Abstract
Latent Epstein-Barr virus (EBV) infection is associated with several lymphoproliferative disorders, including posttransplant lymphoma, Hodgkin's disease, and Burkitt's lymphoma, as well as nasopharyngeal carcinoma (NPC). Twenty-nine microRNAs (miRNAs) have been identified that are transcribed during latent infection from three clusters in the EBV genome. Two of the three clusters of miRNAs are made from the BamHI A rightward transcripts (BARTs), a set of alternatively spliced transcripts that are highly abundant in NPC but have not been shown to produce a detectable protein. This study indicates that while the BART miRNAs are located in the first four introns of the transcripts, processing of the pre-miRNAs from the primary transcript occurs prior to completion of the splicing reaction. Additionally, production of the BART miRNAs correlates with accumulation of a spliced mRNA in which exon 1 is joined directly to exon 3, suggesting that this form of the transcript may favor production of miRNAs. Sequence variations and processing of pre-miRNAs to the mature form also may account for various differences in miRNA abundance. Importantly, residual intronic pieces that result from processing of the pre-miRNAs were detected in the nucleus. The predicted structures of these pieces suggest there is a bias or temporal pattern to the production of the individual pre-miRNAs. These findings indicate that multiple factors contribute to the production of the BART miRNAs and to the apparent differences in abundance between the individual miRNAs of the cluster.
Epstein-Barr virus (EBV) is associated with multiple human cancers of both lymphoid and epithelial cell origins, including Burkitt's lymphoma (BL), Hodgkin's disease, posttransplant lymphoma, and nasopharyngeal carcinoma (NPC) (21, 32, 41). EBV also efficiently transforms B lymphocytes in vitro into permanent lymphoblastoid cell lines (LCLs) (29, 31). Initial studies of viral transcription in NPC revealed striking differences in comparison with transcription in LCLs and BL cell lines (19, 20, 24, 34). In NPC, the regions that encoded the viral nuclear proteins EBNAs 2, 3A, 3B, and 3C were not transcribed while the right terminal region including the BamHI A fragment was abundantly transcribed (21, 34). Subsequent studies determined that these differences reflected three types of latent EBV infection. In type III latency, which is characteristic of LCLs, the EBNA2 and EBNA3 proteins were expressed, while in type II latency found in NPC, these EBNAs were not expressed (21). In addition in NPC, the BamHI A RNAs (BamHI A rightward transcripts [BARTs]) were very abundant and could be detected by Northern blotting (13, 17). In contrast, in LCLs, the BARTs were expressed at very low levels, as detected by Northern blotting, but could be detected by using reverse transcription-PCR (RT-PCR) to detect some of the specific splices (1, 8, 13, 18, 35). Several open reading frames (ORFs) have been identified within the differently spliced BARTs (1, 8, 18, 35, 37). Recombinant proteins representing the ORFs have interesting properties; however, the protein products have not been identified in infected cells (12, 14, 18, 22, 25, 38).
EBV was the first human virus shown to encode microRNAs (miRNAs) that mapped to the BHRF1 and BART regions of the genome (30). These miRNAs were initially identified in the prototype laboratory strain, B95-8, which has a 12-kb deletion that spans the BART sequences (33). A subsequent study identified and cloned an additional 14 miRNAs produced from the BART transcripts in the BC1 cell line, a cell line that is coinfected with EBV and Kaposi sarcoma herpesvirus (KSHV) (6). This study analyzed expression of the miRNAs in additional lymphoid cell lines and the C15 NPC xenograft and C666.1 NPC cell line. The BART miRNAs were abundant in C15 and C666.1 and expressed at lower levels in BC1 and the Jijoye BL cell line (6). Other lymphoid cell lines had barely detectable expression of some of the miRNAs. This study also revealed that the BHRF1 miRNAs were actually expressed from the primary EBNA transcript that is only produced in lymphocytes with type III latency (6). In addition, a computer analysis identified nine additional miRNAs within the BART that are located at various positions between the initial 16 miRNAs (16).
In this study, expression of the BART miRNAs was evaluated in multiple lymphoid and epithelial cell lines. The data indicate that the levels of expression of the miRNAs are linked in part to the transcription of the BART mRNA. Interestingly, stable residual pieces of the intron were detected in the nucleus of cells that express the miRNAs. Characterization of these residual pieces indicated that the miRNAs are produced from a large initial transcript prior to splicing and that a specific spliced form of the transcript favors production of miRNAs. The specific residual intron pieces remaining after the processing of the miRNAs suggest that the individual miRNAs in the transcript are produced with different efficiencies. These findings indicate that mRNA abundance and structure regulate the production of the EBV miRNAs.
MATERIALS AND METHODS
Cell culture and reagents.
The EBV-positive BL cell lines Jijoye, Namalwa, Mutu 1, and Mutu 3, the EBV-negative adenocarcinoma cell line AdAH and the EBV-positive adenocarcinoma cell line NPC-KT, the EBV-positive and KSHV-positive PEL line BC1 and the KSHV-positive PEL cell line BCBL1, the EBV-positive multiple myeloma cell line IM9, and the EBV-positive epithelial cell line C666.1 were grown at 37°C in RPMI 1640 (Gibco) supplemented with 10% fetal bovine serum (Sigma) and antibiotics. The gastric carcinoma cell line AGS infected with the green fluorescent protein (GFP)-tagged Akata EBV strain was maintained in F-12 medium (Gibco) supplemented with 10% fetal bovine serum and antibiotics and 500 μg/ml G418 (Gibco). NPC tumors C15, C17, and C18 were serially passaged in nude mice and have been described previously (3).
RNA preparations.
Total RNA was prepared using TRIzol reagent (Invitrogen) from cell pellets or pulverized tumor tissue. Cell fractionations were as follows: cells were lysed for 10 min on ice in a mixture of 10 mM Tris (pH 7.4), 50 mM KCl, 5 mM MgCl2, and 0.05% NP-40 and then centrifuged at 900 × g for 1 min at 4°C. The supernatant was designated “cytosol.” The pellet was washed in a mixture of 150 mM NaCl, 1.5 mM MgCl2, 10 mM Tris (pH 7.4), and 0.65% NP-40 and then centrifuged at 900 × g for 1 min at 4°C. The resulting pellet was designated the “nuclear fraction.” RNA was prepared from each fraction using TRIzol.
Northern blotting.
Northern gels to screen for miRNAs were prepared from precast 15% urea-Tris-borate-EDTA (TBE) gels (Bio-Rad). RNA (5 μg) was mixed with 2× TBE-urea sample buffer (Bio-Rad), heated at 70°C for 4 min, quick cooled on ice, and then run at 30 mA in 1× TBE. A 72-mer oligonucleotide and a 20-mer oligonucleotide were used as markers. The gels were stained in ethidium bromide for visualization and then transferred to Hybond N+ (GE Biosciences) at 65 V in 1× TBE for 1 h. After UV cross-linking, the membranes were hybridized at 37°C with end-labeled antisense oligonucleotides to the miRNAs or U6 for a control using ExpressHyb solution (Clontech) as directed.
To resolve the mRNA structure and stable intron structure, 10 μg of total RNA, nuclear RNA, or cytosolic RNA was run on 1% agarose gels using NorthernMax denaturing gel buffer (Ambion), NorthernMax MOPS (morpholinepropanesulfonic acid) gel running buffer (Ambion), and RNA Millennium size markers with formamide (Ambion) as directed. The gels were transferred in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium phosphate) to Hybond N+ (GE Biosciences), UV cross-linked, and hybridized with 32P-labeled antisense riboprobes at 68°C in UltraHyb (Ambion) as directed.
Probes.
DNA sequences corresponding to the BamHI A coordinates noted in Table 1 and Fig. 6 were PCR amplified at 40 cycles using Sure Start Taq (Stratagene), gel purified (Qiaquick gel extraction kit; Qiagen), restriction digested (NEB), and cloned with EcoRI/HindIII into pGEM3Z (Promega). Antisense riboprobes were generated from linearized plasmids using Sp6 polymerase (Promega) as previously described (35). An antisense riboprobe for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was synthesized from the pTRI-GAPDH human antisense control template (Ambion). The riboprobes were purified over G-50 Quick columns (Roche).
TABLE 1.
Primers used for PCR and hybridizationa
| Name | Location | Sequence |
|---|---|---|
| miRNA probe oligonucleotides | ||
| BART3-3p | 139145L | ACACCTGGTGACTAGTGGTGCG |
| BART4 | 139249L | AGCACACCAGCAGCATCAGGTC |
| BART1-3p | 139409L | AGACATAGTGGATAGCGGTGCTA |
| BART5 | 139698L | CGATGGGCAGCTATATTCACCTT |
| BART6 | 140093L | TCTAAGGCTAGTCCGATCCCCG |
| BART18 | 145984L | CTGTATAGGAAGTGCGAACTTG |
| BART7 | 146496L | CCCTGGACACTGGACTATGAT |
| BART8 | 146793L | CTGTACAATCTAGGAAACCGT |
| BART9 | 147018L | CTACGGGACCCATGAAGTGTTA |
| BART10 | 147378L | ACAGCCAACTCCATGGTTATGT |
| BART12 | 147958L | AAACCACACCAAACACCACAGGA |
| BART13 | 149585L | TCAGCCGTCCCTGGCAAGTTAC |
| BART14 | 148765L | TGTAAATCGGCAGCGTAGGGTA |
| Promoter usage | ||
| P2 (DH) | 138203R | CACAGCAGCACAATAGAAGCAC |
| P2 | 138081R | CTCGATCCGAGAGACCGACT |
| P1 | 138356R | GCCATTTTTCAAGCTGCTAAACC |
| BART RT primers | ||
| 1R | 138356R | Same as P1 |
| 1aL | 139513L | CGGCACAGATTAGAAGATGA |
| 1bR | 140641R | CTACCTGCCCCTCTAATCTG |
| 1bL | 140625L | TTCTAGAAACACGTGTCCCG |
| 2R | 146302R | GCTGATAAGTGCTGCGTCCACT |
| 2L | 146268L | CGTCCAAATTCAGCTCA |
| 3L | 149659L | ATTCCCGGGGAAGGTGTGTC |
| BamHI A sequencing primers | ||
| 138356R | Same as P1 | |
| 138895R | CGGAATATGCAAGTGCATCTT | |
| 139001R | CTATAGGTCCTACCGGAGCT | |
| 139566R | TTTCCTTGATAGAGACACAA | |
| 139711L | TCCAGCGATACGTCGATGGG | |
| 140216L | TAGTGAGCCAGGCTTTCTAT | |
| 140764L | TGGGTCCTGAGACCCAAAGGGACAG | |
| 145931R | ATTGTTGCCGTTGAAAGACG | |
| 146161R | TTTTTTGCAAATTGGCCTTA | |
| 146302R | Same as 2R | |
| 146552L | CGTACTGGCCACACTAAACA | |
| 147281R | AGACAGGCGGCGCATTGGAA | |
| 147378L | ACAGCCAACTCCATGGTTATGT | |
| 148881L | AGCAGTCGCATGGCGGGAGTAT | |
| 149659L | Same as 3L | |
| Other oligonucleotides for probes | ||
| U6 | ATATGGAACGCTTCACGAATT | |
| Ex3 | 149659L | ATTCCCGGGGAAGGTGTGTC |
| Ex1 | 138419L | TCCATGTTGCTGAGGAGCTT |
| Primers for 5′ RACE | ||
| a | 146420L | CATGGTCCAAGTAGGTCACC |
| b | 146394L | CTACTGCCTTATGCAGCCCACCAA |
| dT-adapter | GACTCGAGTCGACATCGAT(17) | |
| Adapter | GACTCGAGTCGACATCG | |
| Primers for cloning pre-miRNAs | ||
| BART3 | 139076R-E | ATCACGAGGAATTCCCTTTGGTGGAAC |
| 139154L-H3 | ATCACGAGAAGCTTCCTCCGGTGACAC | |
| BART1 | 139340R-E | ATCACGAGGAATTCCCCGTGGGGGGTCTTAG |
| 139417L-H3 | ATCACGAGAAGCTTCCCGGGCGAGACATAG | |
| BART6 | 140016R-E | ATCACGAGGAATTCTGACCTTGTTGGTAC |
| 140107L-H3 | ATCACGAGAAGCTTTGGCCTTGAGTTAC | |
| BART18 | 145942R-E | ATCACGAGGAATTCTGAAAGACGGGTG |
| 146040L-H3 | ATCACGAGAAGCTTCGACATTATCGAG | |
| BART7 | 146425R-E | ATCACGAGGAATTCTCCAGTGTCCTGATC |
| 146510L-H3 | ATCACGAGAAGCTTTCCGAGTGCACTGTC | |
| BART14 | 148731R-E | ATCACGAGGAATTCCAGGGGTGGCCGGTAC |
| 148815L-H3 | ATCACGAGAAGCTTCAGGTCGCGCGTCCAG | |
| Primers for cloning introns | ||
| 1-1a | 138501R-E | ATCACGAGGAATTCGGATCAGGGTATGTAAAC |
| 138931L-H3 | ATCACGAGAAGCTTGCCCCTACTGGTTAGAAAG | |
| 1a-1b | 140198R-E | ATCACGAGGAATTCTAGAAAGCCTGGCTCACTAT |
| 140527L-H3 | ATCACGAGAAGCTTCACTGCCATAAGTCCTCATG | |
| 1b-2 | 145320R-E | ATCACGAGGAATTCAGTATCCAGTTCTAGAAG |
| 146000L-H3 | ATCACGAGAAGCTTATACAAGGCTTTAACACT | |
| 2-3 | 148819R-E | ATCACGAGGAATTCCTCTTCGGAAACGCCAAC |
| 149550L-H3 | ATCACGAGAAGCTTGTGGGCATGACCCTATTG | |
| Primers for cloning exons | ||
| 1 | 138361R-E | ATCACGAGGAATTCTTTTCAAGCTGCTAAACCAC |
| 138479L-H3 | ATCACGAGAAGCTTTCACATGAAGAGGCTAGTGC | |
| 1a | 139454R-E | ATCACGAGGAATTCCCCACCGAACCAGGCATGAT |
| 139535L-H3 | ATCACGAGAAGCTTGGTGGTCATGTTTCCCTCCA | |
| 1b | 140565R-E | ATCACGAGGAATTCTTCTATCAGAATAACAGGGG |
| 140664L-H3 | ATCACGAGAAGCTTCATACAGATTAGAGGGGCAG | |
| 2 | 146233R-E | ATCACGAGGAATTCATGCCGTTGAACGTGTCACT |
| 146310L-H3 | ATCACGAGAAGCTTCTTATCAGCGCTGGAGGCAC | |
| 3a | 149591R-E | ATCACGAGGAATTCTCCACGGAGACTCGGACGTA |
| 149696L-H3 | ATCACGAGAAGCTTCGCCCCCATTTCTAAAGTCA | |
| 3b | 149914R-E | ATCACGGAGAATTCTATGAACAGGCTGCGCGGAT |
| 150050L-H3 | ATCACGAGAAGCTTACGGCAGCCGCTACTTTACG | |
| 4 | 150261R-E | ATCACGAGGAATTCTCCTGAACGACGAG |
| 150340L-H3 | ATCACGAGAAGCTTTCGACGAGCTCCG | |
| 5a | 155278R-E | ATCACGAGGAATTCCAGCAGGATGCGC |
| 155335L-H3 | ATCACGAGAAGCTTATCCTGCGTCCTC | |
| 5b | 156528R-E | ATCACGAGGAATTCGGCATGTTGCTGAGCGAGCT |
| 156709L-H3 | ATCACGAGAAGCTTGCAGCCTGGTCTCTGGTTTC | |
| 6 | 156865R-E | ATCACGAGGAATTCTGGAAGTTGTACTC |
| 156913L-H3 | ATCACGAGAAGCTTCCTCCAACGTCTTTG | |
| 7a | 158629R-E | ATCACGAGGAATTCTCCGTGTGATTCTCCCGCGT |
| 158726L-H3 | ATCACGAGAAGCTTCCTTTCCGAGTCTGCGAGCT | |
| 7b | 159800R-E | ATCACGAGGAATTCTGCGCCGTGAAGG |
| 160192L-H3 | ATCACGAGAAGCTTCAGAGGAGGGCGC | |
| Primers for cloning BART cDNAs | ||
| 138353R | GTTGCCATTTTTCAAGCTGCTAAACCACGAGTGG | |
| 160086L | CTCTCGGAGCTCCTCTATTGGAACTCTGGC |
Primers are numbered at the 5′ end according to the EBV genome sequence AJ507799 and extend in the direction indicated (R, rightward; L, leftward). The EBV-specific nucleotides are shown in boldface in the primers containing restriction sites.
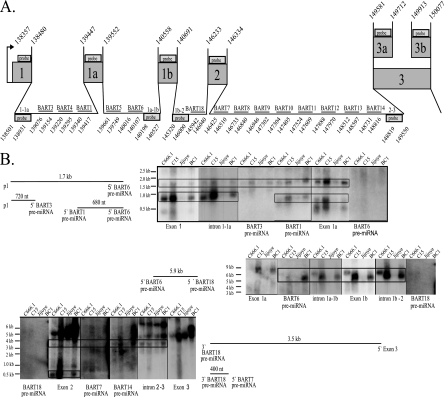
FIG. 6.
Detection of stable intronic RNA structures after miRNA processing. (A) Schematic of the 5′ BART region with the genomic coordinates of the exons, introns, BART pre-miRNAs, and probes used in the Northern analysis. The exon and intron probes are indicated by boxes with the 5′ and 3′ coordinates denoted. The pre-miRNA probes are indicated by horizontal lines with the 5′ and 3′ ends denoted. (B) Northern analysis and diagrams of the potential stable RNA structures in the tumor-derived cell lines resulting from miRNA processing. Total RNA was hybridized to antisense riboprobes representing exons, introns, and pre-miRNAs. Size markers are noted. The sizes and projected endpoints of the stable RNA structures are shown.
Antisense oligonucleotides to the mature miRNAs or U6 were 32P end labeled with polynucleotide kinase (NEB) and purified over G-25 Quick spin columns (Roche).
Sequencing.
The DNA sequence of the BamHI A region including the miRNAs was determined at the UNC-CH Genome Analysis Facility using Applied Biosystems genetic analyzers with the primers noted in Table 1 from PCR fragments amplified and gel purified from DNA isolated from the cell lines and xenographs using the DNeasy kit (Qiagen).
RT-PCR.
Total RNA (200 ng) was used as template for RT-PCR using the primers listed in Table 1 and the Brilliant quantitative RT-PCR core reagent kit (Stratagene) as directed by the manufacturer. The resulting products were resolved on agarose gels, and staining density was quantified using ImageJ. The resulting product bands were also gel purified (Qiaquick gel extraction kit; Qiagen) and sequenced.
cDNA cloning.
Poly(A) RNA was purified from total RNA from the xenograph C15 using Oligotex (Qiagen) according to the manufacturer's instructions. Ten micrograms of poly(A) RNA was used as a template in an oligo(dT)-primed RT reaction using OmniScript reverse transcriptase (Qiagen) as directed. This cDNA was used as a template for full-length cDNA amplification using primers specific for exon 1 (138353R) and exon 7 (160086L) of the BARTs. PCR was performed with 5 μl of a 1:20 cDNA dilution in a 100-μl final volume using the Expand Long PCR system (Roche) for 30 cycles of denaturing at 94°C for 1 min, annealing at 62°C for 1 min, and extension at 68°C for 10 min. The resulting PCR products were size selected on an agarose gel and purified with a Qiaquick gel extraction kit (Qiagen) as directed. The size-selected fragments (0.3 to 1 kb, 1 to 2 kb, and >2 kb) were kinase treated using polynucleotide kinase (NEB) and blunt ligated into pGEM3Z. Colonies were screened with an antisense oligonucleotide to exon 1.
5′ RACE.
Rapid amplification of cDNA ends (RACE) was performed as follows. To determine the 5′ ends of the stable residual pieces resulting from miRNA processing, BC1 nuclear RNA (200 ng) was primed with intron-specific primers (primer a) for first-strand cDNA synthesis using Omniscript (Qiagen) as directed (Table 1). Following cDNA synthesis, the RNA template was subsequently digested with RNase H and the cDNA was purified over a Qiaquick column (Qiagen). The cDNA was poly(A) tailed with terminal transferase (NEB). The resulting dA-tailed cDNA was diluted 1/5 and used as substrate for PCR with nested leftward intronic primer b as denoted in Table 1 and an adapter-dT rightward primer. The PCR was performed under the following conditions: denaturing at 94°C for 5 min, annealing at 48°C for 2 min, and extension at 72°C for 40 min, followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 2 min. The resulting PCR was run on a 1.5% gel, and the resulting band was gel purified (Qiagen gel extraction kit) and sequenced.
Nucleotide sequence accession numbers.
The EBV coordinates listed correspond to EBV genome sequence AJ507799, in which the Raji sequences have been inserted into the B95-8-deleted EBV genome (11). The following BamHI A coordinates of the genomes were sequenced from the cell lines and xenographs and were deposited in GenBank under the corresponding accession numbers: for C15 138361 to 140753, 145320 to 148024, and 148181 to 148850, EU828625; for C17 138381 to 140185, 145931 to 147761, 147821 to 148118, and 148390 to 148850, EU828626; for C18 138384 to 140160, 145931 to 147761, 147831 to 148153, and 148391 to 148850, EU828627; for C666.1 139030 to 140216, 145931 to 147761, 147843 to 148200, and 148378 to 148836, EU828629; for BC1 139036 to 140216 and 145931 to 148850, EU828630; for IM9 138391 to 140170, 145931 to 148030, and 148360 to 148850, EU828628; for Jijoye 139037 to 139300, 139311 to 140216, 145931 to 148307, and 148501 to 148850, EU828631; for Mutu 139041 to 140216, 145931 to 148030, and 148431 to 148850, EU828632; and for Nam 139035 to 140216 and 145931 to 148850, EU828633.
RESULTS
Detection of miRNAs.
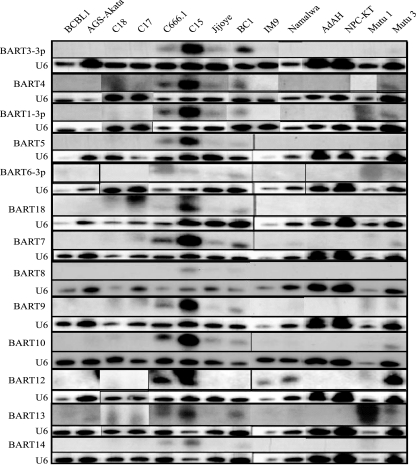
The BART miRNAs were originally shown to be expressed at a higher level in epithelial cells, including the C15 NPC xenograft tumor and NPC cell line C666.1. Some B cells such as BC1 and Jijoye also expressed the miRNAs at readily detectable levels (6). To determine if expression of the miRNAs was related to cell type and to identify expression of the miRNAs in EBV-infected epithelial and lymphoid cells, probes were prepared for the BART1-3p, -3-3p, -4, -5, -6-3p, -18, -7, -8, -9, -10, -12, -13, and -14 miRNAs and hybridized to RNA preparations from the infected epithelial cells AGS-Akata, C18, C17, C666.1, C15, and NPC-KT and from infected lymphoid cells Jijoye, BC1, IM9, Namalwa, Mutu 1, and Mutu 3 (Fig. 1). The EBV-negative epithelial cell line AdAH and the KSHV-positive/EBV-negative lymphoid cell line BCBL1 were included as negative controls. Loading was assessed by hybridization to the U6 snRNA. All of the miRNAs were detected in the C15 NPC tumor, and all but the BART8 miRNA were detected in the C666.1 NPC cell line. Lower levels were detected in the BC1 and Jijoye lymphoid lines. The miRNAs were not detectable in the EBV-infected NPC-KT or AGS-Akata epithelial cell lines. These findings indicate that expression of the miRNAs is not characteristic of a specific cell type. In an interesting study that indicated that the EBV BART miRNAs could regulate expression of LMP1, some of the miRNAs were detected in an EBV-infected derivative of the AGS cell line (AGS-BXI) (28). However, the AGS cell line analyzed in this study was infected with GFP-Akata EBV. The absence of miRNAs in the AGS cell line analyzed here may be due to the difference in the strain of EBV used to infect the AGS cell line. Surprisingly, BART12 miRNA was readily identified in several lymphoid cell lines, including IM9, Namalwa, and Mutu, that did not have detectable expression of the other miRNAs (Fig. 1).
FIG. 1.
Analysis of the BART miRNA expression in the tumor-derived cells and cell lines. Shown is Northern analysis of selected EBV BART miRNAs in total RNA samples derived from the indicated cell lines and tumors. The BCBL1 cell line served as the negative control, and U6 RNA served as a loading control.
Structures of the BART RNAs.
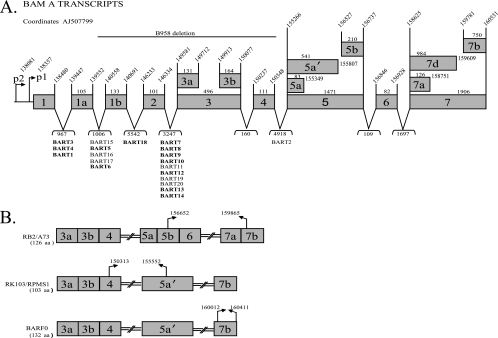
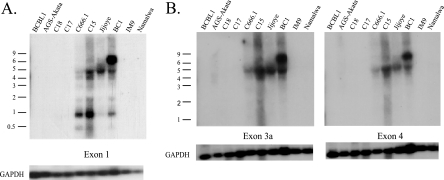
Previous studies that cloned large numbers of EBV miRNAs from RNA libraries and the variable detection of miRNAs by Northern blotting suggest that there must be additional factors regulating the abundance of particular miRNAs in a given cell line (Fig. 1) (6, 26). Considering the variety of mRNAs transcribed from the BART region, it is possible that the BART miRNA precursors are produced from a specific transcript(s). It has been difficult to study NPC, as cell lines that retain the EBV genome cannot readily be established from the primary tumors and do not usually grow in nude mice (32). Three NPC xenografts were established (C15, C17, and C18) that can be passaged in nude mice (3). The C15 tumor has the same pattern of expression as EBV-positive NPC primary tumors, while both C17 and C18 do not express most EBV genes (4, 17, 35). One cell line, C666.1, has been established from NPC that has retained the EBV genome (9). The BARTs were first identified in the C15 tumor, and multiple RNAs of different sizes can be identified by Northern blotting (13, 14, 35). Subsequent studies have identified various structures using cDNA cloning and PCR amplification (Fig. 2A) (35, 37). The exact structures of the multiple BARTs of different sizes are not known. Several splicing patterns were detected in size-selected RNAs with the predominant 4.8-kb RNA containing the spliced form that would produce the RB2 ORF (Fig. 2B) (35). Two full length cDNAs were successfully cloned in a cDNA library. One cDNA began at the P1 promoter, was spliced directly from exon 1 to exon 3, and contained exons 4, 5, 6, and 7. This is thought to represent the predominant 4.8-kb mRNA, however, this RNA only contains approximately 2 kb. The second cDNA had additional splices within exons 5 and 7 (36). Additional structures containing exon 1, 1a, 1b, or 2 have been obtained (7, 11, 35-37). The variation in structure at the 5′ end of the RNA is revealed by hybridization with a probe representing exon 1 (Fig. 3A). In contrast, fewer RNAs are identified by hybridization with probes for exon 3 or exon 4, with the major RNA of approximately 4.8 kb in size detected in the NPC cell line C666.1, the NPC xenograft C15, the KSHV/EBV-coinfected cell line BC1, and the Jijoye BL cell line and with a larger 7.0-kb RNA detected in BC1 (Fig. 3B). The relative abundance of the BART RNAs was similar to that previously shown with readily detectable expression in C666.1, C15, Jijoye, and BC1 (6). The BARTs were not detectable by Northern blotting in two additional NPC xenografts, C17 and C18, or the EBV-infected gastric cancer cell line AGS-Akata and in the IM9 and Namalwa lymphoid cell lines.
FIG. 2.
Structure of the EBV BARTs and the genomic location of the BART miRNAs. (A) Schematic of the promoter, exon, and intron structures of composite BART cDNAs cloned from the xenograph C15 with the corresponding EBV coordinates (AJ507799) and the location of the BART miRNAs. The boldface miRNAs were analyzed in this study. The bp sizes of exons and introns are noted. (B) Putative proteins encoded by the ORFs of the BART cDNAs with the positions of the start and stop codons indicated and the amino acid length of each ORF.
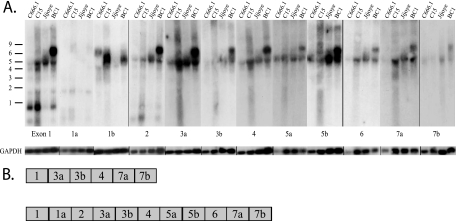
FIG. 3.
Analysis of the BART mRNA expression and 5′ structure in tumor-derived cells and cell lines. (A) Total RNA samples from the indicated cell lines and tumors were hybridized to an antisense riboprobe to exon 1. GAPDH mRNA expression was used as a loading control. Size markers are indicated. (B) Total RNA samples were hybridized to antisense riboprobes to exons 3a and 4. GAPDH mRNA expression was used as a loading control. Size markers are indicated.
To further characterize BART expression in C15, C666.1, BC1, and Jijoye, probes representing the known exons were hybridized to Northern blots (Fig. 4). The BC1 cell line uniquely expressed a larger, approximately 7.0-kb RNA. The 7.0-kb mRNA and 4.8-kb mRNA both hybridized to probes from exons 1, 2, 3, 4, 5, 6, and 7 but not to exon 1a or 1b (Fig. 4A). The molecular size of an RNA with these exons is less than 2.0 kb, and the additional sequences present in the 7.0-kb RNA in BC1 are unknown. A 4.0-kb mRNA was detected in C666.1, C15, and BC1 that hybridized to exons 1, 3, and 4 but not to exon 1a, 1b, or 2 (Fig. 4A). These exons add up to less than 0.8 kb. Previous studies have identified both the 5′ and 3′ ends of the BART RNAs, and the reasons for the disparity between cDNA sequence and mRNA size have not been determined but may reflect aberrant gel migration.
FIG. 4.
Variation in the BART mRNA structure. (A) Total RNA samples were hybridized to antisense riboprobes to the BamHI A exons. GAPDH mRNA expression was used as a loading control. Size markers are indicated. (B) Structure of full-length cDNAs cloned from the xenograph C15.
In support of the hybridization patterns, two distinct spliced forms were identified in C15 by cDNA cloning of products that were amplified using primers specific for exon 1 to exon 7 (Fig. 4B). One cDNA included exons 1, 1a, 2, 3a, 3b, 4, 5a, 6, 7a, and 7b, while a second included exons 1, 3a, 3b, 4, 7a, and 7b. Other smaller forms spliced directly to exon 7 or included exon 2 or various forms of exon 3. In all of the clones, 43% spliced from exon 1 to exon 3, while 18% spliced from exon 1 to exon 2. These data confirm that the BART mRNAs differ in their retention of specific exons with variation in the presence of exons 1a, 1b, 2, and 5. These cDNA sequences suggest that some of the mRNAs that splice directly from exon 1 to exon 3 also do not include exons 5 or 6. Thus, the 7.0- and 4.8-kb mRNAs which contain exon 5 would produce the RB2/A73, Rk103/RPMS1, and BARF0 ORFs (Fig. 2). The 4.0-kb mRNA which did not contain exon 5 would not contain any of these larger ORFs.
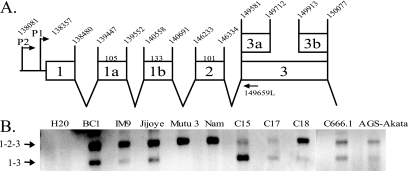
To confirm expression of BART RNAs that do not contain exons 1a, 1b, and 2 and to determine if promoter usage correlated with the expression of the BART miRNAs, the RNA was amplified using primers from the previously identified P1 (bp 138357) and P2 (bp 138081) promoters to exon 3 at bp 149581 (Fig. 5A) (7). Although the BART RNAs were not detected on Northern blots of C17, C18, IM9, or Namalwa, BART transcription can be detected by PCR amplification. Two discrete species initiating from P1 were detected with variable abundance (Fig. 5B). Discrete bands initiating from the P2 promoter were not readily detectable (data not shown). This may reflect greater heterogeneity of transcripts initiating from P2 (7). The products were excised from the gel and sequenced. The top 300-bp band contained exons 1, 2, and 3, while the lower 200-bp fragment contained exons 1 and 3. Products containing exon 1a or 1b were not obtained. The fragment containing exons 1, 2, and 3 was detected in all of the cell lines and was the primary product in IM9, Mutu 3, Namalwa, and C18. In contrast, the fragment containing exons 1 and 3 and lacking exon 2 was the most abundant form in C15. Both forms were detected in BC1, C666.1, and Jijoye, with low levels in IM9, C17, and AGS-Akata (Fig. 5C). The detection of the 4.0-kb mRNA and the spliced product from exon 1 to exon 3 correlated with those samples in which the expression of the miRNAs was more abundant: C15, BC1, and C666.1. This suggests that miRNAs are produced from an mRNA that splices out a large intron spanning the sequences between exons 1 and 3.
FIG. 5.
Differential splicing at the 5′ end of the mRNAs in tumor-derived cells. (A) Schematic of the promoter and exons of the 5′ end of the BART mRNA with the genomic coordinates noted. (B) RT-PCR analysis using primers specific for P1 and exon 3. The splicing structures of the products are denoted from sequencing the gel-purified bands.
Stable intron pieces are detectable after miRNA processing.
Stable residual pieces of the intron after Drosha processing have been identified for the human miR-30 miRNA and for the EBV BHRF1, -2, and -3 miRNAs, and their detection correlated with expression of the miRNAs (2, 10, 40). The BART miRNAs are located between exons 1 and 1a, 1a and 1b, 1b and 2, and 2 and 3 (Fig. 6A). To identify RNA species that could represent residual introns, RNA blots were hybridized to single-stranded RNA probes representing exons, specific introns, and pre-miRNAs (Fig. 6A and Table 1). The probes were prepared by cloning the sequences indicated in Table 1, and the bp coordinates of the probes are denoted beneath the boxes marked “probe” in Fig. 6A. The introns are designated by the exons that flank them as introns 1-1a, 1a-1b, 1b-2, and 2-3. Several pieces of RNA that hybridized to specific probes were identified in those cell lines and tumors that produced the miRNAs (Fig. 6B). An abundant RNA of approximately 720 nucleotides (nt) was detected with probes representing exon 1 and the 5′ sequences of intron 1-1a. This RNA did not hybridize to the pre-miRNA for the BART3 miRNA. The size of this RNA and its hybridization to exon 1 and intron 1-1a suggest that it is a residual piece of RNA that would initiate at the P1 promoter and is not spliced at exon 1 but is processed at 5′ end of the BART3 pre-miRNA (Fig. 6B). A second 1.7-kb RNA was detected that hybridized to exon 1 and intron 1-1a but also hybridized to the BART3 pre-miRNA, the BART1 pre-miRNA, and exon 1a but not to the BART6 pre-miRNA. This RNA could also be a cleavage product of an unspliced transcript through exon 1 to the BART6 pre-miRNA. The detection of two residual RNAs that terminate at either the BART3 pre-miRNA or the BART6 pre-miRNA suggests that either the BART6 and BART3 miRNAs are not both processed simultaneously from the same individual RNA or the 5′ cleavage for the BART6 miRNA occurs first with subsequent production of the miRNAs between the BART3 and BART6 miRNAs.
The initial 5′ cleavage of BART6 miRNA is also suggested by the detection of two additional RNA hybridization patterns (Fig. 6B). One RNA of approximately 680 nt hybridized to the BART1 pre-miRNA and exon 1a but did not hybridize to the BART6 pre-miRNA. An additional 5.9-kb RNA was also detected that hybridized to the BART6 pre-miRNA, the probe representing the sequences at the 3′ end of intron 1a-1b, exon 1b, and sequences immediately 5′ to the BART18 miRNA but not to the BART18 pre-miRNA. This suggests that both BART6 and BART18 miRNAs are processed from the same intron.
A second large 3.5-kb RNA hybridized to exon 2, BART7 pre-miRNA, BART14 pre-miRNA, and a probe representing the 3′ sequences of intron 2-3a, but not to the BART18 pre-miRNA or exon 3. This piece would represent an RNA from which the BART18 miRNA was processed but was contiguous to exon 3 without processing of BART7, -8, -9, -10, -11, -12, -13, or -14 miRNA. A small 400-nt RNA that included sequences 3′ to BART18 miRNA and exon 2 but not the BART7 pre-miRNA would represent the residual piece after processing of the BART18 and BART7 pre-miRNAs.
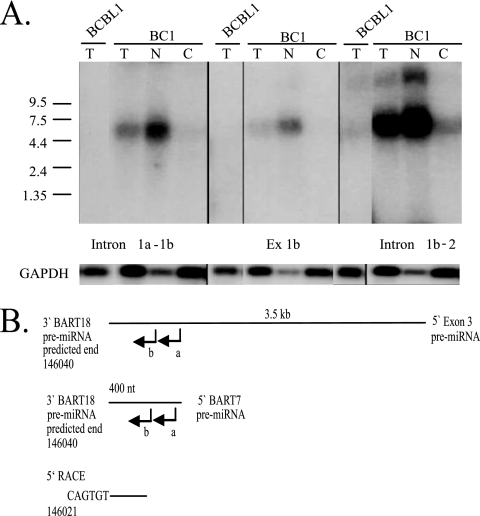
The production of pre-miRNAs occurs within the nucleus and the pre-miRNAs are subsequently translocated to the cytoplasm. Therefore, the residual pieces formed from processing the BART introns into pre-miRNAs should be present only within the nucleus. To determine the location of these RNAs, the BC1 cell line was separated into nuclear and cytoplasmic fractions (Fig. 7A). Hybridization with the cytoplasmic marker GAPDH revealed a slight contamination of the nuclear fraction by cytoplasm. The probes intron 1a-1b, exon 1b, and intron 1b-2 had identified a 5.9-kb residual piece (Fig. 6B). Hybridization with these probes to RNA from fractionated cells identified this RNA piece in the total RNA and strongly within the nuclear fractions (Fig. 7A). The low levels that were detected in the cytoplasmic fraction are likely due to a small amount of nuclear RNA that separated with the cytoplasmic fractions (Fig. 7A). The presence of the 5.9-kb RNA within the nuclear fraction indicates that these RNAs likely represent residual RNA after pre-miRNA processing.
FIG. 7.
Detection of the stable intronic RNA in fractionated BC1 cells and determination of the 5′ end of stable RNAs after miRNA processing. (A) Detection of stable intronic RNA in nuclear RNA. The BC1 cell line was separated into nuclear and cytoplasmic fractions. Northern blots were prepared with total (T), nuclear (N), and cytoplasmic (C) RNA and probed with antisense riboprobes representing introns and exon 1b. An antisense riboprobe to GAPDH mRNA was used as an indicator of fraction purity as well as a loading control. Size markers are noted. BCBL1 RNA was used as a negative control. (B) 5′ RACE of the stable 400-nt and 3.5-kb RNAs identified in Fig. 6B. Leftward intron-specific primer (a) was used to prime cDNA synthesis. cDNA was tagged with oligo(dT) and amplified using an oligo(dT) adapter primer and the nested leftward intron-specific primer (b).
To confirm the structures suggested by the hybridizations, 5′ RACE was performed. The potential 5′ end of the 3.5-kb and 400-nt RNAs was identified using nested primers immediately 5′ to the BART7 pre-miRNA and an oligo(dT) adapter primer (Fig. 7B). The sequence of the amplified fragment terminated at bp 146021 within the predicted BART18 pre-miRNA. BART18 was not cloned but was identified by sequence analysis and predicted to extend to bp 146040.
The detection of these residual pieces that span the sequences from P1 to exon 3 and contain exons 1, 1a, 1b, and 2 indicates that the BART miRNAs are produced from an RNA originating from the P1 promoter prior to any splicing throughout the region spanning exon 1 through exon 3. The specific residual pieces also suggest that certain miRNAs are processed more efficiently from this primary transcript, perhaps accounting for the sometimes large differences in expression seen between individual miRNAs.
Impaired processing in a subset of cell lines and NPC tumors.
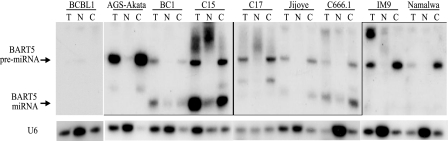
The Jijoye cell line has abundant BART transcripts and detectable residual intronic pieces. However, the much lower expression of the BART miRNAs suggests that there may be impairment in processing of the pre-miRNAs. To identify very low levels of expression and identify potential blocks in processing, AGS-Akata, BC1, and C15 were fractionated into nuclear and cytoplasmic fractions, hybridized to the BART1-3p, 3-3p,- 4, -5, -7, -9, and -12 miRNAs, and exposed to film for extended times. The pre-miRNAs for the BART4 and -7 miRNAs were not detected in C15, BC1, C666.1, and Jijoye, although the processed miRNAs were readily detectable (data not shown). This suggests that these pre-miRNAs are efficiently processed. In contrast, the pre-miRNAs for the BART1-3p, -3-3p, -5, and -9 miRNAs could be detected in those samples that expressed the miRNAs. The variable levels of the pre-miRNAs may reflect differences in the efficiency of recognition and processing by the Dicer complex. These pre-miRNAs were also detected in some samples with low or undetectable miRNA expression, including AGS-Akata, IM9, and Namalwa (Fig. 1 and data not shown). The pre-miRNAs for BART1, -5, -9, and -12 were the only pre- or processed miRNAs that were detectable in the C17 and C18 tumors. Similar bands were not detected in fractionated BCBL1 cells with the BART miRNA probes, indicating that they do not cross-hybridize to cellular miRNAs. The BART5 pre-miRNA that is the Drosha product could be detected in all cell lines and tumors, including those that had low or undetectable expression of the mature miRNAs, such as AGS-Akata, C17, IM9, and Namalwa (Fig. 8 and data not shown). This finding indicates that the cell lines that do not express the miRNAs do have functional Drosha that produces some of the pre-miRNAs. Hybridization with the U6 snRNA indicated significant enrichment in the nuclear fraction with some contamination in the cytoplasmic fraction. However, the BART5 pre-miRNA was detected in the cytoplasmic fraction but not nuclear fraction of AGS-Akata, C17, IM9, and Namalwa (Fig. 8). Thus, this detection did not reflect contamination of the cytoplasmic fraction with nuclear RNA as the pre-miRNAs were not present in the nucleus of these cell lines. This finding indicates that potential differences in nuclear export were not responsible for the impaired miRNA expression.
FIG. 8.
Impaired processing of the BART5 miRNA. Northern analysis of the BART5 miRNA in total, nuclear, and cytosolic RNA samples from the indicated tumor-derived cells and cell lines and the detection of the pre-miRNA and the processed miRNA forms. The nuclear U6 RNA was used as a loading control as well as an indicator of cell fraction integrity.
Sequence variation within the pre-miRNAs.
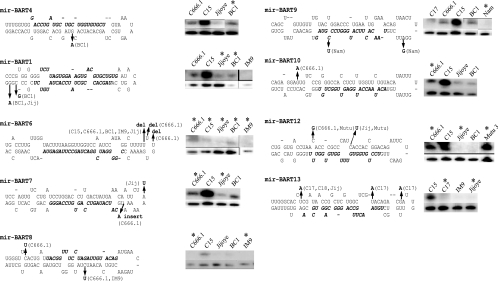
Previous studies have revealed that sequence variation of a single base pair can impair or inhibit processing of miRNAs (15). As the different miRNAs were detected at various levels between the various samples, the sequences of the pre-miRNAs were determined and compared to the EBV sequence database that represents B95-8 sequences with sequences from the Raji cell line that span the B95-8 deletion. The sequence differences and the positions of the changes in the pre-miRNAs are indicated in Fig. 9. The levels of expression of miRNAs in the samples with sequence changes are compared to those of samples without changes, with U6 included as a loading control (Fig. 9). BC1 had a sequence change within the BART4 pre-miRNA, and BART4 miRNA was detectable at levels comparable to those in C666.1 that did not have this change. In addition, the BART4 miRNA was the most frequently cloned miRNA from BC1, suggesting that this sequence change does not affect expression (6). BC1 and Jijoye had changes in the subsequent miRNA (BART1), which is 3′ to the BART4 miRNA. However, the BART1-3p miRNA was expressed in BC1 at levels comparable to C15 and C666.1 that did not have this sequence change. The BART6 miRNA had several deletions within the proposed hairpin loop, with a deletion of a U in C15, C666.1, BC1, IM9, and Jijoye and with two additional deletions of U in C666.1. These sequence changes in BART6 pre-miRNA were detected in many of samples and may be consistent polymorphisms. However, it was difficult to detect BART6 miRNA, and only three clones of BART6 miRNA were originally obtained (6). Thus, these changes within the computer-predicted hairpin possibly impede processing of this pre-miRNA. The BART7 pre-miRNA had an insert of an A in C666.1 and a change from A to U in Jijoye but was expressed at comparable levels in C666.1 and BC1 and at low levels in Jijoye. In the BART8 pre-miRNA, C666.1 had two changes of C to U and IM9 shared one of the changes. BART8 miRNA was difficult to detect in most samples and was not detected in C666.1. However, all of the other BART miRNAs were readily detectable in C666.1; therefore, these changes may affect BART8 miRNA expression (Fig. 1). Two changes in the BART9 pre-miRNA were detected in Namalwa, a cell line that does not produce the BART miRNAs. Interestingly, one of these sequence changes was the only sequence variation detected within the actual miRNA. A single change was detected within BART10 in C666.1; however, C666.1 expressed this miRNA at levels higher than those in Jijoye or BC1. The BART12 pre-miRNA had sequence changes in C666.1, Mutu, and Jijoye but was readily detected in C666.1, C15, and Mutu 3, indicating that these changes did not affect processing. The C17 tumor had multiple changes in the BART13 pre-miRNA and did not express this miRNA; however, the BART miRNAs could not be detected in C17 (Fig. 1). These data indicate that with the exception of the BART6 and BART8 miRNAs, the sequence variation in the BART pre-miRNAs analyzed here apparently does not affect their production. Sequence changes were also detected in the BART15 miRNA in C15 (T→G, bp 139561) and C17 and C18 (T→C, bp 139545), the BART17 miRNA in C666.1 (C→T, bp 139946), and the BART19 miRNA in C15 and BC1 (G→A, bp 148238) and Namalwa (C→T, bp 148223), which may affect their processing and expression; however, these miRNAs were not screened in this study.
FIG. 9.
Sequence variation within the BART miRNAs (mir-BART) and analysis of miRNA expression. The sequence of the BART pre-miRNAs is shown with the mature miRNA indicated in italics. Sequence changes are denoted in boldface with the corresponding cell line indicated. Expression of the miRNA in cell lines with and without sequence changes was determined by Northern blotting and is shown in the top band. Hybridization to the small nuclear U6 RNA is included as a loading control and is shown in the lower band. Asterisks denote cell lines with sequence changes.
DISCUSSION
The data presented in this study reveal that several factors contribute to production of the EBV BART miRNAs. A major factor that has been previously shown is that the detection and apparent abundance of the miRNAs largely reflect the presence and abundance of the BART RNAs (6). Several promoters have been identified for these RNAs, and the factors that regulate the transcription of these RNAs have been identified (7, 11, 35). The P1 promoter has been shown to be the predominant start site; however, transcripts containing further upstream sequences have been identified, suggesting multiple start sites (35). The promoter regions are possibly regulated by SP1, ets, Jun family members, IRF (interferon regulatory factor) proteins, c-myc, and C/EBP proteins. Interestingly, one study that assessed methylation in this region showed that the sequences downstream of exon 1 were not methylated (11). This suggested that additional important genetic functions exist in this region. The transcription through exon 1 and the downstream region likely reflects that this region is the active template for the miRNAs (11).
BART miRNA expression did not correlate with cell type in this study since some epithelial and lymphoid lines had strong expression, while others had little to no expression. This data suggests that the regulation of BART miRNA expression is not specific for cell type or disease state (26). However, the BART RNAs are not readily detectable in lymphoid cell lines, with the notable exception of the Jijoye cell line, which has relatively high levels of BART transcription. Although the BART RNAs are relatively abundant and comparable to those in C666.1 or BC1, the miRNAs are present at considerably lower levels in Jijoye. One potentially important difference in the BART transcripts in the Jijoye cell line is that the 4.0-kb mRNA that lacked exons 1a, 1b, 5, and 6 was not detected. In addition, the primary PCR product in Jijoye contained exons 1, 2, and 3. These findings suggest that the miRNAs are poorly produced in Jijoye as the mRNA lacking exons 1a, 1b, and 2 is expressed at low levels. Thus, a second level of regulation of BART miRNA production is the splicing pattern of the mRNAs. It will be of interest to determine what governs the processing of the internal 1a, 1b, and 2 exons.
Algorithms have been developed to identify sequences that contribute to intron/exon retention (39). However, significant differences in the exon and intron sequences were not detected between cells that did or did not splice at these exons. Notably, C15, C666.1, C17, IM9, and C18 have common amino acid changes in intron 1-1a, and all cell lines studied had similar amino acid changes in intron 1a-1b. Sporadic changes were observed in intron 2-3, yet none of the sequence changes correlated with differential splicing of the BART exons (data not shown). Interestingly, a comprehensive study of the KSHV miRNAs and the structure of the mRNAs that function as the primary pre-miRNAs indicated that the KSHV miRNAs are also produced from a large intron produced by alternate splicing (5). One of these mRNAs would be the template for kaposin and the miRNAs, while the second would contain the ORFs for the 71, 72, and 73 proteins. The structure of the BART RNAs also suggests that one form of the mRNAs contains the intron that is the template for miRNAs while another contains the large ORFs.
The link between the large intron and the production of miRNAs is also indicated by the detection of stable, residual pieces of this intron. These residual RNAs hybridize to exons 1a, 1b, and 2 and to probes representing intron sequences immediately 5′ or 3′ to the clusters of miRNAs. The complexity of the RNAs detected on Northern blots probed with exon 1 likely reflects both the complex structure of the mRNAs and the residual RNAs produced after pre-miRNA cleavage. Importantly, these residual RNAs were detected in the nuclear fraction, indicating that they are produced by pre-miRNA processing. It is possible that different miRNAs are formed from differently spliced RNAs. Perhaps the smaller intron that is produced from an RNA containing exon 1a, 1b, or 2 is a template for some of the miRNAs. This could explain why IM9, which only contained RNA with exon 2, only produced BART12 miRNA.
Considering the complexity of the RNAs detected with multiple probes, it is likely that other residual pieces are produced that were not identified. However, the retention of several pre-miRNAs but not others within these pieces may indicate that some of the miRNAs are preferentially processed. The BART18 pre-miRNA probe did not detect any residual RNAs, although RNAs were detected that hybridized to probes immediately 5′ and 3′ to this RNA. This suggests that this pre-miRNA is efficiently processed. The processing at the 3′ end of the BART18 pre-miRNA was confirmed by 5′ RACE for two of the residual fragments. Preferential processing is also suggested by the residual fragments that contain the BART6 pre-miRNA. Two RNAs were detected that would represent the pieces after cleavage at the 5′ end of the BART6 pre-miRNA. One included the BART1 pre-miRNA through BART6, and the second included exon 1 to BART6. The ability to detect these RNAs, which are likely the most abundant of the residual pieces, may be due to preferential processing of some pre-miRNAs or may indicate a temporal sequence of cleavage.
The detection of exon 1 sequences in two of the RNAs suggests that the miRNAs are produced from the intron prior to processing of the exons. Studies of cellular miRNA synthesis have suggested that intronic miRNAs are processed from the intron prior to splicing (23). This would require that the miRNAs are produced after the exon borders are tethered together but are not cleaved. The residual pieces detected in this study indicate that either the intron lariat has been cleaved at the 3′ end but not the 5′ end of the intron or that the primary pre-miRNA has an additional 5′ splice that produces the intron containing the identified exon 1 (42).
Another potential regulatory level for miRNA synthesis is the Dicer processing to produce the miRNAs. With the exception of miRNA 5, detection of the pre-miRNAs was variable. The pre-miRNAs for BART1, -3, and -5 could be detected in cell lines that produced the miRNAs, while the precursors for BART4 and -7 were not detected. This finding suggests the Drosha products vary in their efficiency of processing by Dicer. In cell lines that did not have detectable mature miRNAs, the pre-miRNAs could be detected for some but not all of the miRNAs. In particular, the pre-miRNAs for BART5, -9, and -12 miRNAs were detected in C17, C18, and IM9. Of these, only the processed BART12 miRNA could be detected in IM9. Thus, there is some regulation of Dicer activity preventing the production of mature miRNAs in these cases. However, in all cases, the pre-miRNAs were detectable in the cytoplasmic fractions, indicating that transport to the cytoplasm is not impaired or regulated distinctly in cell lines without miRNAs.
The further study of viral miRNAs is likely to increase our understanding of factors that regulate their synthesis. The genesis of cellular miRNAs is complicated by uncertainties in coding and noncoding regions and multiple mRNAs whose structures are not known. Many potential cellular miRNAs have been predicted by sequence, and their positions with regard to known protein coding sequences have been determined (27). These analyses have revealed that approximately 25% of cellular miRNAs are within the introns of known coding mRNAs, and half of these are within introns greater than 5 kb. The large BART intron that is the template for the miRNAs is approximately 11 kb. Splicing at exon 1a or 1b would result in introns approximately 1 kb, while splicing at exon 2 would produce a 3-kb intron. This larger intron could serve at the template for the BART7 to -14 miRNAs. In the lymphoid cell lines IM9, Namalwa, and Mutu, where the predominant form was spliced to include exon 2, only BART12 miRNA, which lies in the middle of this intron, was identified. Thus, the production of miRNAs in these cell lines may be impeded by the splicing reaction at exon 2.
The data presented here indicate several factors contribute to expression of the BART miRNAs. These include the abundance of the BART RNAs and specific splicing patterns. The residual RNAs produced by BART pre-miRNA processing have been partially characterized here by their hybridization to exons and introns. It will be of interest to further identify the exact 5′ and 3′ ends of these RNAs and other residual RNAs. The further study of these RNAs is likely to provide new information on the temporal production of the pre-miRNAs and other factors that contribute to their biogenesis.
Acknowledgments
This study was supported by grant CA32979 from the National Institutes of Health.
Footnotes
Published ahead of print on 9 July 2008.
REFERENCES
- 1.Brooks, L. A., A. L. Lear, L. S. Young, and A. B. Rickinson. 1993. Transcripts from the Epstein-Barr virus BamHI A fragment are detectable in all three forms of virus latency. J. Virol. 673182-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buck, A. H., J. Santoyo-Lopez, K. A. Robertson, D. S. Kumar, M. Reczko, and P. Ghazal. 2007. Discrete clusters of virus-encoded microRNAs are associated with complementary strands of the genome and the 7.2-kilobase stable intron in murine cytomegalovirus. J. Virol. 8113761-13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Busson, P., G. Ganem, P. Flores, F. Mugneret, B. Clausse, B. Caillou, K. Braham, H. Wakasugi, M. Lipinski, and T. Tursz. 1988. Establishment and characterization of three transplantable EBV-containing nasopharyngeal carcinomas. Int. J. Cancer 42599-606. [DOI] [PubMed] [Google Scholar]
- 4.Busson, P., R. McCoy, R. Sadler, K. Gilligan, T. Tursz, and N. Raab-Traub. 1992. Consistent transcription of the Epstein-Barr virus LMP2 gene in nasopharyngeal carcinoma. J. Virol. 663257-3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai, X., and B. R. Cullen. 2006. Transcriptional origin of Kaposi's sarcoma-associated herpesvirus microRNAs. J. Virol. 802234-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai, X., A. Schafer, S. Lu, J. P. Bilello, R. C. Desrosiers, R. Edwards, N. Raab-Traub, and B. R. Cullen. 2006. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, H., J. Huang, F. Y. Wu, G. Liao, L. Hutt-Fletcher, and S. D. Hayward. 2005. Regulation of expression of the Epstein-Barr virus BamHI-A rightward transcripts. J. Virol. 791724-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, H. L., M. L. Lung, J. S. T. Sham, D. T. K. Choy, B. E. Griffin, and M. E. Ng. 1992. Transcription of the BamH1 A region of the EBV genome in NPC tissues and B cells. Virology 191193-201. [DOI] [PubMed] [Google Scholar]
- 9.Cheung, S. T., D. P. Huang, A. B. Hui, K. W. Lo, C. W. Ko, Y. S. Tsang, N. Wong, B. M. Whitney, and J. C. Lee. 1999. Nasopharyngeal carcinoma cell line (C666-1) consistently harbouring Epstein-Barr virus. Int. J. Cancer 83121-126. [DOI] [PubMed] [Google Scholar]
- 10.Cullen, B. R. 2004. Transcription and processing of human microRNA precursors. Mol. Cell 16861-865. [DOI] [PubMed] [Google Scholar]
- 11.de Jesus, O., P. R. Smith, L. C. Spender, C. Elgueta Karstegl, H. H. Niller, D. Huang, and P. J. Farrell. 2003. Updated Epstein-Barr virus (EBV) DNA sequence and analysis of a promoter for the BART (CST, BARF0) RNAs of EBV. J. Gen. Virol. 841443-1450. [DOI] [PubMed] [Google Scholar]
- 12.Fries, K. L., T. B. Sculley, J. Webster-Cyriaque, P. Rajadurai, R. H. Sadler, and N. Raab-Traub. 1997. Identification of a novel protein encoded by the BamHI A region of the Epstein-Barr virus. J. Virol. 712765-2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilligan, K., H. Sato, P. Rajadurai, P. Busson, L. Young, A. Rickinson, T. Tursz, and N. Raab-Traub. 1990. Novel transcription from the Epstein-Barr virus terminal EcoRI fragment, DIJhet, in a nasopharyngeal carcinoma. J. Virol. 644948-4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilligan, K. J., P. Rajadurai, J.-C. Lin, P. Busson, M. Abdel-Hamid, U. Prasad, T. Tursz, and N. Raab-Traub. 1991. Expression of the Epstein-Barr virus BamHI A fragment in nasopharyngeal carcinoma: evidence for a viral protein expressed in vivo. J. Virol. 656252-6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottwein, E., X. Cai, and B. R. Cullen. 2006. A novel assay for viral microRNA function identifies a single nucleotide polymorphism that affects Drosha processing. J. Virol. 805321-5326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grundhoff, A., C. S. Sullivan, and D. Ganem. 2006. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA 12733-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hitt, M. M., M. A. Allday, T. Hara, L. Karran, M. D. Jones, P. Busson, T. Tursz, I. Ernberg, and B. E. Griffin. 1989. EBV gene expression in an NPC related tumor. EMBO J. 82639-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karran, L., Y. Gao, P. R. Smith, and B. E. Griffin. 1992. Expression of a family of complementary-strand transcripts in Epstein-Barr virus-infected cells Proc. Natl. Acad. Sci. USA 898058-8062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kerr, B. M., A. L. Lear, M. Rowe, D. Croom-Carter, L. S. Young, S. M. Rookes, P. H. Gallimore, and A. B. Rickinson. 1992. Three transcriptionally distinct forms of Epstein-Barr virus latency in somatic cell hybrids: cell phenotype dependence of virus promoter usage. Virology 187189-201. [DOI] [PubMed] [Google Scholar]
- 20.Kieff, E., and D. Liebowitz. 1990. Epstein-Barr virus and its replication, p. 1889-1920. In B. N. Fields et al. (ed.), Fields virology, 2nd ed. Raven Press, New York, NY.
- 21.Kieff, E., and A. B. Rickinson. 2001. Epstein-Barr virus and its replication, p. 2511-2573. In B. N. Fields, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, S. E. Straus, and D. M. Knipe (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 22.Kienzle, N., T. B. Sculley, L. Poulsen, M. Buck, S. Cross, N. Raab-Traub, and R. Khanna. 1998. Identification of a cytotoxic T-lymphocyte response to the novel BARF0 protein of Epstein-Barr virus: a critical role for antigen expression. J. Virol. 726614-6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, Y. K., and V. N. Kim. 2007. Processing of intronic microRNAs. EMBO J. 26775-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.King, W., A. L. Thomas-Powell, N. Raab-Traub, M. Hawke, and E. Kieff. 1980. Epstein-Barr virus RNA. V. Viral RNA in a restringently infected, growth-transformed cell line. J. Virol. 36506-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kusano, S., and N. Raab-Traub. 2001. An Epstein-Barr virus protein interacts with Notch. J. Virol. 75384-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Landgraf, P., M. Rusu, R. Sheridan, A. Sewer, N. Iovino, A. Aravin, S. Pfeffer, A. Rice, A. O. Kamphorst, M. Landthaler, C. Lin, N. D. Socci, L. Hermida, V. Fulci, S. Chiaretti, R. Foa, J. Schliwka, U. Fuchs, A. Novosel, R. U. Muller, B. Schermer, U. Bissels, J. Inman, Q. Phan, M. Chien, D. B. Weir, R. Choksi, G. De Vita, D. Frezzetti, H. I. Trompeter, V. Hornung, G. Teng, G. Hartmann, M. Palkovits, R. Di Lauro, P. Wernet, G. Macino, C. E. Rogler, J. W. Nagle, J. Ju, F. N. Papavasiliou, T. Benzing, P. Lichter, W. Tam, M. J. Brownstein, A. Bosio, A. Borkhardt, J. J. Russo, C. Sander, M. Zavolan, and T. Tuschl. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 1291401-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, S. C., P. Tang, and W. C. Lin. 2007. Intronic microRNA: discovery and biological implications. DNA Cell Biol. 26195-207. [DOI] [PubMed] [Google Scholar]
- 28.Lo, A. K., K. F. To, K. W. Lo, R. W. Lung, J. W. Hui, G. Liao, and S. D. Hayward. 2007. Modulation of LMP1 protein expression by EBV-encoded microRNAs. Proc. Natl. Acad. Sci. USA 10416164-16169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller, G., and M. Lipman. 1973. Release of infectious Epstein-Barr virus by transformed marmoset leukocytes. Proc. Natl. Acad. Sci. USA 70190-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pfeffer, S., M. Zavolan, F. A. Grasser, M. Chien, J. J. Russo, J. Ju, B. John, A. J. Enright, D. Marks, C. Sander, and T. Tuschl. 2004. Identification of virus-encoded microRNAs. Science 304734-736. [DOI] [PubMed] [Google Scholar]
- 31.Pope, J. H., M. K. Horne, and W. Scott. 1968. Transformation of foetal human keukocytes in vitro by filtrates of a human leukaemic cell line containing herpes-like virus. Int. J. Cancer 3857-866. [DOI] [PubMed] [Google Scholar]
- 32.Raab-Traub, N. 2002. Epstein-Barr virus in the pathogenesis of NPC. Semin. Cancer Biol. 12431-441. [DOI] [PubMed] [Google Scholar]
- 33.Raab-Traub, N., T. Dambaugh, and E. Kieff. 1980. DNA of Epstein-Barr virus. VIII. B95-8, the previous prototype, is an unusual deletion derivative. Cell 22257-267. [DOI] [PubMed] [Google Scholar]
- 34.Raab-Traub, N., R. Hood, C.-S. Yang, B. Henry II, and J. S. Pagano. 1983. Epstein-Barr virus transcription in nasopharyngeal carcinoma. J. Virol. 48580-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sadler, R. H., and N. Raab-Traub. 1995. Structural analyses of the Epstein-Barr virus BamHI A transcripts. J. Virol. 691132-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith, P. 2001. Epstein-Barr virus complementary strand transcripts (CSTs/BARTs) and cancer. Semin. Cancer Biol. 11469-476. [DOI] [PubMed] [Google Scholar]
- 37.Smith, P. R., O. de Jesus, D. Turner, M. Hollyoake, C. E. Karstegl, B. E. Griffin, L. Karran, Y. Wang, S. D. Hayward, and P. J. Farrell. 2000. Structure and coding content of CST (BART) family RNAs of Epstein-Barr virus. J. Virol. 743082-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thornburg, N. J., S. Kusano, and N. Raab-Traub. 2004. Identification of Epstein-Barr virus RK-BARF0-interacting proteins and characterization of expression pattern. J. Virol. 7812848-12856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, Z., M. E. Rolish, G. Yeo, V. Tung, M. Mawson, and C. E. Burge. 2004. Systemic identification and analysis of exonic splicing silencers. Cell 119831-845. [DOI] [PubMed] [Google Scholar]
- 40.Xing, L., and E. Kieff. 2007. Epstein-Barr virus BHRF1 micro- and stable RNAs during latency III and after induction of replication. J. Virol. 819967-9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Young, L., C. Alfieri, K. Hennessy, H. Evans, C. O'Hara, K. C. Anderson, J. Ritz, R. S. Shapiro, A. Rickinson, E. Kieff, et al. 1989. Expression of Epstein-Barr virus transformation-associated genes in tissues of patients with EBV lymphoproliferative disease. N. Engl. J. Med. 3211080-1085. [DOI] [PubMed] [Google Scholar]
- 42.Zhou, H., and K. Lin. 2008. Excess of microRNAs in large and very 5′ biased introns. Biochem. Biophys. Res. Commun. 368709-715. [DOI] [PubMed] [Google Scholar]