Abstract
The mitochondrial genome of trypanosomes is composed of thousands of topologically interlocked circular DNA molecules that form the kinetoplast DNA (kDNA). Most genes encoded by the kDNA require a posttranscriptional modification process called RNA editing to form functional mRNAs. Here, we show that alternative editing of the mitochondrial cytochrome c oxidase III (COXIII) mRNA in Trypanosoma brucei produces a novel DNA binding protein, alternatively edited protein 1 (AEP-1). AEP-1 localizes to the region of the cell between the kDNA and the flagellum and purifies with the tripartite attachment complex, a structure believed to physically link the kDNA and flagellar basal bodies. Expression of the DNA binding domain of AEP-1 results in aberrant kDNA structure and reduced cell growth, indicating that AEP-1 is involved in the maintenance of the kDNA. Perhaps most important, our studies show a gain of function through an alternatively edited mRNA and, for the first time, provide a link between the unusual structure of the kDNA and RNA editing in trypanosome mitochondria.
The kinetoplast of trypanosomes represents more than 10% of the total cellular DNA and is organized as a unique topological structure containing approximately 50 maxicircles (∼20 kb) and 5,000 minicircles (∼1 kb) catenated into a single network (reviewed in reference 13). The kinetoplast DNA (kDNA) network is localized to a region of the mitochondrion immediately adjacent to the basal bodies of the flagellum and is physically linked to the flagellum by the tripartite attachment complex (TAC). The TAC is composed of filaments extending from the proximal end of the basal bodies to the cytosolic face of the outer mitochondrial membrane and of unilateral filaments located between the inner mitochondrial membrane and the kDNA network (17). Replication of the kDNA is restricted to a discrete S phase during which all minicircles and maxicircles are faithfully replicated (reviewed in reference 12). The kDNA network increases in size until the sister kDNA networks are segregated prior to cell division (25). While the precise mechanism of kDNA segregation is unknown, the physical association of the kDNA to the flagellum suggests that the TAC plays a critical role in kinetoplast maintenance (8). Until recently none of the proteins composing the TAC had been identified. Using an RNA interference library to screen for proteins involved in kDNA replication or segregation, a protein encoded by a nuclear gene was identified, p166, that is a component of the TAC (26). Knockdown in the expression of p166 had little effect on kDNA replication but resulted in cells with enlarged kDNA networks that failed to segregate.
The kDNA maxicircles encode rRNAs and components of the electron transport chain and the ATP synthase; however, many of these genes lack complete coding information, and posttranscriptional editing is necessary to produce mRNAs for these essential mitochondrial proteins (1, 10). It is commonly believed that RNA editing, in trypanosomes and other organisms, evolved to correct these missense primary transcripts (22). The mechanism of trypanosome mRNA editing has been extensively studied and is carried out by an enzyme cascade consisting of endonuclease cleavage, uridine insertion or deletion, and RNA ligation (1, 10, 24). These activities are associated with multiple high-molecular-weight complexes, editosomes, that display protein microheterogeneity and mRNA substrate specificity (4, 18). The coding information for RNA editing is found in small guide RNAs (gRNAs) that interact with their cognate mRNAs to direct precise uridine insertion or deletion (2, 19). The gRNAs are primarily encoded by the kDNA minicircles, and recent estimates of minicircle gRNA gene diversity suggest that the number of unique gRNAs greatly exceeds the number needed to direct the editing of the conventional mitochondrial mRNAs (14, 16). We have proposed that the unexpected diversity of gRNA sequences is a powerful means to diversify the mitochondrial gene pool in order to produce novel proteins (14-16). Our laboratory has previously identified an alternatively edited mRNA of the cytochrome c oxidase III (COXIII) gene as well as the corresponding gRNA responsible for the alternative editing pattern. Using two independently raised antibodies against the N terminus of the alternatively edited COXIII transcript, we were able to show expression of this mRNA into a protein we named AEP-1 (15). Alternative editing of the COXIII mRNA leads to a transcript that is edited in the 3′ region and remains preedited in the 5′ region. Through alternative editing of one site between the preedited and edited region, the two reading frames of the 3′ and 5′ region are fused to one contiguous open reading frame (AEP-1). The resultant predicted protein contains the C-terminal 155 amino acids of COXIII forming five membrane-spanning domains and a unique N-terminal domain of 59 amino acids. This protein resides in a high-molecular-weight complex in Trypanosoma brucei mitochondrial membranes; here, we show by homology modeling that the N-terminal domain is positioned in the mitochondrial matrix. In this paper we also investigate the function of alternatively edited protein 1 (AEP-1). We show that the majority of AEP-1 localizes at the kDNA in vivo and within the purified TAC. We have developed reverse genetic analysis of AEP-1 mutants by recoding the AEP-1 gene for expression from the nucleus with an N-terminal mitochondrial localization signal. We found that nuclear expression of a mitochondrially targeted, truncated AEP-1 in a fusion protein with green fluorescent protein (GFP) results in a dominant negative phenotype of decreased cell growth and aberrant kDNA structure. To further investigate the function of AEP-1, we expressed the unique N-terminal 59 amino acids in Escherichia coli and found that it binds DNA, suggesting its involvement in association of the kDNA network to the mitochondrial membrane. Based on these findings, we propose that AEP-1 is a kDNA maintenance factor that is encoded by a maxicircle gene, and we show for the first time a gain of biological function in trypanosomes through alternative editing.
MATERIALS AND METHODS
Immunofluorescence microscopy.
Cultured bloodstream T. brucei (5 × 107 cells/ml) was washed in ice-cold phosphate-buffered saline (PBS) with 1% (wt/vol) glucose, fixed with 1% paraformaldehyde in PBS for 5 min at 4°C, spread onto precleaned microscopic slides, and air dried. Cells were subsequently permeabilized with methanol at −20°C for 5 min. Cells were blocked in 2% native goat serum in PBS and incubated with the primary antibody. The following antibodies were used: polyclonal rabbit AEP-1 (15) at a dilution of 1:3,000, monoclonal mouse anti-trypanosome alternative oxidase ([TAO] 1:400; a kind gift from M. Chaudhuri, Nashville, TN), and monoclonal mouse BBA4 (1:10; a kind gift from K. Gull, Oxford University, United Kingdom). Primary antibodies were incubated with cells for 20 min, followed by two washes with 2% native goat serum in PBS and incubation with the appropriate secondary antibody for 20 min at a 1:1,000 dilution. After incubation with the secondary antibody, cells were washed twice with PBS and then covered with the 4′,6′-diamidino-2-phenylindole (DAPI) containing the antifade reagent ProlongGold (Molecular Probes). Images were acquired using an Axioobserver Z1 equipped with an Axiocam MRm camera controlled by Axiovision, version 4.6, software. Quantitation of fluorescence images was done using the densitometry measurement tool from the Axiovision, version 4.6, software.
Transmission electron microscopy.
Cells were harvested at mid-log phase and washed two times with PBS. Cells were fixed for 1 h at 25°C in a buffer containing 2.5% glutaraldehyde, 4% paraformaldehyde, 5 mM CaCl2, and 100 mM cacodylate (pH 7). After two PBS washes, cell were postfixed with 1% OsO4 in PBS for 1 h at 25°C. Dehydration in a 50, 70, 90, and 100% ethanol series was followed by incubation in propylene oxide (two times for 10 min each). The cell pellet was embedded in Epon resin, and ultrathin sections were prepared. Images were taken on a Philips/FEI Technai 20 (FEI Co., The Netherlands) or a Jeol 100 CX transmission electron microscope (JEOL Ltd.).
Flow cytometry.
Flow cytometry was done as described previously (7). In brief, cells were fixed and permeabilized in 1% Triton X-100, 40 mM citric acid, 20 mM sodium phosphate, pH 7.0, and 200 mM sucrose for 5 min at 25°C. After the addition of MgCl to a final concentration of 87.5 mM in PBS, cells were incubated for 3 h at 37°C with 0.5 mg/ml RNase in a phosphate buffer, pH 7.0, at a final concentration of 120 mM. Propidium iodide (50 μg/ml) in 1.12% sodium citrate was added to the cells to a final concentration of 20 μg/ml and incubated for 30 min at 25°C. Cells were analyzed using a FACSCalibur instrument (Becton Dickinson, NJ). Only individual cells were analyzed. Data analysis was done using the FlowJo software (Treestar, Inc.).
Molecular modeling.
Helices 3, 4, and 5 of APE-1 were modeled with a high degree of confidence with Swiss Model (21) using bovine COXIII (Protein Data Bank identifier 1v54, chain C) as a template. Helices 1 and 2 of AEP-1 are topologically equivalent to the corresponding helices 3 and 4 in bovine COXIII but could not be modeled by Swiss Model due to sequence divergence. Still, TMHMM (11) predicts these transmembrane helices and their topology in AEP-1 with high likelihood, so they were interactively modeled using the bovine structure as a template, with the caveat that their spatial relationship to one another, as well as to the last three helices, is not necessarily as depicted. No other transmembrane helices are predicted in AEP-1.
Construction of a dominant negative cell line.
The unique 60-amino-acid sequence of AEP-1 was recoded for optimal expression from T. brucei nuclear machinery and purchased from Genelink (Hawthorne, NY). Primers were designed against recoded AEP-1 containing a 14-amino-acid mitochondrial import sequence. The forward primer contained a HindIII linker (5′-ATGTTTCGTCGTTGCTTTCC-3′), and the reverse primer contained an SbfI linker (5′-AAGAAGAAGACGGGGAC-3′). AEP-1 was cloned into the HindIII/Sbf1 site of pTuBeGFP vector. Translation of this transcript yielded a chimeric protein of AEP-1 fused with enhanced GFP containing a mitochondrial import signal. pTuBeGFP vector was digested with XbaI and XhoI and transfected into bloodstream form TREU667 trypanosomes. Cells were cloned and selected against 2.5 μg/ml of phleomycin.
Expression of recombinant AEP-1.
AEP-1 was amplified with NdeI and BamHI linkers and cloned into pET16B vector (Novagen EMD, San Diego, CA) and transformed into BL21(DE3) cells. AEP-1 was induced at an optical density of 0.1 with 0.5 mM isopropyl-β-d-thiogalactopyranoside at 25°C for 4 h. Cells were harvested after induction and frozen at −20°C. Cells were lysed in buffer A (300 mM NaCl, 50 mM NaH2PO4, pH 8.0, 10 mM imidazole, 2 mM phenylmethylsulfonyl fluoride, 1 tablet of Roche cocktail protease inhibitor), 1% Triton X-100, 1 mM lysozyme, RNase A (10 μg/ml), and DNase I (5 μg/ml) for 1 h at 4°C. The sample was centrifuged for 30 min at 10,000 rpm. The pellet was collected and resuspended in buffer A containing 2% Triton X-100 and placed over a Ni-nitrilotriacetic acid agarose column. The column was washed three times with buffer A containing 20 mM imidazole. Recombinant AEP-1 was eluted from the column using 300 mM NaCl, 50 mM NaH2PO4 (pH 8.0), 250 mM imidazole, and 1% Triton X-100. Samples were dialyzed overnight at 4°C against 20 mM HEPES (pH 7.9), 60 mM KCl, 1 mM EDTA, and 1 mM dithiothreitol. Samples were collected after dialysis and concentrated using Millipore Centriplus (molecular mass cutoff, 3.5 kDa; Millipore).
Mobility shift.
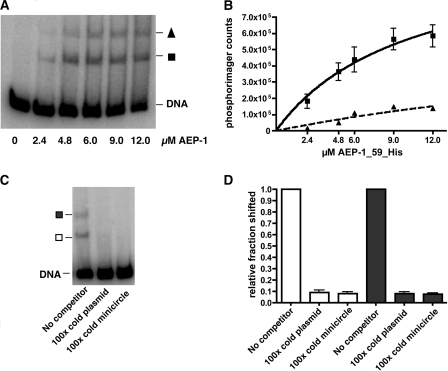
Minicircle probe (233 bp) was generated by PCR from a cloned minicircle sequence. Maxicircle probe (211 bp) was generated by PCR (see Table S1 in the supplemental material) from kDNA. Probes were gel purified prior to 32P end labeling. In Fig. 6, a total of 5 fmole of 32P-labeled minicircle or maxicircle probe was added to the indicated amount of recombinant AEP-1 in 20 mM HEPES (pH 7.9), 60 mM KCl, 1 mM EDTA, and 1 mM dithiothreitol and incubated for 1 h in a total volume of 10 μl at 24°C. The entire reaction was resolved on a 4% polyacrylamide native gel at 30 mA for 2 h at room temperature. Quantitation was done using a phosphorimager (Molecular Dynamics).
FIG. 6.
Recombinant AEP-1(1-59)-His binds DNA. (A) 32P-labeled minicircle DNA was incubated with different concentrations of recombinant AEP-1(1-59)-His and then separated on a native polyacrylamide gel (4%). Two mobility-shifted products are designated with a triangle (slow migrating) and a square (more rapidly migrating). (B) Quantitation of three independent gel shift assays using a phosphorimager. The curve was fitted using the following equation: Y = Bmax × X(Kd + X), where X is the concentration of AEP-1(1-59)-His, Bmax is the maximal binding, and Kd is the concentration of AEP-1(1-59)-His required to reach half-maximal binding. Triangles and squares indicate the mobility-shifted complexes identified in panel A. (C) Competition of binding using AEP-1(1-59)-His (4.8 μM) with 32P-labeled minicircle DNA, 100× cold minicircle DNA, and 100× cold plasmid DNA as competitors and subsequent separation on a native polyacrylamide gel (4%). The mobility-shifted products are indicated with a filled square (slow migrating) and an open square (more rapidly migrating). (D) Quantitation of competition expressed as the fraction shifted after the addition of competitor compared to the result without competitor. Open bars correspond to the more rapidly migrating AEP-1/DNA complex (open square in panel C), and the closed bars correspond to the slower-migrating AEP-1/DNA complex (filled square in panel C).
RESULTS
Orientation of AEP-1 in the mitochondrial membrane.
The AEP-1 mRNA consists of both preedited and edited sequence and contains a 214-amino-acid open reading frame (Fig. 1A). It encodes a protein with a unique, arginine-rich hydrophilic amino-terminal domain fused to five membrane-spanning domains of COXIII in the carboxy-terminal portion of the protein. The mRNA encoding AEP-1 is present in both bloodstream and procyclic (data not shown) T. brucei organisms, suggesting that the protein encoded is not a component of the developmentally regulated mitochondrial oxidative pathways. In order to determine the orientation of AEP-1 in the mitochondrial membrane, we built a homology model using the bovine COXIII crystal structure (Fig. 1B). We were able to model the three C-terminal transmembrane helices with great confidence. Two additional transmembrane helices that are equivalent to the corresponding bovine helices were predicted with high confidence using membrane topology prediction software (TMHMM) (11); however, these helices could not be modeled accurately using Swiss Model (21) due to sequence divergence. Based on our homology model, the arginine-rich N terminus of AEP-1 faces the matrix side of the inner mitochondrial membrane and therefore retains the expected membrane topology of fully edited COXIII.
FIG. 1.
Orientation of AEP-1 in the mitochondrial membrane. (A) Primary amino acid sequence of AEP-1. Secondary structure elements are depicted as seen in panel B. The N-terminal region (magenta, residues 1 to 51) is followed by five transmembrane-spanning regions (I and II, residues 52 to 95 and 95 to 121, respectively; III to V, residues 128 to 145, 149 to 171, and 178 to 200, respectively). Between the transmembrane-spanning regions are short loops (black bars). (B) Homology model of AEP-1 using bovine COXIII (Protein Data Bank identifier 1v54, chain C) as a template. Transmembrane helices modeled with great confidence are shown in cyan. In green are the transmembrane helices predicted by TMHMM that are equivalent to the corresponding bovine helices, but these could not be modeled accurately using Swiss Model due to sequence divergence. The N terminus lacks any identifiable structural homolog and is modeled as a random coil. Based on the homology model with the bovine COXIII, the N terminus of AEP-1 faces the matrix side of the inner mitochondrial membrane (gray).
Localization of AEP-1 to the TAC.
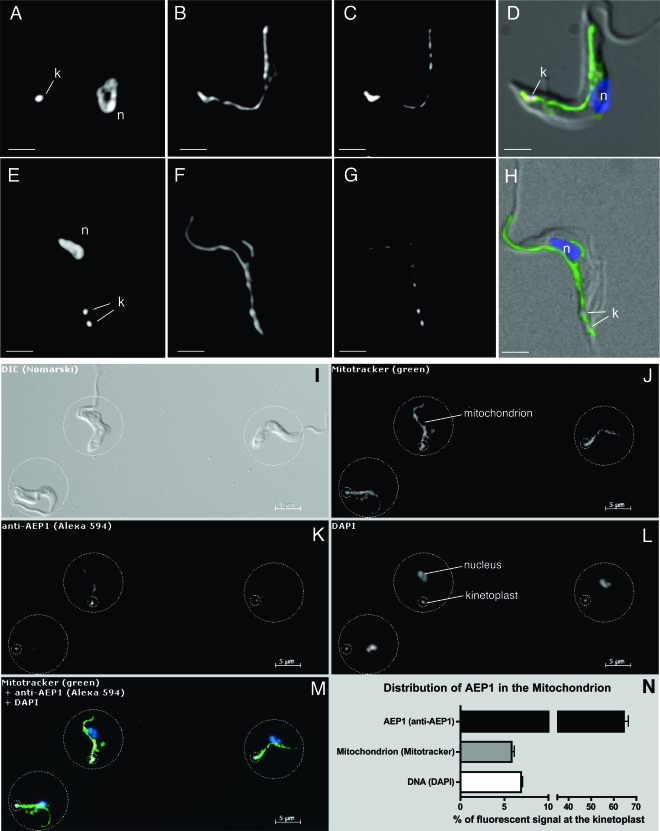
Since AEP-1 is encoded by the maxicircle, we anticipated it would be found exclusively in the mitochondrion, and previous studies showed that AEP-1 fractionated with mitochondrial membranes (15). Further investigation of the cellular localization of AEP-1 by immunofluorescence microscopy with anti-AEP-1 and the fluorescent marker MitoTracker confirmed that AEP-1 is present in the mitochondrion of bloodstream developmental stage T. brucei (Fig. 2A to D). We also found AEP-1 in the mitochondrion of the procylic developmental stage of T. brucei (data not shown). Despite the detection of AEP-1 throughout the mitochondrion, its distribution was nonuniform, with significantly greater anti-AEP-1 signal found in the kinetoplast region of the mitochondrion (Fig. 2A to D). Cells with two kinetoplasts show localization of AEP-1 at both (Fig. 2E to H). To better evaluate the distribution of AEP-1, we did a rigorous quantitative immunofluorescence analysis of AEP-1 in bloodstream T. brucei (Fig. 2I to M). Relative to the distribution of MitoTracker, we found that approximately 65% of AEP-1 is localized to the kinetoplast region that comprises only 6% of the total volume of the mitochondrion. The remaining AEP-1 is distributed throughout the mitochondrion (Fig. 2I to M). Since AEP-1 is localized to the region of the mitochondrion adjacent to the kDNA network, we investigated whether AEP-1 remains associated with the kinetoplast and flagellum by using conditions used to isolate the TAC (17). When the TAC is isolated by nonionic detergent extraction and depolymerization of subpellicular microtubules, AEP-1 remains stably associated while an abundant mitochondrial membrane protein in the bloodstream T. brucei, the TAO, is efficiently removed (Fig. 3A to D; see Fig. S1A to H in the supplemental material).
FIG. 2.
Analysis of the distribution of AEP-1 in bloodstream form trypanosomes using immunofluorescence microscopy. (A to H) Immunofluorescence microscopy of whole cells using the DNA stain DAPI (A and E), the mitochondrial marker MitoTracker (B and F), and an antibody to AEP-1 (C and G). Panels D and H show the merged images of panels A to C and panels E to G, respectively, using artificial coloring (blue, A and E; green, B and F; red, C and G) with a differential interference contrast image. (I to N) Fluorescence microscopy showing the distribution of AEP-1 in the mitochondrion of bloodstream form T. brucei. Shown is a representative field of T. brucei cells following incubation with MitoTracker FM green that was subsequently fixed and stained for AEP-1. (I) Differential interference contrast (DIC) image of bloodstream form T. brucei. (J) Fluorescence image (excitation, 490 nm; emission, 516 nm) of the same cells showing MitoTracker signal. (K) Fluorescence image of the same cells (excitation, 590 nm; emission, 617 nm) stained with anti-AEP-1 antibody. (L) Cells stained with DAPI. (M) Overlay of images J to L using artificial colors: green, MitoTracker; red, AEP-1; blue, DAPI. (N) Quantitation of signal distribution over 38 individual cells. Relative distribution of the individual channels between the large and small circles was measured, and the average and standard deviations are displayed in the graph. The small circle (diameter, 2 μm) was centered at the kinetoplast. The large circle was chosen to contain the entire signal from a given cell in each channel. The positions of the kinetoplast (k) and nucleus (n) are indicated.
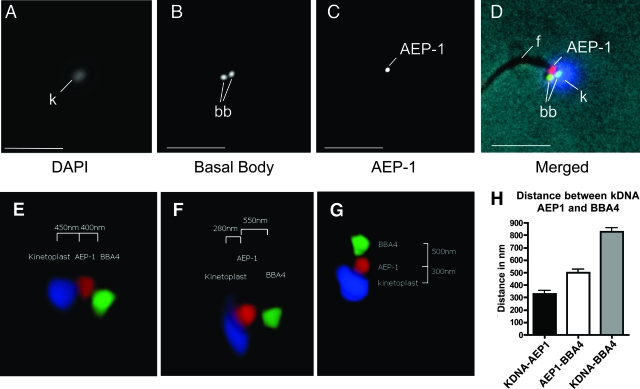
FIG. 3.
Localization of AEP-1 to the TAC. (A to D) Immunofluorescence microscopy of isolated flagella using DAPI, a basal body antibody, and an antibody to AEP-1. Panel D shows the merged image using artificial coloring (blue, A; green, B; red, C) with the phase-contrast image. The positions of the flagellum (f), basal bodies (bb), and kDNA (k) are indicated. Bar, 2.5 μm. (E to G) Three-dimensional reconstruction of the TAC complex from purified flagella. Three representative images show the three-dimensional relationship of the basal body (green), AEP-1 (red), and the kDNA (blue). (H) Bar graph of average distances (n = 7) between basal body, AEP-1, and the kDNA.
In order to gain a more detailed representation of the orientation of AEP-1 in the TAC, we prepared three-dimensional reconstructions of deconvoluted images of TAC preparations stained for DNA (DAPI) and reacted with antibodies against AEP-1 and a basal body protein (BBA4) (Fig. 3E to G). The three-dimensional imaging allows us to orient the kDNA network, AEP-1, and the basal bodies within the TAC. These studies show that AEP-1 is juxtaposed to the kDNA network; the average distance from the AEP-1 to the kDNA is 320 (± 20) nm while the distance from the kDNA to the basal body is 810 (± 20) nm (Fig. 3E to H). The localization of AEP-1 to the zone between the kDNA and the basal bodies of the flagellum is consistent with intact cell imaging using anti-AEP-1 and the basal body-specific antibody BBA4 (data not shown). Because of the DNA binding properties of AEP-1 we describe later, it is important to note that in these three-dimensional reconstructions, AEP-1 is not distributed throughout the kDNA but is restricted to the surface of the kDNA network adjacent to the flagellum (Fig. 3E to G).
Expression of truncated AEP-1 from the nucleus.
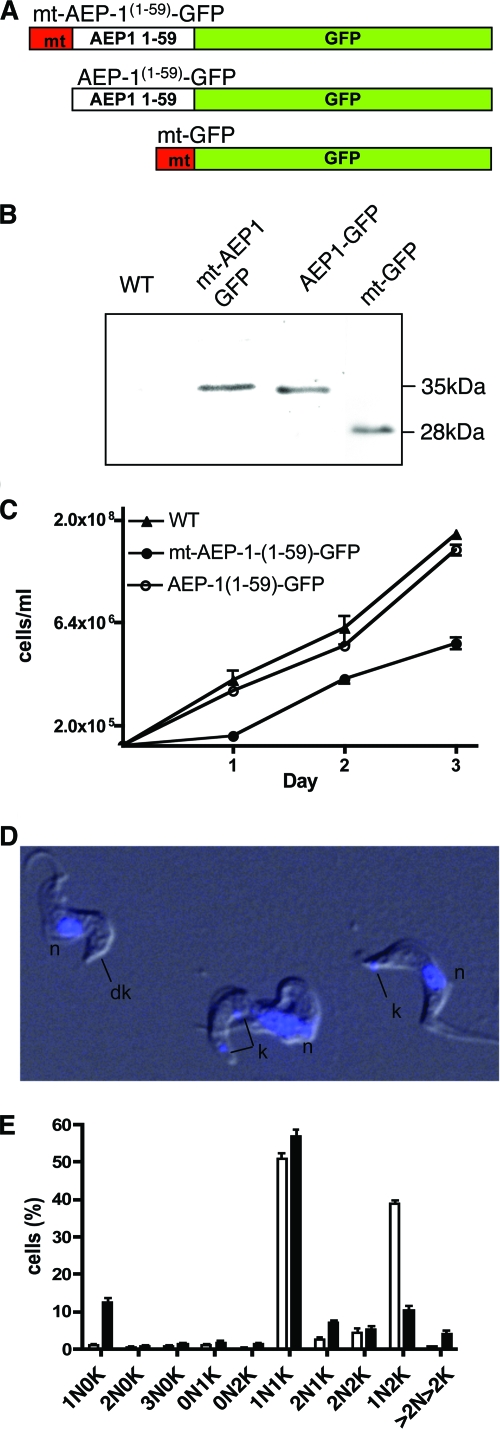
To investigate the function of AEP-1, we reasoned that expression of the unique amino-terminal 59 amino acids, without the multiple membrane-spanning regions of COXIII, might compete with the endogenous AEP-1 and lead to a dominant negative phenotype. The mitochondrial AEP-1 gene was recoded to match the nuclear codon usage, allowing translation on cytosolic ribosomes, and the gene then expressed the truncated AEP-1 ectopically from the nucleus with a mitochondrial targeting sequence (Fig. 4; see Fig. S2A in the supplemental material). A construct encoding a 14-amino-acid mitochondrial targeting sequence, the first 59 amino acids of AEP-1, and a C-terminal fusion to enhanced GFP [mt-AEP-1(1-59)-GFP] was cloned into a trypanosome expression vector (Fig. 4A). Control constructs lacking the mitochondrial targeting sequence [AEP-1(1-59)-GFP] or with the mitochondrial targeting sequence fused directly to GFP (mt-GFP) were also cloned into the trypanosome expression vector (Fig. 4A).
FIG. 4.
Expression of a truncated AEP-1 produces a dominant negative phenotype. (A) Representation of the AEP-1(1-59)-GFP construct with and without the N-terminal mitochondrial targeting sequence (mt; red) and a GFP (green) with the mitochondrial targeting sequence. (B) Western blot analysis of total protein extract from 1 × 105 wild-type (WT) and transfected cells with anti-GFP. (C) Growth of transfected and WT cells. (D) Overlay of differential interference contrast image and DAPI-stained image of AEP-1(1-59)-GFP-transfected cells (n, nucleus; k, kinetoplast; dk, dyskinetoplast) (for the full field, see Fig. S4 in the supplemental material). (E) Histogram from seven independent experiments. A total of 1,037 wild-type cells and 887 mt-AEP-1(1-59)-GFP-transfected cells were analyzed for numbers of nuclei (N) and kinetoplasts (K) using DAPI staining. Cells are classified as, for example, 1N0K, indicating one nucleus and no kinetoplast.
Expression of each protein, mt-AEP-1(1-59)-GFP, AEP-1(1-59)-GFP, and mt-GFP, was confirmed by Western blotting using an antibody to GFP (Fig. 4B). A 35-kDa band reacts with the antibody corresponding to the mt-AEP-1(1-59)-GFP fusion protein containing the mitochondrial targeting sequence, AEP-1, and GFP. The anti-GFP reactive band is absent in nontransfected cells. Cells transfected with AEP-1(1-59)-GFP, lacking the mitochondrial targeting sequence, also express the fusion protein as did trypanosomes transfected with mt-GFP (27 kDa) (Fig. 4B).
Phenotype of cells expressing truncated AEP-1.
Trypanosomes ectopically expressing mt-AEP-1(1-59)-GFP show a growth defect, resulting in an increased cell doubling time (6.45 ± 0.63 h versus 12.2 ± 0.65 h) leading to a 10-fold reduction in cell number after 72 h in culture. Cells expressing AEP-1(1-59)-GFP or mt-GFP show wild-type growth (Fig. 4C). Consistent with the observed growth defect, cells expressing mt-AEP-1(1-59)-GFP have an altered cell cycle compared with wild-type T. brucei, with an increase in G1 phase cells (60% versus 40%) and a decrease of cells in G2/S phase (12% versus 30%) (see Fig. S3 in the supplemental material). The expression of mt-AEP-1(1-59)-GFP also results in the unexpected phenotype of impaired kinetoplast segregation. The mt-AEP-1(1-59)-GFP cells have a 10-fold increase in the number of dyskinetoplastic cells lacking kinetoplast staining (∼13%) as well as an increase in cells with more than two kinetoplasts (Fig. 4D and E; see Fig. S4 in the supplemental material).
Localization of mt-AEP-1(1-59)-GFP.
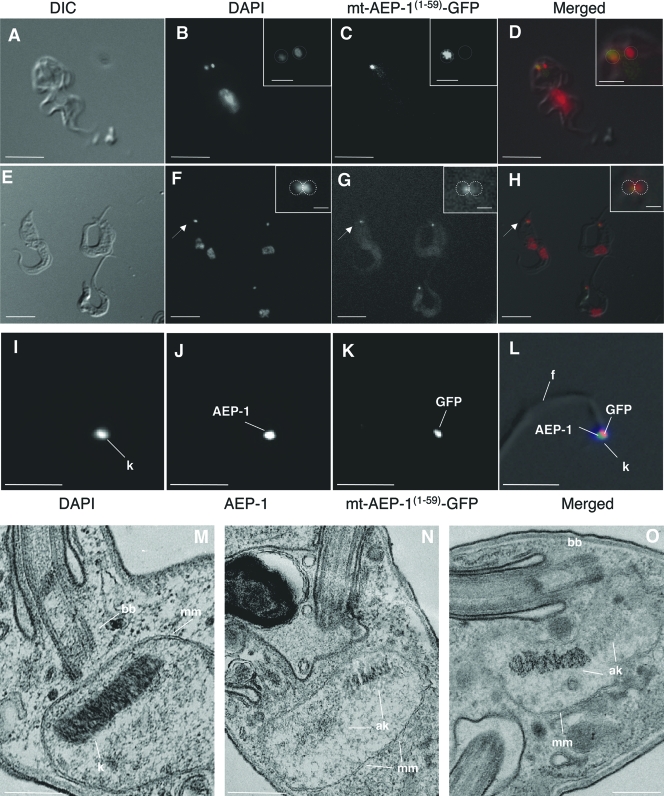
The observed effect of mt-AEP-1(1-59)-GFP expression on cell growth and kinetoplast number suggests that mt-AEP-1(1-59)-GFP could be competing for endogenous AEP-1 within the kinetoplast of the transfected cells. To investigate this possibility, we followed the localization of mt-AEP-1(1-59)-GFP in T. brucei by fluorescence microscopy. In stable transfectants the mt-AEP-1(1-59)-GFP localizes at the kinetoplast (Fig. 5A to H). The truncated AEP-1 (mt-AEP-1(1-59)-GFP) was also stably associated with the purified TAC and colocalized with endogenous AEP-1 (Fig. 5I to L). In dividing cells, with multiple kinetoplasts, mt-AEP-1(1-59)-GFP localization was restricted to one of the sister kinetoplasts, and this kinetoplast always appeared as a less distinct DAPI-stained granule, suggesting that the organization of the kDNA network might be affected by association with the truncated AEP-1(1-59)-GFP (Fig. 5A to D). In dividing cells that had not segregated the sister kinetoplasts, the morphology of the elongated kinetoplast suggested that AEP-1(1-59)-GFP was associated with the portion of the kinetoplast with more diffuse DAPI staining, suggesting a less ordered state (Fig. 5E to H, Insets).
FIG. 5.
T. brucei expressing mt-AEP-1(1-59)-GFP has altered kinetoplast morphology. (A and E) Differential interference contrast (DIC) microscopy of mt-AEP-1(1-59)-GFP trypanosomes. (B and F) DAPI-stained cells. (C and G) GFP signal from the same cells shown in panels B and F. (D and H) Overlay using artificial colors for DAPI (red) and GFP (green). Insets show an enlargement of the kinetoplast region of the cells indicated by an arrow. Bar, 10 μm (large frames) and 1 μm (insets). (I to L) Immunofluorescence microscopy of isolated flagella using DAPI, an AEP-1 antibody, and an antibody to GFP. Panel L shows the merged image of panels I to K using artificial coloring (blue, I; green, J; red, K) with the DIC image. Bar, 2.5 μm. (M to O) Transmission electron microscopy images of thin sections from wild type (M) and mt-AEP-1(1-59)-GFP-transfected cells (N and O). Bar, 200 nm. k, kinetoplast; mm, mitochondrial membrane; bb, basal body; ak, aberrant kinetoplast.
To ensure that the growth and morphological changes associated with mt-AEP-1(1-59)-GFP expression were a consequence of mitochondrial localization of the truncated AEP-1, we examined cells transfected with AEP-1(1-59)-GFP (i.e., lacking the mitochondrial targeting sequence). These cells express the fusion protein (Fig. 4B), but the protein is restricted to the cytoplasm, as judged by GFP fluorescence (see Fig. S2C in the supplemental material). We also tested whether the kinetoplast localization of mt-AEP-1(1-59)-GFP was due to GFP sequences. Cells transfected with the mitochondrion-targeted GFP, without AEP-1 sequences, express the protein (Fig. 4B), and fluorescence microscopy showed that while the GFP signal was mitochondrial, there was no discrete localization to the kinetoplast region (see Fig. S2B in the supplemental material).
In order to further investigate the morphology of the kinetoplast in mt-AEP-1(1-59)-GFP trypanosomes, cells were examined by transmission electron microscopy. Sections through the kinetoplast of wild-type T. brucei show a highly ordered structure of tightly arranged DNA filaments localized to the region of the mitochondrion adjacent to the basal bodies of the flagellum (Fig. 5M). As previously described, stained fibers and unilateral filaments are localized in the region between the kDNA network and the inner mitochondrial membrane, and detailed morphological and cytochemical analyses suggest that there are distinct subdomains within this region (8, 17). Transmission electron microscopy of mt-AEP-1(1-59)-GFP-transfected cells reveals an altered kDNA structure not found in wild-type T. brucei (Fig. 5N and O). Alterations in kDNA organization are most often seen in cells with elongated kDNA discs, consistent with late stages in kDNA replication. It generally results in a disordered appearance to a portion of the kDNA network, with DNA filaments extending up to 300 nm from the kDNA disc (Fig. 5N and O). Taken together, these data suggest that endogenous AEP-1 is normally associated with the kDNA and that expression of the unique N-terminal 59 amino acids, without transmembrane domains of AEP-1, produces a dominant negative phenotype of altered kDNA organization.
AEP-1 binds DNA.
To better understand how AEP-1 contributes to the organization and segregation of the kDNA network, we expressed the unique N-terminal 59 amino acids of AEP-1 as a His-tagged protein in E. coli (data not shown). Because of the unusual amino acid composition of this region of AEP-1 (11 arginine residues; 18%), the ability to bind DNA was examined (Fig. 6A and B). In an electrophoretic mobility shift assay, recombinant AEP1(1-59)-His10 binds both minicircle and maxicircle DNA with approximately equal affinity (Fig. 6B; see Fig. S5A and B in the supplemental material). Two shifted products were detected in these assays, with the more rapidly migrating complex forming at lower concentrations of AEP-1. Detailed analysis of the AEP-1/DNA complexes is under way, but it is likely that the slower-migrating complex is an oligomeric product of AEP-1 bound to DNA. Competition studies with unlabeled minicircle, maxicircle, or plasmid DNA suggest that AEP1(1-59)-His10 binding is not specific for kDNA (Fig. 6C and D; see Fig. S5A to D in the supplemental material). These results suggest that AEP-1 is a membrane-anchored, DNA binding protein localized to the flagellum/kinetoplast segregation apparatus. Remarkably, the properties of AEP-1 are the result of alternative editing of COXIII mRNA. Membrane anchoring is provided by the portion of AEP-1 that is identical to COXIII, and the DNA binding activity is encoded by the unedited portion of the AEP-1 mRNA. These results suggest that alternative mRNA editing plays an important role in generation of protein diversity in trypanosomes.
DISCUSSION
RNA editing in trypanosome mitochondria is an essential process for the production of mRNAs encoding conventional proteins involved in energy production. Recently, it has been shown that COXIII and several other mitochondrial mRNA can be alternatively edited to produce novel open reading frames (14, 15). We have previously shown that one alternative edited mRNA is translated in the mitochondrion of T. brucei and that this protein is associated with mitochondrial membranes (15). In this paper, we investigated the function of AEP-1 and found it to be necessary for the maintenance of the kDNA.
AEP-1 is a kinetoplast-associated membrane protein.
The mRNA encoding AEP-1 contains approximately 180 nucleotides of preedited COXIII joined by a short junction sequence of alternatively edited RNA to the fully edited bona fide COXIII sequence. The predicted protein sequence for AEP-1 contains the C-terminal five transmembrane segments of COXIII and a hydrophilic N-terminal region of approximately 50 amino acids (Fig. 1A). Based on the crystal structure of bovine COXIII, we modeled the T. brucei AEP-1 with a high degree of confidence; we showed the overall orientation of the COXIII transmembrane segments and positioned the hydrophilic N-terminal domain in the mitochondrial matrix (Fig. 1B). This model is consistent with physical properties of AEP-1 indicating that it is an integral, inner mitochondrial membrane protein (15).
AEP-1 is a component of the TAC.
We have shown that the majority of AEP-1 localizes to the portion of the mitochondrion of T. brucei containing the kDNA (Fig. 2). The replication and segregation of the kDNA present several physical challenges. To maintain a high level of sequence diversity for gRNA-encoding genes, each minicircle must be faithfully replicated and segregated during cell division (12). Minicircle replication is unique and requires the cyclical release of individual minicircles, followed by replication and reattachment to the growing kDNA network. To ensure that each minicircle is replicated, the replication machinery preferentially detaches covalently closed, unreplicated minicircles and attaches gapped, newly replicated minicircles (12). At the end of S phase, all the minicircles and maxicircles have replicated, and the sister kinetoplasts segregate. The separation of the sister kDNA networks and the mechanism of segregation at cell division are poorly understood but are likely mediated by the association of the basal bodies of the flagellum with the kDNA networks through the TAC. The first protein component of the TAC, p166, was recently identified (26). Fluorescence microscopy and immuno-electron microscopy showed that this nucleus-encoded protein is largely associated with the TAC but that approximately one-third was in the mitochondrial matrix. This distribution is similar to that we observed for AEP-1; ∼65% of the AEP-1 associated with the kDNA, and the remaining protein was distributed throughout the mitochondrion (Fig. 2). The presence of a predicted mitochondrial targeting signal on p166 suggests that it is assembled into the TAC from the matrix side of the inner mitochondrial membrane. It is possible that both p166 and AEP-1 might represent precursor molecules that have not yet assembled into the TAC but may associate in a partially assembled complex.
Direct analysis of AEP-1 and other components of the AEP-1 mitochondrial membrane complexes will provide important insight into both the function of AEP-1 and the TAC. These studies will require the development of protein purification methods that will allow the sequencing of mitochondrially encoded proteins, which have been difficult to analyze by conventional mass spectrometry methods.
Functional analysis of AEP-1.
The fact that AEP-1 is part of the TAC suggests that it might be involved in the maintenance of the kDNA network. In contrast to p166, functional analysis of trypanosome mitochondrion-encoded proteins is complicated by the lack of mitochondrial transfection methods. To circumvent this limitation, we developed a novel strategy to examine the function of a mitochondrially encoded gene. Since short N-terminal signal sequences can direct the import of nucleus-encoded mitochondrial proteins from the cytoplasm into the mitochondrial matrix, we reasoned that AEP-1, or portions of AEP-1, could be recoded for efficient nuclear transcription, cytosolic translation, and mitochondrial import. By expressing the N terminus of AEP-1 from the nucleus, we observed a dominant negative phenotype of decreased cell growth, aberrant kDNA segregation, and disruption of the kDNA structure (Fig. 4 and 5). The phenotype is consistent with the immunofluorescence localization of the truncated AEP-1 protein at the kinetoplast in vivo. The retention of the truncated AEP-1 in the TAC complex after detergent extraction (Fig. 5I to L) indicates strong interaction of the protein with one of the TAC components. In the absence of the membrane-anchoring domains, this component is likely to be the kDNA. This model is supported by the DNA binding properties of the recombinant N-terminal domain of AEP-1 (Fig. 6).
The sheer size and complexity of the trypanosome kDNA network may have contributed to the evolution of the TAC; however, replication and segregation of less complex mitochondrial genomes in other organisms have also been shown to be membrane associated and mediated through association with cytoplasmic cytoskeletal elements (5). While the DNA binding activity of AEP-1 is associated with a unique N-terminal domain encoded by preedited COXIII mRNA, the C-terminal transmembrane domains of COXIII may be important in membrane anchoring of the TAC. This is somewhat reminiscent of the dual function of mitochondrial metabolic proteins, including aconitase and pyruvate dehydrogenase, in the packaging and maintenance of mitochondrial genomes in Saccharomyces cerevisiae and other eukaryotes (3, 6). The localization and orientation of AEP-1 in the mitochondrial membrane with its N terminus facing the kinetoplast in the mitochondrial matrix, together with the ability of the N-terminal domain to bind DNA in vitro and the dominant negative phenotype in vivo, point toward a function as a kinetoplast maintenance factor.
Alternative mRNA editing generates protein diversity.
Alternative splicing of mRNAs has long been recognized as a mechanism to generate complexity in multicellular eukaryotes by expanding the coding potential for many genes (9). The most extreme example of how alternative splicing can produce different mRNAs from the same gene is seen in the transcripts from the Drosophila melanogaster gene for an axon receptor, Dscam (20). Alternative splicing of Dscam transcripts can produce 38,016 different mRNAs. The tissue and developmental regulation of alternative splicing also contribute to the diversity of gene expression in many organisms (23).
RNA editing bears superficial similarities to RNA splicing. Both are posttranscriptional and result in mature mRNA sequences that differ from the gene. RNA editing of transcripts for the trypanosome mitochondrial NADH dehydrogenase, cytochrome c reductase, COX, and the ATP synthase are incomplete, and editing is necessary to form complete open reading frames for these conventional mitochondrial proteins. Although we have not formally shown the existence of AEP-1 by mass spectrometry, we now present very strong evidence that alternative editing produces a protein with novel functions, indicating that RNA editing provides a powerful evolutionary force for the generation of protein diversity and functional adaptation in these organisms.
Supplementary Material
Acknowledgments
We are grateful to Keith Gull for antibodies to the basal body, Minu Chaudhuri for antibodies to TAO, John Shields for excellent assistance with electron microscopy, Justin Widener and members of the Hajduk laboratory for discussion, and Justin Graham for initial expression of AEP1(1-59)-His10.
This work was supported by a grant from the National Institutes of Health (AI21401).
Footnotes
Published ahead of print on 7 July 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Benne, R., J. Van den Burg, J. P. Brakenhoff, P. Sloof, J. H. Van Boom, and M. C. Tromp. 1986. Major transcript of the frameshifted CoxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 46819-826. [DOI] [PubMed] [Google Scholar]
- 2.Blum, B., N. Bakalara, and L. Simpson. 1990. A model for RNA editing in kinetoplastid mitochondria: “guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell 60189-198. [DOI] [PubMed] [Google Scholar]
- 3.Bogenhagen, D. F., Y. Wang, E. L. Shen, and R. Kobayashi. 2003. Protein components of mitochondrial DNA nucleoids in higher eukaryotes. Mol. Cell Proteomics 21205-1216. [DOI] [PubMed] [Google Scholar]
- 4.Carnes, J., J. R. Trotter, N. L. Ernst, A. Steinberg, and K. Stuart. 2005. An essential RNase III insertion editing endonuclease in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 10216614-16619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, X. J., and R. A. Butow. 2005. The organization and inheritance of the mitochondrial genome. Nat. Rev. Genet. 6815-825. [DOI] [PubMed] [Google Scholar]
- 6.Chen, X. J., X. Wang, B. A. Kaufman, and R. A. Butow. 2005. Aconitase couples metabolic regulation to mitochondrial DNA maintenance. Science 307714-717. [DOI] [PubMed] [Google Scholar]
- 7.Dvorak, J. A. 1993. Analysis of the DNA of parasitic protozoa by flow cytometry, p. 191-204. In J. H. Hyde (ed.), Protocols in molecular parasitology. Methods in molecular biology, vol. 21. Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 8.Gluenz, E., M. K. Shaw, and K. Gull. 2007. Structural asymmetry and discrete nucleic acid subdomains in the Trypanosoma brucei kinetoplast. Mol. Microbiol. 641529-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graveley, B. R. 2001. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 17100-107. [DOI] [PubMed] [Google Scholar]
- 10.Horvath, A., E. A. Berry, and D. A. Maslov. 2000. Translation of the edited mRNA for cytochrome b in trypanosome mitochondria. Science 2871639-1640. [DOI] [PubMed] [Google Scholar]
- 11.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305567-580. [DOI] [PubMed] [Google Scholar]
- 12.Liu, B., Y. Liu, S. A. Motyka, E. E. Agbo, and P. T. Englund. 2005. Fellowship of the rings: the replication of kinetoplast DNA. Trends Parasitol. 21363-369. [DOI] [PubMed] [Google Scholar]
- 13.Lukes, J., H. Hashimi, and A. Zikova. 2005. Unexplained complexity of the mitochondrial genome and transcriptome in kinetoplastid flagellates. Curr. Genet. 48277-299. [DOI] [PubMed] [Google Scholar]
- 14.Ochsenreiter, T., M. Cipriano, and S. L. Hajduk. 2008. 2008. Alternative mRNA editing is extensive and may contribute to mitochondrial protein diversity in trypanosomes. PLoS ONE 3e1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ochsenreiter, T., and S. L. Hajduk. 2006. Alternative editing of cytochrome c oxidase III mRNA in trypanosome mitochondria generates protein diversity. EMBO Rep. 71128-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ochsenreiter, T., and S. L. Hajduk. 2007. Function of RNA editing in trypanosomes, p. 181-197. In H. U. Göringer (ed.), RNA editing. Nucleic acids and molecular biology, vol. 20. Springer, Heidelberg, Germany. [Google Scholar]
- 17.Ogbadoyi, E. O., D. R. Robinson, and K. Gull. 2003. A high-order trans-membrane structural linkage is responsible for mitochondrial genome positioning and segregation by flagellar basal bodies in trypanosomes. Mol. Biol. Cell 141769-1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Panigrahi, A. K., N. L. Ernst, G. J. Domingo, M. Fleck, R. Salavati, and K. D. Stuart. 2006. Compositionally and functionally distinct editosomes in Trypanosoma brucei. RNA 121038-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pollard, V. W., M. E. Harris, and S. L. Hajduk. 1992. Native mRNA editing complexes from Trypanosoma brucei mitochondria. EMBO J. 114429-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmucker, D., J. C. Clemens, H. Shu, C. A. Worby, J. Xiao, M. Muda, J. E. Dixon, and S. L. Zipursky. 2000. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell 101671-684. [DOI] [PubMed] [Google Scholar]
- 21.Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 313381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simpson, L., O. H. Thiemann, N. J. Savill, J. D. Alfonzo, and D. A. Maslov. 2000. Evolution of RNA editing in trypanosome mitochondria. Proc. Natl. Acad. Sci. USA 976986-6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stetefeld, J., and M. A. Ruegg. 2005. Structural and functional diversity generated by alternative mRNA splicing. Trends Biochem. Sci. 30515-521. [DOI] [PubMed] [Google Scholar]
- 24.Stuart, K. D., A. Schnaufer, N. L. Ernst, and A. K. Panigrahi. 2005. Complex management: RNA editing in trypanosomes. Trends Biochem. Sci. 3097-105. [DOI] [PubMed] [Google Scholar]
- 25.Woodward, R., and K. Gull. 1990. Timing of nuclear and kinetoplast DNA replication and early morphological events in the cell cycle of Trypanosoma brucei. J. Cell Sci. 9549-57. [DOI] [PubMed] [Google Scholar]
- 26.Zhao, Z., M. E. Lindsay, A. R. Chowdhury, D. R. Robinson, and P. T. Englund. 2008. p166, a link between the trypanosome mitochondrial DNA and flagellum, mediates genome segregation. EMBO J. 27143-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.