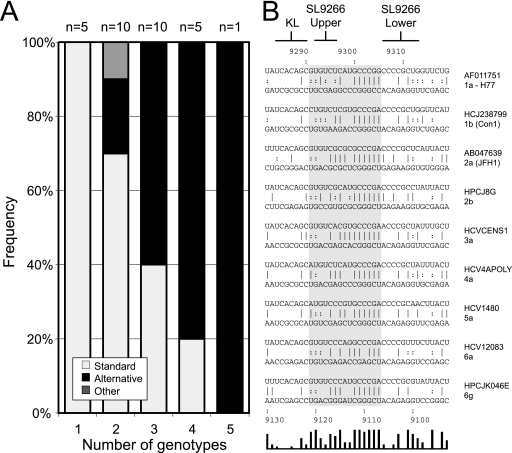
Exchange of SL9266 with the analogous region of other genotypes of HCV. (A) The SL9266 nucleotide sequence is shown (left) together with the nucleotide differences introduced by exchange with the sequences from a range of genotypes indicated. At the top and emphasized with a dark shaded box is the kissing loop interaction between the terminal loop of SL9266 and the 3′ NCR (8). At the bottom and highlighted by a pale shaded box is the predicted interaction between SL9266 and upstream sequences centered around nt 9110. Underlined nucleotides in the SL9266 or upstream sequences indicate the third base “wobble” position of codons. The upper and lower duplexes that form SL9266 are indicated by horizontal joined brackets (see also Fig. 1C). Nucleotides underlined in the alternative genotype sequences retain the ability to form these duplexes. Nucleotides in bold type within the dark shaded box retain (or acquire) the potential to base pair with the upstream sequence. The phenotypes (+, growth; −, no growth) and genotypes (genotype 1b, the parental positive control [+ive]) are shown to the right of the SL9266 nucleotide sequences. The NS5B amino acid sequences altered by exchange of SL9266 with the analogous region from other genotypes is indicated on the right-hand side of the panel. (B) G418 selection assay of SL9266 substitutions for the sequences from the genotypes indicated (genotypes 1a, 2b, 3b, 4a, 5a, 6a, and 6g).
In vitro RNA transcription and replicon analysis.
One microgram of ScaI-linearized replicon cDNA was used as the template for the production of RNA in vitro using a T7 MEGAscript kit (Ambion) according to the manufacturer's instructions. RNA was purified using RNeasy (Qiagen), the integrity of the RNA was confirmed by agarose gel electrophoresis, and the RNA was quantified spectrophotometrically.
Huh7 cells were transfected by electroporation. Briefly, 400 μl of trypsinized, washed Huh7 cells at 1 × 107 cells/ml in phosphate-buffered saline (PBS) was mixed with 5 μg in vitro-transcribed RNA in a prechilled 4-mm cuvette, pulsed once (25 milliseconds, 250 V, 950 μF, square wave) using a Bio-Rad Gene Pulser Xcell unit, and transferred into 100-mm dishes with 10 ml of DMEM P/S added. After 24 h of culture at 37°C, the medium was replaced with medium supplemented with 500 μg/ml G418 (Geneticin, G418 sulfate; Invitrogen), and the medium was changed at 2- to 3-day intervals for the duration of the selection period. G418-resistant colonies were washed with PBS, fixed with 4% formaldehyde, and visualized with Giemsa stain after about 3 weeks.
Luciferase-encoding replicon RNA (10 μg) was transfected into Huh7 cells as described previously and transferred into 20 ml of DMEM P/S, and 4 ml was placed in five wells of a six-well dish. At each time point (4, 24, 48, and 72 h posttransfection), cells in one well were washed with PBS, lysed with 0.5 ml Glo lysis buffer (Promega) and stored frozen before analysis using the Bright-Glo luciferase assay system (Promega) and quantified on a Turner TL-20 luminometer.
RESULTS
Synonymous substitutions within SL9266 define a cis-acting replication element.
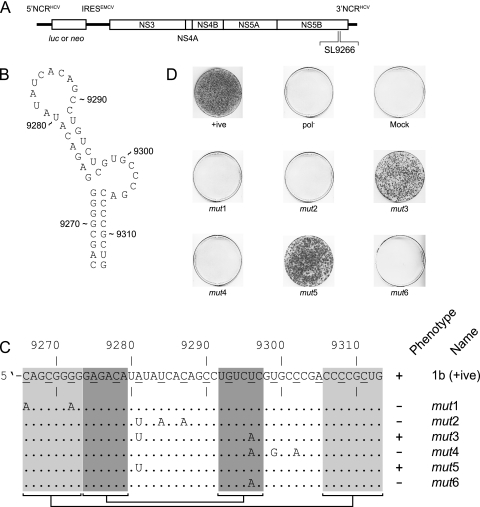
We investigated the role of base pairing within the SL9266 stem-loop structure by introducing a limited number of nucleotide substitutions to the region. For each of the six mutants, designated SL9266mut1 to SL9266mut6 and hereafter referred to as mut1 to mut6, respectively, the modifications were at synonymous sites and were generated in a neomycin-encoding subgenomic replicon (Fig. 1). MFOLD analysis (data not shown) indicated that the mutations introduced in mut1, mut3, mut4, and mut6 probably disrupted the predicted structure of SL9266, but that introduction of the mutations in mut2 and mut5 had no structural consequences, being restricted to the unpaired terminal loop region. RNA generated in vitro was transfected into Huh7 cells and analyzed in a G418 transduction assay. Two of the six mutants analyzed, mut3 and mut5, generated colony numbers consistent with replication levels at, or near, that of the positive control. The remaining four mutants (mut1, mut2, mut4, and mut6) failed to yield significant numbers of colonies following transfection and neomycin selection (Fig. 1D). Of these, substitutions involving the 5′ side of the lower duplex in SL9266 (mut1 [Fig. 1C]) or the terminal loop (mut2) were lethal, presumably reflecting a requirement for stable base pairing in the former or interaction with the 3′ X tail in the latter, and were in agreement with the results of several other published studies (8, 16, 36). Substitution of A9281U (mut5) alone did not impair replication, again consistent with other studies (see A68C in Fig. 7 of reference 36), and appeared to complement the otherwise lethal substitution of U9296A (compare mut3, mut5, and mut6 in Fig. 1C and D). Although the nonviable phenotype of mut4 could probably be ascribed to the U9296A mutation disrupting the upper duplex of SL9266, two other substitutions in this mutant were located in the unpaired 3′ bulge loop (Fig. 1B and C). A potential functional role for this region of SL9266, also hinted at by the currently unexplained lack of viability of replicons bearing mutations of C9303 and/or A9305 (designated C90A and A92G in Fig. 7 of reference 36), prompted us to investigate additional features of SL9266 and possible interactions of the unpaired regions of the CRE with flanking RNA sequences.
FIG. 1.
SL9266 is a cis-acting replication element in hepatitis C virus. (A) The genetic organization of the hepatitis C subgenomic replicon expressing either a luciferase reporter gene or neomycin selection marker is shown, together with an indication of the location of SL9266 in the region encoding the C terminus of NS5B. EMCV, encephalomyocarditis virus. (B) The thermodynamically predicted structure of SL9266. (C) Genetic analysis of synonymous mutations introduced to subgenomic replicons. The sequence of SL9266 is shown with the third “wobble” position of each triplet underlined. Underneath the top sequence, the locations of individual mutations (mut1 to mut6) are shown, together with their phenotype (+, growth; −, no growth) after G418 selection. The shaded boxes joined by horizontal brackets and lines indicate the duplex regions (lower [pale shading] and upper [dark shading]) of SL9266. (D) The phenotypes of SL9266 neomycin-encoding replicon mutants mut1 to mut6 in a G418 selection assay. +ive, positive control; pol−, defective polymerase negative control.
FIG. 7.
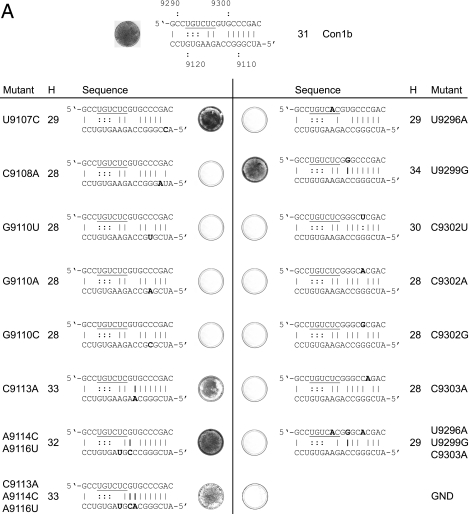
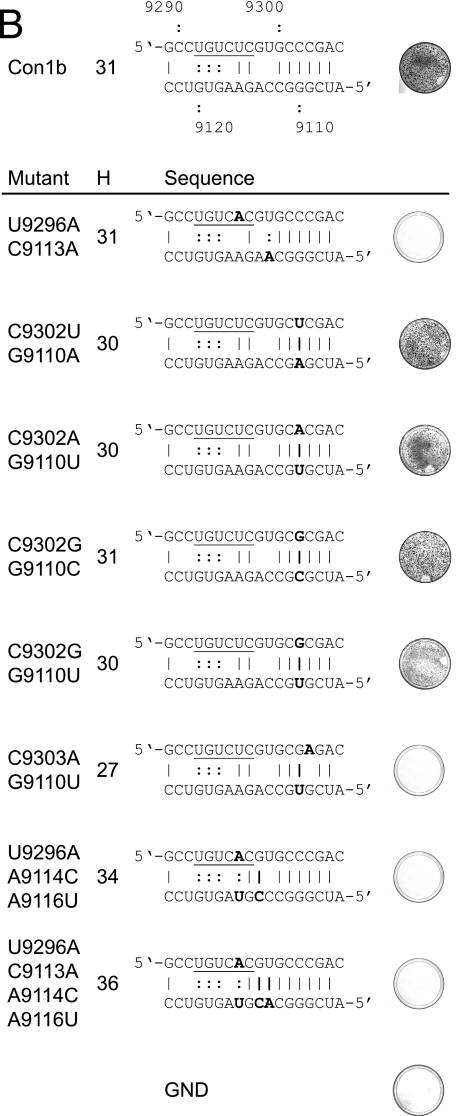
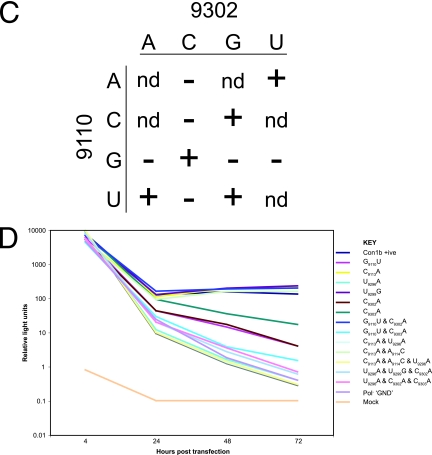
Mutational analysis of the alternative interaction of sequences within SL9266. (A) Phenotypes of neomycin-encoding replicons containing mutations within the upstream region (nt 9107 to 9121; left panel) or within the sequences that form part of SL9266 (nt 9291 to 9305; right panel). For each named mutant, a photograph of a stained dish after G418 selection is shown next to the sequence indicating the impact on the alternative interaction predicted bioinformatically. For consistency with other figures, the upstream sequence is the lower sequence depicted. Substitutions are indicated in bold type, as are additional or changed hydrogen bonding interactions. The total number of hydrogen bonds that could form between the sequences shown are indicated in the column labeled H. The regions of the SL9266 sequence that form the 3′ side of the upper duplex of SL9266 are underlined. The positive-control replicon is shown at the top of the figure. GND indicates a control replicon containing active site mutations within the NS5B polymerase (see Materials and Methods). (B) Phenotypes of neomycin-encoding replicons containing substitutions in both upstream and SL9266 sequences. (C) Summary of changes made at nt 9110 and 9302. A plus sign indicates a replication phenotype similar to that of a positive control, a minus sign indicates no apparent replication, and nd indicates that the change was not done. (D) Replication phenotypes of luciferase-encoding subgenomic replicons bearing mutations at nucleotides 9110, 9113, 9114, 9296, 9299, 9302, 9303, and combinations thereof. The average of two or three independent repeats at each time point are plotted. Con1b +ve, Con1b, the parental positive-control replicon; Pol−, defective-polymerase replicon.
RNA secondary structure prediction.
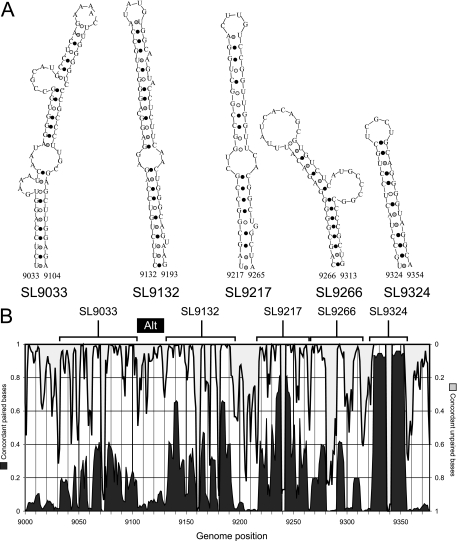
Previous comparative analysis of minimum free energy structures of the NS5B region of HCV revealed a series of evolutionarily conserved stem-loops spanning the terminal 700 bases of the coding sequence (Fig. 2A) (16, 31, 32, 36). Using an automated method (StructureDist) to quantify the frequencies of concordant and discordant pairings at individual sites for pairwise comparison of structure predictions for each sequence (31), substantial variability in the degree of conservation of the stem-loops was found between HCV genotypes (Fig. 2B). Similar variability was observed within sets of approximately equally energetically favored structure predictions for individual sequences (data not shown). The most highly conserved predicted stem-loop was SL9324, while SL9266 was the least conserved. Lack of conservation of the latter structure was unexpected and relevant to the investigation of its demonstrated role as a CRE (8, 16, 36).
FIG. 2.
Stem-loop structures in the NS5B-encoding region of HCV. (A) Predicted RNA secondary structures in the terminal 350 bases of the HCV coding sequence (in NS5B). Structures were numbered according their position in the H77 reference sequence, using standard nomenclature for stem-loops (see Materials and Methods). (B) Frequencies of concordant pairing (left-hand y axis) predictions and predicted unpaired bases (right-hand y axis) at each nucleotide position (x axis) in pairwise comparisons of the most energetically favored RNA structures predicted by MFOLD (38) for a set of 150 sequences representative of HCV genotypes 1 to 6. Frequencies were compiled using StructureDist v.1.3 (31). The location of each of the five predicted stem-loop structures is indicated above the graph. The location of the alternative upstream paired region is indicated as a black bar labeled Alt.
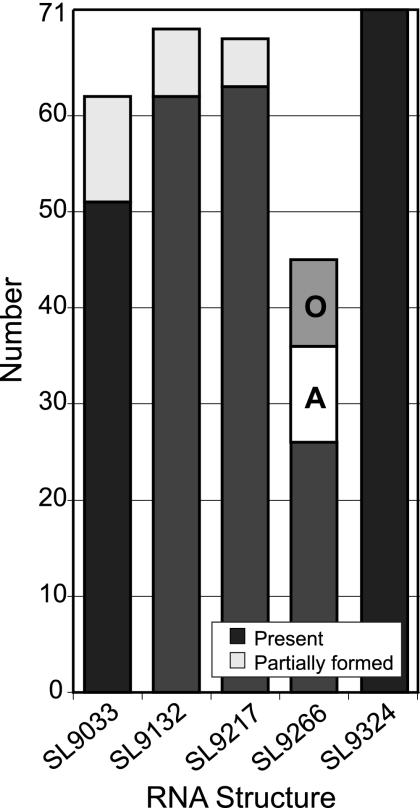
To investigate whether there were alternative RNA structures or pairings underlying this observed lack of conservation of SL9266, RNA structures were predicted for 26 NS5B nucleotide sequences, with each representing different (up to four) subtypes within the six genotypes of HCV using the program SFOLD. This generates a statistical sample of secondary structures from the Boltzmann ensemble of RNA secondary structures using Turner free energy rules (7). The relatedness of structures to each was determined using the Diana method for cluster analysis followed by calculation of the Calinski and Harabasz index to determine the optimal number of centroids for which consensus structures can be calculated, as previously described (6). Each sequence submitted to SFOLD analysis generated between two and six centroids, whose consensus structure predictions were compared to the previously described RNA structure for the NS5B region (Fig. 3). Despite the variability in RNA pairings between centroids for individual sequences of the wide range of genotypes analyzed, four of the five stem-loops were frequently (SL9033, SL9132, and SL9217) or invariably (SL9324) found among sampled structures, generally containing equivalent pairings to the predicted structures for HCV genotype 1a (black filled boxes) or pairings restricted to bases around the terminal loop (gray filled boxes). However, consistent with the more variable structure predictions in this region visualized by StructureDist (Fig. 2B), only around a third (26) of the 71 consensus structures of the centroids contained pairings that matched those of SL9266 (Fig. 3). Alternative structures for this region frequently retained the pairing of the terminal stem and loop (bases 9274 to 9297, but with a partially overlapping longer range pairing of bases forming the bulge loop and part of the 3′ lower stem of SL9266 [bases 9296 to 9306]) with upstream predicted unpaired regions in NS5B. In approximately one half of these alternative structures (labeled A in Fig. 3), bases between 9296 and 9306 formed a duplex with the predicted unpaired bases between structures SL9033 and SL9132 (bases 9106 to 9123). Analogous alternative conformations were found in predicted structures for all six genotypes of HCV and frequently alternated with the standard structure in the Boltzmann ensemble for individual sequences. Similar frequencies of standard and alternative pairings were observed when longer sequences spanning position 8301 to the 3′ NCR were analyzed by SFOLD (data not shown).
FIG. 3.
SFOLD analysis of HCV NS5B sequences. Numbers of consensus structures in 72 centroids generated by SFOLD from a total of 26 HCV NS5B sequences (positions 9001 to 9377) corresponding to standard stem-loop structures (Fig. 2A) (filled black) or containing partial structure (filled gray). The frequencies of alternative pairings of the 3′ side of SL9266 to upstream sequences are shown by the Alternative (A) and Other (O) boxes.
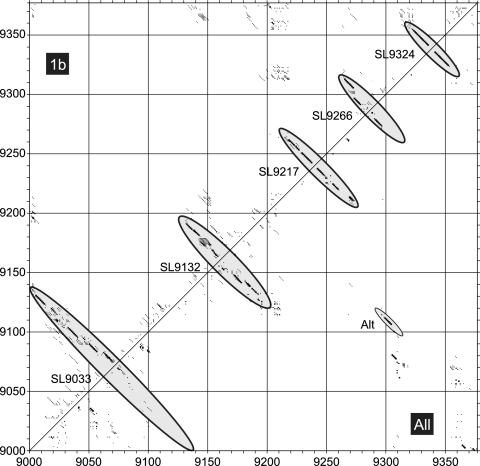
StructureDist and SFOLD use free energy minimization algorithms (e.g., MFOLD) to predict candidate RNA structures. Given the poor resolution of RNA structure in the HCV CRE, we used an independent, non-energy-minimizing algorithm that makes better use of the substantial comparative sequence information available for HCV (14, 24). The method, implemented as PFOLD, combines an explicit evolutionary model of RNA sequences with a probabilistic model for secondary structures. A stochastic context-free grammar is used to produce a prior probability distribution of RNA structures. For the analysis, a set of 40 NS5B sequences between positions 9001 and 9377 from genotype 1b and further sets of 20 sequences from genotypes 1, 2, 3, 4, and 6 containing as diverse a range of subtypes as possible, were analyzed individually and in combination by PFOLD (Fig. 4 and 5). For the set of genotype 1b sequences, pairing predictions corresponded to those of the standard structure, with robust prediction of SL9266 (upper left half in Fig. 4) and the four other stem-loops predicted for NS5B. Similar results were obtained for pairing predictions of alignments of each genotype individually (Fig. 5A). Intriguingly, analyzing the combined data set of all five genotypes produced a distinct pairing for the HCV CRE corresponding to the alternative pairing found by SFOLD (lower right, labeled Alt in Fig. 4). By analyzing alignments of each combination of two, three, and four genotypes, a relationship was found between sequence diversity and frequency of detection of standard and alternative RNA structures (Fig. 5A). Representative comparisons of duplexes formed in the alternative pairing for a range of HCV genotypes are shown in Fig. 5B. The region of maximum potential interaction (positions 9121 to 9107 with positions 9291 to 9305, see the bar graph at the bottom of Fig. 5B) can be divided into two areas, a less well conserved region (on the left in Fig. 5B) involving sequences already implicated in forming the 3′ side of the upper duplex of SL9266 and a highly conserved block of five nucleotides centered around positions 9110 and 9302. To functionally test the relevance of the predicted alternative pairing, we undertook further mutagenesis studies.
FIG. 4.
PFOLD analysis of HCV NS5B sequences. Coordinates (dot plot) of pairing predictions for consensus structures predicted for alignments of HCV genotype 1b sequences (top left) or HCV genotypes 1 to 6 (bottom right) using PFOLD. The size of the dot depicts the reliability of the pairing prediction. The positions of standard predicted structures and base pairing forming the alternative RNA structure (Alt) are shown as gray filled ellipses.
FIG. 5.
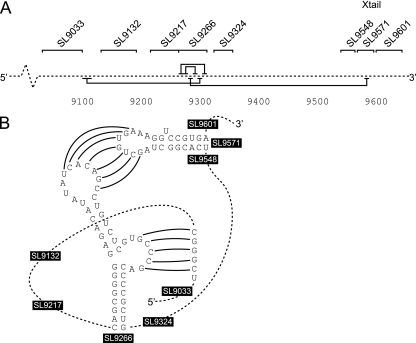
Alternative interactions of SL9266 sequences in a range of HCV genotypes. (A) Frequencies of RNA structure prediction by PFOLD corresponding to the standard model or containing the alternative pairing. The x axis records the number of different genotypes in each alignment; the numbers above the bars records the number of different genotype combinations tested by PFOLD. For example, there are 10 possible combinations of the five genotypes tested, all of which were analyzed, and these results are presented in the second column (the column with 2 for the number of genotypes) of the graph. (B) Comparison of duplexes formed in the alternative pairing for representative sequences of HCV genotypes 1 to 6. Genomic numbering for upstream and downstream bases is shown at the top and bottom of the figure, respectively. The locations of known interactions of genotype 1b SL9266 are indicated at the top of the figure; KL indicates the location of sequences forming a kissing loop interaction with the 3′ X tail (8), and SL9266 Upper and SL9266 Lower indicate the 3′ side of the upper and lower duplexes of SL9266. The gray block highlights the area of maximal conserved base pairing (nucleotides 291 to 9305 and 9121 to 9107; indicated in a simple bar chart at the bottom of the figure, each bar representing a single nucleotide in the aligned sequences) forming the predicted alternative interaction of sequences within SL9266 and the upstream region.
Substitution of SL9266 with the analogous region of alternate genotypes.
Of the two previously defined interactions of SL9266, one is local, forming the interrupted base pairing of the CRE (16, 36), whereas the second is long-range, involving an interaction with the X tail SL2 (8). Within SL9266, the nucleotides in the terminal loop that base pair with the 3′ NCR are very highly conserved (8). Similarly, sequences occupying the bulge loop of SL9266 are highly conserved, whereas those forming the upper and lower duplexes show more variability. This accounts for the different levels of conservation of base pairing between the left- and right-hand sides of the interaction depicted with the upstream sequences depicted in Fig. 5B. Assuming SL9266 folds similarly in each genotype of HCV, we reasoned that replacement of SL9266 in the subgenomic replicon (genotype 1b) with the analogous structures from other HCV genotypes might allow us to determine whether just some or all of the sequences between positions 9291 and 9305 were also involved in the alternative pairing we predict.
Using a BsmBI-based cassette system (see Materials and Methods), we precisely replaced the regions between nucleotides 9266 and 9312 with complementary oligonucleotides corresponding to the analogous sequences of other genotypes of HCV. Inevitably, due to the sequence variation inherent in HCV, this strategy resulted in changes to the encoded NS5B polypeptide sequence (Fig. 6A). All modifications were made in a neomycin-expressing replicon that, in parallel with appropriate controls, was independently transfected into Huh7 cells and selected with G418. Of the eight substitutions made, five were tolerated well, generating approximately equivalent colony numbers to the positive control after G418 selection. The remaining three substitutions of genotypes 3b (Tr), 4a (ED43), and 6g (JK046) produced markedly reduced colony numbers, indicating that the modifications introduced within SL9266 were incompatible with replication.
It seemed unlikely that the differences in the replication phenotypes of the chimeric replicons were due to introduction of incompatible residues into the NS5B polypeptide, with the possible exception of the genotype 3b (Tr) sequence. The latter contains two amino acid substitutions (G558N and P569S; Fig. 6A) not present in the other sequences analyzed. In the remaining genotype swaps, amino acid substitutions were restricted to just three residues of NS5B, with both viable and nonviable chimeric replicons containing the same changes, implying that they alone do not account for the phenotype. For example, the replication-deficient replicon containing genotype 4a (ED43) sequences has substitutions at positions 556, 564, and 566; of these, S556G is in genotype 2b (HCJ8), L564M is in genotype 5a (EUH1480), and R566H is in genotype 1a (HP-H), all of which are replication competent. Therefore, unless particular combinations of these changes are deleterious, it seemed probable that the poor replication of genotypes 6g (JK046) and 4a (ED43) must be mediated at the level of RNA, either by disruption of an RNA-RNA interaction, or alteration of a sequence motif bound by a cellular or viral protein(s).
Replication competence of the chimeric replicon did not correlate directly with either invariant or covariant (underlined in Fig. 6A) base pairing within the upper duplex region of SL9266 or the covariation within the alternative interaction with the upstream sequence (in bold type in Fig. 6A). For example, genotype 1a (HP-H) and 4a (ED43) replicons were identical to the control 1b replicon in the upper duplex of SL9266, but only the former could replicate. Similarly, the genotype 6g (JK046) replicon contains two compensating changes in the upper duplex but cannot replicate, whereas genotype 6a (EUHK2) and 5a (EUH1480) replicons had the same covariance in the upper duplex and were replication competent. Within the region forming the bulge loop of SL9266, none of the chimeras changed the highly conserved 5′-GCCCG motif. However, of the six that contained variation within this region of SL9266 (namely, genotype 1a [GLA], genotype 1a [HP-H], genotype 2b [HCJ8], genotype 4a [ED43], genotype 6a [EUHK2], and genotype 6g [JK046]), two of the nonviable chimeras with genotype 4a (ED43) and genotype 6g (JK046) lacked any covariant changes within this region, whereas the genotype 1a (GLA), genotype 1a (HP-H), and genotype 6a (EUHK2) chimeras all contained at least one covariant substitution that could be involved in base pairing to the upstream sequence (highlighted in bold type in Fig. 6A). All chimeras also introduced covariant changes at C9291 (to A or G), the 5′ nucleotide within the SL9266 sequence that could pair with U9121 (Fig. 5B and 6A), though there was not a correlation between the viability of the replicon and the particular substitution at this position.
Results obtained with the chimeric replicons suggested that the RNA-RNA interactions within SL9266 and the proposed alternative upstream pairing were nontrivial. We therefore specifically examined the upstream interaction in a more focused manner by further site-directed mutagenesis.
Critical interactions between SL9266 and the upstream sequence.
Mutations were introduced singly or in combination into SL9266 or the upstream sequence located around nt 9110. In each instance, substitutions were selected to leave the encoded NS5B polypeptide unchanged, thereby excluding the possibility that the resulting phenotype was due to the introduction of an incompatible amino acid into the virus polymerase. The majority of the mutations introduced were within the SL9266 subterminal bulge loop or the complementary sequence around nt 9110, though additional changes were also made in the sequences implicated in forming the 3′ side of the upper duplex in SL9266. These, or the complementary changes 3′ to nt 9110, were designed to test the extent of the alternative interaction proposed by our bioinformatic analysis.
In the upstream sequence (Fig. 7A, left-hand panel), substitutions at C9108 and G9110 were incompatible with replication, whereas substitution of U9107C, C9113A, or a combination of the changes at A9114C and A9116U, also in combination with C9113A, were tolerated well. Within the sequences that contribute to the upper duplex or bulge loop of SL9266 (Fig. 7A, right-hand panel), substitutions of U9296A, alone or in combination with U9299G and C9303A, prevented replication. This phenotype is presumably attributable to the change at U9296 which disrupts the stability of the upper duplex. Of the other single substitutions constructed, only U9299G had no impact on replication, with changes of C9302 and C9303 all preventing colony formation in the G418 transduction assay.
Mutations in the upstream and SL9266 regions were also combined to test whether complementary substitutions could restore the replication phenotype to resemble that of the parental replicon (Fig. 7B). In addition, combinations of substitutions were introduced to determine the influence of increasing the potential hydrogen bonding between the upstream region and SL9266 sequences. Of the combinations constructed, four that restored the predicted ability to base pair G9110 and C9302 all generated significant numbers of G418-resistant colonies after transduction and selection. The demonstration that individual substitutions of G9110 or C9302 that disrupted the predicted base pairing prevented replication, whereas all but one in which duplex formation could occur (summarized in Fig. 7C) were replication competent, provides strong support for the interaction of these regions. Double substitution of nucleotides C9110U and C9303A did not restore replication capacity. Furthermore, all combinations of mutations that included U9296A were incapable of replicating (Fig. 7B); this included substitutions at nt 9113, 9114, and 9116, the addition of which significantly increased the potential for hydrogen bonding between the upstream and SL9266 sequences. This result suggested that disruption of the upper duplex of SL9266 by U9296A could not be compensated for by strengthening the predicted interaction with upstream sequences.
The majority of mutations constructed in the neomycin-encoding replicon were also rebuilt into a replicon carrying a luciferase reporter gene. Huh7 cells were transfected, and a time course experiment of luciferase activity over 3 days was performed (Fig. 7D). Of those tested, the mutants could be divided into three broadly defined groups. With the exception of single mutations involving nucleotides G9110, C9302, or C9303, all the replicons harboring mutations that prevented replication in the G418 colony-forming assay (Fig. 7A) exhibited a phenotype similar to that of the negative control (which lacks an NS5B active site). This group included replicons with the mutation of U9296A, the double mutations of C9113A plus U9296A, and all the triple mutations tested. In contrast, replicons that had generated colony numbers similar to that of the parental 1b replicon (positive control) generated luciferase activities indistinguishable from the parental luciferase-encoding replicon. These included C9113A, U9299G, and the double mutant A9114C A9116U. Significantly, this group also included the double mutant G9110U C9302A (Fig. 7D). The final group had intermediate phenotypes, exhibiting a steady decline of luciferase activity over the second and third day of the time course experiment but at a lower rate than that of the replicons that resembled the defective-polymerase negative control. Although we tested only a limited representative range of substitutions predicted to be involved in the highly conserved (Fig. 4 and 5B) upstream interaction, it was notable that all those exhibiting an intermediate phenotype were from this group. This included G9110U, C9302A, and C9303A (Fig. 7D). One explanation for this could be an increase in RNA stability. However, since this phenotype was observed only in mutants in which the RNA structure was destabilized, we suspect that the enhanced translation may be explained by some factor other than an increase in RNA stability.
DISCUSSION
Many viral proteins are multifunctional, for example controlling aspects of the virus replication cycle and the intracellular milieu. Increasingly, studies are demonstrating that the virus genome also has multiple functions, particularly in the small RNA and DNA viruses where coding capacity is limited. In the case of the small positive-strand RNA viruses, the genome must act as a template for both translation and replication. At least on the input genome, before a pool of progeny genomes have been generated, these are mutually exclusive processes. In certain examples, additional functions ascribed to the RNA genome include subversion of the innate immune response, temporal and spatial control of the replication process, and encapsidation (12, 28, 35, 37). Functional specificity is provided by the evolutionary conservation of binding determinants, often in a structural context. The accurate prediction of stem-loop and higher-order structures therefore provides primary information on key functional domains of the virus genome.
Well-established thermodynamic methods to predict two-dimensional RNA structure (e.g., MFOLD; see references 20 and 38) exist; we have extended these methods and implemented them in the program StructureDist to extract the additional information present in large data sets of related sequences. Using this and an alternative thermodynamic approach, SFOLD (7), we investigated structures in the terminal 700 nt of the HCV coding region, an area of the genome in which we had previously identified at least five well-conserved stem-loop elements (31). One of the five structures predicted, an interrupted stem-loop starting at nt 9266 (SL9266) shown in previous studies to be a cis-acting replication element, was only poorly predicted. An alternative nonthermodynamic method (PFOLD [see references 14 and 24]) robustly predicted SL9266 in genotype 1, but analysis of all six genotypes of HCV indicated a hitherto unsuspected interaction of sequences within SL9266 and a region located approximately 200 nt in a 5′ direction (Fig. 4).
The finding of poor RNA structure conservation of the HCV replication element among alternative structures showing similar folding free energies (StructureDist and SFOLD), may arise from either an incorrect structure prediction for the HCV CRE using thermodynamic methods or because there is more than one (metastable) RNA structure in this region. The evidence that the alternative folding better accommodates sequence variability between genotypes using PFOLD even though the standard structure was predicted for individual genotypes provides further evidence for possible alterations in RNA structure in this genome region. Unfortunately, none of the structure prediction methods are able to incorporate tertiary RNA structure interactions, such as pseudoknots or kissing loop interactions, in predicted structure models. These interactions may have significant stabilizing or destabilizing influences on the two predicted structures for the HCV CRE. Variability in prediction outcomes in this study may therefore result from incomplete prediction of potential pairings in this region of the HCV genome.
We investigated the relevance of the two predicted conformations of SL9266 to HCV replication by site-directed mutagenesis of a subgenomic replicon encoding either a neomycin resistance marker or luciferase reporter gene. The definition of SL9266 as a functional CRE was supported by limited site-directed mutagenesis (Fig. 1C and D and Fig. 7A and B). Disruption of the lower duplex (in mut1) or the sequences (mut2) implicated in the “kissing loop” interaction with SL2 (now SL9571 [see reference 15]) in the 3′ X tail prevented replication in agreement with the published results of other studies (8, 16, 36). Three of the mutants (mut3, mut4, and mut6) had substitutions of U9296A, a substitution that in our more extensive mutagenic analysis (Fig. 7) was always incompatible with replication. However, our results suggest that the additional presence of A9281U (compare mut3, mut5, and mut6 in Fig. 1C and D) could somehow compensate for the otherwise lethal substitution of U9296A. Our present understanding of SL9266, together with knowledge of interactions of SL9266 with the 3′ untranslated region or the upstream sequences demonstrated here, does not explain how substitution of 9281 (unpaired in the terminal loop of SL9266) compensates for a mutation that destabilizes the upper duplex of the CRE.
More extensive modification of SL9266 was achieved by substituting the entire structure with the analogous region of other genotypes of HCV. These modifications were intended to allow the distinction between the importance of interactions within the SL9266 structure and those involving more distant sequences. Of the representative genotypes chosen, the sequence variation was unevenly distributed within the SL9266 structure, presumably reflecting evolutionary conservation of certain features. Significantly, all of the introduced sequences were invariant between nucleotides 9284 and 9290 (inclusive) in the terminal loop, thereby excluding the possibility that the resulting phenotypes of the chimeric replicons were due to disruption of the “kissing loop” interaction with SL9571 in the 3′ NCR (8). Other regions of significant conservation existed within the 3′ side of the bulge loop (nt 9300 to 9304) and the central region of each of the two duplexes on either side of the bulge loop. Unsurprisingly, considering the predicted structure of SL9266, there was good evidence for covariation within the region (underlined in Fig. 6A), in particular at nt 9267/9312 and 9275/9296. All but one of the chosen sequences included an A9281U substitution, and all also carried a change at nt 9291 that created the potential to interact with U9121 in the upstream region. The resulting phenotypes of replicons in the G418 transduction assay (Fig. 6B) indicated that there was a good correlation between the overall level of retained base pairing—both within SL9266 and between SL9266 and the upstream sequence around position 9110—and viability of the chimeric replicon. Chimeras either generated good numbers of colonies, broadly equivalent in number to the unmodified replicon, or very limited numbers of G418-resistant colonies; the latter phenotype is consistent with the introduced mutation being grossly suboptimal for replication, with the appearance of a limited number of colonies due to the acquisition of one or more compensatory mutations that restore replicative capacity. These are considered nonviable without the adaptive changes. The nonviable chimeras exhibited only 43% (genotype 6g [JK046]), 40% (genotype 4a [ED43]), or 30% (genotype 3b [Tr]) covariation, whereas all viable chimeras contained >50% covariation. For example, 70% of the 10 nucleotide changes between the genotype 1b parental replicon and the genotype 6a (EUHK2) chimera were covariant—5 within duplex regions of SL9266, at nt 9267, 9268, 9275, 9296 and 9311, and a further 2, at nt 9291 and 9299, with regard to the upstream alternative interaction proposed here. Although based on a limited sample size, these results suggest that both the SL9266 CRE and the interaction of SL9266 sequences with the upstream region were important for replication. These studies also demonstrated that there was no absolute requirement for a U at nt 9296; the viable chimeric replicons with genotypes 2b (HCJ8), 5a (EUH1480), and 6a (EUHK2) all had a substitution at nt 9296 but also carried a covariant change at nt 9275 that retained the base pairing in the upper stem of SL9266 (Fig. 6A). However, base pairing of nt 9275/9296, for example in genotype 6g (JK046), was alone not sufficient for replication. In this chimera, encoding an NS5B polypeptide identical to that encoded by the viable genotype 1a HP-H construct (Fig. 6A), it is presumed that the overall reduced level of conserved base pairing within SL9266 and between the bulge loop of SL9266 and the upstream sequences rendered the chimera nonviable.
Despite demonstrating that replicons chimeric for the SL9266 CRE exhibiting divergence of ∼20% in this region were still replication competent, the distribution of substitutions within the replaced sequence meant that further site-directed mutagenesis was required to determine the contribution of individual nucleotides to the predicted RNA-RNA interactions with the upstream region. Individual substitutions of U9107, C9113, and U9299 were not detrimental to replicon activity, whether determined by luciferase activity or the generation of G418 resistance (Fig. 7). Of these, C9113 and U9299 are juxtaposed in the predicted long-range interaction but are not complementary in the majority of sequences. In contrast, a possible base pair between nt 9107 and 9305 is highly conserved but apparently not necessary for replication (see U9107C in Fig. 7A and Fig. 5B). Although substitution of nt 9107 had no apparent effect, modification of A9305 in isolation in a previous study (A92G/C/U in Fig. 7 of reference 36) generated a wild-type phenotype when the A was converted to C, reduced colony numbers when it was changed to U, and no colonies when it was converted to G. This suggests qualitative differences between the potential A-U or G-U pairing of nt 9107/9305 or, more likely, that nt 9305 is possibly involved in another RNA or protein interaction that has yet to be defined.
Although covariation of nt 9275/9296 (Fig. 6A) could be accommodated without destroying replication, all individual substitutions of A9296 or combinations of mutations that included a change of A9296 were incapable of replicating (Fig. 7A and B). This included the combination of A9296U with substitutions at nt 9113, 9114, and 9116. The latter were designed to increase potential hydrogen bonding between sequences within SL9266 and the upstream region. We interpret this to mean that additional bonding between these more distant regions cannot compensate for disruption of the upper duplex of SL9266.
The remaining substitutions involved the highly conserved 5-nt 5′-GCCCG motif occupying the subterminal bulge loop of SL9266 and the perfect complementarity to a 5′-CGGGC sequence centered on nt 9110. Individual synonymous substitutions in both regions, of C9108A, of G9110 to U, A, or C, and of C9303A or C9302 to U, A, or G all prevented colony formation in the G418 transduction assay. Of these, only C9302U was predicted to retain any capacity to base pair with the upstream region. Interestingly, despite using standardized transfection conditions as with the chimeric SL9266 exchanges, point mutations in this region did not generate any colonies in our assays. Although not tested, this implies these mutants were incapable of generating revertant colonies under G418 selection. We went on to investigate the effect of substitutions in both parts of the predicted interacting sequence. In every case, dual mutations that restored the potential for base pairing between positions 9110 and 9302 resulted in a replication-competent phenotype (Fig. 7B and C). Individually, both nt 9110 and 9302 were substituted for each possible alternative nucleotide, indicating no sequence specificity at either position. It was perhaps surprising therefore that the single substitution of C9302U, which left a potential interaction with G9110, was incapable of replicating when a G9110A C9302U double mutant was viable. This strongly implies that a canonical Watson-Crick pairing may be essential in this position to ensure the interaction of the two interacting regions. This conclusion is supported by the results of analysis of a large data set of divergent HCV sequences, corresponding to available complete genome sequences of all six genotypes of HCV, in which none were identified with a G-U at this position (the distribution was 12% A-U and 88% G-C; data not shown). The requirement to retain synonymous substitutions prevented an individual mutation being introduced to restore complementarity between nt 9303 and 9109 (which, respectively, form the first and second nucleotides in codons coding for arginine and serine).
Our results strongly support a long-range interaction between highly conserved sequences located in the subterminal bulge loop of SL9266 and a similarly conserved upstream region around nt 9110 that is not implicated in any evolutionary conserved RNA structure. Additional supporting data for the importance of this interaction comes from the study by Friebe et al., who constructed a G9300A substitution (designated bulge-G→A [see reference 8]) in a replicon with a duplication of SL9266 sequences and the flanking regions within the 3′ NCR. This substitution rendered the replicon nonviable and because G9300 was now noncoding, this could not be attributed to a defect in NS5B. In one construct, P1-ins3.2 (8), SL9266 alone was duplicated in the 3′ NCR of a replicon bearing synonymous substitutions that disrupted the native SL9217, SL9266, and SL9324 structures in the NS5B coding region. Although this replicon exhibited 10- to 15- fold-lower replication activity than the wild type did, it implies that the distance separating sequences around nt 9110 and the complementary functional SL9266 sequences are not absolutely critical for replication.
The data available from our analysis and reinterpretation of previous studies of SL9266 (8, 16, 36) cannot unequivocally demonstrate whether formation of SL9266 and either or both of the upstream and downstream interactions are mutually exclusive events or could occur simultaneously. A number of scenarios are possible; the rather weak (as evidenced by the poor bioinformatic prediction) SL9266 structure could be stabilized by interaction with either or both sequences around nt 9110 and SL9571 to form a complex extended pseudoknot containing four duplexed regions. Alternatively, interaction of sequences normally not paired within SL9266 with the 3′ NCR and the 9110 region could destabilize or prevent formation of SL9266, thereby forming a molecular switch capable of adopting at least two conformations. Intermediates between these two examples, separately involving the 3′ NCR or the upstream region, are also possible. Further mutagenic and functional studies will be needed to distinguish between these various possibilities. Considering the available data, we currently favor a model in which SL9266 interacts, at least some of the time, with both the upstream and downstream sequences to form an extended pseudoknot structure, as illustrated in Fig. 8. In our model, we define the upstream interaction as involving complementarity between 5′-CGGGC and 5′-GCCCG sequences centered on nt 9110 and 9302, respectively. Good evidence to support this interpretation includes the primary involvement of single-stranded regions of SL9266 in the long-range interactions. Furthermore, the phenotype exerted by the majority of substitutions introduced to SL9266 in this and previous studies can be interpreted as affecting either SL9266 per se or one or other of the long-range interactions. Sequences within the region from nt 9108 to 9112/9300 to 9304 are highly conserved; of 192 divergent HCV sequences analyzed, all exhibited G9109 to C9303 and C9112 to G9300 pairings. There was a single, presumably unpaired, variant of C9108 to A9304, the remainder being C9108 to G9304, and another singleton of G9111 to U9301, with all others in the data set being G to C pairs at this position (data not shown). The variation of nt 9110 and 9302 is listed above. This conservation of Watson-Crick pairings presumably explains the inhibition of replication mediated by the C9303A substitution constructed by You and colleagues (for their substitutions of C90, see reference 36). Overall, there is less variation or covariation in the unpaired regions of SL9266, compared with the lower and upper duplexes of the stem-loop (36; data not shown). The lack of covariation in the pentanucleotide motif forming the upstream interaction described here is presumably a consequence of the juxtaposition of the third base “wobble” position of the codons in these regions; almost all variation is restricted to substitution of a G9110-C9302 pair by an A-U pair in genotype 6 sequences.
FIG. 8.
Proposed structure of a complex pseudoknot in hepatitis C virus. (A) The solid black horizontal lines above and below a linear representation of the HCV genome (broken line) indicate the interactions involved in formation of SL9266 (above) and the long-range interactions (below) with sequences located 5′ and 3′ to SL9266. The positions of evolutionarily conserved stem-loop structures in the NS5B coding region and the X tail in the 3′ NCR are also indicated. (B) Schematic of a complex pseudoknot involving SL9266 and long-range interactions between the subterminal bulge loop and sequences centered on nucleotide 9110 and the SL9266 terminal loop and complementary sequences in SL9571.
Many viruses are known to possess pseudoknots that contribute essential functions during the replication cycle. In most viruses, pseudoknots located within coding regions are primarily involved in translational control, in particular −1 frameshifting (2). However, there is no evidence for such a role in HCV, and the previously demonstrated positional independence of SL9266 would argue strongly against any such function. Instead, it seems likely that the RNA structure forming SL9266, together with interactions of the unpaired loop sequences of SL9266 and both upstream and downstream regions, has one of more functions in genome replication. Precedents exist in bacteriophages, several plant viruses, and some animal RNA viruses. The first identified pseudoknot, the tRNA-like sequence (TLS) of turnip yellow mosaic virus (27), has multiple functions, including recruitment of a nucleotidyltransferase for genome completion and genome circularization (or at least juxtaposition of the 5′ and 3′ ends) probably via interaction with eIF1a and consequent enhancement of translation. The TLS is also implicated in the switch from translation of the input genome to replication by competitive binding with newly synthesized viral polymerase and may also have a role in late replication functions, such as encapsidation (4, 11, 21, 22). Genome circularization by the TLS is probably protein mediated, but long-range RNA-RNA interactions that form pseudoknots can critically influence the global folding of RNA. Such interactions form the core of the ribosome (reviewed in reference 2) and are also known to occur in virus genomes. In bacteriophage Qβ, a pseudoknot spanning 1.2 kb of the genome recruits the 3′ end of the genome to the internally bound viral replicase (13). Similarly, recruitment of the replicase to the 3′ end of porcine reproductive and respiratory syndrome virus requires a long-range (∼300-nt) pseudoknot (33).
Considering the important role in replication of the complex pseudoknot proposed here, it is perhaps unsurprising that the RNA structures in the 3′ end of the HCV coding region (3) and SL9266, forming the core of the pseudoknot, interact with NS5B in in vitro assays (16). Although further investigation is required to define the function(s) of this complex RNA structure in the translation and replication of the HCV genome, our demonstration of important 5′ interactions with the subterminal bulge loop of SL9266 provides a structural basis on which these studies can be based.
Supplementary Material
Acknowledgments
We thank R. Bartenschlager for the neomycin-encoding replicon and GlaxoSmithKline for the luciferase-encoding replicon.
We thank the Medical Research Council for financial support (D.J.E. and P.S.) and MRC/GlaxoSmithKline for a CASE Ph.D. studentship (to R.M.E.) for V.A.
Footnotes
Published ahead of print on 9 July 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 2901972-1974. [DOI] [PubMed] [Google Scholar]
- 2.Brierley, I., S. Pennell, and R. J. Gilbert. 2007. Viral RNA pseudoknots: versatile motifs in gene expression and replication. Nat. Rev. Microbiol. 5598-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng, J. C., M. F. Chang, and S. C. Chang. 1999. Specific interaction between the hepatitis C virus NS5B RNA polymerase and the 3′ end of the viral RNA. J. Virol. 737044-7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi, Y. G., and A. L. N. Rao. 2003. Packaging of brome mosaic virus RNA3 is mediated through a bipartite signal. J. Virol. 779750-9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clerte, C., and K. B. Hall. 2006. Characterization of multimeric complexes formed by the human PTB1 protein on RNA. RNA 12457-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding, Y., C. Y. Chan, and C. E. Lawrence. 2005. RNA secondary structure prediction by centroids in a Boltzmann weighted ensemble. RNA 111157-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding, Y., and C. E. Lawrence. 2003. A statistical sampling algorithm for RNA secondary structure prediction. Nucleic Acids Res. 317280-7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friebe, P., J. Boudet, J. P. Simorre, and R. Bartenschlager. 2005. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J. Virol. 79380-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friebe, P., V. Lohmann, N. Krieger, and R. Bartenschlager. 2001. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 7512047-12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukushi, S., M. Okada, T. Kageyama, F. B. Hoshino, K. Nagai, and K. Katayama. 2001. Interaction of poly(rC)-binding protein 2 with the 5′-terminal stem loop of the hepatitis C-virus genome. Virus Res. 7367-79. [DOI] [PubMed] [Google Scholar]
- 11.Giege, R. 1996. Interplay of tRNA-like structures from plant viral RNAs with partners of the translation and replication machineries. Proc. Natl. Acad. Sci. USA 9312078-12081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han, J.-Q., H. L. Townsend, B. K. Jha, J. M. Paranjape, R. H. Silverman, and D. J. Barton. 2007. A phylogenetically conserved RNA structure in the poliovirus open reading frame inhibits the antiviral endoribonuclease RNase L. J. Virol. 815561-5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klovins, J., V. Berzins, and J. van Duin. 1998. A long-range interaction in Qbeta RNA that bridges the thousand nucleotides between the M-site and the 3′ end is required for replication. RNA 4948-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knudsen, B., and J. Hein. 1999. RNA secondary structure prediction using stochastic context-free grammars and evolutionary history. Bioinformatics 15446-454. [DOI] [PubMed] [Google Scholar]
- 15.Kuiken, C., C. Combet, J. Bukh, I. T. Shin, G. Deleage, M. Mizokami, R. Richardson, E. Sablon, K. Yusim, J. M. Pawlotsky, and P. Simmonds. 2006. A comprehensive system for consistent numbering of HCV sequences, proteins and epitopes. Hepatology 441355-1361. [DOI] [PubMed] [Google Scholar]
- 16.Lee, H., H. Shin, E. Wimmer, and A. V. Paul. 2004. cis-Acting RNA signals in the NS5B C-terminal coding sequence of the hepatitis C virus genome. J. Virol. 7810865-10877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a.Lemon, S. M., C. Walker, M. J. Alter, and M. Yi. 2007. Hepatitis C virus, p. 1253-1304. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 17.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309623-626. [DOI] [PubMed] [Google Scholar]
- 18.Lohmann, V., F. Körner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285110-113. [DOI] [PubMed] [Google Scholar]
- 19.Luo, G., S. Xin, and Z. Cai. 2003. Role of the 5′-proximal stem-loop structure of the 5′ untranslated region in replication and translation of hepatitis C virus RNA. J. Virol. 773312-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Markham, N. R., and M. Zuker. 2005. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 33W577-W581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuda, D., and T. W. Dreher. 2004. The tRNA-like structure of turnip yellow mosaic virus RNA is a 3′-translational enhancer. Virology 32136-46. [DOI] [PubMed] [Google Scholar]
- 22.Matsuda, D., S. Yoshinari, and T. W. Dreher. 2004. eEF1A binding to aminoacylated viral RNA represses minus strand synthesis by TYMV RNA-dependent RNA polymerase. Virology 32147-56. [DOI] [PubMed] [Google Scholar]
- 23.McMullan, L. K., A. Grakoui, M. J. Evans, K. Mihalik, M. Puig, A. D. Branch, S. M. Feinstone, and C. M. Rice. 2007. Evidence for a functional RNA element in the hepatitis C virus core gene. Proc. Natl. Acad. Sci. USA 1042879-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pedersen, J. S., I. M. Meyer, R. Forsberg, P. Simmonds, and J. Hein. 2004. A comparative method for predicting and folding RNA secondary structures within protein-coding regions. Nucleic Acids Res. 324925-4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pietschmann, T., A. Kaul, G. Koutsoudakis, A. Shavinskaya, S. Kallis, E. Steinmann, K. Abid, F. Negro, M. Dreux, F. L. Cosset, and R. Bartenschlager. 2006. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl. Acad. Sci. USA 1037408-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reusken, C. B., T. J. Dalebout, P. Eerligh, P. J. Bredenbeek, and W. J. Spaan. 2003. Analysis of hepatitis C virus/classical swine fever virus chimeric 5′NTRs: sequences within the hepatitis C virus IRES are required for viral RNA replication. J. Gen. Virol. 841761-1769. [DOI] [PubMed] [Google Scholar]
- 27.Rietveld, K., R. van Poelgeest, C. W. A. Pleij, J. H. Van Boom, and L. Bosch. 1982. The tRNA-like structure at the 3′ terminus of turnip yellow mosaic virus RNA. Differences and similarities with canonical tRNA. Nucleic Acids Res. 101929-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sasaki, J., and K. Taniguchi. 2003. The 5′-end sequence of the genome of Aichi virus, a picornavirus, contains an element critical for viral RNA encapsidation. J. Virol. 773542-3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simmonds, P., A. Tuplin, and D. J. Evans. 2004. Detection of genome-scale ordered RNA structure (GORS) in genomes of positive-stranded RNA viruses: implications for virus evolution and host persistence. RNA 101337-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spangberg, K., and S. Schwartz. 1999. Poly(C)-binding protein interacts with the hepatitis C virus 5′ untranslated region. J. Gen. Virol. 801371-1376. [DOI] [PubMed] [Google Scholar]
- 31.Tuplin, A., D. J. Evans, and P. Simmonds. 2004. Detailed mapping of RNA secondary structures in core and NS5B-encoding region sequences of hepatitis C virus by RNase cleavage and novel bioinformatic prediction methods. J. Gen. Virol. 853037-3047. [DOI] [PubMed] [Google Scholar]
- 32.Tuplin, A., J. Wood, D. J. Evans, A. H. Patel, and P. Simmonds. 2002. Thermodynamic and phylogenetic prediction of RNA secondary structures in the coding region of hepatitis C virus. RNA 8824-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verheije, M. H., R. C. L. Olsthoorn, M. V. Kroese, P. J. M. Rottier, and J. J. M. Meulenberg. 2002. Kissing interaction between 3′ noncoding and coding sequences is essential for porcine arterivirus RNA replication. J. Virol. 761521-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, S., and K. A. White. 2007. Riboswitching on RNA virus replication. Proc. Natl. Acad. Sci. USA 10410406-10411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.You, S., D. D. Stump, A. D. Branch, and C. M. Rice. 2004. A cis-acting replication element in the sequence encoding the NS5B RNA-dependent RNA polymerase is required for hepatitis C virus RNA replication. J. Virol. 781352-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu, L., and L. Markoff. 2005. The topology of bulges in the long stem of the flavivirus 3′ stem-loop is a major determinant of RNA replication competence. J. Virol. 792309-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 313406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.