Abstract
The human genome encodes over 500 microRNAs (miRNAs), small RNAs (19 to 26 nucleotides [nt]) that regulate the expressions of diverse cellular genes. Many cellular processes are altered through a variety of mechanisms by human cytomegalovirus (HCMV) infection. We asked whether HCMV infection leads to changes in the expression of cellular miRNAs and whether HCMV-regulated miRNAs are important for HCMV replication. Levels of most miRNAs did not change markedly during infection, but some were positively or negatively regulated. Patterns of miRNA expression were linked to the time course of infection. Some similarly reregulated miRNAs share identical or similar seed sequences, suggesting coordinated regulation of miRNA species that have shared targets. miRNAs miR-100 and miR-101 were chosen for further analyses based on their reproducible changes in expression after infection and on the basis of having predicted targets in the 3′ untranslated regions (3′-UTR) of genes encoding components of the mammalian target of rapamycin (mTOR) pathway, which is important during HCMV infection. Reporter genes that contain the 3′-UTR of mTOR (predicted targets for miR-100 and miR-101) or raptor (a component of the mTOR pathway; predicted site for miR-100) were constructed. Mimics of miR-100 and miR-101 inhibited expression from the mTOR construct, while only miR-100 inhibited the raptor construct. Together, miR-100 and miR-101 reduced mTOR protein levels. While the miR-100 and miR-101 mimics individually modestly inhibited production of infectious progeny, much greater inhibition was achieved with a combination of both (33-fold). Our key finding is that HCMV selectively manipulates the expression of some cellular miRNAs to help its own replication.
MicroRNAs (miRNAs) are small (∼21-nucleotide [nt]) RNA species that are expressed from specialized genes and have important roles in the regulation of cellular gene expression, including the regulation of development, the differentiation of hematopoietic stem cells, apoptosis, and the development of cancer (reviewed in references 16, 25, and 26). miRNA-mediated gene regulation is related to cellular defenses that are mediated via very similar mechanisms (small interfering RNA) but target exogenous mRNAs, such as those expressed by viruses. The human genome contains perhaps 500 distinct miRNA genes (1, 33). miRNAs are initially expressed as 5′-capped and polyadenylated RNA polymerase II transcripts (4, 31). They are expressed either as individually regulated genes or as clusters of miRNAs that are expressed and then processed from a single primary transcript that might contain several miRNAs (22, 29, 49). After cytoplasmic processing to the ∼21-nt single-stranded mature form by the enzyme Dicer, miRNAs associate with the RNA-induced silencing complex. miRNA-mediated RNA interference is manifest as reduced levels of translation from the targeted mRNAs. This translational silencing comes in two forms: (i) by inhibiting protein synthesis after binding via incomplete base pairing to the 3′ untranslated regions (3′-UTR) of target mRNAs, and (ii) by binding to mRNAs with perfect complementarity, which leads to cleavage of the targeted mRNA. One important consequence of miRNA-mediated inhibition of gene expression via imperfect base pairing is that individual miRNAs can potentially regulate many cellular targets. Further, individual genes can be targeted by multiple miRNAs.
There are connections between viruses and the miRNA world. Simian virus 40, human immunodeficiency virus type 1 (HIV-1), herpes simplex virus type 1, Marek's disease virus, murine cytomegalovirus, human cytomegalovirus (HCMV), Epstein-Barr virus, and human herpesvirus 8 encode sets of miRNAs (3, 12, 18, 40, 42, 44, 48, 53). HCMV encodes at least 12 miRNAs that are expressed as immediate early or early viral genes in infected cells; their abundance increases for at least 72 h after infection (12, 18, 41). Similarly, a herpes simplex virus type 1 viral miRNA encoded upstream of the latency-associated transcript also persisted at high levels until late in infection (9). Thus, the miRNA machinery remains operational in cells infected by these viruses. While functions have yet not been ascribed to most of the virally encoded miRNAs, HCMV-miR-UL112 inhibits NK cell killing by reducing major histocompatibility complex class I chain-related molecule B protein levels (46), the simian virus 40 miRNA plays a role in preventing the immune recognition of infected cells by reducing the production of some viral proteins (47), several human herpesvirus 8-encoded viral miRNAs target cellular genes that may be relevant to pathogenesis (45), and an HIV-1-encoded viral miRNA suppresses nef gene expression (40).
In addition to encoding miRNAs, viruses can interact with host miRNAs or affect their regulation. Nuclear export of miRNA precursors is inhibited by the adenovirus VA1 noncoding RNA (35). Accumulation of primate foamy virus type 1 is inhibited by a cellular miRNA (miR-32), and the virus encodes a protein that suppresses miRNA silencing in mammalian and plant cells (30). In addition, a cellular miRNA (miR-122) that targets the 5′ end of the hepatitis C virus genome facilitates viral replication (24). Transfection of HeLa cells with HIV-1 leads to a substantial alteration in the expression of cellular miRNAs, with many of them being downregulated (54). Moreover, HIV-1 infection downregulates cellular miRNAs that repress viral growth (50).
HCMV exerts diverse and profound effects on the regulation of host cell metabolism, including altering levels of cellular transcripts, perturbing the cell cycle, and inhibiting infection-triggered apoptosis (reviewed in reference 39), and the machinery for miRNA biosynthesis and activity is functional in HCMV-infected cells. Thus, we asked whether HCMV infection results in altered expression of cellular miRNA species. We found that levels of most miRNA species do not change dramatically during infection, but several species were markedly up- or downregulated, indicating that HCMV infection leads to specific changes in the expression of some cellular miRNAs. Further, we identified cellular targets for two of the virally regulated miRNAs and found that synthetic mimics of these miRNAs can inhibit viral replication.
MATERIALS AND METHODS
Cells and virus.
MRC-5, HeLa, and 293T cells were obtained from the American Type Culture Collection (Manassas, VA) and maintained in Earle's modified Eagle's medium containing 10% fetal bovine serum (HyClone, Logan, UT), 2 mM l-glutamine, 0.1 mM nonessential amino acids, and 1 mM sodium pyruvate. T-BACwt (a gift from Hua Zhu) is a derivative of the HCMV Towne strain that has several nonessential genes (US1 through half of US12) replaced with a gene encoding a green fluorescence protein and was used in all experiments (38). Virus stocks were prepared in MRC-5 cells infected with T-BACwt at a multiplicity of infection (MOI) of 0.01, fresh culture medium was added when the cells showed >90% cytopathic effects, and the cells were harvested after 3 to 4 days. Harvested cells were resuspended in a thrice-autoclaved mixture of 50% fat-free milk and 50% culture medium and then frozen in aliquots at −80°C. Viral titers were determined by a plaque-forming assay done in triplicate in 48-well flat-bottom plates (Corning Incorporated, Corning, NY).
RNA preparation.
MRC-5 cells were infected at an MOI of 2 in 100-mm cell culture dishes (Becton Dickinson, Franklin Lakes, NJ). Total RNA was extracted at 6, 24, 48, and 96, or 120 h postinfection (hpi) with Trizol reagent (Invitrogen, Carlsbad, CA). Briefly, the culture medium was removed and the cells were lysed with 6 ml of Trizol reagent and then frozen at −80°C. The RNA phase was partitioned with 6 ml of chloroform, precipitated with 3 ml of isopropyl alcohol, washed, and resuspended in DNase/RNase-free sterile water and then stored at −80°C. RNA quality was verified by electrophoresis in agarose gels.
miRNA microarray.
miRNA microarray analysis was performed as described previously (34), using an updated version of the chip (human and mouse miRNA 11K version 2 chip). Arrays included 40-mer oligomers with sequences corresponding to 250 human miRNAs and their precursors known at the time this work was initiated. The chips also included a series of well-tested control probes with sequences corresponding to human tRNA and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) that served as positive controls and numerous random sequences that served as negative controls. Each probe was printed in quadruplicate on activated Amersham CodeLink slides (Amersham, Piscataway, NJ). Five micrograms of total RNA was labeled with biotin by reverse transcription (RT), and the labeled cDNA was hybridized to the printed chips. Bound sequences were detected with a streptavidin-Alexa647 conjugate. After processing, data were collected using an Axon 4000B scanner and the Genepix Pro 6.0 software package (Molecular Devices, Sunnyvale, CA).
miRNA data analysis.
Microarray data were analyzed with BRB-ArrayTools (version 3.4_Beta_1a, National Cancer Institute, Rockville, MD). In the initial data filtering, spots were excluded if the minimum fluorescence intensity was ≤10 or if there was insufficient agreement among the quadruplicate spots. The mean fluorescence intensities (MFI) of the quadruplicate spots were log2 transformed and then normalized using the median-centering array procedure. miRNAs were excluded if one of the expression values was less than 20% or had a 1.5-fold change in either direction from the averaged median value (34). For each miRNA for which data of sufficient quality were available, array intensities from each time point after HCMV infection were compared with those of mock-infected MRC-5 cells.
qRT-PCR.
Short RNAs (≤200 nt) were separated from total RNA preparations (described above) by use of the mirVanaTM miRNA isolation kit (Ambion, Austin, TX) according to the manufacturer's directions, beginning with 50 μg of total RNA that had been treated with 10 units of DNase at 37°C for 30 min. Levels of miRNA expression at different times after HCMV infection were determined by quantitative RT-PCR (qRT-PCR) from 10-ng aliquots of short RNA by using the mirVanaTM qRT-PCR miRNA detection kit (Ambion, Austin, TX) and primers for specific miRNAs (Applied Biosystems, Foster City, CA).
miRNA target verification.
Fragments containing the 3′-UTR of mTOR and raptor that contain predicted targets of miR-100 and/or miR-101 were cloned from MRC-5 cells by use of the following primers, each of which contains a NotI site (italicized): 5′-GCGGCCGCAGATGTGCCCATCACGTT-3′ and 5′-GCGGCCGCTGATGTCATTTATTGGCACA-3′ (mammalian target of rapamycin [mTOR], NM_004958), and 5′-GCGGCCGCCCTGCTACTCGCTTTTGTC-3′ and 5′-GCGGCCGCTTTCCCGAATTTCCAGTGTC-3′ (raptor, NM_020761). The mTOR primers amplify a 927-bp fragment located 11 to 937 bp downstream of the stop codon, and the raptor primers amplify a 406-bp fragment located 143 to 557 bp downstream of the stop codon. Cloned sequences were confirmed and the fragments were transferred to the NotI site in the 3′ multiple cloning sequence of pHygEGFP (BD Biosciences, Palo Alto, CA), which is located downstream of the stop codon of a gene encoding a fusion protein of hygromycin and enhanced green fluorescent protein (EGFP) to produce pHygEGFP-mTOR and pHygEGFP-raptor. HeLa cells were cotransfected with reporter genes and mimics of miR-100, miR-101, or a negative-control miRNA mimic (Dharmacon, Lafayette, CO) by use of Lipofectamine 2000 (Invitrogen, Carlsbad, California) as described previously (10). At 15 to 24 h after transfection, digital pictures of the cells were taken with a Leica (DM IRB) UV microscope system with a Q-Imaging camera at 100× magnification, and EGFP expression levels were determined using Image-Pro Plus software (version 6.1).
HCMV growth in the presence of miR-100 and miR-101 mimics.
MRC-5 cells were grown in 48-well plates (Becton Dickinson, Franklin Lakes, NJ) to 40 to 50% confluence and transfected using Lipofectamine 2000 with miR-100, miR-101, or a negative-control miRNA mimic. Two days after transfection, the cells were serum starved for 48 h and then infected with T-BACwt (MOI of 2). Four days after infection, virus titers in culture supernatants were determined by standard serial dilution on MRC-5 cells.
Immunoblot analyses.
Cells from six-well plates were collected and lysed on ice in lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% Triton X-100 [vol/vol], 10% glycerol [vol/vol], and 1% protease inhibitor cocktail). After clarification by centrifugation, 100 μg of protein lysate was separated in a gradient (4 to 20%) polyacrylamide-sodium dodecyl sulfate gel and then transferred to a membrane (Immobilon-P; Millipore Corp., Billerica, MA). The membrane was preincubated in blocking buffer containing 5% fat-free milk in Tris-buffered saline containing 0.1% Tween 20 (pH 7.6). The membrane was probed with a rabbit anti-mTOR antibody (2972; Cell Signaling Technology, Inc., Beverly, MA) overnight at 4°C and then with a horseradish peroxidase-conjugated anti-rabbit secondary antibody for 1 h at room temperature with chemiluminescent detection (Millipore Corp.). Bands were scanned and then quantified using Image-Pro Plus software (version 6.1).
Statistical analysis.
The two-tailed Student's t test was used to analyze the effects of miRNA mimics on reporter assays and viral replication. The samples were treated as having equal variances (43, 52).
RESULTS
Based on the abundant observations that HCMV employs a variety of mechanisms to regulate cellular processes, we hypothesized that (i) cellular miRNAs play a significant role in CMV biology and (ii) HCMV regulates cellular miRNA populations, collectively or individually.
miRNA microarray analysis. (i) Experimental design.
To gain an overview of the impact of HCMV infection on host cellular miRNA expression, MRC-5 cells that had been confluent for 3 days were infected at an MOI of 2 with HCMV strain T-BACwt. In this way, we excluded the effects of cell replication and ensured the synchronous infection of the metabolically synchronized cells (2, 55). The HCMV strain we used has an integrated GFP gene so that infection efficiency can be conveniently monitored, as infected cells show green fluorescence from 48 hpi. Two independent experiments were done in which total RNA samples were extracted from mock-infected cells and cells that had been infected for 6, 24, and 48 h plus a 96-h time point in experiment 1 and a 120-h time point in experiment 2, thus enabling analysis of early and late stages of infection. The rationale for this design was that in previous global analyses of host cellular gene expression, the number of affected cellular genes increased with time after HCMV infection, and groups of genes associated with different cellular activities, such as innate immune responses and cell cycle regulation, were affected at different times after infection (2, 21).
The miRNA microarray chips used in this study are well established and have been used in many studies of cancer and cellular differentiation (5, 7, 8, 13, 17, 20, 34, 51, 52). The array results passed the standard blank, negative, and positive quality controls.
(ii) Similar results in experiments 1 and 2.
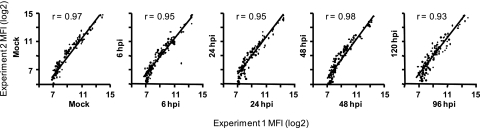
Results of two independent experiments (experiment 1 and experiment 2) at each time point were compared to evaluate their reproducibility. For the scatterplots shown in Fig. 1, the analysis was restricted to probes that met the quality control criteria; because of the variability in the data for spots with very low fluorescence signals, signals of ≤64 fluorescence units are not represented in the scatterplots. These species are considered to be negative for expression in our experiments; probes for 153 of the 250 miRNAs were negative at all time points in both experiments. There was generally good agreement between the two experiments. The correlation coefficients (r) were >0.9 at all time points. The similarity between results obtained at the 96-hpi and 120-hpi time points (r = 0.93) is consistent with the cells being in the late phase of the replication cycle. Because of the high agreement between the two experiments, further analyses were based on experiment 2.
FIG. 1.
Comparison of results from two independent miRNA microarray experiments. RNA was prepared from MRC-5 fibroblasts mock infected or infected with HCMV T-BACwt (MOI of 2) at 6, 24, and 48 hpi plus 96 hpi (experiment 1) or 120 hpi (experiment 2). These RNAs were used as probes on miRNA microarrays, as described in Materials and Methods. Data that passed the quality control criteria were compared with the BRB-ArrayTools software.
(iii) miRNA expression after HCMV infection.
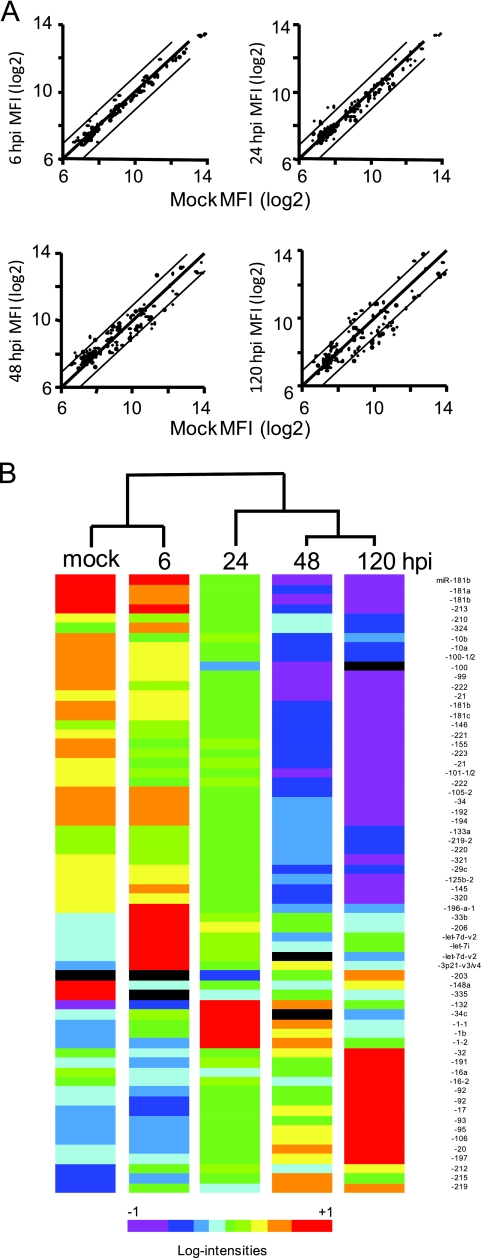
The scatterplots in Fig. 2A are comparisons of the expressions of individual miRNAs at each time point with mock-infected cells. Two general observations are apparent from these plots: (i) there is no global unidirectional change in miRNA expression, and (ii) the expression levels of individual miRNAs may be unaffected by infection or may be positively or negatively regulated after infection. Table 1 shows the changes relative to levels for mock-infected cells for a set of miRNAs for which high-quality data (as defined in Materials and Methods) were available from every time point. In the comparison with mock-infected cells at the 6-hpi time point, the miRNA expression profiles are very similar; only one cellular miRNA was upregulated, but none were downregulated by more than twofold. At 24 hpi, three cellular miRNA species were upregulated by more than twofold and none were downregulated (one of the miRNAs was targeted by probes for two precursor forms; thus, it shows up in the plot as two points). Especially manifest at 48 hpi and 120 hpi is a progressive change in the expression of individual miRNAs relative to what was seen for mock-infected cells, such that the differences in expression levels of most miRNAs are readily visible. In total, 2 and 8 miRNA species were upregulated by more than twofold at 48 and 120 hpi, respectively, and 22 and 24 miRNAs were downregulated by more than twofold at 48 and 120 hpi, respectively. In many instances, changes of 20% to 30% were statistically significant at a P value of <0.01, and all of the changes of at least twofold were significant at that or higher levels (Table 1). qRT-PCR was used to confirm the microarray data for selected miRNAs; as shown in Table 2, the two methods were in good agreement.
FIG. 2.
Impact of HCMV infection on the expression profile of cellular miRNA. (A) Comparison of miRNA expression levels at different times after HCMV infection with levels in mock-infected cells. (B) Cluster analyses and heat map showing the expression levels of probes representing miRNAs with high-quality data available for every time point. The heat map is based on center-normalized MFI for each miRNA probe. Black bars denote probes for which signal intensities were below the level of quantitation.
TABLE 1.
Human miRNA expression patterns after HCMV infectiona
| miRNA IDb | Mock MFI (± SD)c | Ratio of infected cell MFI to mock MFI atd:
|
|||
|---|---|---|---|---|---|
| 6 hpi | 24 hpi | 48 hpi | 120 hpi | ||
| miRNAs with ≥2-fold decrease | |||||
| -10p | 3,022 ± 249 | 0.8** | 0.7*** | 0.4** | 0.6* |
| -21 | 4,286 ± 274 | 1.1 | 0.9* | 0.5*** | 0.4** |
| -29p | 2,366 ± 128 | 1.2 | 0.8* | 0.5*** | 0.5** |
| -34p | 1,112 ± 37 | 1 | 0.7* | 0.5*** | 0.4*** |
| -99p | 7,208 ± 809 | 1 | 0.8** | 0.4*** | 0.5** |
| -100 | 9,842 ± 632 | 1 | 0.7 | 0.4** | 0.1*** |
| -101p | 2,558 ± 171 | 1 | 0.8* | 0.4*** | 0.4*** |
| -105 | 1,412 ± 135 | 1.1 | 0.7* | 0.4** | 0.4*** |
| -125p | 9,903 ± 173 | 1.2* | 0.8 | 0.6*** | 0.5** |
| -133 | 1,127 ± 73 | 1.1 | 0.9* | 0.6** | 0.5** |
| -145p | 2,442 ± 184 | 1.2 | 0.8* | 0.5** | 0.5** |
| -146p | 873 ± 56 | 1.1 | 0.9 | 0.5** | 0.5** |
| -155p | 4,035 ± 252 | 0.9** | 0.8** | 0.4** | 0.4** |
| -181p | 2,445 ± 114 | 1.1 | 0.7** | 0.4** | 0.3*** |
| -181 | 5,051 ± 425 | 1 | 0.6** | 0.3** | 0.3** |
| -181p | 1,943 ± 49 | 1.1 | 0.8 | 0.5*** | 0.4** |
| -192 | 1,844 ± 219 | 1 | 0.7*** | 0.5** | 0.4* |
| -194 | 963 ± 46 | 1 | 0.7** | 0.5** | 0.4** |
| -213p | 4,350 ± 250 | 1.1 | 0.6*** | 0.4*** | 0.3** |
| -221p | 24,951 ± 982 | 1 | 0.9 | 0.5*** | 0.5** |
| -222p | 31,697 ± 589 | 0.9 | 0.7* | 0.4*** | 0.4*** |
| -223p | 2,847 ± 51 | 0.9 | 0.8* | 0.5*** | 0.4*** |
| -320 | 1,970 ± 145 | 1.2 | 0.8** | 0.5** | 0.4** |
| -321 | 3,449 ± 259 | 1.1 | 0.9* | 0.6** | 0.4** |
| -335 | 1,170 ± 70 | 1 | 0.6** | 0.5** | 0.5** |
| miRNAs with no changes of >2-fold (up or down) | |||||
| -let-7p | 944 ± 53 | 1.9* | 1.1 | 0.8* | 1 |
| -16 | 856 ± 69 | 1.1 | 1 | 0.9 | 1.4 |
| -32p | 927 ± 54 | 1.1 | 1 | 1.1 | 1.4*** |
| -34 | 311 ± 15 | 1.2 | 1.1 | 0.9* | 0.8* |
| -93p | 955 ± 43 | 1 | 1.2* | 1.1 | 1.9** |
| -148 | 331 ± 110 | 1 | 0.9 | 0.8 | 1.1 |
| -191p | 2,137 ± 119 | 1 | 1.1 | 1 | 1.7*** |
| -196 | 1,481 ± 272 | 1.5* | 0.8 | 0.7 | 0.7 |
| -206p | 1,146 ± 91 | 1.8* | 1.3** | 1 | 1 |
| -210p | 819 ± 48 | 1.1 | 0.9 | 0.7* | 0.6** |
| -212p | 1,537 ± 48 | 1.9* | 1.7 | 1.3 | 1.8 |
| -215p | 271 ± 30 | 1.4*** | 1.4* | 1.6* | 1.5* |
| -220p | 1,896 ± 113 | 1.2* | 1 | 0.6** | 0.6** |
| -324 | 596 ± 58 | 1.3 | 0.9* | 0.7* | 0.6** |
| miRNAs with ≥2-fold increase | |||||
| -1p | 302 ± 11 | 1.5* | 2.1* | 1.4* | 1.1 |
| -1 | 227 ± 5 | 1.3 | 2.7*** | 1.6 | 1.2 |
| -17p | 1,008 ± 58 | 1 | 1.2* | 1.3 | 2.3*** |
| -20p | 792 ± 78 | 1 | 1.1 | 1.2* | 2.4** |
| -33p | 453 ± 24 | 2.2* | 1.2 | 0.9 | 1 |
| -92p | 13,751 ± 616 | 0.9* | 1.1 | 1.1 | 2** |
| -95p | 1,745 ± 136 | 1 | 1.2 | 1.2 | 2** |
| -106p | 807 ± 56 | 1.1 | 1.2* | 1.2 | 2.1** |
| -132p | 404 ± 38 | 1.4* | 2.7** | 2** | 1.7* |
| -197p | 1,287 ± 167 | 1.2 | 1.1 | 1.1 | 2* |
| -219 | 5,463 ± 689 | 1.8** | 1.5* | 1.9* | 2* |
| -203p | 196 ± 15 | 1.1 | 1.7* | 2.3** | 3.5*** |
Array data for human miRNAs with high-quality data (as defined in Materials and Methods) at every time point. Data from a representative probe are shown if similar results were obtained with multiple probes for the same miRNA species.
miRNA names are for human miRNAs as listed in the Sanger miRNA registry. The suffix “p” denotes species for which the array probe targeted the precursor form of the miRNA.
MFI and SD of mock-infected cells are shown to give baselines for interpreting the change levels.
At each time point, the MFI for each probe was compared with the corresponding value from mock-infected cells with the two-tailed Student t test, considering the samples as paired. *, P ≤ 0.01; **, P ≤ 0.001; ***, P ≤ 0.0001.
TABLE 2.
Comparison of microarray and qRT-PCR results
| miRNA and testa | Fold change atb:
|
|||
|---|---|---|---|---|
| 6 hpi | 24 hpi | 48 hpi | 120 hpi | |
| miR-223 | ||||
| Microarray | 0 | 0 | 3 | 3 |
| qRT-PCR | 0 | 0 | 2 | 3 |
| miR-101 | ||||
| Microarray | 0 | 0 | 3 | 4 |
| qRT-PCR | 0 | 0 | 8 | 8 |
| miR-181 | ||||
| Microarray | 0 | 0 | 4 | 7 |
| qRT-PCR | 0 | 2 | 2 | 4 |
| miR-100 | ||||
| Microarray | 0 | 0 | 3 | 34 |
| qRT-PCR | 0 | 0 | 4 | 16 |
Short RNAs (≤200 nt) isolated from the same RNA preparations used in microarray analysis were reverse transcribed and then analyzed by qRT-PCR.
The change is the ratio of threshold cycles (CT), CTmock to CTinfected, for each time point. Results are shown from a subset of downregulated miRNAs.
To compare the expression patterns of individual miRNAs over the time course of the experiment, we did a cluster analysis based on the MFI for 64 probes for which high-quality data were available for every time point (Fig. 2B). From the dendrogram and the associated heat maps, it is apparent that changes in miRNA expression levels link to the infection time course. The heat maps reveal coherent temporal linkages from time point to time point. Thus, the mock and 6-hpi time points are closely related to each other, as are the 48- and 120-h time points, with the 24-h time point being related to the 48- and 120-h time points. From the heat maps, persistent and transient effects on the expression levels of individual miRNAs are apparent. The expression levels of some species progressively changed from high to low over the experimental time course; conversely, the expression levels of some species changed from low to high. The expression levels of some species transiently increased, especially at the 6- and 24-h time points.
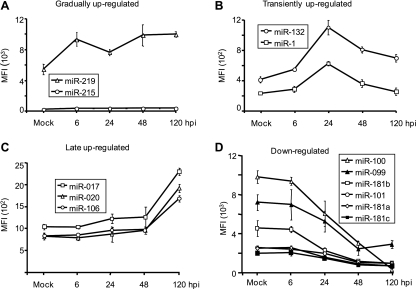
Specific examples of the types of changes mentioned above are shown in Fig. 3. Cellular miRNAs were upregulated in three general patterns: persistent increases beginning from 6 hpi (Fig. 3A), transient increases in expression that peaked at 24 hpi (Fig. 3B), and late increases (Fig. 3C). Some miRNAs were downregulated, with their levels decreasing until 48 hpi with little change thereafter (Fig. 3D).
FIG. 3.
Patterns of changes in cellular miRNA expression after HCMV infection. Cellular miRNAs were upregulated in three general patterns: gradual upregulation over the time course of infection (A), transient upregulation that peaked at 24 hpi (B), and late increases (C). There was only one pattern for downregulated cellular miRNAs (D).
Interestingly, some miRNAs with the same seed sequence (residues 2 to 8) had similar patterns of changes in expression after HCMV infection. This included miR-99 and miR-100, which have identical seed sequences and have only a single-nucleotide difference elsewhere. In addition, of the nine miRNAs upregulated at 120 hpi, four (miR-17, -20, -93, and -106) have identical seed sequences and only 2- to 3-nt sequence differences elsewhere (11). These miRNAs are encoded on different chromosomes. Thus, some groups of unique miRNA species that have similar targets are coordinately regulated after infection.
(iv) Selection of miRNAs for functional analyses.
After the initial data quality filtering described above, we applied a series of additional filters to identify the miRNA species whose expression levels were most profoundly and reproducibly changed after HCMV infection. This involved identifying miRNAs for which there was at least a twofold change in the expression level (up or down), MFI of ≥1,024 fluorescence units at least one time point, and concordance between experiments 1 and 2. Species that passed these filters are miR-21, -99, -100, -101, -155, -181, -213, -222, -223, and -320 (downregulated) and miR-17, -20, -106, and -219 (upregulated). Interestingly, many of these species have been identified by others as playing possible roles in cell differentiation or oncogenesis (reviewed in references 16, 25, and 26), and some are known to target cellular regulators that may be important during infection.
miR-100 and miR-101 interact with components of the mTOR pathway.
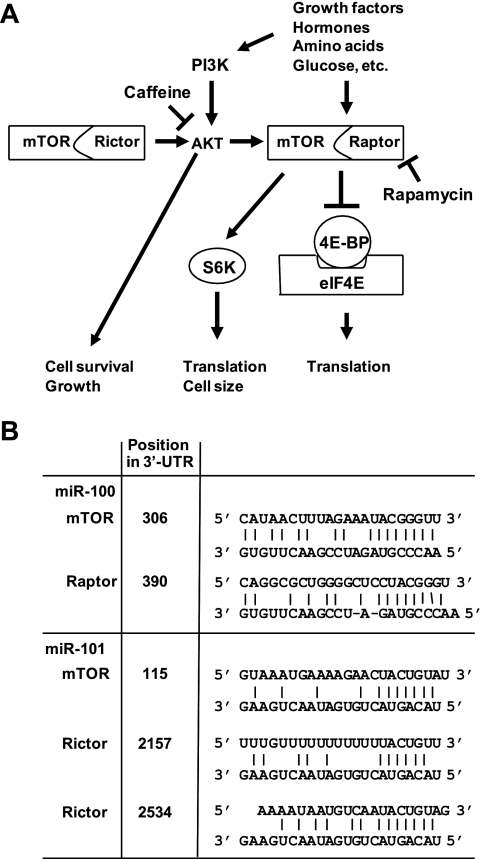
For the miRNA species that had the most consistent and significant changes in expression following HCMV infection, we examined lists of targets predicted by MIRANDA and TargetScan (19, 32). miR-100 and miR-101 have predicted targets on the mTOR pathway, which is important in regulating the translation of capped mRNAs, cell size, cell growth, cell cycle, cell survival, and cytoskeletal organization (37) (Fig. 4A). Specifically, miR-100 and miR-101 each have a predicted target in the mTOR 3′-UTR. In addition, miR-100 has a predicted target in the 3′-UTR of raptor, which is a partner of mTOR, and miR-101 has two predicted targets in the 3′-UTR of another mTOR partner, rictor (Fig. 4B).
FIG. 4.
Predicted targets of miR-100 and miR-101 in the 3′-UTR of members of the mTOR pathway. (A) Schematic of the mTOR signaling pathway. mTOR functions by partnering with either raptor or rictor and controls mRNA translation or cell growth and survival, respectively. Rapamycin inhibits the function of the mTOR/raptor complex, while the function of Akt, the only known target of the mTOR/rictor complex, can be inhibited by caffeine. PI3K, phosphatidylinositol 3-kinase. (B) Predicted targets. For each duplex, the upper sequence is from the predicted target and the lower sequence is the mature form of the indicated miRNA. For mTOR at least, the predicted target sequences are highly conserved among diverse mammalian species.
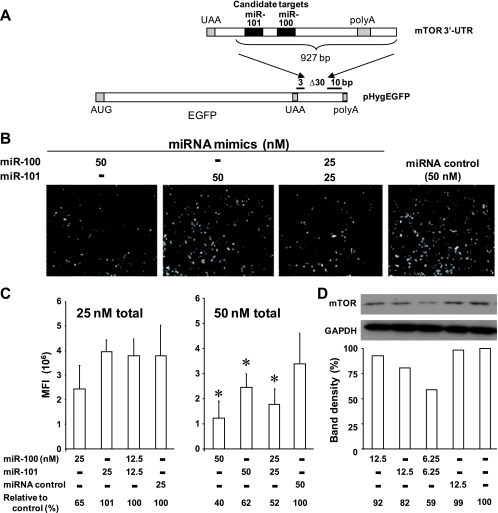
To determine whether miR-100 and miR-101 interact with components of the mTOR pathway, we constructed reporter genes containing segments of the 3′-UTR from the mTOR and raptor genes downstream of the EGFP reading frame (pHygEGFP-mTOR and pHygEGFP-raptor) (Fig. 5A and 6A). Cells transfected with the reporter genes express EGFP, and the effects of miRNAs on EGFP expression can be assessed by comparing fluorescence intensities in the presence or absence of synthetic miRNA mimics or a negative-control miRNA mimic (Fig. 5B). The miRNA mimics are able to be processed by Dicer and produce stable and active silencing of their cognate targets.
FIG. 5.
miR-100 and miR-101 interact with the mTOR 3′-UTR and downregulate mTOR expression. (A) A segment of the 3′-UTR of the mTOR mRNA (927 nt) containing predicted targets for miR-100 and miR-101 was inserted downstream of the EGFP reading frame of pHygEGFP. (B and C) Inhibition of pHygEGFP-mTOR reporter gene activity in HeLa cells that were cotransfected with the reporter gene and mimics of miR-100 and/or miR-101. (B) Fluorescence micrographs. (C) Graphs of fluorescence intensities from the same experiment. Values are the means ± standard deviations (SD) of triplicate determinations. (D) Reduced mTOR protein levels after transfection of cells with mimics of miR-100 and/or miR-101. Chemiluminescence images were scanned, band densities were normalized relative to GAPDH levels, and the data were plotted as percentages relative to GAPDH levels in mock-transfected cells. *, P value of <0.05 in comparison with the control.
FIG. 6.
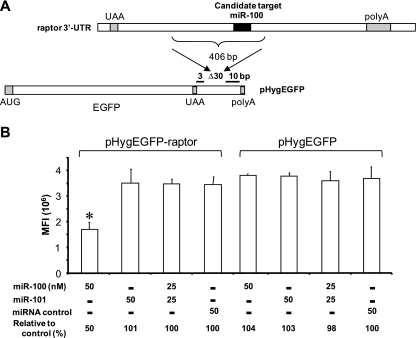
miR-100 interacts with the raptor 3′-UTR. (A) Part (406 nt) of the raptor 3′-UTR containing a predicted miR-100 target was inserted downstream of the EGFP reading frame of pHygEGFP. (B) Expression levels of pHygEGFP-raptor and the parental pHygEGFP reporter in HeLa cells cotransfected with mimics of miR-100, miR-101, or the negative-control miRNA mimic. Values are the means ± SD of triplicate determinations. *, P value of <0.05 in comparison with the control.
For pHygEGFP-mTOR, the cloned region contains predicted targets for miR-100 and miR-101 (Fig. 5A). At 25 nM, the miR-100 mimic alone inhibited EGFP expression by 35%, while the miR-101 mimic alone had no effect. At 50 nM, the miR-100 mimic alone inhibited EGFP expression by 60%, and the miR-101 mimic alone inhibited it by 38%; the combination of 25 nM of each of the miR-100 and miR-101 mimics (50 nM total) inhibited EGFP by 48% (Fig. 5C). The raptor 3′-UTR fragment contains a predicted target for miR-100 but has none for miR-101. Consistent with this, in experiments performed in parallel to those for which results are shown in Fig. 5B and C, miR-100 inhibited pHygEGFP-raptor by about 50%, but neither miR-101 nor the negative-control miRNA mimic had an effect (Fig. 6B). In addition, the miR-100 and miR-101 mimics had no effect on the parental EGFP vector, which lacks the mTOR and raptor 3′UTR (Fig. 6B). We extended this to an analysis of the effects of miR-100 and miR-101 on the expression of the mTOR protein in 293T cells. As shown in Fig. 5D, a combination of the two miRNAs (6.25 nM of each) reduced mTOR expression by 41% relative to what was seen for the transfection control, an effect greater than that seen for either of the mimics alone at 12.5 nM; the control miRNA had a negligible effect. Note that the levels of miRNA mimics required to achieve silencing of the EGFP reporter constructs (Fig. 5A to C) and mTOR protein expressed at its native levels (Fig. 5D) are not directly comparable, due to the high level of reporter gene transcripts expressed from the HCMV immediate early promoter in the transfected HeLa cells and the relatively low level of mTOR expressed from its native transcript in 293T cells and possibly to differences in silencing resulting from differences in the overall structure of the mTOR 3′UTR reporter transcript versus that of the native mTOR transcript. To summarize, two of the miRNAs reregulated after HCMV infection interact specifically with the 3′-UTR of at least two members of the mTOR pathway and can reduce the expression of mTOR at the protein level.
miR-100 and miR-101 inhibit HCMV replication.
We tested the hypothesis that if levels of a cellular miRNA are reduced following HCMV infection, their absence might be helpful for viral replication. Therefore, by supplementing intracellular levels of the miRNA by exogenous addition of the miRNA, viral replication might be suppressed.
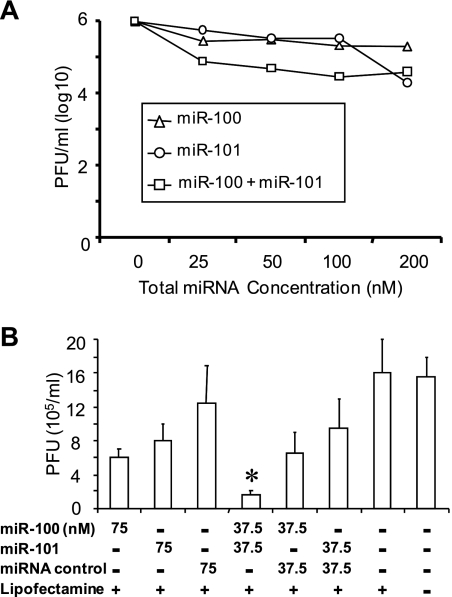
First, we directly compared miR-100 and miR-101. Based on work from Kudchodkar and colleagues, subconfluent human fibroblasts (28) were transfected with the mimics of miR-100 and miR-101 individually and as equimolar mixtures (10). Two days after transfection, the cells were serum starved for two more days and then infected with the T-BACwt virus at an MOI of 2. Virus titers in the culture supernatant on the fourth day after infection are shown in Fig. 7A. Individual mimics reduced virus titers by about one-half of a log at concentrations from 25 to 100 nM. Equimolar mixes of the two mimics were much more potent (>10-fold reduction) than individual mimics at the same total molar concentration. Inhibition by the 25 nM mixture (12.5 nM of each species) was >4-fold greater than that in the presence of 100 nM of either species alone. These inhibitory effects were verified and extended in an independent experiment that included a commercial miRNA mimic as a negative control (Fig. 7B). In this experiment, miR-100 and miR-101, either alone or in combination, reduced the amount of infectious HCMV in the culture supernatants at 4 days after infection to levels similar to those shown in Fig. 7A. Importantly, the miRNA mimic negative control had no significant effect on HCMV production when used either alone or in combination with miR-100 or miR-101. Thus, the inhibitory effects of the combination of miR-100 and miR-101 are specific. Further, we saw no difference in GFP expression from the recombinant virus in the presence or absence of the miRNA mimics (data not shown), indicating that the mimics do not adversely affect viral entry or the early stages of gene expression.
FIG. 7.
Inhibition of HCMV replication by miR-100 and miR-101. (A) Effects of miR-100 and miR-101, alone or in combination, on HCMV replication. (B) Confirmation of the specificity of miR-100 and miR-101 inhibition of HCMV replication. A control miRNA mimic had no effect on HCMV replication or on the inhibitory effects of miR-100 and miR-101. HCMV titers are expressed in PFU/ml ± the SD of triplicate samples. *, P value of <0.05 in comparison with the control.
The combinatorial effect may be the product of the two mimics together targeting common targets and/or each having different targets that together are important for the production of infectious progeny. The enhancement of inhibitory effects seen with the equimolar mixtures of the two mimics indicates that the induction of cellular antiviral mechanisms such as interferon does not explain the observed inhibition of viral replication.
DISCUSSION
Previous studies of herpesviruses and miRNAs have focused on virally encoded miRNAs. Here we examined the connection between infection and global patterns of cellular miRNA expression. The major observations from this work are that HCMV infection results in the altered regulation of cellular miRNAs, that mTOR pathway components are targeted by infection-regulated miRNAs, and that infection-regulated miRNAs target genes important in viral replication. This illustrates a novel mode of viral regulation of cellular processes.
HCMV infection leads to altered regulation of cellular miRNAs.
HCMV affects the regulation of many cellular processes (reviewed in reference 39). In some cases, these changes are directed by the virus for its advantage and others are cellular defense responses to infection. Here, we found that HCMV infection leads to altered regulation of cellular miRNAs. Given the number of genes that can be regulated by individual miRNAs and the number of miRNAs expressed in cells and tissues, this greatly expands the range of possible virus-host regulatory interactions. The complexity is underscored by there being no uniform global pattern of regulation; rather, it appears that individual (or groups of) miRNAs are independently regulated, some positively and some negatively. Persistent and transient effects were seen, and changes in miRNA expression profiles linked to the time course of infection. Interestingly, many of the virally regulated miRNAs are predicted to target important biological pathways and have altered regulation in some cancers and some states of cellular differentiation.
Our results contrast with what has been observed for some plant viruses, which encode proteins that cause global inhibition of the miRNA system (6, 36). The altered patterns of cellular miRNA we observed for HCMV-infected cells are similar in some respects to changes seen for cells infected with HIV in that there was both up- and downregulation of cellular miRNAs (50). Most of the miRNAs affected by HCMV are different from those affected by HIV. In contrast to HIV-infected cells, in HeLa cells that were transfected with HIV-1, 43% of miRNA species were downregulated, with few being upregulated (54). It will be important to learn the mechanisms of regulation of miRNA levels, which could be at the stages of transcription, maturation, and/or degradation. The temporal patterns of regulation indicate that for HCMV, viral genes of more than one kinetic class are involved.
Biological effects of reregulated miRNAs.
Because of their target degeneracy, it is a major challenge to identify the full spectrum of regulatory targets of individual miRNAs (23, 27). We made the novel observations that mimics of miR-100 and miR-101 downregulate the expression of a reporter gene containing in its 3′-UTR the 3′-UTR of either mTOR (silenced by mimics of miR-100 and miR-101) or raptor (silenced only by the miR-100 mimic), and that these miRNAs can suppress mTOR protein levels. mTOR and raptor are components of the mTOR protein translation initiation regulatory pathway, which is important for cell size (15) and processes that include protection from cell death (37), cell cycle progression (14), and HCMV replication under some conditions (28). It will be important to determine whether miR-100 and miR-101 regulate the mTOR pathway in infected cells and to identify other cellular and/or viral genes that might be involved in this process.
A key observation was that some infection-regulated miRNAs can influence virus production. By themselves, the miR-100 and miR-101 mimics had only modest effects on virus yield (∼70% reduction at 50 nM of either mimic); together, the inhibitory effect was amplified (33-fold reduction in the presence of 25 nM of each mimic). When virally vectored small interfering RNAs, each containing two distinct short hairpin RNAs with perfect sequence matches in either the raptor or the rictor genes, were delivered prior to HCMV infection, both of the targeted genes were downregulated and viral replication was inhibited, with the inhibition from targeting rictor being greater than that seen from targeting raptor (28). As rictor also has two candidate targets for miR-101, our results suggest that the combination of mir-100 and miR-101 may inhibit the mTOR pathway more completely than the targeting of either mTOR or raptor alone. Other miRNAs that also contribute to this process may be found.
Potential roles for miRNA during HCMV infection.
The vast diversity of miRNA regulatory targeting and the diverse patterns of the regulation of cellular miRNAs during infection suggest that miRNA might be involved in the regulation of cells at any and potentially at every step of HCMV replication, including manipulating the initially infected target cell at the portal of entry, creating an environment suitable for establishing latency, reactivating cells from latency, and causing end organ disease. Some cellular miRNAs are already known to be important for virus replication (24, 30, 50). Similar to our result for HCMV, the addition of exogenous miRNA miR-17/92, whose expression levels were substantially decreased after HIV infection, inhibited HIV growth (50). It remains to be seen which components of the altered regulation of miRNA expression in infected cells are cellular responses to infection and which are done by the virus for its purposes.
Our study has some limitations that will need to be addressed in future studies. We did not assess the roles in infection of most of the miRNAs whose expression is altered after infection. The miRNA microarrays we used do not contain probes for every known miRNA; thus, it is possible that HCMV infection affects the expression of a list of miRNAs longer than that enumerated here. The virus may interact with miRNA regulatory pathways differently in cells in other metabolic states, in different cell types, or in human tissues. Genes present in wild-type viruses but absent in the laboratory strain we used may have additional effects on cellular miRNA expression.
Acknowledgments
We thank Pamela Berk for technical assistance, Subhendu Das for helpful discussions, Naoki Inoue for helpful comments on the manuscript, and the staff of the Lerner Research Institute Imaging and Flow Cytometry Cores for valuable assistance.
Footnotes
Published ahead of print on 2 July 2008.
REFERENCES
- 1.Berezikov, E., V. Guryev, J. van de Belt, E. Wienholds, R. H. Plasterk, and E. Cuppen. 2005. Phylogenetic shadowing and computational identification of human microRNA genes. Cell 12021-24. [DOI] [PubMed] [Google Scholar]
- 2.Browne, E. P., B. Wing, D. Coleman, and T. Shenk. 2001. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J. Virol. 7512319-12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burnside, J., E. Bernberg, A. Anderson, C. Lu, B. C. Meyers, P. J. Green, N. Jain, G. Isaacs, and R. W. Morgan. 2006. Marek's disease virus encodes microRNAs that map to meq and the latency-associated transcript. J. Virol. 808778-8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai, X., C. H. Hagedorn, and B. R. Cullen. 2004. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 101957-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calin, G. A., M. Ferracin, A. Cimmino, G. Di Leva, M. Shimizu, S. E. Wojcik, M. V. Iorio, R. Visone, N. I. Sever, M. Fabbri, R. Iuliano, T. Palumbo, F. Pichiorri, C. Roldo, R. Garzon, C. Sevignani, L. Rassenti, H. Alder, S. Volinia, C. G. Liu, T. J. Kipps, M. Negrini, and C. M. Croce. 2005. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 3531793-1801. [DOI] [PubMed] [Google Scholar]
- 6.Chapman, E. J., A. I. Prokhnevsky, K. Gopinath, V. V. Dolja, and J. C. Carrington. 2004. Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev. 181179-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ciafre, S. A., S. Galardi, A. Mangiola, M. Ferracin, C. G. Liu, G. Sabatino, M. Negrini, G. Maira, C. M. Croce, and M. G. Farace. 2005. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem. Biophys. Res. Commun. 3341351-1358. [DOI] [PubMed] [Google Scholar]
- 8.Cimmino, A., G. A. Calin, M. Fabbri, M. V. Iorio, M. Ferracin, M. Shimizu, S. E. Wojcik, R. I. Aqeilan, S. Zupo, M. Dono, L. Rassenti, H. Alder, S. Volinia, C. G. Liu, T. J. Kipps, M. Negrini, and C. M. Croce. 2005. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA 10213944-13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui, C., A. Griffiths, G. L. Li, L. M. Silva, M. F. Kramer, T. Gaasterland, X. J. Wang, and D. M. Coen. 2006. Prediction and identification of herpes simplex virus 1-encoded microRNAs. J. Virol. 805499-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Das, S., Y. Skomorovska-Prokvolit, F. Z. Wang, and P. E. Pellett. 2006. Infection-dependent nuclear localization of US17, a member of the US12 family of human cytomegalovirus-encoded seven-transmembrane proteins. J. Virol. 801191-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dostie, J., Z. Mourelatos, M. Yang, A. Sharma, and G. Dreyfuss. 2003. Numerous microRNPs in neuronal cells containing novel microRNAs. RNA 9180-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunn, W., P. Trang, Q. Zhong, E. Yang, C. van Belle, and F. Liu. 2005. Human cytomegalovirus expresses novel microRNAs during productive viral infection. Cell. Microbiol. 71684-1695. [DOI] [PubMed] [Google Scholar]
- 13.Felli, N., L. Fontana, E. Pelosi, R. Botta, D. Bonci, F. Facchiano, F. Liuzzi, V. Lulli, O. Morsilli, S. Santoro, M. Valtieri, G. A. Calin, C. G. Liu, A. Sorrentino, C. M. Croce, and C. Peschle. 2005. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc. Natl. Acad. Sci. USA 10218081-18086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fingar, D. C., C. J. Richardson, A. R. Tee, L. Cheatham, C. Tsou, and J. Blenis. 2004. mTOR controls cell cycle progression through its cell growth effectors S6K1 and 4E-BP1/eukaryotic translation initiation factor 4E. Mol. Cell. Biol. 24200-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fingar, D. C., S. Salama, C. Tsou, E. Harlow, and J. Blenis. 2002. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 161472-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garzon, R., M. Fabbri, A. Cimmino, G. A. Calin, and C. M. Croce. 2006. MicroRNA expression and function in cancer. Trends Mol. Med. 12580-587. [DOI] [PubMed] [Google Scholar]
- 17.Garzon, R., F. Pichiorri, T. Palumbo, R. Iuliano, A. Cimmino, R. Aqeilan, S. Volinia, D. Bhatt, H. Alder, G. Marcucci, G. A. Calin, C. G. Liu, C. D. Bloomfield, M. Andreeff, and C. M. Croce. 2006. MicroRNA fingerprints during human megakaryocytopoiesis. Proc. Natl. Acad. Sci. USA 1035078-5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grey, F., A. Antoniewicz, E. Allen, J. Saugstad, A. McShea, J. C. Carrington, and J. Nelson. 2005. Identification and characterization of human cytomegalovirus-encoded microRNAs. J. Virol. 7912095-12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffiths-Jones, S., R. J. Grocock, S. van Dongen, A. Bateman, and A. J. Enright. 2006. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 34D140-D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He, H., K. Jazdzewski, W. Li, S. Liyanarachchi, R. Nagy, S. Volinia, G. A. Calin, C. G. Liu, K. Franssila, S. Suster, R. T. Kloos, C. M. Croce, and A. de la Chapelle. 2005. The role of microRNA genes in papillary thyroid carcinoma. Proc. Natl. Acad. Sci. USA 10219075-19080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hertel, L., and E. S. Mocarski. 2004. Global analysis of host cell gene expression late during cytomegalovirus infection reveals extensive dysregulation of cell cycle gene expression and induction of pseudomitosis independent of US28 function. J. Virol. 7811988-12011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Houbaviy, H. B., M. F. Murray, and P. A. Sharp. 2003. Embryonic stem cell-specific microRNAs. Dev. Cell 5351-358. [DOI] [PubMed] [Google Scholar]
- 23.John, B., A. J. Enright, A. Aravin, T. Tuschl, C. Sander, and D. S. Marks. 2004. Human microRNA targets. PLoS Biol. 2e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jopling, C. L., M. Yi, A. M. Lancaster, S. M. Lemon, and P. Sarnow. 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 3091577-1581. [DOI] [PubMed] [Google Scholar]
- 25.Jovanovic, M., and M. O. Hengartner. 2006. miRNAs and apoptosis: RNAs to die for. Oncogene 256176-6187. [DOI] [PubMed] [Google Scholar]
- 26.Kluiver, J., B. J. Kroesen, S. Poppema, and A. van den Berg. 2006. The role of microRNAs in normal hematopoiesis and hematopoietic malignancies. Leukemia 201931-1936. [DOI] [PubMed] [Google Scholar]
- 27.Krutzfeldt, J., M. N. Poy, and M. Stoffel. 2006. Strategies to determine the biological foundation of microRNAs. Nat. Genet. Suppl. 38S14-S19. [DOI] [PubMed] [Google Scholar]
- 28.Kudchodkar, S. B., Y. Yu, T. G. Maguire, and J. C. Alwine. 2006. Human cytomegalovirus infection alters the substrate specificities and rapamycin sensitivities of raptor- and rictor-containing complexes. Proc. Natl. Acad. Sci. USA 10314182-14187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lagos-Quintana, M., R. Rauhut, J. Meyer, A. Borkhardt, and T. Tuschl. 2003. New microRNAs from mouse and human. RNA 9175-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lecellier, C. H., P. Dunoyer, K. Arar, J. Lehmann-Che, S. Eyquem, C. Himber, A. Saib, and O. Voinnet. 2005. A cellular microRNA mediates antiviral defense in human cells. Science 308557-560. [DOI] [PubMed] [Google Scholar]
- 31.Lee, Y., M. Kim, J. Han, K. H. Yeom, S. Lee, S. H. Baek, and V. N. Kim. 2004. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 234051-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewis, B. P., I. H. Shih, M. W. Jones-Rhoades, D. P. Bartel, and C. B. Burge. 2003. Prediction of mammalian microRNA targets. Cell 115787-798. [DOI] [PubMed] [Google Scholar]
- 33.Lim, L. P., M. E. Glasner, S. Yekta, C. B. Burge, and D. P. Bartel. 2003. Vertebrate microRNA genes. Science 2991540. [DOI] [PubMed] [Google Scholar]
- 34.Liu, C. G., G. A. Calin, B. Meloon, N. Gamliel, C. Sevignani, M. Ferracin, C. D. Dumitru, M. Shimizu, S. Zupo, M. Dono, H. Alder, F. Bullrich, M. Negrini, and C. M. Croce. 2004. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc. Natl. Acad. Sci. USA 1019740-9744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu, S., and B. R. Cullen. 2004. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and microRNA biogenesis. J. Virol. 7812868-12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mallory, A. C., B. J. Reinhart, D. Bartel, V. B. Vance, and L. H. Bowman. 2002. A viral suppressor of RNA silencing differentially regulates the accumulation of short interfering RNAs and micro-RNAs in tobacco. Proc. Natl. Acad. Sci. USA 9915228-15233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mamane, Y., E. Petroulakis, O. LeBacquer, and N. Sonenberg. 2006. MTOR, translation initiation and cancer. Oncogene 256416-6422. [DOI] [PubMed] [Google Scholar]
- 38.Marchini, A., H. Liu, and H. Zhu. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J. Virol. 751870-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mocarski, E. S., T. Shenk, and R. F. Pass. 2007. Cytomegaloviruses, p. 2701-2772. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, PA.
- 40.Omoto, S., M. Ito, Y. Tsutsumi, Y. Ichikawa, H. Okuyama, E. A. Brisibe, N. K. Saksena, and Y. R. Fujii. 2004. HIV-1 nef suppression by virally encoded microRNA. Retrovirology 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pfeffer, S., A. Sewer, M. Lagos-Quintana, R. Sheridan, C. Sander, F. A. Grasser, L. F. van Dyk, C. K. Ho, S. Shuman, M. Chien, J. J. Russo, J. Ju, G. Randall, B. D. Lindenbach, C. M. Rice, V. Simon, D. D. Ho, M. Zavolan, and T. Tuschl. 2005. Identification of microRNAs of the herpesvirus family. Nat. Methods 2269-276. [DOI] [PubMed] [Google Scholar]
- 42.Pfeffer, S., M. Zavolan, F. A. Grasser, M. Chien, J. J. Russo, J. Ju, B. John, A. J. Enright, D. Marks, C. Sander, and T. Tuschl. 2004. Identification of virus-encoded microRNAs. Science 304734-736. [DOI] [PubMed] [Google Scholar]
- 43.Richardson, B. A., and J. Overbaugh. 2005. Basic statistical considerations in virological experiments. J. Virol. 79669-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Samols, M. A., J. Hu, R. L. Skalsky, and R. Renne. 2005. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi's sarcoma-associated herpesvirus. J. Virol. 799301-9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samols, M. A., R. L. Skalsky, A. M. Maldonado, A. Riva, M. C. Lopez, H. V. Baker, and R. Renne. 2007. Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathogens 3e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stern-Ginossar, N., N. Elefant, A. Zimmermann, D. G. Wolf, N. Saleh, M. Biton, E. Horwitz, Z. Prokocimer, M. Prichard, G. Hahn, D. Goldman-Wohl, C. Greenfield, S. Yagel, H. Hengel, Y. Altuvia, H. Margalit, and O. Mandelboim. 2007. Host immune system gene targeting by a viral miRNA. Science 317376-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sullivan, C. S., and D. Ganem. 2005. A virus-encoded inhibitor that blocks RNA interference in mammalian cells. J. Virol. 797371-7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sullivan, C. S., A. T. Grundhoff, S. Tevethia, J. M. Pipas, and D. Ganem. 2005. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature 435682-686. [DOI] [PubMed] [Google Scholar]
- 49.Tanzer, A., and P. F. Stadler. 2004. Molecular evolution of a microRNA cluster. J. Mol. Biol. 339327-335. [DOI] [PubMed] [Google Scholar]
- 50.Triboulet, R., B. Mari, Y. L. Lin, C. Chable-Bessia, Y. Bennasser, K. Lebrigand, B. Cardinaud, T. Maurin, P. Barbry, V. Baillat, J. Reynes, P. Corbeau, K. T. Jeang, and M. Benkirane. 2007. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science 3151579-1582. [DOI] [PubMed] [Google Scholar]
- 51.Volinia, S., G. A. Calin, C. G. Liu, S. Ambs, A. Cimmino, F. Petrocca, R. Visone, M. Iorio, C. Roldo, M. Ferracin, R. L. Prueitt, N. Yanaihara, G. Lanza, A. Scarpa, A. Vecchione, M. Negrini, C. C. Harris, and C. M. Croce. 2006. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 1032257-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weber, F., R. E. Teresi, C. E. Broelsch, A. Frilling, and C. Eng. 2006. A limited set of human microRNA is deregulated in follicular thyroid carcinoma. J. Clin. Endocrinol. Metab. 913584-3591. [DOI] [PubMed] [Google Scholar]
- 53.Yao, Y., Y. Zhao, H. Xu, L. P. Smith, C. H. Lawrie, A. Sewer, M. Zavolan, and V. Nair. 2007. Marek's disease virus type 2 (MDV-2)-encoded microRNAs show no sequence conservation with those encoded by MDV-1. J. Virol. 817164-7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yeung, M. L., Y. Bennasser, T. G. Myers, G. Jiang, M. Benkirane, and K. T. Jeang. 2005. Changes in microRNA expression profiles in HIV-1-transfected human cells. Retrovirology 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu, H., J. P. Cong, G. Mamtora, T. Gingeras, and T. Shenk. 1998. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 9514470-14475. [DOI] [PMC free article] [PubMed] [Google Scholar]