Abstract
Alphavirus replicase protein nsP1 has multiple functions during viral RNA synthesis. It catalyzes methyltransferase and guanylyltransferase activities needed in viral mRNA capping, attaches the viral replication complex to cytoplasmic membranes, and is required for minus-strand RNA synthesis. Two temperature-sensitive (ts) mutations in Semliki Forest virus (SFV) were previously identified within nsP1: ts10 (E529D) and ts14 (D119N). Recombinant viruses containing these individual mutations reproduced the features of the original ts strains. We now find that the capping-associated enzymatic activities of recombinant nsP1, containing ts10 or ts14 lesions, were not ts. The mutant proteins and polyproteins also were membrane bound, mutant nsP1 interacted normally with the other nonstructural proteins, and there was no major defect in nonstructural polyprotein processing in the mutants, although ts14 surprisingly displayed slightly retarded processing. The two mutant viruses were specifically defective in minus-strand RNA synthesis at the restrictive temperature. Integrating data from SFV and Sindbis virus, we discuss the domain structure of nsP1 and the relative positioning of and interactions between the replicase proteins. nsP1 is suggested to contain a specific subdomain involved in minus-strand synthesis and interaction with the polymerase nsP4 and the protease nsP2.
The alphaviruses are plus-strand RNA viruses whose genome of approximately 11.5 kb encodes four nonstructural (ns) proteins, nsP1 to nsP4, in its 5′ end region. The structural protein genes, located at the 3′ end of the genome, are translated from a subgenomic mRNA generated by internal initiation on the complementary minus-strand template. In the case of Semliki Forest virus (SFV), the ns proteins are initially produced as a single large polyprotein, P1234, of 2,432 amino acid residues, which is processed to the final products in a carefully controlled sequential order (43). The transient processing intermediate P123 together with nsP4 is responsible for minus-strand synthesis, whereas completely processed nsPs in mature, stable replication complexes are active only in genomic and subgenomic plus-strand synthesis (17, 21, 37, 47).
Many temperature-sensitive (ts) strains of Sindbis virus (SIN) and SFV have been isolated and used in studies of RNA synthesis, of the processing and intracellular transport of viral proteins, and of the maturation of virus particles (for clarity, SIN ts mutants will be designated herein with the prefix SIN, whereas SFV ts mutants will be shown without a prefix). The ts strains have yielded several important insights into the different stages of viral RNA synthesis (reviewed in references 15 and 39). SIN ts6 causes a rapid cessation of all RNA synthesis when cells or cell extracts are shifted to the nonpermissive temperature, indicative of a defect in the polymerase (5, 16). Thus, nsP4 was genetically identified as the polymerase subunit a long time ago, but only very recently was the first direct demonstration of nsP4 as the alphavirus RNA-dependent RNA polymerase achieved (41). So far, both genetics and biochemistry have struggled in revealing the exact roles of nsP3, although several pieces of the puzzle are emerging. nsP3 participates in the formation of replication complexes and in the synthesis of minus-strand RNA (6, 47). nsP3 is phosphorylated in the carboxy-terminal tail region (44), and the conserved amino-terminal domain of the protein is capable of binding poly(ADP-ribose) with high affinity (8).
Early on, analysis of RNA synthesis intermediates led to the suggestion that a replicase protein regulates subgenomic RNA synthesis by directly and reversibly binding to the subgenomic promoter (31). This regulatory factor was later revealed to be nsP2, as many mutants causing defects in subgenomic RNA production are found in nsP2 and particularly in the carboxy-terminal domain of the protein (40). A curious feature of several of the nsP2 ts mutants is that when shifted to the restrictive temperature late in infection, they can reactivate minus-strand RNA synthesis in the mature replication complexes (32). This behavior has not been demonstrated for the wild-type replication complex under any circumstances, but it suggests that nsP2 is the central regulator of RNA synthesis and that the general organizations of the early and late replication complexes are similar. The N-terminal domain of nsP2 carries out the first of the viral RNA capping reactions, the RNA triphosphatase reaction (42). The N-terminal domain is also an NTPase which uses the same active site as triphosphatase (4). The NTPase activity fuels the RNA helicase activity of nsP2 (11). The carboxy-terminal domain carries out the highly regulated site-specific processing of the ns polyprotein (13, 43).
The only SIN ts mutant mapping to nsP1, SIN ts11, is specifically defective in minus-strand synthesis at the restrictive temperature (33, 46). This defect is due to a substitution, A348T, in nsP1 (12), indicating that nsP1 is specifically involved in the regulation of minus-strand production. Biochemical studies with recombinant proteins have shown that nsP1 possesses guanine-7-methyltransferase and guanylyltransferase activities needed in viral RNA capping (1, 20, 27). nsP1 has affinity to cytoplasmic membranes and especially to the inner surface of the plasma membrane; the essential membrane binding activity is mediated primarily by an amphipathic helix that can bind to negatively charged phospholipids (3, 38). It should be noted that only a fraction of the nsPs is present in the active replication complexes, which are located on the cytoplasmic surfaces of endosomes and lysosomes. In addition to this common site, each of the proteins has a specific individual localization pattern, which is independent of those of the other nsPs: nsP1 on the inner side of the plasma membrane, nsP2 in the nucleus, nsP3 in cytoplasmic aggregates, and nsP4 dispersed in the cytoplasm (15, 30, 43).
We have recently mapped those SFV ts mutants that display a significant overall phenotype in RNA synthesis to the individual ns proteins (23). Similar to what is seen for SIN, many of the mutations mapped to the largest protein, nsP2, and the first biochemical studies carried out with purified proteins containing ts mutations allowed the examination of the connections between the multiple functions of nsP2 and a direct comparison of biochemical and genetic data (4). Two individual mutations in SFV nsP1, D119N and E529D, were found to be responsible for the ts phenotypes of ts14 and ts10, respectively (23). Here we have characterized these two mutants in temperature upshift experiments in cell culture. We have also characterized the enzymatic properties, localizations, and protein-protein interactions of nsP1 proteins containing these substitutions. We found that both mutants were specifically defective in minus-strand RNA synthesis, whereas the RNA capping activities, membrane binding, and interaction with nsP3 and nsP4 were not affected by the mutations.
MATERIALS AND METHODS
Viruses, recombinant clones, and plasmids.
Wild-type SFV (generated from the infectious cDNA clone pSP6-SFV4) and the recombinant viruses containing individual ts mutations in nsP1 (SFons10 and SFots14) or nsP2 (SFots4) were derived and propagated as described previously (23). Wild-type SIN (heat-resistant strain) and mutant SIN ts11 virus have been described previously (46).
nsP1 genes with ts10 and ts14 mutations from the infectious cDNA clones pSP6-SFV4-ts10 and pSP6-SFV-ts14 (23) were PCR amplified using specific primers with NcoI and HindIII adaptors, treated with these enzymes, and ligated into the bacterial expression vector pBAT4-nsP1 (20) digested with the same enzymes, resulting in plasmids pBAT4-nsP1-ts10 and pBAT4-nsP1-ts14, respectively. For in vitro translation, the two genes were cloned under the T7 promoter in the vector pTSF1, a derivative of pGEM3, giving vectors pTSF1-ts10 and pTSF1-ts14. pBAT-nsP1-ts10 and pBAT-nsP1-ts14 were digested with NcoI and HindIII and fragments ligated into pcDNA4/TO (Invitrogen) and treated with AflII, Klenow fragment, and HindIII, resulting in mammalian expression plasmids pcDNA4/TO-nsP1-ts10 and pcDNA4/TO-nsP1-ts14. pcDNA4/TO-nsP3 for the expression of nsP3 was constructed after PCR amplification of nsP3 with suitable primers. The ts10 and ts14 mutations were transferred from the infectious clones to pTSF-P12 vector for in vitro translation of P12 (25) as StuI-SacI and EcoRV-DraIII fragments, respectively. The ts4 mutation (M781T in nsP2 [40])-containing region was PCR amplified and transferred to pTSF-P12 as a KpnI-BglII fragment. The SIN ts11 mutation (12) was generated by site-directed mutagenesis in pBAT-SINnsP1 vector for the bacterial expression of SIN nsP1.
To obtain plasmids expressing protease-inactivated P123 of SFV (designated P12CA3) from the cytomegalovirus (CMV) promoter, the luciferase-coding region from plasmid pRL-CMV (Promega) was replaced by the region encoding P12CA3, derived from a T7 promoter-based expression vector (25). The resulting clone was designated pCMV-P12CA3. The clones expressing P12CA3 with ts10 and ts14 mutations were constructed by the replacement of restriction fragments from mutation-containing infectious cDNA clones (23).
In vitro translation.
In vitro translation of P12 proteins was carried out with the T7 TNT rabbit reticulocyte lysate system (Promega) according to the manufacturer's protocol. Reaction mixtures (10 μl) containing 10 μCi of [35S]methionine (GE Healthcare) and 0.5 μg of plasmid DNA were incubated at 30°C for 30 min, after which the translation was stopped by adding cycloheximide to a final concentration of 1 mM. Samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and phosphorimaging, and quantification was performed with Tina 2.09c software. The background was subtracted and kinetic constants were calculated using the following first-order reaction equations for P12 → nsP1 plus nsP2.
For P12 decomposition, the following equation was used:
 |
where c0 is the initial value for the amount of P12 (t = 0 min) and cx is the value for the amount of P12 at time point x (t = x min).
For nsP2 formation, the following equation was used:
 |
where c∞ is the final value for the amount of nsP2 (t = ∞ min) and cx is the value for the amount of nsP2 at time point x (t = x min).
Calculation of the average constant was performed in Microsoft Excel using a linear trend line for the equation x = f(y), where x is time (min) and y is the corresponding ln.
Expression of nsP1.
To produce active recombinant nsP1, the expression plasmids were transformed into the Escherichia coli BL21(DE3) strain (Novagen). Cells were grown in the presence of ampicillin (100 μg/ml) in LB medium at 37°C until the optical density at 600 nm of the culture reached 0.6. The culture was then transferred to 15°C, and protein expression was induced by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside; final concentration, 500 μM). After 20 h of incubation, the cells were collected by centrifugation, washed in buffer containing 15 mM Tris (pH 8) and 140 mM NaCl, and resuspended in 1/30 the original volume in lysis buffer (50 mM Tris [pH 7.5], 50 mM NaCl, 10% glycerol, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride). The cell suspension was passed twice through a French press at a cell pressure of 10,000 lb/in2. The lysate was centrifuged at 15,000 × g at 4°C for 15 min, and the resulting supernatant (S15) was used in further experiments.
Methyltransferase and guanylyltransferase assays.
Guanine-7-methyltransferse activities of the recombinant proteins in S15 were assayed by use of a buffer containing 50 mM Tris (pH 6.95), 4 mM MgCl2, 2 mM DTT, 10 μM S-adenosylmethionine (AdoMet), 1 μCi of S-adenosyl-[methyl-3H]methionine, and 10 mM GTP in a 25-μl volume for 30 min. The reactions were stopped on ice by adding an equal volume of 0.2% SDS in 20 mM EDTA, pH 7.5. The labeled reaction products were isolated in small DEAE Sepharose columns prepared in Pasteur pipettes and quantitated by liquid scintillation counting (20). Covalent guanylate complex formation was assayed in a buffer containing 50 mM Tris (pH 7.5), 2 mM MgCl2, 5 mM DTT, 10 mM KCl, 100 μM AdoMet, 5 μCi of [α-32P]GTP in a 20-μl volume for 20 min at 28°C or 39°C. The reactions were stopped by adding SDS to a final concentration of 2% and boiling for 2 min. Samples were subjected to SDS-PAGE and phosphorimaging, and quantification was performed with Tina 2.09c software.
Cells, transfection, and immunofluorescence.
BHK-21 cells were cultivated as described by Salonen et al. (30). Lipofectamine 2000 (Invitrogen) was used as a transfection reagent according to the manufacturer's instructions. For immunofluorescence, BHK cells were grown on coverslips in 35-mm dishes. Cells were transfected with pcDNA4/TO or pCMV-P12CA3 constructs, followed by incubation for 16 h at 39°C. Cells were fixed with 4% paraformaldehyde, permeabilized with Triton X-100, and stained with rabbit anti-nsP1 antiserum (18), followed by treatment with secondary antibodies conjugated with Alexa568 (Molecular Probes). Labeled cells were analyzed with the Nikon Eclipse TE2000-U confocal microscopy system.
Interactions between nsP1 and nsP3.
BHK cells (106) were coelectroporated with 3 μg of each of the two plasmids (pcDNA4/TO-nsP1 and pcDNA4/TO-nsP3) and incubated for 24 h at 28°C or 39°C. Cells were lysed in 1 ml of buffer containing 50 mM Tris (pH 8), 150 mM NaCl, 1% NP-40, 5 mM EDTA, and protease inhibitor cocktail (Roche). Clarified lysate was collected after 15 min of centrifugation at 15,000 × g. Half of the lysate was incubated with anti-nsP3 antiserum (guinea pig [18]), followed by incubation with protein A-Sepharose beads equilibrated in phosphate-buffered saline (PBS) containing 0.2% bovine serum albumin. The beads were washed four times with lysis buffer, the samples were denatured by SDS and boiled for 5 min, and one-fifth of the lysate was applied to SDS-PAGE gels. Western blotting of the resulting proteins was performed using rabbit anti-nsP1 antiserum.
Protein labeling and immunoprecipitation.
BHK cells in 35-mm dishes (approximately 106 cells/well) were infected at 28°C with 100 PFU of each virus (23) per cell, incubated for 5.5 h at 28°C, and then transferred to 39°C. Cells were washed once with prewarmed PBS, supplied with methionine-free medium, incubated for 30 min at 39°C, and pulsed with 50 μCi/plate of [35S]methionine for 15 min (10 min in experiments with short chase times). Chase periods were done with medium containing excesses of cold methionine and cysteine (final concentrations, 10 mM and 5 mM, respectively). For immunoprecipitation under denaturing conditions, cells were lysed in 1% SDS followed by boiling. One-tenth of each lysate was used for immunoprecipitation with polyclonal rabbit antisera as described previously (18). For native immunoprecipitation, cells were collected by being scraped in PBS and were centrifuged at 250 × g for 10 min; were resuspended in buffer containing 10 mM NaCl, 10 mM Tris (pH 8.0), and protein inhibitor cocktail (Roche); were swollen for 15 min on ice; and were homogenized with a Dounce homogenizer. Postnuclear supernatant was collected by centrifugation at 500 × g for 10 min. All of the clarified lysate was used for immunoprecipitation with polyclonal rabbit antisera as described previously (30). The immunoprecipitates were analyzed by SDS-PAGE and phosphorimaging.
RNA labeling and quantitation of minus strands.
BHK cells in 35-mm dishes (for overall RNA synthesis) or in 60-mm dishes (for minus-strand synthesis) were infected with 25 PFU/cell and then incubated in complete medium containing 20 mM HEPES, pH 7.4, at 30°C until the time of upshift to 40°C or harvesting. At the times indicated for each experiment, cells on 35-mm dishes were labeled for a 1-h period with 1 ml [3H]uridine (50 μCi/ml [3H]uridine and 20 μg/ml actinomycin D), those on 60-mm dishes were so labeled with 200 μCi/ml, and then cells were harvested. The medium was removed, and 5% LiDS in LET buffer (0.1 M LiCl, 1 mM EDTA, 10 mM Tris, pH 8.0) containing 200 μg/ml proteinase K was added. The cells were harvested approximately at 2.5 × 106 cells/ml (0.7 ml per 35-mm dish). The lysate was passed four times through a 27-gauge needle to shear DNA. Duplicate or triplicate samples equivalent to 50,000 cells were precipitated with trichloroacetic acid to measure total incorporation.
To analyze minus-strand synthesis, the lysates were extracted two or three times with phenol at pH 4.3 and then two times with chloroform-isoamyl alcohol; the aqueous phase was adjusted to 0.2 M LiCl or 0.3 M sodium acetate, and ethanol (2 volumes) was used to precipitate the nucleic acids at −20°C overnight. The precipitate was collected by centrifugation, washed with 70% ethanol, dried, and resuspended in 0.2 ml of STE (0.1 M NaCl, 1 mM EDTA, and 10 mM Tris, pH 7.4). The samples were subjected to digestion with limited RNase A and chromatography on CF-11 columns as described previously (32). Briefly, the NaCl concentration was adjusted to 0.3 M, and 25 μl of 1 μg/ml RNase A was added for a 15-min incubation at room temperature. Immediately thereafter, the solution was adjusted back to STE by the addition of Tris-EDTA and 35% ethanol. The samples were applied to washed CF-11 columns, tubes were rinsed with 1 ml of 35% ethanol-STE, and the wash was applied to the column, which was then allowed to drain. Then columns were washed with 15 ml of 35% ethanol-STE, followed by 15 ml of 15% ethanol-STE. The replicative forms (RFs) were eluted with 9 ml/column of STE directly into siliconized tubes. Then, 0.5 ml of 4 M LiCl, 100 μg of purified yeast tRNA at 20 mg/ml, and 20 ml of absolute ethanol were added per tube. After thorough mixing, precipitation was carried out at −20°C overnight, followed by centrifugation at 10,000 rpm for 1 h at 4°C. The pellets were dried and resuspended in 200 μl of nuclease-free water. Minus-strand RNA was measured by a nuclease protection assay, which determines the amount of heat-denatured labeled RF RNA that was protected from RNase digestion (5 μg RNase A per ml) by hybridization to an approximately 100-fold excess of unlabeled plus-strand RNA.
RESULTS
It was shown previously that the ts10 and ts14 strains have mutations in nsP1 (amino acid changes E529D and D119N, respectively), which cause a ts phenotype in SFV RNA replication. The ts10 strain was demonstrated to have an RNA± phenotype, with 6% RNA synthesis remaining at 39°C, whereas the ts14 strain showed a clear RNA-negative phenotype, with very low RNA synthesis at the restrictive temperature. A shift to 39°C after 5 h of incubation at 28°C did not cause any reduction in 26S or 42S RNA synthesis, indicating that once synthesized at the permissive temperature, the mature replicase complex is capable of functioning normally at the restrictive temperature (23). Since several functions are known for nsP1, characterization of the specific defect(s) in the mutants was expected to illuminate the roles of nsP1 in RNA replication.
Polyprotein processing.
As one of the mutations (ts10) mapped very close to the nsP1/nsP2 border (position −9 relative to the cleavage site), we considered the possibility of a cleavage defect at this site. The 1/2 site is cleaved relatively slowly to permit the existence of the minus-strand-synthesizing replication complex, and the cleavage takes place in cis (43). We therefore examined the cleavage efficiency of P12 translated in vitro. The wild-type P12 polyprotein was translated for 30 min and the synthesis was stopped by cycloheximide at this point, at which reasonable amounts of P12 have accumulated but most of the protein remains uncleaved. The time course of cleavage was monitored by gel electrophoresis and autoradiography (Fig. 1A). First-order reaction kinetics were measured both for the decomposition of P12 and for the formation of nsP2 from quantification of the [35S]methionine-labeled protein bands. Cleavage experiments at different temperatures showed that there is a very inefficient cleavage of P12 at 39°C (Fig. 1B). This is also the case when purified recombinant protease is used in cleavage studies, showing that the protease is more sensitive to temperature in vitro than it is in infected cells (4). Therefore, we used 22°C as a permissive temperature and 32°C and 36°C as restrictive temperatures and compared the processing efficiencies of mutant polyproteins with the wild-type P12 processing efficiency in vitro. A well-characterized ts protease activity mutant located within the protease domain of nsP2, ts4 (4, 40), was used as a control. As expected, P12-ts4 had greatly reduced kinetic constant values at 32°C and 36°C compared with the wild-type P12 (Fig. 1C). In contrast, for P12-ts10 and P12-ts14, the first-order kinetic constant did not differ significantly from the wild-type P12 data (Fig. 1C).
FIG. 1.
Processing of wild-type and mutant polyproteins. (A) Polyprotein P12 was synthesized in a cell-free translation system in the presence of [35S]methionine at 30°C. Aliquots were taken at the indicated times and analyzed by SDS-PAGE and autoradiography. In a parallel experiment, P12 synthesis was stopped after 30 min with 1 mM cycloheximide, and the processed samples were collected at the indicated time points and analyzed similarly. Arrows indicate the positions of nsPs and their precursor P12 polyproteins. Note that both nsP1 and P12 give rise to a double band due to aberrant initiation also at the second in-frame methionine. (B) Processing kinetics of wild-type P12 in vitro at different temperatures (temp). The protein amounts used in the calculations were measured during the cycloheximide treatment as done for panel A. The y axis represents the first-order kinetic constant in min−1. (C) Comparison of the first-order kinetic constants for the processing of wild-type and mutant P12 proteins at different temperatures. (D) Polyprotein processing in cells infected with SFowt, SFons10, and SFots14. Proteins were labeled with [35S]methionine for 10 min at 39°C. The sample from one parallel plate was extracted immediately after the pulse, while the samples from the other plates were chased with an excess of unlabeled methionine for 5 min or 15 min before extraction. The samples were immunoprecipitated with the indicated combinations of antibodies against the ns proteins (either anti-nsP1 plus anti-nsP2 [1+2] or anti-nsP3 plus anti-nsP4 [3+4]) and analyzed by SDS-PAGE and fluorography to visualize all the nsPs. The virus strain, sample (pulse, 5-min chase, or 15-min chase), and antibody combinations used for immunoprecipitation are indicated at the top, and the positions of proteins and precursors are marked on the right.
Polyprotein processing in virus-infected cells was studied in pulse-chase experiments with [35S]methionine at 39°C as described in Materials and Methods. Recombinant virus containing the ts4 mutation was again included as a known protease-defective control. The wild-type virus, SFowt, showed almost complete processing of the labeled polyprotein after a 60-min chase, whereas in SFots4-infected cells, P1234, P123, P12, and P34 polyprotein precursors accumulated, and out of the individual proteins only nsP4 could be detected even after the 60-min chase (data not shown). Using shorter chase times revealed that the cleavage of the polyprotein in SFons10-infected cells occurred exactly as during SFowt infection, whereas in SFots14-infected cells the processing of P1234 was slightly slower, and the precursors were more strongly visible than for SFowt (Fig. 1D). Nevertheless, all of the individual ns proteins could also be clearly detected already after the pulse. Thus, the processing defect of SFots14 is very mild compared to what was seen for the nsP2 mutants previously examined (4), and although the defect was reproducibly observed and was verified in two completely independently derived virus clones, its significance is not easy to estimate.
Enzymatic activities of nsP1.
To study the enzymatic activities of nsP1 involved in RNA capping, the genes coding for nsP1-ts10 and nsP1-ts14 were cloned into the pBAT4 expression vector and expressed in E. coli as recombinant proteins. E. coli transformed with the empty pBAT4 plasmid was used as a control in all the experiments, and exactly the same procedures were performed with the four samples, designated as mock, nsP1-wt, nsP1-ts10, and nsP1-ts14. Western blotting with anti-nsP1 antibodies was used in these experiments to verify the equal expression levels of the proteins (data not shown).
Guanine-7-methyltransferase activity was measured in bacterial lysates clarified by a short centrifugation (2) with a 30-min reaction time. The temperature dependence of the wild-type nsP1 methyltransferase was not as marked as that of the protease (Fig. 2A). Although optimal activity was found around 35°C, the permissive and restrictive temperatures of virus growth, 28°C and 39°C, respectively, could easily be used in the assays. All three proteins, namely, nsP1-ts10, nsP1-ts14, and nsP1-wt, showed equal and strong methyltransferase activities both at 28°C and 39°C (Fig. 2B). Under these conditions, the reactions continue linearly for at least 2 hours (20), indicating that the enzyme is not present at saturating levels.
FIG. 2.
Enzymatic activities of nsP1 at different temperatures. Wild-type and mutant proteins produced in bacteria were assayed as explained in Materials and Methods. (A) Guanine-7-methyltransferase activity of nsP1-wt. Incorporation of [3H]methyl (in cpm) from AdoMet to 7-methyl-guanosine at different temperatures (temp) is shown; error bars indicates standard deviation. (B) Guanine-7-methyltransferase activities of nsP1-wt, nsP1-ts10, and nsP1-ts14 at 28°C and 39°C. The reactions were done in triplicate; error bars indicate standard deviation. In mock cells, no nsP1 was expressed. (C) Formation of nsP1-guanylate complex. S15 fractions of E. coli lysates containing the indicated SFV or SIN proteins were incubated with [α-32P]GTP at 28°C and 39°C. Following the reaction, proteins were separated by SDS-PAGE and visualized by phosphorimaging. nsP1 expressed in E. coli always yields a major product, which represents the full-length protein, and a minor, shorter product, which arises due most likely to premature translation termination. Both products are enzymatically active (2). Abbreviations: wt, nsP1-wt; ts10, nsP1-ts10; ts14, nsP1-ts14.
The other RNA capping reaction catalyzed by nsP1 is guanylyltransferase, which can be measured by covalent complex formation between nsP1 and 7-methyl-GMP (1, 2). Again in this reaction, nsP1-ts10, nsP1-ts14, and nsP1-wt showed equally strong activities, which were somewhat higher at 28°C than at 39°C for all three proteins (Fig. 2C). The activity of the bacterially produced SIN nsP1 mutant ts11 was also compared with that of the SIN wild-type control under the same conditions. Both SIN proteins showed equal covalent complex formation activities at both temperatures (Fig. 2C). These results strongly suggest that defects in the RNA capping reactions are not involved in the ts phenotypes of these alphavirus nsP1 mutants.
Plus-strand RNA synthesis.
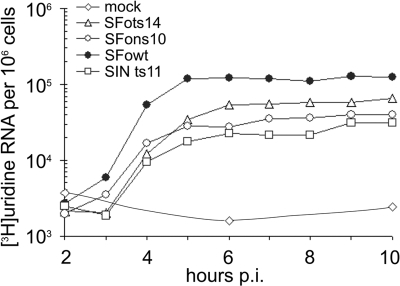
We next turned our attention directly to RNA synthesis in virus-infected cells. We first studied RNA synthesis at the permissive temperature (30°C was used in these experiments) to compare the mutant and wild-type viruses. Plus-strand synthesis (measured as cpm incorporated to viral RNA per hour of labeling) increased early and then reached a constant, maximum rate, in a pattern that is typical for alphavirus infection. The SFV nsP1 mutants resembled SIN ts11, another nsP1 mutant, in showing a maximum rate that was ∼30 to 50% of the wild-type SFV rate (Fig. 3). There were no ts defects in genome or 26S mRNA synthesis either at 30°C or after the shift to 40°C (Table 1), indicating that the mutants are not defective in 26S RNA synthesis. At the higher temperature, genomic RNA synthesis was increased relative to subgenomic RNA synthesis for the wild type and for the mutants, as reported previously for both SFV and SIN (15).
FIG. 3.
Plus-strand synthesis by wild-type SFV and the nsP1 ts mutants. Cells infected with the indicated viruses were labeled with [3H]uridine in the presence of actinomycin D for 1 h. The labeling was terminated at the indicated time points by cell lysis and RNA extraction. Incorporated label was measured by scintillation counting. The total RNA synthesis is taken to represent plus-strand RNA synthesis, since >90% of total viral RNA synthesis at all time points is plus-strand RNA synthesis.
TABLE 1.
Ratio of the synthesis of genome and 26S RNA at 30°C and after shift to 40°C
| Virus | Molar ratio (genome/26S RNA synthesis) at:
|
40°C/30°C ratio | |
|---|---|---|---|
| 30°C | shift to 40°C | ||
| SFowt | 0.33 | 1.3 | 3.9 |
| SFons10 | 0.21 | 0.76 | 3.6 |
| SFots14 | 0.38 | 1.41 | 3.7 |
| SIN ts11 | 0.43 | 0.9 | 2.1 |
Minus-strand RNA synthesis.
Minus-strand synthesis at 30°C was efficient early in infection: ∼40% of [3H]uridine-labeled RF RNA was in minus strands each hour, indicating that ∼80% of the minus strands present were newly made during the previous hour. Both wild-type SFV (Fig. 4D) and the nsP1 ts mutants (Fig. 4A to C) showed a cessation of minus-strand synthesis by 6 h postinfection (p.i.) (Fig. 4A to C). The shift of cells to 40°C at 3 h p.i. or 4 h p.i. resulted in a rapid cessation of minus-strand synthesis by SFV ts10 and ts14 that was similar to the rapid shutoff by SIN ts11 (Fig. 4A to C). Wild-type SFV did not show a rapid shutoff when shifted to 40°C, but instead the rate of its normal minus-strand cessation was slightly increased at the higher temperature (Fig. 4D). This finding argues that the nsP1 mutants SFV ts10 and ts14 are ts specifically for minus-strand synthesis.
FIG. 4.
Minus-strand synthesis by the nsP1 ts mutants and wild-type SFV. Cells were labeled with [3H]uridine as described in Materials and Methods, and the RFs were isolated. The percentage of labeled minus-strand RNA was determined by an RNase protection assay (see Materials and Methods). The percentage of RF RNA cpm in minus strands was determined for SFons10 (A), SFots14 (B), SIN ts11 (C), and SFowt (D); incubations at 30°C (○) and then shift after 3 h to 40°C (▪), shift after 4 h to 40°C (▴), and shift after 3 h to 40°C in medium containing cycloheximide (•) were done.
Shift to 40°C at 3 h p.i. and incubation in a medium containing cycloheximide (100 μg/ml) led to a rapid cessation of minus-strand synthesis by SFV ts10 and ts14 and SIN ts11 (Fig. 4A to C). The rate of loss was similar to that seen for temperature shift alone, although for ts10 the temperature-mediated loss was slightly slower and less complete, which may reflect the RNA± nature (23) of the original mutant. This indicates that shift to 40°C prevented any (or with ts10 most) further production of nsP1 proteins functional in minus-strand RNA synthesis, since it was equivalent to conditions that prevented the actual synthesis of nascent nsP1 proteins. These results further support a conclusion that the SFV nsP1 mutants have ts defects in functions required for minus-strand synthesis that would include promoter recognition or initiation.
Protein localization and protein-protein interactions.
We were also interested in examining whether the ts mutations would affect nsP1 localization or protein-protein interactions in a temperature-dependent manner. First, the localization of nsP1 was studied by use of BHK cells transiently transfected with derivatives of the pcDNA4/TO vector containing the CMV promoter. Both the wild type and the two mutant proteins showed predominant association with the plasma membrane (Fig. 5A, left), as described before for nsP1 (19, 38). Second, since polyproteins mediate the membrane association and localization of the replication complexes during infection, we studied the effect of the nsP1 mutations on the localization of P123, which was rendered uncleavable through a protease active-site mutation. The wild-type P12CA3 and the two mutants showed indistinguishable stainings of intracellular vesicles (Fig. 5A, right), some of which have been shown to be of endolysosomal origin, reflecting the actual localization of replication complexes in infected cells (30). The uncleaved nature of the polyprotein was verified by Western blotting (data not shown). No temperature-dependent (28°C versus 39°C) difference in the localization of nsP1 or polyproteins for any of the derivatives used in this assay was seen, and the experiments were therefore carried out mainly at 39°C (Fig. 5A). Third, we studied the interactions between nsP1 and nsP3 proteins by coexpressing these proteins in BHK cells at 28°C or 39°C. It was found that both nsP1-ts10 and nsP1-ts14 showed a clear interaction with nsP3, similar to what was seen for nsP1-wt (Fig. 5B), indicating that this interaction, which has been shown to be crucial for correct replicase complex formation (30), is not temperature dependent for ts10 and ts14.
FIG. 5.
Interactions of wild-type and mutant nsP1 with cellular and viral components. (A) Localization of nsP1 (left) or uncleavable polyprotein P12CA3 (right) derivatives in transfected BHK cells at 39°C. The cells were stained with antibodies against nsP1. (B) Interaction of nsP1 with nsP3 at different temperatures. BHK cells were coelectroporated with the plasmids expressing nsP1 and nsP3 at 28°C or 39°C. The cell lysates were incubated with guinea pig anti-nsP3 antiserum, followed by incubation with protein A-Sepharose beads and immunoprecipitation. The precipitated samples were denatured and analyzed by SDS-PAGE followed by Western blotting using rabbit anti-nsP1 antiserum. The analyzed mutant and wild-type proteins are indicated at the top, the position of nsP1 is marked on the left, and the temperatures used in the experiments are shown at the bottom. (C) Interaction of nsP1 with viral proteins in infected cells at 39°C. Proteins were labeled with [35S]methionine and chased with an excess of cold methionine for 60 min. The samples were immunoprecipitated (IP) with anti-nsP1 antibodies under only a native condition (left panel) or with a combination of anti-nsP1 and anti-nsP4 antibodies under denaturing conditions (right panel) and analyzed by SDS-PAGE and fluorography. The virus strains are indicated at the top, and the positions of nsPs are marked on the left.
Finally, we studied the interactions of the ns proteins in infected cells by pulse-chase labeling. The cells were labeled for 15 min starting 30 min after the shift to the restrictive temperature of 39°C in order to examine only those protein-protein interactions which take place under restrictive conditions. After a 60-min chase, which allows complete polyprotein processing, cell extracts were prepared and subjected to immunoprecipitation. Under the native condition, antibodies against nsP1 precipitated nsP1 as well as some of the other nsPs, most notably significant quantities of nsP4 (Fig. 5C). For control purposes, immunoprecipitation under denaturing conditions was performed with a combination of the nsP1 and nsP4 antisera, which revealed both proteins as expected (Fig. 5C). No differences were noticed between cells infected with mutant virus and those with wild-type virus, suggesting that nsP1 interacts normally with nsP4 and that ns protein complexes are formed under conditions, which are restrictive for ts10 and ts14 minus-strand RNA synthesis.
DISCUSSION
We have here analyzed in detail the effects of two ts mutations mapping to SFV nsP1 by use of both biochemical assays performed in vitro and RNA synthesis and protein interaction studies with mammalian cells. The RNA capping-related enzymatic functions of nsP1, the guanine-7-methyltransferase and guanylyltransferase activities, were not affected by the ts mutations in nsP1, as assayed at different temperatures (Fig. 2). The same result was obtained with the otherwise previously characterized SIN nsP1 mutant ts11. It was also shown that the mutant SFV proteins interacted with membranes and localized properly. These known direct functions of nsP1 are linked with each other, since the membrane binding of nsP1 both determines its subcellular localization (38) and is a prerequisite for its enzymatic functions (3). Furthermore, it appeared that the mutant nsP1s formed proper interactions with the other nsPs, as detected by immunoprecipitation (Fig. 5). In infected cells, the major interaction partner of nsP1 seems to be the core polymerase nsP4 (Fig. 5C) (18). Upon pairwise expression of the nsPs, an interaction of nsP1 and nsP3 is also observed (Fig. 5B) (30). The only major defect found for ts10 and ts14 was an inability to synthesize minus strands, a defect which appeared very rapidly upon shift to the nonpermissive temperature (Fig. 4).
It is interesting to note that none of the three known alphavirus ts mutations in nsP1 cause a defect in the RNA capping activities of the protein. This contrasts with the situation of nsP2, where ts mutations often specifically affect the protease and NTPase functions (4). Since the capping activities of nsP1 are essential for RNA replication (45), they are in principle susceptible targets for ts mutations. Although multiple ts mutants have been mapped to each of the alphavirus nsPs (12, 15, 23), the mutational screens are very far from saturating. Still, it seems to be the case that ts mutations are more frequently located in nsP2 than in any of the other nsPs and that all the known functions of nsP2 are affected, often in various combinations, by the ts mutations. The molecular mechanisms leading to a ts phenotype are poorly understood, but ts mutations can occur both in the core of the protein and in surface residues involved in protein-protein interactions, e.g. (28). Since nsP2 centrally regulates the cleavages of nsPs and also the different steps of RNA synthesis, the tight packing of functions and interactions in nsP2 may lead to a higher number of ts mutations. Alternatively, it is possible that some protein domains are for structural reasons intrinsically more prone to ts mutations than others.
The three ts mutations of nsP1 cause very similar defects in minus-strand synthesis (Fig. 4), yet they are located in different parts of the primary sequence of the protein (Fig. 6). The overall structure of nsP1 remains unknown and due to the membrane-bound nature of the protein it is a difficult problem to approach, but some clues have emerged from numerous biochemical and genetic studies. It was suggested some time ago that certain parts of nsP1 fold in a pattern resembling the then-recently-determined general AdoMet-dependent methyltransferase structures (2). The conserved general structure of methyltransferases is an example of a Rossman fold, a central β sheet of seven strands flanked by α helices. It consists of two halves, in which the “first” half (usually but not always N terminal in the primary sequence) contains the AdoMet binding site and the “second” half is mostly involved in binding the methyl acceptor substrate (GTP in the case of nsP1) (24). The positioning of the first half within alphavirus nsP1 is strongly supported by secondary structure prediction and by mutational and cross-linking studies which pinpointed two conserved acidic residues (D64 and D90), located at the ends of two β-strands, essential for AdoMet binding (Fig. 6) (2). Mutations in SIN altering the affinity of nsP1 toward AdoMet also map to the same area (residues 87 and 88) (26, 34). The location of the GTP binding site is less well defined, but the mutant selection using drugs which lower cellular GTP levels suggests that residues S23 and V302 are involved (35). During the last few years, the first structures of enzymes specifically involved in the methylation of the guanine 7 position in mRNA cap structures have become available (7, 9) In contrast to many other AdoMet-dependent methyltransferases, the guanine-7-methyltrasferases contain very prominent insertions of other subdomains in the loops of the core Rossman fold. The fact that several deletion studies of nsP1 have failed to define enzymatically active subdomains of the protein (2, 3, 19, 45) may support a complex domain structure for nsP1 in which structurally continuous domains are assembled by the coming together of distantly located primary sequence elements.
FIG. 6.
Schematic representation of nsP1. The amino acids and their positions are given according to the SFV sequence, except when indicated otherwise with the prefix SIN. Locations of conserved and functionally important residues are shown. Sequence alignments for the regions surrounding the ts10 and ts14 mutations are provided for representatives of the principal groups of alphaviruses. VEE indicates Venezuelan equine encephalitis virus and SDV indicates sleeping disease virus (salmonid alphavirus).
So far, specific mutations affecting the binding of enzymatic substrates have been found to be located within the first ∼310 amino acids of nsP1. This area then is likely to contain the entire methyltransferase-like fold, which uniquely in the alphavirus-like group also contains an associated guanylyltransferase activity dependent on methylation (1). We propose that the ts14 mutation is located within an insertion which is not part of the core methyltransferase fold. Sequence alignment of alphavirus nsP1 with the related plant virus sequences reveals that the plant viruses lack a short stretch of sequence surrounding the ts14 site (2), supporting a structural insertion. The SIN ts11 region seems to be located after the core methyltransferase area. We suggest that the ts11 region forms a specific subdomain of nsP1 which is necessary for minus-strand RNA synthesis and not directly involved in RNA capping functions. The ts14 region could be involved in the same function with the ts11 region and could even be part of the same subdomain. One could hypothesize that the recognition of the already formed cap structure, which is present only in plus-strand RNAs, by these regions of nsP1 would be important for the initiation of minus-strand synthesis. We favor an alternative hypothesis that although the SFV nsP1 ts mutants appear capable of forming strong interactions with the other nsPs which are detected by immunoprecipitation, the mutations disturb crucial functional or regulatory aspects of these protein-protein interactions.
The most probable direct interaction partner for nsP1 during minus-strand RNA is core polymerase nsP4. Other genetic studies strongly support such a functional interaction between nsP1 and nsP4. The proper N-terminal residue of nsP4 is required for RNA synthesis, but certain mutations of this residue can be suppressed by mutations in nsP1. The suppressors characterized are located at residue 349 next to the SIN ts11 mutation at residue 348 (Fig. 6) (36). Other mutations nearby, probably in the same subdomain of nsP1, i.e., N374H and N374I in SIN, suppress the minus-strand synthesis defect of a polymerase altered at residue 183 in nsP4 (10).
Another functional interaction partner for nsP1 could be nsP2. Although the nsP1-nsP2 interaction appears to be biochemically weak, as it is not readily observed by immunoprecipitation, this does not preclude an important contact between the proteins. Interestingly, the ts14 mutation caused a slight polyprotein-processing defect in infected cells (Fig. 1D), which could be symptomatic of a problem in interacting with the protease. According to the generally accepted model of alphavirus RNA synthesis, in which the polyprotein P123 (with nsP4) is mainly responsible for minus-strand synthesis (21, 37, 47), slower processing of P123 should if anything stabilize the early polymerase and increase minus-strand synthesis. Yet, ts14 displays a strong and immediate reduction in minus-strand synthesis, indicating that altered processing is not the primary cause of this defect. The ts10 mutation is located at position −9 relative to the nsP1/nsP2 cleavage site. The mutation site is outside of the short stretch of residues specifically recognized by the protease, which extends upstream approximately to position −5 (22), and accordingly it did not cause a defect in processing (Fig. 1D). In contrast, mutation I538T within the recognition sequence (position −3 relative to the 1/2 site) in a SIN strain resulted in faster processing of the nsPs and accelerated the growth of the virus, due to the earlier expression of structural protein mRNA from the subgenomic promoter (14). Despite its more distal location from the cleavage site, the ts10 site would still be placed near nsP2 and could be involved in another protein-protein interaction with nsP2 which does not affect proteolysis.
It is interesting to note that the N terminus of nsP4, crucially important for RNA synthesis as described above, also represents a protease cleavage site. The relative positions of the alphavirus nsPs within the replication complex are not known, but it is known that both cleavages 3/4 and 1/2 can occur in cis within the same polyprotein, whereas the 2/3 cleavage must take place in trans (13, 43). During minus-strand synthesis, the 3/4 cleavage has already occurred and the 1/2 cleavage is yet to take place. One possibility is that the overall configuration of the polyprotein stays the same during all these stages: the bulk of nsP4 remains in the same general area where it was during the 3/4 cleavage and the body of nsP1 is poised near the position which enables the subsequent 1/2 cleavage. After each proteolytic cleavage, the actual substrate amino acid stretch naturally needs to be displaced from the protease active site in order to permit subsequent cleavages, but this could be accomplished by relatively minor movement of the flexible cleavage regions themselves. Assuming that the overall relative configuration of the nsPs is maintained (e.g., by nsP1-nsP4 and other protein-protein interactions), then nsP1 (an N-terminal cleavage product) and nsP4 (a C-terminal cleavage product) would flank the nsP2 protease on two sides, which would enable a three-way interaction of the proteins.
In conclusion, the ts mutations so far discovered in alphavirus nsP1 do not affect the essential enzymatic or protein localization functions of nsP1 but instead appear to perturb a delicate functional interaction within the replication complex which is required for minus-strand RNA synthesis. In the future, structural studies will help in interpreting the effects of some of the ts mutations, as is already the case for the protease domain of nsP2 (29). Structural studies could also define the strong interaction sites between the nsPs. However, the replication complex during different functional stages of RNA synthesis must be capable of making and breaking multiple weak protein-protein and protein-RNA interactions. Genetic studies will continue to be essential in understanding the many functions of the alphavirus nsPs and in pinpointing the protein regions involved in those functions.
Acknowledgments
This work was supported by European Union 5th Framework Programme project SFvectors, by grant 067575 from The Wellcome Trust, and by Academy of Finland grant 211121.
Footnotes
Published ahead of print on 2 July 2008.
REFERENCES
- 1.Ahola, T., and L. Kääriäinen. 1995. Reaction in alphavirus mRNA capping: formation of a covalent complex of nonstructural protein nsP1 with 7-methyl-GMP. Proc. Natl. Acad. Sci. USA 92507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahola, T., P. Laakkonen, H. Vihinen, and L. Kääriäinen. 1997. Critical residues of Semliki Forest virus RNA capping enzyme involved in methyltransferase and guanylyltransferase-like activities. J. Virol. 71392-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahola, T., A. Lampio, P. Auvinen, and L. Kääriäinen. 1999. Semliki Forest virus mRNA capping enzyme requires association with anionic membrane phospholipids for activity. EMBO J. 183164-3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balistreri, G., J. Caldentey, L. Kääriäinen, and T. Ahola. 2007. Enzymatic defects of the nsP2 proteins of Semliki Forest virus temperature-sensitive mutants. J. Virol. 812849-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barton, D. J., S. G. Sawicki, and D. L. Sawicki. 1988. Demonstration in vitro of temperature-sensitive elongation of RNA in Sindbis virus mutant ts6. J. Virol. 623597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dé, I., C. Fata-Hartley, S. G. Sawicki, and D. L. Sawicki. 2003. Functional analysis of nsP3 phosphoprotein mutants of Sindbis virus. J. Virol. 7713106-13116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De la Peña, M., O. J. P. Kyrieleis, and S. Cusack. 2007. Structural insights into the mechanism and evolution of the vaccinia virus mRNA cap N7 methyl-transferase. EMBO J. 264913-4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Egloff, M. P., H. Malet, A. Putics, M. Heinonen, H. Dutartre, A. Frangeul, A. Gruez, V. Campanacci, C. Cambillau, J. Ziebuhr, T. Ahola, and B. Canard. 2006. Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J. Virol. 808493-8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fabrega, C., S. Hausmann, V. Shen, S. Shuman, and C. D. Lima. 2004. Structure and mechanism of mRNA cap (guanine-N7) methyltransferase. Mol. Cell 1377-89. [DOI] [PubMed] [Google Scholar]
- 10.Fata, C. L., S. G. Sawicki, and D. L. Sawicki. 2002. Modification of Asn374 of nsP1 suppresses a Sindbis virus nsP4 minus-strand polymerase mutant. J. Virol. 768641-8649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gomez de Cedrón, M., N. Ehsani, M. L. Mikkola, J. A. García, and L. Kääriäinen. 1999. RNA helicase activity of Semliki Forest virus replicase protein NSP2. FEBS Lett. 44819-22. [DOI] [PubMed] [Google Scholar]
- 12.Hahn, Y. S., E. G. Strauss, and J. H. Strauss. 1989. Mapping of RNA- temperature-sensitive mutants of Sindbis virus: assignment of complementation groups A, B, and G to nonstructural proteins. J. Virol. 633142-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hardy, W. R., and J. H. Strauss. 1989. Processing the nonstructural polyproteins of Sindbis virus: nonstructural proteinase is in the C-terminal half of nsP2 and functions both in cis and in trans. J. Virol. 634653-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heise, M. T., J. W. Laura, D. A. Simpson, C. L. Kristen, A. Bernard, R. B. Meeker, and R. E. Johnston. 2003. An attenuating mutation in nsP1 of the Sindbis-group virus S.A.AR86 accelerates nonstructural protein processing and up-regulates viral 26S RNA synthesis. J. Virol. 771149-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kääriäinen, L., and T. Ahola. 2002. Functions of alphavirus nonstructural proteins in RNA replication. Prog. Nucleic Acid Res. Mol. Biol. 71187-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keränen, S., and L. Kääriäinen. 1979. Functional defects of RNA-negative temperature-sensitive mutants of Sindbis and Semliki Forest viruses. J. Virol. 3219-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, K. H., T. Rumenapf, E. G. Strauss, and J. H. Strauss. 2004. Regulation of Semliki Forest virus RNA replication: a model for the control of alphavirus pathogenesis in invertebrate hosts. Virology 323153-163. [DOI] [PubMed] [Google Scholar]
- 18.Kujala, P., A. Ikäheimonen, N. Ehsani, H. Vihinen, P. Auvinen, and L. Kääriäinen. 2001. Biogenesis of the Semliki Forest virus RNA replication complex. J. Virol. 753873-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laakkonen, P., T. Ahola, and L. Kääriäinen. 1996. The effects of palmitoylation on membrane association of Semliki Forest virus RNA capping enzyme. J. Biol. Chem. 27128567-28571. [DOI] [PubMed] [Google Scholar]
- 20.Laakkonen, P., M. Hyvönen, J. Peränen, and L. Kääriäinen. 1994. Expression of Semliki Forest virus nsP1-specific methyltransferase in insect cells and in Escherichia coli. J. Virol. 687418-7425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemm, J. A., T. Rümenapf, E. G. Strauss, J. H. Strauss, and C. M. Rice. 1994. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J. 132925-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lulla, A., V. Lulla, K. Tints, T. Ahola, and A. Merits. 2006. Molecular determinants of substrate specificity for Semliki Forest virus nonstructural protease. J. Virol. 805413-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lulla, V., A. Merits, P. Sarin, L. Kääriäinen, S. Keränen, and T. Ahola. 2006. Identification of mutations causing temperature-sensitive defects in Semliki Forest virus RNA synthesis. J. Virol. 803108-3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin, J. L., and F. M. McMillan. 2002. SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Curr. Opin. Struct. Biol. 12783-793. [DOI] [PubMed] [Google Scholar]
- 25.Merits, A., L. Vasiljeva, T. Ahola, L. Kääriäinen, and P. Auvinen. 2001. Proteolytic processing of Semliki Forest virus-specific non-structural polyprotein by nsP2 protease. J. Gen. Virol. 82765-773. [DOI] [PubMed] [Google Scholar]
- 26.Mi, S., R. Durbin, H. V. Huang, C. M. Rice, and V. Stollar. 1989. Association of the Sindbis virus RNA methyltransferase activity with the nonstructural protein nsP1. Virology 170385-391. [DOI] [PubMed] [Google Scholar]
- 27.Mi, S., and V. Stollar. 1991. Expression of Sindbis virus nsP1 and methyltransferase activity in Escherichia coli. Virology 184423-427. [DOI] [PubMed] [Google Scholar]
- 28.Mondal, K., A. G. Dastidar, G. Singh, S. Madhusudhanan, S. L. Gande, K. VijayRaghavan, and R. Varadarajan. 2007. Design and isolation of temperature-sensitive mutants of Gal4 in yeast and Drosophila. J. Mol. Biol. 370939-950. [DOI] [PubMed] [Google Scholar]
- 29.Russo, A. T., M. A. White, and S. J. Watowich. 2006. The crystal structure of the Venezuelan equine encephalitis alphavirus nsP2 protease. Structure 141449-1458. [DOI] [PubMed] [Google Scholar]
- 30.Salonen, A., L. Vasiljeva, A. Merits, J. Magden, E. Jokitalo, and L. Kääriäinen. 2003. Properly folded nonstructural polyprotein directs the Semliki Forest virus replication complex to the endosomal compartment. J. Virol. 771691-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sawicki, D. L., L. Kääriäinen, C. Lambek, and P. J. Gomatos. 1978. Mechanism for control of synthesis of Semliki Forest virus 26S and 42S RNA. J. Virol. 2519-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sawicki, D. L., and S. G. Sawicki. 1993. A second nonstructural protein functions in the regulation of alphavirus negative-strand RNA synthesis. J. Virol. 673605-3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sawicki, D. L., S. G. Sawicki, S. Keränen, and L. Kääriäinen. 1981. Specific Sindbis virus-coded function for minus-strand RNA synthesis. J. Virol. 39348-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scheidel, L. M., R. K. Durbin, and V. Stollar. 1989. SVLM21, a Sindbis virus mutant resistant to methionine deprivation, encodes an altered methyltransferase. Virology 173408-414. [DOI] [PubMed] [Google Scholar]
- 35.Scheidel, L. M., and V. Stollar. 1991. Mutations that confer resistance to mycophenolic acid and ribavirin on Sindbis virus map to the nonstructural protein nsP1. Virology 181490-499. [DOI] [PubMed] [Google Scholar]
- 36.Shirako, Y., E. G. Strauss, and J. H. Strauss. 2000. Suppressor mutations that allow Sindbis virus RNA polymerase to function with nonaromatic amino acids at the N-terminus: evidence for interaction between nsP1 and nsP4 in minus-strand RNA synthesis. Virology 276148-160. [DOI] [PubMed] [Google Scholar]
- 37.Shirako, Y., and J. H. Strauss. 1994. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J. Virol. 681874-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spuul, P., A. Salonen, A. Merits, E. Jokitalo, L. Kääriäinen, and T. Ahola. 2007. Role of the amphipathic membrane binding peptide of Semliki Forest virus replicase protein nsP1 in membrane association and virus replication. J. Virol. 81872-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suopanki, J., D. L. Sawicki, S. G. Sawicki, and L. Kääriäinen. 1998. Regulation of alphavirus 26S mRNA transcription by replicase component nsP2. J. Gen. Virol. 79309-319. [DOI] [PubMed] [Google Scholar]
- 41.Thal, M. A., B. R. Wasik, J. Posto, and R. W. Hardy. 2007. Template requirements for recognition and copying by Sindbis virus RNA-dependent RNA polymerase. Virology 358221-232. [DOI] [PubMed] [Google Scholar]
- 42.Vasiljeva, L., A. Merits, P. Auvinen, and L. Kääriäinen. 2000. Identification of a novel function of the alphavirus capping apparatus: RNA 5′-triphosphatase activity of nsP2. J. Biol. Chem. 27517281-17287. [DOI] [PubMed] [Google Scholar]
- 43.Vasiljeva, L., A. Merits, A. Golubtsov, V. Sizemskaja, L. Kääriäinen, and T. Ahola. 2003. Regulation of the sequential processing of Semliki Forest virus nonstructural polyprotein. J. Biol. Chem. 27841636-41645. [DOI] [PubMed] [Google Scholar]
- 44.Vihinen, H., T. Ahola, M. Tuittila, A. Merits, and L. Kääriäinen. 2001. Elimination of phosphorylation sites of Semliki Forest virus replicase protein nsP3. J. Biol. Chem. 2765745-5752. [DOI] [PubMed] [Google Scholar]
- 45.Wang, H.-L., J. O'Rear, and V. Stollar. 1996. Mutagenesis of the Sindbis virus nsP1 protein: effects on methyltransferase activity and viral infectivity. Virology 217527-531. [DOI] [PubMed] [Google Scholar]
- 46.Wang, Y.-F., S. G. Sawicki, and D. L. Sawicki. 1991. Sindbis virus nsP1 functions in negative-strand RNA synthesis. J. Virol. 65985-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang, Y.-F., S. G. Sawicki, and D. L. Sawicki. 1994. Alphavirus nsP3 functions to form replication complexes transcribing negative-strand RNA. J. Virol. 686466-6475. [DOI] [PMC free article] [PubMed] [Google Scholar]