Abstract
Parainfluenza virus 5 (PIV5) is a prototypical paramyxovirus. The V/P gene of PIV5 encodes two mRNA species through a process of pseudotemplated insertion of two G residues at a specific site during transcription, resulting in two viral proteins, V and P, whose N termini of 164 amino acid residues are identical. Previously it was reported that mutating six amino acid residues within this identical region results in a recombinant PIV5 (rPIV5-CPI−) that exhibits elevated viral protein expression and induces production of cytokines, such as beta interferon and interleukin 6. Because the six mutations correspond to the shared region of the V protein and the P protein, it is not clear whether the phenotypes associated with rPIV5-CPI− are due to mutations in the P protein and/or mutations in the V protein. To address this question, we used a minigenome system and recombinant viruses to study the effects of mutations on the functions of the P and V proteins. We found that the P protein with six amino acid residue changes (Pcpi−) was more efficient than wild-type P in facilitating replication of viral RNA, while the V protein with six amino acid residue changes (Vcpi−) still inhibits minigenome replication as does the wild-type V protein. These results indicate that elevated viral gene expression in rPIV5-CPI− virus-infected cells can be attributed to a P protein with an increased ability to facilitate viral RNA synthesis. Furthermore, we found that a single amino acid residue change at position 157 of the P protein from Ser (the residue in the wild-type P protein) to Phe (the residue in Pcpi−) is sufficient for elevated viral gene expression. Using mass spectrometry and 33P labeling, we found that residue S157 of the P protein is phosphorylated. Based on these results, we propose that phosphorylation of the P protein at residue 157 plays an important role in regulating viral RNA replication.
Parainfluenza virus 5 (PIV5), formerly known as simian virus 5, is a prototypical paramyxovirus in the Rubulavirus genus of Paramyxovirinae (4). The paramyxovirus family includes many important human and animal pathogens, such as Sendai virus, mumps virus, human parainfluenza viruses 1, 2, 3, and 4, Newcastle disease virus, measles virus, and emerging viruses, such as Hendra virus and Nipah virus (19). Paramyxoviruses contain nonsegmented negative-stranded RNA genomes, which are encapsidated by nucleocapsid protein (NP). The gene order within the paramyxovirus genomes is 3′-NP-P(V/W/C)-M-F-(SH)-HN-L-5′, where genes in parenthesis are not found in all species (reviewed in reference 19). The viral RNA-dependent RNA polymerase of paramyxoviruses minimally consists of two proteins, phosphoprotein (P) and the large (L) polymerase protein (11). The L proteins of paramyxoviruses have masses of 220 to 250 kDa. They have the capacity to initiate, elongate, and terminate transcription. In addition, they have the capacity to insert nontemplated G residues at selected sites within viral mRNAs during transcription and to add cap structures to the 5′ ends of viral transcripts. While the L protein of the P-L complex of viral RNA-dependent RNA polymerase is thought to contain polymerase activity, the P protein is thought to be a regulatory protein without intrinsic enzymatic activity. The P protein of paramyxoviruses is phosphorylated (hence the name phosphoprotein). The phosphorylation of P in nonsegmented negative-stranded RNA virus is thought to play a critical role in virus RNA synthesis (19). Mutations within P of vesicular stomatitis virus that selectively affect viral RNA replication or viral mRNA transcription have been identified, suggesting that the protein plays an important role in regulating viral RNA replication and viral mRNA transcription (7). However, the roles of phosphorylation of paramyxovirus P proteins in viral RNA synthesis have been less defined. Numerous reports utilizing minigenome systems indicate that mutating putative phosphorylation sites within P proteins affects their activities (6, 10). Unexpectedly, when these mutations are incorporated into virus genomes, no effect on virus RNA synthesis has been observed, leading to the theory that phosphorylation of the P protein does not have a role in viral RNA synthesis (17, 18, 26). Recently studies using recombinant PIV5 expressing an additional copy of the P gene suggest that the P protein may have a role in limiting induction of host cell antiviral responses (9, 13). However, it is not clear whether the putative role of the PIV5 P protein in preventing host innate immune responses is direct or indirect due to the P protein's role in viral RNA synthesis.
The V/P gene of PIV5 can be transcribed into two species of mRNA in about equal amounts through a process of pseudotemplated nucleotide insertion, in which two G residues are inserted at a specific location during viral RNA transcription. The faithful transcription of the gene results in a population of mRNAs encoding the V protein, whereas insertion of the nontemplated two G residues results in a population of mRNAs encoding the P protein. The V protein (222 amino acid residues) and the P protein (392 amino acid residues) have identical N termini (164 amino acid residues) (39). The V protein is a structural component of PIV5 virions (∼350 molecules per virion) and is multifunctional (30). The V protein C-terminal domain contains seven cysteine residues, resembling a zinc finger domain, and binds atomic zinc (25, 30, 36, 39). It interacts with soluble NP (32), and the N-terminal domain binds RNA through a basic region (21). The PIV5 V protein interacts with a cellular protein (DDB1). This interaction requires the presence of the C-terminal domain of the V protein (20). Expression of the PIV5 V protein slows down the cell cycle in a manner that is dependent on the C-terminal region of the protein (20). Coexpression of DDB1 can partially restore the changes in the cell cycle caused by V (20). The V protein of PIV5 can cause degradation of the STAT1 protein, an essential regulator of interferon (IFN) signaling, through a proteasome-mediated pathway in human cells but not in mouse cells (8). It has been shown that V, DDB1, Cul4A, STAT1, and STAT2 form a complex, which is essential for V-mediated STAT1 degradation, and the V protein has an E3 ubiquitin ligase activity (28, 40). In addition to preventing IFN signaling, the V protein can inhibit beta IFN (IFN-β) production through an IRF-3-dependent pathway (15, 31) and block interleukin 6 (IL-6) expression in virus-infected cells (24). Recently it was shown that the V protein interacts with MDA-5 and that this interaction plays an important role in blocking activation of IFN-β production (1). The V protein is also known to play an essential role in blocking apoptosis in virus-infected cells. While the exact mechanism is not clear, it is thought that the V protein blocks endoplasmic reticulum stress-induced apoptosis and this blockage function is independent of its involvement with IFN pathways (38). Using a minigenome system developed for PIV5 free of vaccinia virus infection, it has been reported that the expression of the V protein inhibits minigenome replication by inhibiting viral RNA synthesis (23). Further investigation of the mechanism indicated that the V protein interacts with Akt1, a serine/threonine kinase, and the V protein likely exerts its influence on viral RNA synthesis via its interaction with Akt1 (37).
A strain of PIV5 causing a neurological disorder in canines was isolated by Evermann et al. (12) and is called canine parainfluenza virus (CPI+). During the course of studying the CPI+ virus, a derivative of CPI+, termed CPI−, was isolated from a dog that was experimentally infected with the CPI+ virus (2). It was found that there are differences among the V/P genes of CPI+, CPI−, and the commonly used lab strain W3A, in addition to differences in other viral genes (4, 5, 35). The CPI− virus has eight amino acid residues in the V/P gene that are different from those of the W3A strain (referred to as the wild type [wt] in this work), and six of them are in the shared region of the V and P proteins. The CPI+ virus has five amino acid residues that are different from those of wt PIV5 in the V/P gene, and three of them are in the shared region of the V and P proteins. Wansley et al. generated recombinant PIV5 based on the W3A strain but containing six amino acid residue changes in the shared region of the V and P proteins, corresponding to the CPI− virus. This virus was termed rPIV5-CPI− (42). They observed that rPIV5-CPI− causes elevated viral gene expression, induces expression of host antiviral response genes, such as IFN-β and IL-6, and induces apoptosis of infected cells compared to the wt W3A strain. Because the six mutations are within the shared region of the V and P proteins, it is not clear whether the phenotypes associated with rPIV5-CPI− are due to mutations in the P protein or mutations in the V protein. To address this question, we used both a vaccinia virus-free minigenome system that we developed and recombinant viruses to study the effects of mutations on the functions of the P and V proteins.
MATERIALS AND METHODS
Plasmids, viruses, and cells.
Plasmids used in this work were constructed using standard molecular cloning techniques. Details of the construction of the plasmids and the computer sequence files of the plasmids are available on request. The plasmid pSMG-RL, which contains a PIV5 minigenome system that complies with the rule of six, was constructed from pMG-RL (23) by adding three nucleotides between the reporter gene and the leader sequence. Mutant P genes encoding V32I, T33I, S157F, S157A, S157D, V32I-T33I, T33I-S157F, or V32I S157F were made from a copy of P from the W3A strain (23) (Table 1) by using four-primer PCR as described previously (14). The P mutants were cloned into the pCAGGS expression vector (27). An L protein with two copies of Flag epitope tags at its C terminus was constructed in the pCAGGS background for expression and detection in coimmunoprecipitation experiments. Plasmids containing full-length viral genomes corresponding to rPIV5-CPI− and rPIV5-CPI+ were generated by introducing six or three substitution mutations, located at amino acid residue positions 26, 32, 33, 50, 102, and 157 or 32, 33, and 157, respectively, in the V/P sharing region of the rPIV5 genome (Table 1) in a plasmid containing the PIV5 genome, pBH276, which contains the wt W3A full-length genome, as previously described (16). The viruses (rPIV5-CPI− and rPIV5-CPI+) were rescued following a procedure described by He et al. and Waning et al. (16, 41). The recovered viruses were plaque purified, and the V/P gene sequence was confirmed by reverse transcription (RT)-PCR sequencing. The viruses were grown in Vero cells and titrated in BHK cells as previously described (16).
TABLE 1.
Amino acid residue substitutions encoded in V/P gene of PIV5a
| Description or strain | Amino acid residue at position:
|
Effectivenessb | |||||
|---|---|---|---|---|---|---|---|
| 26 | 32 | 33 | 50 | 102 | 157 | ||
| wt | Y | V | T | L | L | S | Normal |
| CPI− | H | I | I | P | P | F | High |
| CPI+ | Y | I | I | L | L | F | High |
| V32I | Y | I | T | L | L | S | Normal |
| T33I | Y | V | I | L | L | S | Normal |
| S157F | Y | V | T | L | L | F | High |
| V32I-T33I | Y | I | I | L | L | S | Normal |
| V32I-S157F | Y | I | T | L | L | F | High |
| T33I-S157F | Y | V | I | L | L | F | High |
| S157A | Y | V | T | L | L | A | High |
| S157D | Y | V | T | L | L | D | High |
Boldfacing indicates residues associated with CPI−.
Effectiveness indicates the ability of the P protein to facilitate viral RNA synthesis.
HeLa and Vero cells were grown in Dulbecco modified Eagle medium (DMEM) (Gibco-BRL) containing 10% fetal calf serum. BHK cells were grown in DMEM containing 10% fetal calf serum and 10% tryptose phosphatase broth. BSR T7 cells were growth in the same medium as BHK cells, with the addition of 400 μg/ml G418 to maintain the expression of T7 RNA polymerase (3). All cell lines were maintained in 100 IU/ml penicillin-100 μg/ml streptomycin and incubated at 37°C in 5% CO2.
Flow cytometry.
BSR T7 or HeLa cells were mock infected or rPIV5, rPIV5-CPI−, or rPIV5-CPI+ infected at a multiplicity of infection (MOI) of 3. The cells were collected at 16 h postinfection (hpi) and fixed with 0.5% formaldyhyde for 2 h. The fixed cells were pelleted by centrifugation and then resuspended in 500 μl of solution of fetal bovine serum (FBS)-DMEM (50:50). The cells were permeabilized in 70% ethanol overnight. The cells were washed once with phosphate buffered saline deficient in Mg2+ and Ca2+ (PBS−) and then incubated with mouse monoclonal anti-hemagglutinin-neuraminidase (HN) antibody in PBS− containing 10% FBS for 30 min at room temperature. The cells were stained with antimouse antibody labeled with phycoerythrin for 30 min at room temperature in the dark and then washed once with PBS− containing 10% FBS. The fluorescence intensity was measured using a flow cytometer.
Transfection and dual luciferase assay.
To measure the activities of the P proteins, a modified minigenome system of PIV5 was used. The BSR T7 cells were seeded in 24-well culture plates. The cells were transfected at 70 to 80% confluence with a total of 0.827 μg of plasmid DNA per well containing pCAGGS-NP, pCAGGS-L, pSMG-R-luc (minigenome plasmid in which the number of nucleotides from the trailer to leader sequence is a multiple of six), pCAGGS-P or -P mutants and/or p-CAGGS-V or pCAGGS-Vcpi− (which has six substitution mutations in the V/P shared region [Table 1]). The total amounts of transfected plasmids were kept constant using pCAGGS-GFP. Plasmids were transfected using Lipofectamine Plus (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. To measure the transfection efficiency in the luciferase assay, an equal amount of the pT7-F-Luc plasmid, which contains a firefly luciferase (F-Luc) reporter gene, was included in the DNA transfection mixture. At 18 to 20 h posttransfection, the cells were lysed in 140 μl passive lysis buffer (Promega). Twenty microliters of lysate from each well was then used in the subsequent dual luciferase assay, according to the manufacturer's protocol (Promega). To examine the transcription activity of P and P mutants, a minigenome with a defective trailer sequence that is active only in transcription was used as described by Lin et al. (23). Minigenome replication and/or transcription was normalized for each sample using dual luciferase activity. The relative luciferase activity is defined as the ratio of R-Luc to F-Luc activities.
Western blot.
An aliquot of the cell lysate from the dual luciferase assay was mixed with an equal volume of 2× protein lysis buffer (60 mM Tris-HCl [pH 6.8], 40% glycerol, 4% sodium dodecyl sulfate [SDS], 3% dithiothreitol [DTT], and a few grains of bromophenol blue) as previously described (29). Samples were resolved in 10 or 15% SDS-polyacrylamide gel electrophoresis (PAGE) and transferred onto a polyvinylidene difluoride membrane. Immunoblotting using antibodies against PIV5 proteins was performed as previously described (22, 38).
Immunoprecipitation.
To examine the expression levels of viral proteins in infected cells, cells (HeLa or BSR T7) were mock infected or infected with PIV5, rPIV5-CPI−, or rPIV5-CPI+ at a MOI of 3 as before. At 16 hpi, the infected cells were starved in DMEM lacking cysteine-methionine for 30 min and then labeled with 35S-Promix (Amersham Life Sciences) (10 μCi/ml) for 3 h. The cells were lysed with whole-cell extract buffer (WCEB) (50 mM Tris-HCl [pH 8], 280 mM NaCl, 0.5% NP-40, 0.2 mM EDTA, 2 mM EGTA, and 10% glycerol) (40), and the aliquots were coimmunoprecipitated using antibodies against PIV5 NP, P, and M. The precipitated proteins were resolved by 10% SDS-PAGE and visualized using a Storm PhosphorImager (Molecular Dynamics Inc., Sunnyvale, CA). To study the interaction of P and P mutants with either NP or L, plasmids encoding P or P mutants with NP or with L containing two copies of Flag epitope tags at the C terminus encoded by the L gene to facilitate its detection were cotransfected into cells. At 20 to 24 h posttransfection, the cells were labeled and immunoprecipitated using anti-P, anti-NP (NP-214) (33), or anti-Flag M2 (Sigma).
Chemical cross-linking.
The P and P mutants were expressed in BSR T7 cells and then labeled with 35S-Promix as before. Chemical cross-linking was performed as described previously (34). Briefly, metabolically labeled cells were washed with PBS− and removed from the dish with PBS− containing 50 μM EDTA. Cells were pelleted by centrifugation and resuspended in PBS−. Cross-linking reactions were carried out in 200-μl aliquots of cell suspension in 0.5% NP-40. One hundred millimolar disuccinimidyl tartrate (Pierce, Rockford, IL) stock solution was dissolved in dimethylsulfoxide and added to the cross-linking reaction mixture so that the final concentration was 1 mM. Cross-linking reactions were incubated for 2 h at 4°C. The cells were lysed, and the proteins precipitations were processed as mentioned earlier. Samples were mixed with an equal volume of 2× protein lysis buffer in the presence or absence of DTT and then resolved by 10% SDS-PAGE.
Phosphorylation of P.
To investigate the phosphorylation level of P and P mutants in the infected cells, BSR T7 cells were mock infected or rPIV5, rPIV5-CPI− and rPIV5 CPI+ infected at a MOI of 2. At 18 to 20 hpi, the cells were starved with either DMEM lacking cysteine-methionine or DMEM lacking phosphate and then labeled with 100 μCi 35S-Promix or 200 μCi [33P]orthophosphate (PerkinElmer), respectively, for 4 h. The immunoprecipitations using anti-P antibody (33) were processed as described above.
Real-time PCR.
Monolayer cells in 6-cm dishes were washed with PBS and inoculated with PIV5, rPIV5-CPI+, or rPIV5-CPI− viruses in DMEM with 1% bovine serum albumin at a MOI of 5 for 1 to 2 h at 37°C. Cells were then washed and incubated in DMEM with 2% FBS at 37°C with 5% CO2. At 0, 4, 8, 12, 16, and 20 hpi, total RNAs were extracted from infected cells using an RNeasy minikit (Qiagen) and eluted in 100 ml RNase-free H2O. Five milliliters of total RNA for each sample was used for the RT reaction with Superscript III reverse transcriptase (Invitrogen) according to the manufacturer's protocol. Oligo(dT)15 was used to measure mRNA levels; oligonucleotide BH191, which hybridizes to the viral RNA of PIV5 within the HN gene, was added for detection of viral RNA levels. Five percent of the cDNA from each sample was then used for every real-time PCR on an ABI 7300 real-time PCR system using Taqman Universal PCR master mix (Applied Biosystems) and custom-made Taqman gene expression assays (Applied Biosystems) for the HN gene. Primers were as follows: forward primer, GGGTACTAGATGTATGGGCAACA; reverse primer, ACGCCGCCATATATTGGAAAGAG; reporter, CCCCGCTTCCTGTTCC, with 6-carboxyfluorescein dye and NFQ quencher. Results were analyzed with the RQ study software program (Applied Biosystems) to obtain threshold cycle values. Relative levels of mRNA and viral RNA at each time point were determined by calculating the change in the threshold cycle, with 0-hpi samples infected with the same virus serving as the calibrators. The ratios of mRNA and viral RNA levels of the same samples were calculated by setting the viral RNA levels as calibrators. Each sample was done in quadruplicate.
Mass spectroscopic analysis.
To determine the phosphorylated residues in the PIV5 P protein, mass spectroscopic analysis was performed. HeLa cells in 10-cm plates were mock or PIV5 infected at a MOI of 3. At 24 hpi, the cells were lysed with WCEB buffer and the P protein was immunoprecipitated by using anti-V5 agarose-conjugated beads (Sigma-Aldrich) for 4 h at 4°C or overnight. The beads were washed three times with WCEB buffer, mixed with 2× protein lysis buffer in the absence of DTT, and then resolved by 10% SDS-PAGE. The band corresponding to the P protein was excised and digested with trypsin (12.5 ng/μl in 25 mM NH4HCO3) at 37°C for 16 h. The digested peptides were further processed for phosphopeptide enrichment using TiO2. Both the enriched fraction and the flowthrough (which contains all of the nonphosphorylated peptides) were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) on a Waters Q-Tof Ultima mass spectrometer at Yale Cancer Center Mass Spectrometry Resource and W.M. Keck Foundation Biotechnology Resource Laboratory, Yale University. All MS/MS spectra were searched using the automated Mascot algorithm against the NCBI database for possible tyrosine (Tyr), threonine (Thr), and serine (Ser) phosphorylation sites.
RESULTS
Elevated viral protein expression and viral genome replication by rPIV5-CPI− and rPIV5-CPI+.
It has been shown that six amino acid residue substitutions within the V/P gene are sufficient to cause elevated viral gene expression (42). To investigate the roles of individual residues in increasing viral gene expression, we have generated a recombinant PIV5 encoding the P protein that has the same three amino acid residue substitutions, relative to the wt P protein sequence, that are found in CPI+ virus (4). We call it rPIV5-CPI+ (Table 1). HeLa cells, a human cell line, and BSR T7 cells, a murine cell line, were infected with PIV5, rPIV5-CPI−, or rPIV5-CPI+. To examine expression of viral proteins in individual cells, HN protein expression was quantified using flow cytometry (Fig. 1A and B). As expected, rPIV5-CPI− caused increased expression of viral protein compared with wt PIV5 infection, consistent with the previous report (42). Interestingly, infection with rPIV5-CPI+ also caused increased expression of viral protein compared with wt PIV5 infection, indicating that three amino acid residue substitutions (V32I, T33I, and S157F) within the shared region of the V and P proteins are sufficient to convert wt PIV5 into a virus that expresses its proteins at higher levels. It is known that wt PIV5 causes degradation of STAT1 in human cells, such as HeLa cells, but not in BHK cells. Since the same results have been obtained in both human HeLa cells and BSR T7 cells, a derivative of BHK cells, it is unlikely that the ability of virus to cause degradation of STAT1 plays a role in the increased expression of viral proteins by rPIV5-CPI− and rPIV5-CPI+. To further confirm the results, immunoprecipitation of viral proteins from infected HeLa or BSR T7 cells was performed. Consistent with flow cytometry data, both rPIV5-CPI− and rPIV5-CPI+ virus infections resulted in a higher level of viral gene expression (Fig. 1C and D). These results also indicate that the increased viral gene expression is not cell type specific.
FIG. 1.
Expression of viral protein and RNA from cells infected with rPIV5-CPI− or rPIV5-CPI+. (A and B) Determining expression of viral protein from cells infected with rPIV5-CPI− or rPIV5-CPI+ using flow cytometry. BSR T7 or HeLa cells were infected with mock, wt PIV5, rPIV5-CPI−, or rPIV5-CPI+ at a MOI of 3. The cells were collected, fixed, and stained with anti-HN antibody at 16 hpi. A flow cytometer was used to gate HN-positive cells, and then the mean fluorescence intensities for HN were measured and graphed. Error bars are standard deviations of means. Panel A shows HeLa cells; panel B shows BSRT7 cells. (C and D) Determining expression of viral protein from cells infected with rPIV5-CPI− or rPIV5-CPI+ using immunoprecipitation. The infected cells were labeled and immunoprecipitated with anti-PIV5 antibodies as described in Materials and Methods. Panel C shows HeLa cells; panel D shows BSR T7 cells. (E to G) Viral RNA synthesis in cells infected with rPIV5-CPI− or rPIV5-CPI+. HeLa cells were infected with wt PIV5, rPIV5-CPI−, or rPIV5-CPI+. At the indicated hours postinfection, cells were lysed and total RNA extracted. Real-time RT-PCR was performed to determine the relative levels of the mRNA or viral RNA. Panels E and F show relative viral RNA and mRNA levels of cells infected with PIV5, rPIV5-CPI−, or rPIV5-CPI+ at different time points, obtained by comparing the RNA level at each time point with that of the 0-hpi sample of the corresponding virus. (G) Ratios of mRNA to viral RNA levels. Error bars are the standard deviations of means.
The increased viral gene expression could potentially be attributed to increased viral RNA replication, since this would increase the amount of viral RNA genome for use as a template during transcription. Another possibility is that increased viral gene expression results from increased RNA transcription. To investigate the mechanism for the increased viral gene expression in rPIV5-CPI− and rPIV5-CPI+, we examined viral RNA synthesis in infected cells. HeLa cells were infected with mock, PIV5, rPIV5-CPI−, or rPIV5-CPI+ at a MOI of 3. At different time points after infection, RNAs were purified from infected cells and measured using a real-time RT-PCR assay. Both rPIV5-CPI− and rPIV5-CPI+ infection resulted in much higher levels of viral RNA genome production, indicating that viral RNA replication increased in cells infected with rPIV5-CPI− or rPIV5-CPI+ compared with that in PIV5-infected cells (Fig. 1E), consistent with the observation that higher viral protein expression levels were observed in cells infected with rPIV5-CPI− or rPlV5-CPI+ than in wt virus-infected cells. While overall levels of viral transcripts are higher in cells infected with rPIV5-CPI− or rPIV5-CPI+ than in PIV5-infected cells, as expected (Fig. 1F), transcription of viral mRNA normalized to the amount of viral genome RNA remains similar among all virus-infected cells, indicating that the increase in activities of Pcpi− and Pcpi+ proteins results from increased RNA genome replication (Fig. 1G).
Elevated minigenome replication in the presence of Pcpi− and Pcpi+.
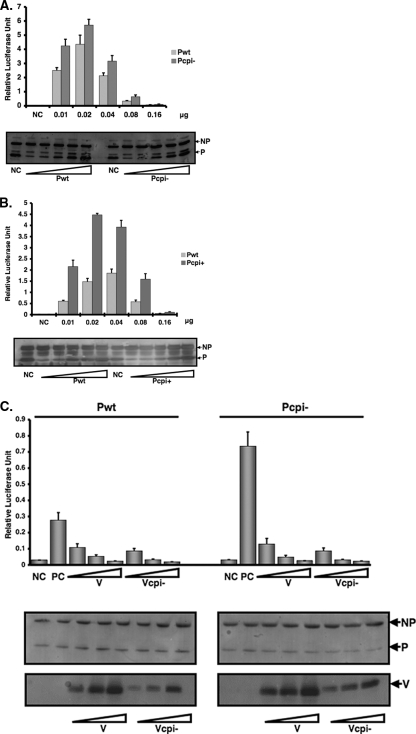
Previously we generated a minigenome system free of vaccinia virus to study viral RNA synthesis of PIV5 (23). To further study the mechanism of increased viral gene expression in cells infected with rPIV5-CPI− or rPIV5-CPI+, we compared the activity of the P protein of rPIV5-CPI− (Pcpi−) and the P protein of rPIV5-CPI+ (Pcpi+) with the P protein of the wt (P or Pwt) in the minigenome system. Because overexpression of the P protein inhibits minigenome replication, an observation that has been reported previously for PIV5 and other systems (23), we used a spectrum of P-protein concentrations to ensure that differences in minigenome expression reflect differences in P-protein activities, not differences in P-protein quantity. As expected, initially, increased expression of the P protein resulted in increased reporter gene expression, and further overexpression of the P protein inhibited minigenome replication (Fig. 2A). Using the same spectrum of protein concentrations, Pcpi− gave rise to higher minigenome reporter expression than wt P, indicating that Pcpi− is more efficient in this system than the wt P protein. Further overexpression of Pcpi− inhibited minigenome replication, similar to the effect observed with the wt P protein. All P protein expression levels in minigenome replication experiments were measured by immunoblotting (Fig. 2A, bottom panel). Using the same approach, Pcpi+ was found to be more efficient for minigenome replication than the wt P protein. The results suggest that a more efficient P protein in rPIV5-CPI− or rPIV5-CPI+ contributes to the increased viral protein expression in cells infected with rPIV5-CPI− or rPIV5-CPI+. This result obtained from the minigenome system is consistent with observations made for virus-infected cells. Thus, three amino acid residues within the identical N-terminal region of the V and P proteins contribute to the increased activity of the P protein of rPIV5-CPI+.
FIG. 2.
Elevated minigenome replication with Pcpi− and Pcpi+. A minigenome plasmid (pSMG-RL) that contains a Renilla luciferase reporter gene described previously (23) was modified to comply with the rule of six as described in the Materials and Methods. A negative-sense minigenome was generated from T7 RNA polymerase transcription in BSR T7 cells. In the presence of NP and L and with type P (Pwt), P from rPIV5-CPI+ (Pcpi+) (A) or P from rPIV5-CPI− (Pcpi−) (C), this negative-sense RNA template is replicated and transcribed to give rise to the reporter gene mRNA, resulting in luciferase activity. The pT7-F-Luc plasmid, which contains an F-Luc reporter gene as a transfection efficiency control, was transfected along with the plasmids. Firefly and Renilla luciferase activities were detected in cell lysates at 18 to 20 h posttransfection, as described in Materials and Methods. The relative luciferase activities are calculated at ratios of Renilla luciferase activity (indicative of minigenome replication) versus F-Luc activity (indicative of transfection efficiency). Due to the quality and amount of plasmids used in the experiments and passages of BSR T7 cells, which affect expression of T7 RNA polymerase, the relative activity fluctuates. To ensure the validity of comparisons among different P proteins, we have used a spectrum of concentrations of the P proteins in the experiments. Cell lysate aliquots from panel A or B were subjected to immunoblotting using anti-NP or anti-P antibody, respectively (as described in Materials and Methods). NC, negative control, containing all plasmids except L encoding plasmid. Error bars are the standard deviations of means from examples containing six replicates for each transfection. (C) Inhibition of minigenome replication by Vcpi−. Increasing amounts of V or Vcpi− expression plasmid were cotransfected with equal amounts of P or Pcpi− expression plasmid in the PIV5 minigenome system as described above. The total amount of DNA transfected was kept constant by using a green fluorescent protein expression plasmid. Cell lysate aliquots were collected and subjected to immunoblotting using anti-NP and anti-P antibodies. NC, negative control, lacking L. Error bars are the standard deviations of means from examples containing six replicates for each transfection.
Vcpi− retains its ability to inhibit minigenome replication.
Previous work from our laboratory demonstrated that the V protein of PIV5 inhibits minigenome replication (23). An alternative explanation for elevated viral gene expression in cells infected with rPIV5-CPI− is that the V protein of rPIV5-CPI− (Vcpi−) loses its ability to inhibit viral RNA synthesis. To test this, we transfected plasmids encoding the V protein or Vcpi− into the minigenome system together with plasmids encoding the P protein or Pcpi-. We found that Vcpi− inhibited minigenome replication (together with either the P protein or Pcpi-) as did the wt V protein (Fig. 2C and D), suggesting that increased viral gene expression in cells infected with rPIV5-CPI− is due to alterations in the P protein and not to alterations in the V protein.
Gene expression from replication-defective minigenome system.
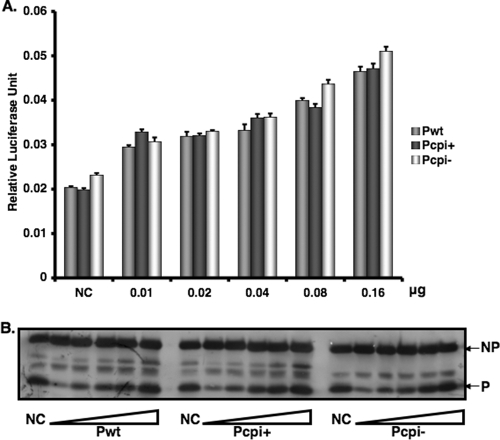
To investigate the mechanism for the more efficient Pcpi− and Pcpi+ proteins, we examined the effects of Pcpi− and Pcpi+ on viral RNA transcription. Previously we generated a minigenome system in which reporter gene expression arises only from viral transcription, not replication, due to mutation in the region that is essential for viral RNA replication (23). Using this transcription-only minigenome system, we found that Pcpi-, Pcpi+, and Pwt all had similar abilities to facilitate viral RNA transcription (Fig. 3), suggesting that the increased activities of the Pcpi− and Pcpi+ proteins comes from a process other than transcription, i.e., viral RNA replication. This result is consistent with those obtained with virus-infected cells (Fig. 1E, F, and G). Interestingly, overexpression of the P protein, Pcpi−, or Pcpi+ did not inhibit viral mRNA transcription, suggesting that the inhibitory effect on viral gene expression observed on overexpression of the P protein reported previously (23) and observed earlier (Fig. 2A and B) depends on viral RNA replication.
FIG. 3.
Transcription activities of P, Pcpi−, and Pcpi+. (A) A replication-deficient minigenome plasmid, pMG-m-R-Luc (23), was transfected into BSR T7 cells together with increasing amounts of plasmid encoding P, Pcpi−, or Pcpi+, along with a plasmid encoding NP and L. Firefly and Renilla luciferase activities were measured in cell lysates 18 to 20 h posttransfection. (B) Cell lysate aliquots were collected and subjected to immunoblotting using anti-NP or anti-P antibody. NC, negative control without L. Error bars are the standard deviations of means from examples containing six replicates for each transfection.
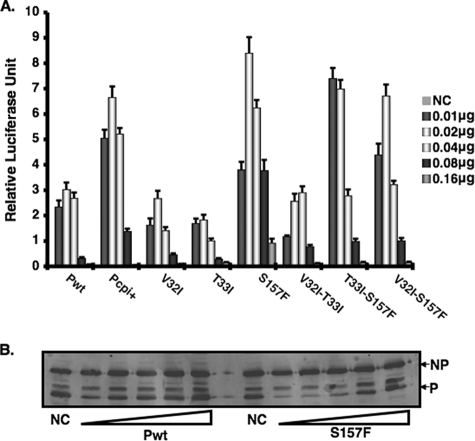
A single change at amino acid residue 157 of the P protein is responsible for elevated viral gene expression.
To further identify the residue that plays a critical role in elevated viral gene expression, we generated a series of P-protein mutations that contain double or single amino acid residue changes at positions 32, 33, and 157 (Table 1). Since the minigenome system reproduces the observation made for virus-infected cells concerning viral RNA synthesis and the minigenome system is free of interference from the V protein, this system was used to identify the amino acid residue that is responsible for increased activity of the P protein. We found that whenever the amino acid residue at position 157 was changed from Ser to Phe (S157F), the P protein had a higher activity than the wt P protein (Fig. 4A), indicating that this residue plays a critical role in modulating P-protein activity. The expression levels of the different viral proteins were similar, as indicated by immunoblotting of the NP and P proteins (Fig. 4B; also data not shown).
FIG. 4.
Activities of mutant P proteins in minigenome system. (A) Increasing amounts of plasmids encoding P and different P mutant proteins that contain double or single substitution mutations at amino acid positions 32, 33, and 157 (Table 1) were transfected along with other plasmids of the PIV5 minigenome system. Replication of the minigenome systems was measured as described in Materials and Methods. (B) Cell lysate aliquots from P and S157F protein-expressing cells were collected and subjected to immunoblotting using anti-NP or anti-P antibody. NC, negative control which lacks L. Error bars are standard deviations of means from examples containing six replicates for each transfection.
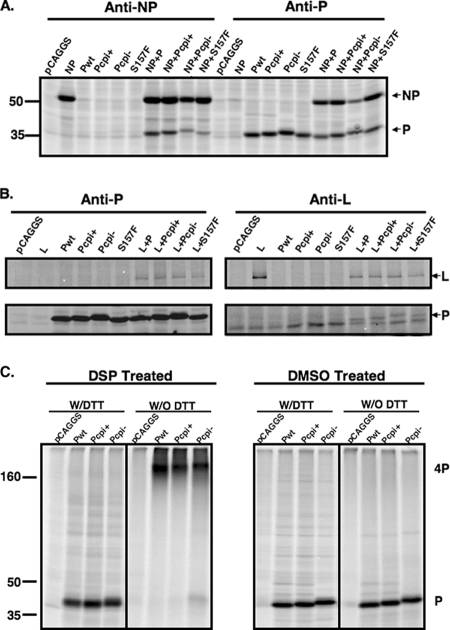
Protein-protein interactions involving the P protein.
To further investigate the mechanism of elevated viral RNA replication of Pcpi−, interactions between P proteins and other viral proteins, as well as those between the P protein and itself, were examined. It is known that the P protein interacts with the NP protein to serve as a chaperone for the NP protein to encapsidate viral RNA genomes, and the P protein also interacts with the L protein to stabilize the L protein. The P protein forms a homo-oligomer, which is essential for its function. To examine whether changes within the P proteins of Pcpi− and Pcpi+ affect their associations with the NP or L protein, coimmunoprecipitation was performed using [35S]Met/Cys-labeled transfected cell lysates. The amounts of NP precipitated by anti-P in Pcpi−transfected cells in Fig. 5A look different. However, the expression level of Pcpi− is much lower than those of the rest. When we adjusted the expression levels of P, the amount of NP precipitated by anti-P in cells transfected with Pcpi− is not obviously different from the rest. Thus, no obvious differences were observed in interactions between NP and P, Pcpi−, or Pcpi+. In addition, no obvious differences were observed in interactions between L and P, Pcpi−, or Pcpi+ (Fig. 5A and B). It is unlikely that interactions between P and NP or interactions between P and L play critical roles in the increased activities of the Pcpi− and Pcpi+ proteins. To examine possible effects on P-protein oligomer formation, we carried out cross-linking experiments. While there was a small amount of the Pcpi− monomeric form in cross-linking experiments, the amount was small and we did not consistently observe it. In addition, the same thing was not observed with Pcpi+. Because Pcpi+ also caused elevated viral gene expression, we conclude that the small amount of the monomeric form of Pcpi− is not significant for its ability to enhance viral gene expression (Fig. 5C). Thus, it is unlikely that homo-oligomer formation contributes to increased RNA replication activities of Pcpi− or Pcpi+.
FIG. 5.
Interactions between P and P mutant with NP or L and oligomer formation of P. (A and B) Coimmunoprecipitation of P and P mutants with either L (with two Flag tags) or NP, respectively. BSR T7 cells were transfected with empty plasmid or plasmids encoding P, Pcpi−, Pcpi+, S157F, and/or L (A) or and/or NP (B). At 24 h posttransfection, the cells were metabolically labeled with 35S-ProMix, lysed, and then subjected to immunoprecipitation with anti-P and anti-NP antibody (A) or anti-Flag antibody for L (B). The precipitates were then resolved in 10% SDS-PAGE and visualized by using a Storm PhosphorImager. (C) P protein oligomer formation. P and P mutants were expressed in BSR T7 cells, metabolically labeled, and then cross-linked using disuccinimidyl tartrate or dimethylsulfoxide as a control. The cells were lysed then subjected to immunoprecipitation with anti-P antibody. The precipitates were mixed with protein lysis buffer in the presence or absence of DTT, resolved in 10% SDS-PAGE, and visualized as described in Materials and Methods.
Decreased phosphorylation of Pcpi− and Pcpi+ proteins.
It is known that the P protein is heavily phosphorylated and its phosphorylation plays a critical role in its function. The Pcpi+ protein contains three residues (V32I, T33I, and S157F) that differ from the wt P protein, and two of them are potential phosphorylation sites in the wt P protein (T33 and S157). To determine whether these sites are involved in phosphorylation of the P protein, cells were infected with mock, PIV5, rPIV5-CPI− or rPIV5-CPI+ and labeled with [35S]Met/Cys or [33P]orthophosphate for 3 to 4 h at 1 dpi. The P proteins were immunoprecipitated and resolved by SDS-PAGE. Interestingly, both the Pcpi− and Pcpi+ proteins had reduced phosphorylation levels (about 70% of that of the wt P protein) (Fig. 6A and B). That the reduction of phosphorylation in Pcpi− is modest is consistent with the notion that the P protein is phosphorylated at multiple sites and that these residues affect phosphorylation only of a subset of these sites. Because rPIV5-CPI− and rPIV5-CPI+ have elevated viral gene expression, the expression levels of P in these viruses are higher than that in wt virus-infected cells.
FIG. 6.
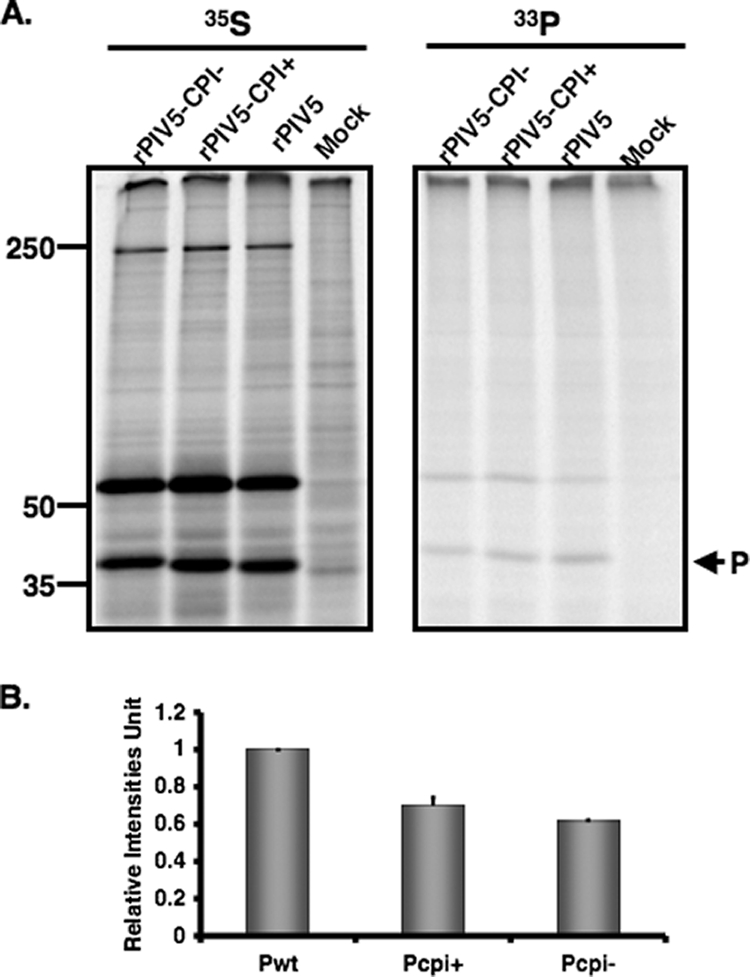
Phosphorylation of P in cells infected with rPIV5-CPI−, rPIV5-CPI+, or PIV5. BSR T7 cells were mock infected or infected with wt PIV5, rPIV5-CPI−, or rPIV5-CPI+ at a MOI of 3. At 18 to 20 hpi, the infected cells were labeled with 35S-ProMix or [33P]orthophosphate for 4 h. The cells were lysed and immunoprecipitated with anti-P antibody. The P proteins were immunoprecipitated and resolved in 10% SDS-PAGE (A). The results of three experiments were averaged and graphed (B). The level of wt P phosphorylation, defined as the intensity of 33P/35S, is set as 1. Error bars are standard deviations of means. The average reduction in phosphorylation of the P protein from three experiments is graphed (P = 0.02).
S157 of P protein is phosphorylated.
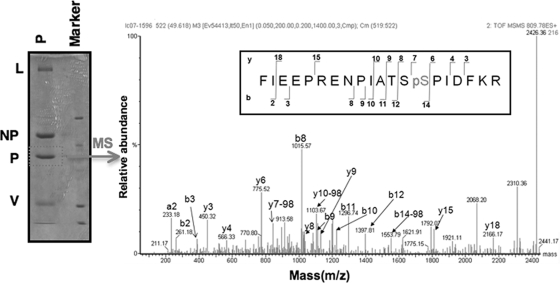
To investigate whether T33 and S157 of the P protein are phosphorylated, we used a web-based program, NetPhos (http://www.cbs.dtu.dk/services/NetPhos/), that predicts phosphorylation sites within a protein. The program predicts that the likelihood of T33 being phosphorylated is 0.025, while the likelihood of S157 being phosphorylated is 0.994 (on a scale from 0 to 1, with 1 being the highest likelihood of phosphorylation). Thus, it is predicted that S157 is phosphorylated. To further investigate this possibility, we purified the P protein from PIV5-infected cells using affinity purification with anti-P antibody. The purified P protein was resolved by SDS-PAGE, and the P protein band was excised and digested with trypsin. The digested peptides were subjected to MS analysis for phosphorylation site determination. The MS detected 71% of the total P protein sequence. A peptide containing S157 was identified in which the S157 position was indeed phosphorylated (Fig. 7). While peptides containing T33 were identified in the MS analysis, no phosphorylation of this residue was detected.
FIG. 7.
Determination of phosphorylation sites within the P protein. HeLa cells were infected with wt PIV5 at a MOI of 3. The P protein was immunoprecipitated from cell lysates using anti-V5-conjugated agarose (Sigma-Aldrich) which recognizes the P protein and resolved by 10% SDS-PAGE as described in Materials and Methods. The band corresponding to the P protein was excised, digested with trypsin, and then processed to enrich phosphopeptides using TiO2. Both the enriched fraction and the flowthrough (which contains all of the nonphosphorylated peptides) were analyzed by LC-MS/MS on a Waters Q-Tof Ultima mass spectrometer. The graph represents the LC-MS/MS product ion spectrum of the parent ion of phosphorylated peptide 144-163 from the trypsin digest of the PIV5 P protein. The yn-98 ion corresponds to the natural loss of H3PO4 from the parent ion. The b- and y-type fragment ions observed are shown with the peptide sequence (insert box). The x axis and y axis show mass-to-charge ratio (m/z) and relative abundance of the ions (% relative intensity), respectively.
To examine the role of phosphorylation at residue S157 in viral RNA synthesis, we constructed P-protein S157A and S157D mutants and examined the activities of these proteins using the minigenome replication system (Fig. 8). As expected, the S157A mutation resulted in higher reporter gene expression than that observed with the wt P protein. Interestingly, the S157D mutation also resulted in higher reporter gene expression than that for the wt P protein.
FIG. 8.
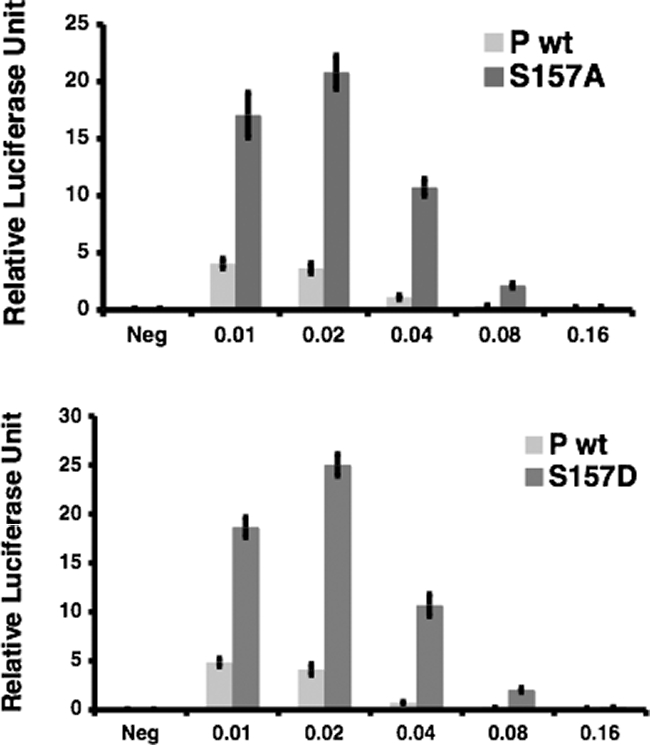
Activities of S157A and S157D proteins. Increasing amounts of plasmids encoding P (wt) or a P mutant (S157A or S157D) were transfected along with other plasmids of the PIV5 minigenome system. Replication of the minigenome systems was measured as described in Materials and Methods. Neg, negative control without L. Error bars are standard deviations of means from examples containing six replicates for each transfection.
DISCUSSION
In this work, we have investigated roles of the P and V proteins in the phenotype of rPIV5-CPI−, which contains six mutations in the shared region of the V and P proteins. One main characteristic of rPIV5-CPI− is that it causes elevated viral gene expression. Because the P protein plays an essential role in virus RNA synthesis and the V protein regulates virus RNA synthesis as well, it was not clear which of these altered proteins contributes to the elevated viral gene expression. It is possible that the P protein in rPIV5-CPI− (Pcpi−) gains new function and/or the V protein in rPIV5-CPI− (Vcpi−) loses its ability to inhibit viral RNA synthesis. We have found that the Pcpi− protein indeed has elevated activity on viral gene expression. We have also found that the Vcpi− maintains its ability to inhibit viral RNA synthesis in the minigenome system. Thus, we conclude that the ability of rPIV5-CPI− to cause elevated viral gene expression is likely due to a P protein with increased ability to facilitate replication of viral RNA.
To study the mechanism of the elevated activity of the Pcpi− protein, we examined viral RNA synthesis (viral RNA and viral mRNA). We have found that rPIV5-CPI− causes elevated viral RNA replication while maintaining similar levels of viral mRNA transcription per viral genome. This result is consistent with our observation that there is no difference between the Pcpi− and P proteins in gene expression from the minigenome system containing a defective trailer sequence in which only viral transcription occurs, whereas Pcpi− enhances reporter gene expression from a fully functional minigenome system. Since we have not found any significant difference among P proteins in their ability to associate with NP or L and to form homo-oligomers, we speculate that the elevated gene expression phenotype may be associated with a host protein. We have mapped the amino acid residue that plays a critical role in the elevated gene expression to serine at position 157. Interestingly, S157 is phosphorylated in the wt P protein. Due to the limitation of methods employed to determine phosphorylation, it is not clear what percentage of P is phosphorylated, nor is it known whether phosphorylation of S157 is dynamic and regulated during the virus replication cycle. Nonetheless, this is the first time a phosphorylation site has been identified in the P protein of PIV5. Furthermore, this is first time that an effect on viral gene expression has been observed after altering the phosphorylation site of P in a paramyxovirus genome. Intriguingly, mutating this phosphorylation site to F results in higher activity for the P protein, and this higher activity is limited to viral RNA replication but not transcription, indicating that this residue plays a critical role in modulating virus RNA replication. The exact mechanism for increased RNA replication with the S157-to-F substitution is not clear. Interestingly, mutating S157 to A or D also results in increased activity for the P protein. It is noteworthy that the context of this S157 is within a Polo-like kinase 1 (PLK1) binding motif (SpSP; the second S is phosphorylated). We speculate that phosphorylation of residue 157 of the P protein is important for its interaction with PLK1 and that PLK1 may play an important role in regulating viral RNA replication. Out of 10 strains of PIV5 examined, 7 have F residues at position 157 instead of the S residue found in the common laboratory strain W3A (4). Further studies on the role of PLK1 will address this interesting possibility.
This increased activity of the P protein may explain some discrepancies in the literature. Previously a recombinant PIV5 lacking the conserved cysteine-rich C terminus (rPIV5VΔC) induced increased IFN-β and IL-6 expression, suggesting that the V protein plays a role in inhibiting expression of IFN-β and IL-6 in infected cells (15, 24). Ecotopic expression of the V protein or the C terminus of the V protein is sufficient to inhibit IFN-β expression (31). Thus, it is thought that the C terminus is required and essential for the V protein to inhibit IFN-β expression. Interestingly, it has also been reported that rPIV5-CPI− induces expression of IFN-β (42). One of the theories for explaining increased IFN-β expression by rPIV5-CPI− is that the V protein (Vcpi−) in the rPIV5-CPI− virus has lost its ability to inhibit IFN-β expression. However, because the mutations of Vcpi− are in the N terminus and the C terminus of Vcpi− is still intact, the result is seemingly inconsistent with reports that the C terminus is required and sufficient for inhibiting IFN-β expression. An alternative explanation is that rPIV5-CPI− induces expression of IFN-β differently from rPIV5VΔC. Our result that Pcpi− is more efficient in facilitating replication of viral RNA is consistent with this hypothesis. It is possible that more efficient Pcpi− protein has an advantage over the wt P protein in allowing more rapid production of viral proteins in infected cells and thus more virus progenies. However, the downside of this increased replication rate may be increased or faster production of viral proteins or RNA that can be detected by host cell innate immune response sensors. This could conceivably lead to enhanced production of IFN early in rPIV5-CPI− infections, before sufficient V protein has been produced to block IFN signaling. Such a scenario is consistent with the observation that the V protein can inhibit IFN-β expression independently of virus infection, such as double-stranded-RNA-induced IFN-β expression (31). Recently it was reported with the studies of rPIV5-CPI− that the P protein may have a role in inhibiting host innate immune responses; however, this putative role of the P protein in limiting host innate immune responses has been observed only in virus-infected cells (9). In this experimental system, when an additional copy of the P gene was inserted into the genome of rPIV5-CPI−- (rPIV5-CPI-/P), expression of IFN-β was reduced in the virus-infected cells. It is possible that this additional copy of the P gene reduces viral gene expression, since overexpression of the P protein inhibits viral gene expression. That Pcpi− is more efficient in facilitating replication of viral RNA is consistent with the theory that synthesizing more viral RNA may result in more-robust innate immune responses. However, the results presented here do not exclude the possibility that the P protein may be able to interrupt innate immune responses through a mechanism that is independent of its role in viral RNA synthesis.
It is well known that overexpression of the P protein results in inhibition of viral RNA synthesis, which is consistent with our observation (Fig. 2 and 4) that using too much plasmid encoding the P protein in the minigenome system reduces viral RNA synthesis (23). Interestingly, increased expression levels did not seem to inhibit reporter gene expression from the minigenome system with a defective trailer sequence, in which only viral RNA transcription is measured. Thus, it appears that overexpression of the P protein inhibits viral RNA replication while having no effect on viral RNA transcription. It is possible that the overexpressed P protein may outcompete the NP protein, preventing it from encapsidating nascent viral RNA genomes, thus inhibiting viral RNA replication. In the case of viral RNA transcription, overexpression of the P protein would have no effect because the NP protein is not needed for functionality of viral mRNA.
Acknowledgments
We thank the members of Biao He's laboratory for helpful discussion and technical assistance. We are grateful to Rick Randal for providing antibody against P.
This work was supported by grants from the National Institute of Allergy and Infectious Diseases to B.H. (AI051372 and K02 AI65795).
Footnotes
Published ahead of print on 9 July 2008.
REFERENCES
- 1.Andrejeva, J., K. S. Childs, D. F. Young, T. S. Carlos, N. Stock, S. Goodbourn, and R. E. Randall. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-β promoter. Proc. Natl. Acad. Sci. USA 10117264-17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baumgartner, W. K., S. Krakowka, A. Koestner, and J. Evermann. 1982. Acute encephalitis and hydrocephalus in dogs caused by canine parainfluenza virus. Vet. Pathol. 1979-92. [DOI] [PubMed] [Google Scholar]
- 3.Buchholz, U. J., S. Finke, and K. K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chatziandreou, N., N. Stock, D. Young, J. Andrejeva, K. Hagmaier, D. J. McGeoch, and R. E. Randall. 2004. Relationships and host range of human, canine, simian and porcine isolates of simian virus 5 (parainfluenza virus 5). J. Gen. Virol. 853007-3016. [DOI] [PubMed] [Google Scholar]
- 5.Chatziandreou, N., D. Young, J. Andrejeva, S. Goodbourn, and R. E. Randall. 2002. Differences in interferon sensitivity and biological properties of two related isolates of simian virus 5: a model for virus persistence. Virology 293234-242. [DOI] [PubMed] [Google Scholar]
- 6.Curran, J. 1998. A role for the Sendai virus P protein trimer in RNA synthesis. J. Virol. 724274-4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das, T., A. K. Pattnaik, A. M. Takacs, T. Li, L. N. Hwang, and A. K. Banerjee. 1997. Basic amino acid residues at the carboxy-terminal eleven amino acid region of the phosphoprotein (P) are required for transcription but not for replication of vesicular stomatitis virus genome RNA. Virology 238103-114. [DOI] [PubMed] [Google Scholar]
- 8.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 739928-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dillon, P. J., and G. D. Parks. 2007. Role for the phosphoprotein P subunit of the paramyxovirus polymerase in limiting induction of host cell antiviral responses. J. Virol. 8111116-11127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dupuy, L. C., S. Dobson, V. Bitko, and S. Barik. 1999. Casein kinase 2-mediated phosphorylation of respiratory syncytial virus phosphoprotein P is essential for the transcription elongation activity of the viral polymerase; phosphorylation by casein kinase 1 occurs mainly at Ser(215) and is without effect. J. Virol. 738384-8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emerson, S. U., and Y.-H. Yu. 1975. Both NS and L proteins are required for in vitro RNA synthesis by vesicular stomatitis virus. J. Virol. 151348-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evermann, J. F., J. D. Lincoln, and A. J. McKiernan. 1980. Isolation of a paramyxovirus from the cerebrospinal fluid of a dog with posterior paresis. J. Am. Vet. Med. Assoc. 1771132-1134. [PubMed] [Google Scholar]
- 13.Gainey, M. D., P. J. Dillon, K. M. Clark, M. J. Manuse, and G. D. Parks. 2008. Paramyxovirus-induced shut off of host and viral protein synthesis: role of the P and V proteins in limiting PKR activation. J. Virol. 82828-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He, B. 1996. A gene expression system based on bacteriophage RNA polymerase and characterization of bacteriophage T7 RNA polymerase. Ph.D. thesis. State University of New York, Brooklyn, NY.
- 15.He, B., R. G. Paterson, N. Stock, J. E. Durbin, R. K. Durbin, S. Goodbourn, R. E. Randall, and R. A. Lamb. 2002. Recovery of paramyxovirus simian virus 5 with a V protein lacking the conserved cysteine-rich domain: the multifunctional V protein blocks both interferon-beta induction and interferon signaling. Virology 30315-32. [DOI] [PubMed] [Google Scholar]
- 16.He, B., R. G. Paterson, C. D. Ward, and R. A. Lamb. 1997. Recovery of infectious SV5 from cloned DNA and expression of a foreign gene. Virology 237249-260. [DOI] [PubMed] [Google Scholar]
- 17.Hu, C., and K. C. Gupta. 2000. Functional significance of alternate phosphorylation in Sendai virus P protein. Virology 268517-532. [DOI] [PubMed] [Google Scholar]
- 18.Hu, C. J., A. Kato, M. C. Bowman, K. Kiyotani, T. Yoshida, S. A. Moyer, Y. Nagai, and K. D. Gupta. 1999. Role of primary constitutive phosphorylation of Sendai virus P and V proteins in viral replication and pathogenesis. Virology 263195-208. [DOI] [PubMed] [Google Scholar]
- 19.Lamb, R. A., and D. Kolakofsky. 2001. Paramyxoviridae: the viruses and their replication, p. 1305-1340. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams and Wilkins, Philadelphia, PA.
- 20.Lin, G. Y., and R. A. Lamb. 2000. The paramyxovirus simian virus 5 V protein slows progression of the cell cycle. J. Virol. 749152-9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin, G. Y., R. G. Paterson, and R. A. Lamb. 1997. The RNA binding region of the paramyxovirus SV5 V and P proteins. Virology 238460-469. [DOI] [PubMed] [Google Scholar]
- 22.Lin, Y., A. C. Bright, T. A. Rothermel, and B. He. 2003. Induction of apoptosis by paramyxovirus simian virus 5 lacking a small hydrophobic gene. J. Virol. 773371-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin, Y., F. Horvath, J. A. Aligo, R. Wilson, and B. He. 2005. The role of simian virus 5 V protein on viral RNA synthesis. Virology 338270-280. [DOI] [PubMed] [Google Scholar]
- 24.Lin, Y., M. Sun, S. M. Fuentes, C. D. Keim, T. Rothermel, and B. He. 2007. Inhibition of interleukin-6 expression by the V protein of parainfluenza virus 5. Virology 368262-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liston, P., and D. J. Briedis. 1994. Measles virus V protein binds zinc. Virology 198399-404. [DOI] [PubMed] [Google Scholar]
- 26.Lu, B., C. H. Ma, R. Brazas, and H. Jin. 2002. The major phosphorylation sites of the respiratory syncytial virus phosphoprotein are dispensable for virus replication in vitro. J. Virol. 7610776-10784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants by a novel eukaryotic vector. Gene 108193-200. [DOI] [PubMed] [Google Scholar]
- 28.Parisien, J. P., J. F. Lau, J. J. Rodriguez, C. M. Ulane, and C. M. Horvath. 2002. Selective STAT protein degradation induced by paramyxoviruses requires both STAT1 and STAT2 but is independent of alpha/beta interferon signal transduction. J. Virol. 764190-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paterson, R. G., and R. A. Lamb. 1993. The molecular biology of influenza viruses and paramyxoviruses, p. 35-73. In A. Davidson and R. M. Elliott (ed.), Molecular virology: a practical approach. IRL Oxford University Press, Oxford, United Kingdom.
- 30.Paterson, R. G., G. P. Leser, M. A. Shaughnessy, and R. A. Lamb. 1995. The paramyxovirus SV5 V protein binds two atoms of zinc and is a structural component of virions. Virology 208121-131. [DOI] [PubMed] [Google Scholar]
- 31.Poole, E., B. He, R. A. Lamb, R. E. Randall, and S. Goodbourn. 2002. The V proteins of simian virus 5 and other paramyxoviruses inhibit induction of interferon-beta. Virology 30333-46. [DOI] [PubMed] [Google Scholar]
- 32.Randall, R. E., and A. Bermingham. 1996. NP:P and NP:V interactions of the paramyxovirus simian virus 5 examined using a novel protein:protein capture assay. Virology 224121-129. [DOI] [PubMed] [Google Scholar]
- 33.Randall, R. E., D. F. Young, K. K. A. Goswami, and W. C. Russell. 1987. Isolation and characterization of monoclonal antibodies to simian virus 5 and their use in revealing antigenic differences between human, canine and simian isolates. J. Gen. Virol. 682769-2780. [DOI] [PubMed] [Google Scholar]
- 34.Russell, R., R. G. Paterson, and R. A. Lamb. 1994. Studies with cross-linking reagents on the oligomeric form of the paramyxovirus fusion protein. Virology 199160-168. [DOI] [PubMed] [Google Scholar]
- 35.Southern, J. A., D. F. Young, F. Heaney, W. K. Baumgartner, and R. E. Randall. 1991. Identification of an epitope on the P and V proteins of simian virus 5 that distinguishes between two isolates with different biological characteristics. J. Gen. Virol. 721551-1557. [DOI] [PubMed] [Google Scholar]
- 36.Steward, M., A. C. R. Samson, W. Errington, and P. T. Emmerson. 1995. The Newcastle disease virus V protein binds zinc. Arch. Virol. 1401321-1328. [DOI] [PubMed] [Google Scholar]
- 37.Sun, M., S. M. Fuentes, K. Timani, D. Sun, C. Murphy, Y. Lin, A. August, M. N. Teng, and B. He. 2008. Akt plays a critical role in replication of nonsegmented negative-stranded RNA viruses. J. Virol. 82105-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun, M., T. A. Rothermel, L. Shuman, J. A. Aligo, S. Xu, Y. Lin, R. A. Lamb, and B. He. 2004. Conserved cysteine-rich domain of paramyxovirus simian virus 5 V protein plays an important role in blocking apoptosis. J. Virol. 785068-5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas, S. M., R. A. Lamb, and R. G. Paterson. 1988. Two mRNAs that differ by two nontemplated nucleotides encode the amino coterminal proteins P and V of the paramyxovirus SV5. Cell 54891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ulane, C. M., and C. M. Horvath. 2002. Paramyxoviruses SV5 and HPIV2 assemble STAT protein ubiquitin ligase complexes from cellular components. Virology 304160-166. [DOI] [PubMed] [Google Scholar]
- 41.Waning, D. L., A. P. Schmitt, G. P. Leser, and R. A. Lamb. 2002. Roles for the cytoplasmic tails of the fusion and hemagglutinin-neuraminidase proteins in budding of the paramyxovirus simian virus 5. J. Virol. 769284-9297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wansley, E. K., and G. D. Parks. 2002. Naturally occurring substitutions in the p/v gene convert the noncytopathic paramyxovirus simian virus 5 into a virus that induces alpha/beta interferon synthesis and cell death. J. Virol. 7610109-10121. [DOI] [PMC free article] [PubMed] [Google Scholar]