Abstract
Dysfunctional telomeres elicit the canonical DNA damage response, which includes the activation of the ATM or ATR kinase signaling pathways and end processing by nonhomologous end joining (NHEJ) or homologous recombination (HR). The cellular response to DNA double-strand breaks has been proposed to involve chromatin remodeling and nucleosome eviction, but whether dysfunctional telomeres undergo chromatin reorganization is not known. Here, we report on the nucleosomal organization of telomeres that have become deprotected through the deletion of the shelterin components TRF2 or POT1. We found no evidence of changes in the nucleosomal organization of the telomeric chromatin or nucleosome eviction near the telomere terminus. An unaltered chromatin structure was observed at telomeres lacking TRF2, which activate the ATM kinase and are a substrate for NHEJ. Similarly, telomeres lacking POT1a and POT1b, which activate the ATR kinase, showed no overt nucleosome eviction. Finally, telomeres lacking TRF2 and Ku70, which are processed by HR, appeared to maintain their original nucleosomal organization. We conclude that ATM signaling, ATR signaling, NHEJ, and HR at deprotected telomeres can take place in the absence of overt nucleosome eviction.
Through their unique structure and composition, specialized nucleoprotein complexes known as telomeres protect the ends of linear chromosomes. Vertebrate telomeres can form so-called t-loops through strand invasion of their long 3′ overhang into the double-stranded region of telomeric TTAGGG repeats (17, 32). Beyond this ultrastructure, the packaging of telomeric DNA in nucleosomal chromatin lends an additional layer of complexity to vertebrate telomeres. Whereas Saccharomyces cerevisiae and Tetrahymena spp. have short, nonnucleosomal telomeres (7, 16, 43), the results of electron microscopy and micrococcal nuclease (MNase) digestion studies have demonstrated the presence of nucleosomes at telomeres in a number of vertebrates, including chickens, rodents, and humans (26, 28, 32, 38). Telomeric chromatin resembles bulk chromatin in its composition, consisting of the canonical core histone components, but the nucleosomal repeat length is short, and the nucleosomal core particle shows hypersensitivity to MNase (26, 28, 38). The results of in vitro studies have shown that nucleosomes assembled on TTAGGG repeats exhibit higher mobility than on other sequences (33). In addition, telomeric chromatin is enriched for heterochromatic marks, such as trimethylation of H3K9 and H4K20, and the results of studies in mice have shown that the loss of such marks correlates with abnormally elongated telomeres (3, 14, 15). Short human telomeres show evidence of an unusual chromatin structure, as deduced from the more diffuse nature of the MNase digestion patterns (38), but the molecular basis of this change is not known.
Shelterin, a six-protein complex unique to chromosome ends, binds TTAGGG repeats within their nucleosomal context and protects telomeres from being recognized as DNA double-strand breaks (DSBs) or other sites of DNA damage (reviewed in reference 12). Telomere dysfunction caused by the inhibition of shelterin or replicative attrition can lead to the activation of the ATM or ATR kinase signaling cascade, resulting in cell cycle arrest and inappropriate repair of telomeres by nonhomologous end joining (NHEJ) or homologous recombination (HR). The DNA damage response at dysfunctional telomeres also results in the presence of DNA damage factors (e.g., 53BP1 and γ-H2AX) at chromosome ends, forming so-called telomere dysfunction-induced foci (TIFs) (37).
The shelterin protein TRF2 binds double-stranded telomeric repeats through its C-terminal Myb/SANT DNA binding domain (5, 6). Its inhibition by either the introduction of a dominant-negative allele or conditional deletion in mouse embryonic fibroblasts (MEFs) activates the ATM kinase at all stages of the cell cycle and results in end-to-end telomere fusions mediated by Ku70- and DNA ligase IV-dependent NHEJ (8, 9, 36, 42). HR at telomeres gives rise to telomere-sister chromatid exchanges (T-SCEs), which are repressed by both TRF2 and Ku70 (9).
On the other hand, the ATR kinase is repressed by POT1, which binds single-stranded TTAGGG repeats (2) and is recruited to telomeres through its interaction with another shelterin component, TPP1 (19, 20, 25, 45). In mice, which contain two POT1 genes (19), deletion of both POT1a and POT1b or POT1a alone leads to the activation of the ATR kinase (25). Like TRF2, the two POT1 proteins also repress HR at telomeres when Ku70 is absent (W. Palm, D. Hockemeyer, and T. de Lange, unpublished data). However, POT1a and -b do not contribute to the repression of NHEJ at telomeres. POT1b has a specific role in limiting the resection of the C-rich telomeric strand, a process that gives rise to the 3′ overhang (19-21). As a result, POT1b-null MEFs have two- to threefold-more single-stranded telomeric DNA.
The cellular machinery that responds to DNA damage executes its functions within the context of chromatin, and the results of several recent studies have suggested that chromatin remodeling and modifications contribute to the recognition and repair of DSBs. Chromatin decondensation at DSBs has been inferred to occur in living cells subjected to DNA-damaging agents, and such conformational changes have been proposed to activate the ATM kinase (1, 24). In addition, nucleosome loss has been observed at sites of induced DSBs in several model systems. In S. cerevisiae, the induction of a single DSB by HO endonuclease leads to histone displacement that depends on the DNA damage sensor MRX and the chromatin remodeling complex INO80 (39, 41). In human cells, break sites introduced by the I-PpoI nuclease exhibit nucleosome disruption that requires NBS1 and ATM (4). Furthermore, several steps in NHEJ or HR are stimulated by one or more of the three major ATP-dependent chromatin remodeling complexes, INO80, Swi/Snf, and RSC (10, 31, 34, 35, 40, 41).
Given that telomere dysfunction activates the canonical response to DSBs, exploring the role of chromatin remodeling at damaged telomeres may lend insight into the mechanisms by which shelterin prevents telomeres from being recognized as DNA damage. For this study, we investigated the nucleosomal organization of telomeres following their deprotection to determine whether the telomeric DNA damage signaling and repair processes involve chromatin remodeling.
MATERIALS AND METHODS
Cell lines.
Deletion of TRF2 was performed in TRF2FLOX/− p53−/−, TRF2FLOX/− p53−/− Lig4−/−, and SV40LT-immortalized TRF2FLOX/− Ku−/− MEFs by infection with Hit&Run-Cre retrovirus as described previously (8, 9). The temperature-sensitive TRF2ts DNA ligase IV-deficient cell line was described previously (23). It contains the TRF2ts allele (mouse TRF2-I468A) in the context of TRF2FLOX/− p53−/− Lig4−/− MEFs from which the endogenous TRF2 allele was deleted with Cre. The cells were grown at the permissive temperature of 32°C and shifted to 37°C to induce the release of TRFts from telomeres.
Conditional deletion of POT1a and/or POT1b from MEFs was performed as previously described (19). SV40LT-immortalized POT1aSTOP/FLOX, POT1bSTOP/FLOX, or POT1aSTOP/FLOX POT1bSTOP/FLOX MEFs were infected with pWzl-Cre retrovirus and selected for 5 days in hygromycin. SV40LT-immortalized POT1aSTOP/FLOX Ku−/− and POT1aSTOP/FLOX POT1bSTOP/FLOX Ku−/− MEFs were infected four times with Hit&Run-Cre at 12-h intervals. All MEFs were cultured in Dulbecco's modified Eagle's medium supplemented with 15% fetal bovine serum, l-glutamine, penicillin/streptomycin, and nonessential amino acids.
MNase digestion of nuclei.
Nuclei were isolated and digested with MNase as described previously (38). Briefly, cells were trypsinized, suspended in growth medium, and harvested by centrifugation in an RT6000 centrifuge at 1,500 rpm for 5 min. Cells were suspended at 2 × 106 cells/ml in buffer A (100 mM KCl, 10 mM Tris [pH 7.5], 3 mM MgCl2, 1 mM CaCl2, 0.5 mM phenylmethylsulfonyl fluoride), washed twice with buffer A, and then resuspended at 5 × 106 cells/ml in buffer A with 0.6% Nonidet P-40 to lyse cells. After gently mixing and incubating on ice for 5 min, nuclei were harvested at 2,000 rpm for 5 min and resuspended in buffer A without NP-40 at 2.5 × 107 cells/ml. Nuclei were homogenized in a dounce with 10 strokes with a tight B-type pestle. Aliquots of 150 μl were digested for 5 min at 30°C with MNase (Roche Diagnostics) at concentrations ranging from 5 to 600 U/ml. Reactions were stopped by adding 1 volume of TEES-protK (10 mM Tris-HCl [pH 7.5], 10 mM EDTA, 10 mM EGTA, 1% sodium dodecyl sulfate [SDS], 50 μg/ml proteinase K) and incubating at 37°C for 2 h to overnight. DNA was extracted with phenol-chloroform, precipitated with isopropanol in the presence of 0.2 M sodium acetate (pH 5.5), and resuspended in 500 μl TE (10 mM Tris, 1 mM EDTA [pH 8.0]).
MNase digestion of DNA in agarose plugs.
Agarose-embedded DNA plugs were prepared as described previously (8). Cells were harvested, resuspended in phosphate-buffered saline at 2 × 106 cells/ml, and mixed in a 1:1 (vol/vol) ratio with 2% agarose. Plugs were digested with 1 mg/ml proteinase K in buffer (100 mM EDTA [pH 8.0], 0.2% sodium deoxycholate, 1% sodium lauryl sarcosine) overnight at 50°C. Plugs were washed extensively with TE, and then equilibrated in buffer A (see above) for 1 h at 4°C. Digestions were performed by incubating plugs for 10 min at 30°C with MNase in buffer A at concentrations ranging from 0 to 8 U/ml in a total volume of 500 μl. Reactions were stopped by adding an equal volume of TEES-protK and incubating overnight at 37°C. The next day, plugs were washed several times with TE and once in water and then incubated overnight with 60 U MboI at 37°C in NEB buffer 3 (New England Biolabs). Following digestion, plugs were loaded on a 1% agarose-0.5× Tris-borate-EDTA gel and fractionated by pulsed-field gel electrophoresis on a Chef-DRII (Bio-Rad) as described previously (8). In-gel detection of single-stranded TTAGGG repeats was performed as previously described (8).
Southern blotting and detection of telomeric DNA.
DNA was electrophoresed in 1% agarose-0.5× Tris-borate-EDTA at 1 V/cm for at least 2 h, followed by raising the voltage to 3 to 4 V/cm until the Orange-G dye in the loading buffer ran off the gel. The DNA was denatured (1.5 M NaCl, 0.5 M NaOH; twice for 30 min), neutralized (3 M NaCl, 0.5 M Tris-HCl, pH 7.0; twice for 30 min), and then transferred to a Hybond-N membrane in 20× SSC (3 M NaCl, 300 mM Na3-citrate, pH 7). The DNA was UV-cross-linked to the membrane, which was then prehybridized in Church mix (0.5 M Na phosphate buffer [pH 7.2], 1 mM EDTA [pH 8], 7% wt/vol SDS, 1% wt/vol bovine serum albumin) for 1 h followed by hybridization with an end-labeled 32P-(CCCTAA)4 oligonucleotide at 50°C for at least 4 h. The membrane was washed, at 55°C, three times for 30 min in 4× SSC and once for 15 min in 4× SSC-0.1% SDS.
Assay for the last nucleosomes.
DNA associated with the last nucleosome was isolated by a variation on the previously described telomere purification protocol (44). Following MNase digestion of nuclei, the recovered DNA fragments were annealed to a biotinylated oligonucleotide in a 50-μl reaction mixture containing ∼40 μg MNase-digested DNA, 0.25 pmol biotin-(CCCTAA)6, 1× SSC, and 1.0% Triton X-100 and incubated for 30 min each at 37°C, 30°C, 25°C, 15°C, 10°C, and 4°C. The mixture was rotated overnight at 4°C with 0.5 mg/ml streptavidin-coupled Dynabeads (Dynal; Invitrogen) which were prewashed with 1× SSC, precoated with 5× Denhardt's solution (0.1% Ficoll 400, 0.1% polyvinylpyrrolidone, 0.1% bovine serum albumin) for 30 min at room temperature, and resuspended in cold 1× SSC.
To isolate the annealed DNA, a magnet (Dynal magnetic-particle concentrator) was used to concentrate the beads, and the supernatant was removed. The beads were washed twice with 150 μl 1× SSC-1% Triton X-100, once with 150 μl 0.2× SSC-1% Triton X-100, and once with 50 μl TE. All washes were performed on ice. The beads were then resuspended in 20 μl TE and heated to 65°C for 10 min. The supernatant was recovered and fractionated on 1% agarose, and the telomeric signal was detected by Southern blot hybridization as described above.
Exonuclease I digestion.
Fragments recovered after MNase digestion were subjected to treatment with Escherichia coli exonuclease I. Approximately 2 μg MNase-treated DNA was incubated with ExoI (1 U/μl; New England Biolabs) in reaction buffer (New England Biolabs) at 37°C overnight. DNA fragments were recovered by phenol-chloroform extraction followed by precipitation with isopropanol in the presence of 0.2 M sodium acetate (pH 5.5).
Construction of model telomeric fragments.
A 226-bp BglII/KpnI fragment containing 25 TTAGGG repeats was excised from pSXneo.25(T2AG3) (18), dephosphorylated with calf intestinal phosphatase, and purified by gel extraction. The BglII end of the double-stranded telomeric fragment was ligated overnight at 16°C to the 5′ phosphorylated oligonucleotide 5′-GATC(T2AG3)12-3′ with T4 DNA ligase. Reaction mixtures lacking either the double-stranded fragment or the single-stranded oligonucleotide were used as controls.
RESULTS
Deletion of TRF2 or POT1a and -b does not disrupt the overall nucleosomal organization of telomeric chromatin.
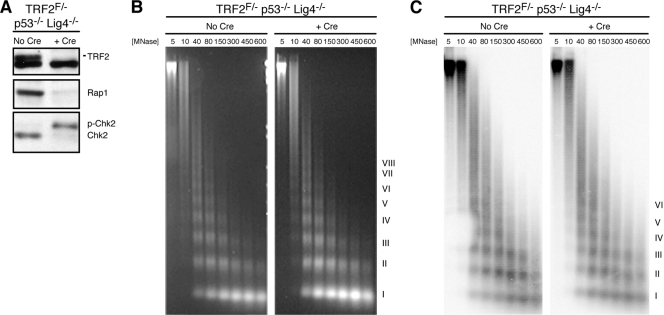
To explore the possibility of chromatin remodeling at telomeres in the setting of ATM kinase activation, we first examined telomeric nucleosomes following Cre-mediated deletion of TRF2 from TRF2FLOX/− p53−/− Lig4−/− MEFs. Due to the absence of p53 and DNA ligase IV, these cells do not undergo cell cycle arrest, nor do they accumulate telomere fusions, thus allowing the assessment of the nucleosomal organization of free dysfunctional telomeres. Nucleosomal chromatin was examined in this and other experiments by digesting nuclei with MNase. Cells were harvested at 90 h after Cre infection, and immunoblots confirmed the loss of TRF2 and its interacting partner Rap1 (Fig. 1A). Furthermore, as expected, the cells showed evidence of ATM kinase signaling based on the phosphorylation of Chk2 (Fig. 1A).
FIG. 1.
No disruption of telomeric chromatin after TRF2 depletion. (A) Immunoblot confirming loss of TRF2 and Rap1 and phosphorylation of Chk2 at 90 h after retrovirus-mediated Hit&Run-Cre expression in TRF2FLOX/− p53−/− Lig4−/− MEFs. The nonspecific band on the TRF2 blot served as a loading control. (B) Bulk nucleosomes in cells with and without TRF2 detected by ethidium bromide staining of DNA from nuclei digested with MNase and fractionated on a 1% agarose gel. (C) Telomeric nucleosomes detected by Southern blot hybridization with a 32P-(CCCTAA)4 probe. Roman numerals represent oligonucleosomes formed by partial digestion. MNase concentrations in U/ml are shown. F, FLOX.
Nuclei from these cells lacking TRF2 were digested with MNase, and the resulting DNA products were fractionated on agarose gels to visualize bulk and telomeric chromatin fragments. The deletion of TRF2 resulted in no obvious change in the MNase sensitivity of bulk nucleosomes, as seen by the results of ethidium bromide staining (Fig. 1B). In cells with and without TRF2, MNase digestion generated the typical nucleosomal ladder with regular periodicity. DNA fragments representing partial digestion products of up to 7 or 8 nucleosomes were seen at low MNase concentrations. In comparison to bulk nucleosomes, telomeric nucleosomes showed a more diffuse ladder of oligonucleosomes (Fig. 1C), as reported previously (38). However, when wild-type and TRF2-deficient cells were compared, no obvious difference in the sensitivity of telomeric chromatin to MNase was observed. The telomeric chromatin maintained the same nucleosomal periodicity in the presence and absence of TRF2. Furthermore, in both cases, the formation of partial products correlated similarly with the MNase concentration, suggesting no difference in the rate of MNase digestion of the dysfunctional telomeres.
While Cre-mediated deletion of TRF2 requires several days before the consequences of telomere dysfunction can be assayed, a recently characterized temperature-sensitive allele of TRF2 allowed us to examine chromatin organization within a shorter time frame after telomere deprotection (23). The TRF2ts allele has a point mutation in the Myb/SANT DNA binding domain (I468A in mouse TRF2) that affects the ability of TRF2 to stably associate with telomeric DNA at the nonpermissive temperature. A DNA ligase IV-deficient cell line carrying TRF2ts was generated by expressing the mouse TRF2ts allele in TRF2FLOX/− p53−/− Lig4−/− MEFs and removing the endogenous TRF2 through Cre-mediated deletion (23). These cells retain telomere protection at 32°C, while incubation at 37°C dislodges TRF2ts from telomeres, leading to a telomere damage response within hours. We compared the MNase sensitivity of telomeric chromatin in TRF2ts cells growing at 32°C to that of cells shifted to 37°C for 3 or 6 h. Immunoblotting confirmed that the temperature shift induced γ-H2AX at 6 h (see Fig. S1A in the supplemental material). Consistent with the data obtained after Cre-mediated deletion of TRF2, the MNase digestion patterns of telomeric chromatin remained unchanged within 3 to 6 h after the removal of TRF2ts from telomeres (data not shown; see Fig. S1B and C in the supplemental material). Thus, there is no overt alteration in the nucleosomal structure of dysfunctional telomeres despite ongoing ATM kinase signaling.
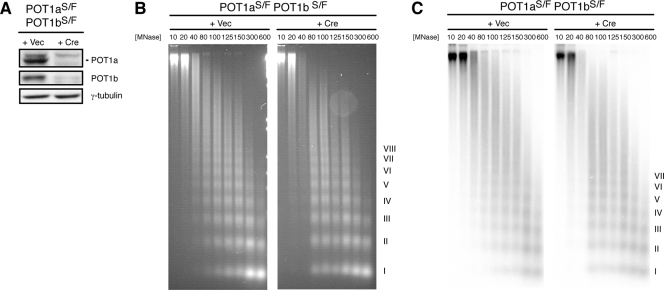
Next, we evaluated the nucleosomal organization at telomeres deprotected by POT1 deletion to determine whether chromatin remodeling accompanies activation of the ATR kinase. The introduction of Cre into POT1aSTOP/FLOX POT1bSTOP/FLOX MEFs resulted in the expected loss of both POT1a and POT1b protein, as confirmed by immunoblots (Fig. 2A). The loss of the POT1 proteins did not affect the MNase sensitivity of the bulk nucleosomes (Fig. 2B) or the telomeric nucleosomes (Fig. 2B and C). Furthermore, similar to the result with TRF2, deletion of both POT1 genes or either POT1a or POT1b alone also did not lead to an obvious change in MNase digestion patterns (Fig. 2B and C; see Fig. S2A and B in the supplemental material).
FIG. 2.
No disruption of telomeric chromatin after POT1 depletion. (A) Immunoblot confirming deletion of POT1a and POT1b after retroviral infection of POT1aSTOP/FLOX POT1bSTOP/FLOX cells with pWzl-Cre followed by 5 days of selection with hygromycin. γ-Tubulin is shown as a loading control. (B) Bulk nucleosomes in cells with and without POT1a and -b detected by ethidium bromide staining of DNA from nuclei digested with MNase and fractionated on 1% agarose gels. (C) Telomeric nucleosomes detected by Southern blot hybridization with a 32P-(CCCTAA)4 probe. Roman numerals represent oligonucleosomes formed by partial digestion. MNase concentrations in U/ml are shown. S/F, STOP/FLOX; Vec, vector; +, present; −, absent.
A novel assay detects nucleosomal organization at the telomere terminus.
Since most telomeres in MEFs are more than 20 kb in length, nucleosome eviction from the telomere end may not be reflected in the MNase digestion patterns of the total telomeric chromatin. Therefore, we devised a novel method for detecting the nucleosomal organization at the telomere terminus, based on the fact that mammalian telomeres end in a 50- to 300-nucleotide, single-stranded tract of TTAGGG repeats, termed the 3′ overhang (11, 27, 29, 46). Although the 3′ overhang is removed from telomeres undergoing NHEJ, the single-stranded TTAGGG repeats are retained when TRF2 is deleted from DNA ligase IV-deficient cells and also persist in POT1-deficient cells. Therefore, the last nucleosome of the dysfunctional telomeres in these settings is expected to be adjacent to the 3′ overhang.
A concern with using the 3′ overhang as a marker for the last nucleosome is that MNase is known to rapidly degrade single-stranded DNA (22). Indeed, when agarose-embedded protein-free genomic DNA was digested with MNase, the single-stranded overhang signal decreased rapidly and at a much faster rate than the total TTAGGG signal representing the duplex telomeric repeats (see Fig. S3A in the supplemental material). In contrast, the telomeric overhang remains largely intact during the MNase treatment of nuclei from wild-type cells, as well as in nuclei lacking either POT1 or TRF2 (see Fig. S3B in the supplemental material; data not shown), indicating that the single-stranded DNA is protected from the nuclease. As discussed below, this protection could be due to strand invasion of the 3′ overhang in the t-loop configuration and/or coating with POT1a and -b.
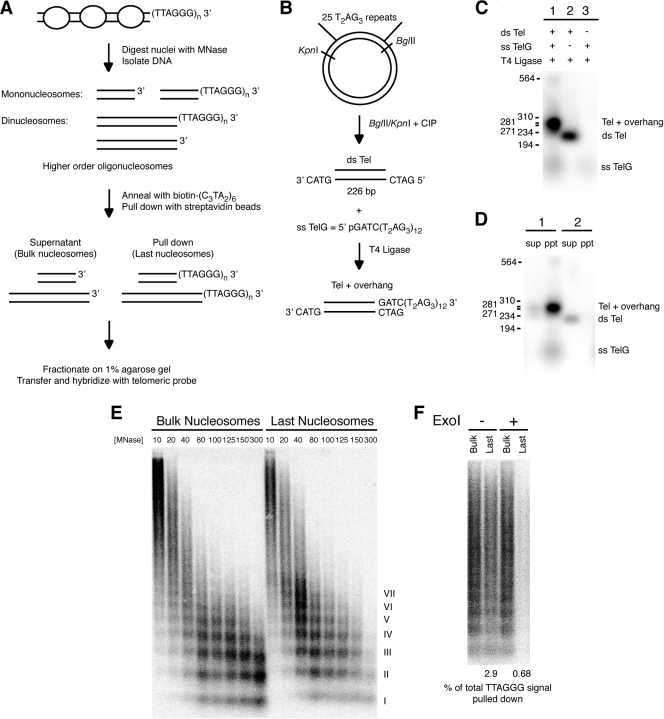
The resistance of the telomeric overhang to MNase digestion of chromatin in nuclei suggested that it should be possible to detect the most terminal nucleosome abutting the 3′ overhang. However, the standard method of detecting DNA fragments bearing a 3′ overhang—in-gel hybridization to native DNA—is not applicable to fragments smaller than ∼500 bp due to their loss during the gel-drying step required for this protocol (data not shown). We therefore developed an alternative method of detecting the MNase product representing the last nucleosome next to the single-stranded TTAGGG repeats (Fig. 3A). Biotinylated oligonucleotides complementary to the 3′ overhang were annealed to the DNA fragments recovered from MNase-treated nuclei; the last nucleosomal fragments could then be precipitated with streptavidin beads. The DNA fragments in the supernatant and precipitate were fractionated on an agarose gel and transferred onto a nitrocellulose membrane, and the telomeric signal was detected with a 32P-(CCCTAA)4 probe.
FIG. 3.
A novel assay for the nucleosomal organization near the telomere terminus. (A) Scheme of the last nucleosome assay. Nuclei are treated with MNase, and the isolated DNA fragments are incubated with a biotinylated oligonucleotide representing the C-rich telomeric DNA strand, biotin-(CCCTAA)6. Magnetic streptavidin beads are used to pull down DNA fragments containing a telomeric (TTAGGG)n 3′ overhang. The supernatant contains fragments of bulk nucleosomes, while the pulldown contains nucleosomal fragments ending at the telomere terminus. Fragments are fractionated on a 1% agarose gel and subjected to Southern blot hybridization with a 32P-(CCCTAA)4 probe. (B) Scheme for the construction of a model telomeric fragment to test the last nucleosome assay. A BglII/KpnI fragment excised from pSXneo.25(T2AG3) (dsTel) was annealed and ligated to a single-stranded telomeric oligonucleotide (ssTel) containing the complementary BglII recognition sequence at its 5′ end. CIP, calf intestinal phosphatase. (C) Southern blot detection of the telomeric signal ligation products of dsTel and ssTel (lane 1), dsTel only (lane 2), or ssTel only (lane 3). (D) Southern blot detection of the telomeric signal after annealing the DNA products from lanes 1 and 2 shown in panel C with biotin-(CCCTAA)6 and separating the supernatant (sup) and pulldown (ppt) following incubation with streptavidin beads. Two percent of the supernatant was loaded next to 50% of the total pulldown. Sizes in base pairs are marked to the left of panels C and D. (E) Detection of the telomeric signal associated with the last nucleosomes in wild-type MEFs. The last nucleosome assay was performed as described above, and the supernatant (2% of total fraction) containing the bulk nucleosomes was loaded in the first eight lanes, while nucleosomal fragments containing the telomeric overhang (50% of total pulldown) were loaded in the last eight lanes. Roman numerals represent oligonucleosomes formed by partial digestion. MNase concentrations in U/ml are shown. (F) Detection of the telomeric signal pulled down by the last nucleosome assay following ExoI treatment of DNA from MNase-digested nuclei. DNA fragments isolated after MNase digestion were mixed and treated with ExoI (300 U in 300 μl), removing the 3′ overhang. ExoI-treated and untreated samples were then subjected to the last nucleosome assay. Two percent of the supernatant (bulk nucleosomes) was loaded next to 50% of the total pulldown (last nucleosomes). Percentage of total TTAGGG signal pulled down was calculated with the formula (2 × pulldown signal)/[(2 × pulldown signal) + (50 × supernatant signal)]. +, present; −, absent.
To verify that this assay could efficiently isolate telomeric fragments containing a G-rich overhang, we constructed a model telomeric fragment by annealing and ligating a BglII/KpnI restriction fragment (dsTel) excised from a plasmid with 25 tandem TTAGGG repeats (pSXneo.25T2AG3) (18) to a single-stranded (TTAGGG)12 oligonucleotide (ssTelG) containing a BglII protrusion at the 5′ end (Fig. 3B). When the products of the ligation reaction were detected by Southern blot for telomeric signal, the major product was a 270- to 280-bp fragment that was absent when either dsTel or ssTelG was excluded from the reaction mixture (Fig. 3C). When this model telomeric fragment was annealed to biotin-(CCCTAA)6 and isolated with streptavidin beads, nearly all of the DNA was pulled down (Fig. 3D). On the other hand, dsTel lacking the 3′ overhang remained in the supernatant, confirming that only fragments containing a G-rich overhang can be isolated by the assay.
We tested this assay for nucleosomes associated with the 3′ overhang on wild-type cells. The pattern of MNase digestion at the terminus largely resembled that of bulk telomeric chromatin (Fig. 3E). Partially digested oligomers of up to 7 or 8 nucleosomes could be visualized. Quantitatively, after normalizing for the relative volumes loaded, the signal intensity of the DNA associated with the last nucleosome accounted for approximately 2 to 4% of the total telomeric signal at low MNase concentrations (Fig. 4B) when the median size of the MNase products was ∼0.9 kb. A maximal yield of ∼3% is expected upon isolation of the terminal fragments from a 30-kb telomere digested into fragments of 1 kb. The observed yield in the last nucleosome assay is therefore within the expected range.
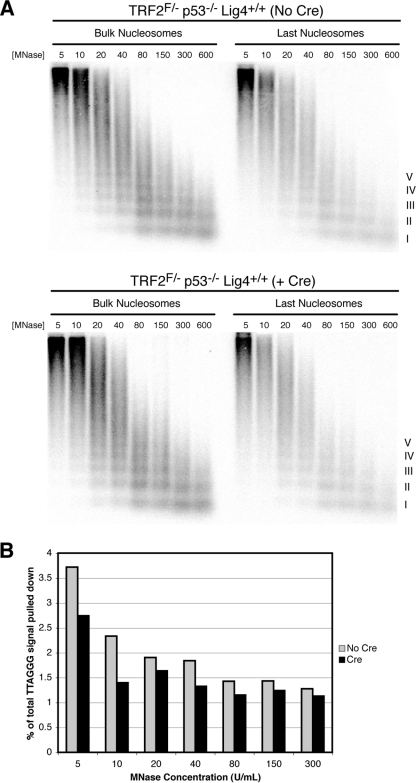
FIG. 4.
Telomere fusion reduces the signal in the last nucleosome assay. (A) MNase sensitivity of bulk and last telomeric nucleosomes assessed by the last nucleosome assay, performed as described in Materials and Methods, of TRF2FLOX/− p53−/− Lig4+/+ cells not infected with Cre (top) or at 120 h following Cre-mediated TRF2 deletion (bottom). Roman numerals represent oligonucleosomes formed by partial digestion. MNase concentrations in U/ml are shown. F, FLOX. (B) Quantitation of the percentage of total TTAGGG signal pulled down in gel shown in panel A.
The results of additional control experiments confirmed the specificity of the assay for the 3′ overhang. When DNA fragments recovered from MNase digestion were treated with the 3′ to 5′ E. coli exonuclease I prior to annealing with the biotinylated oligonucleotide, the percentage of total telomeric signal pulled down by the assay decreased by 50 to 70% (Fig. 3F). Furthermore, when the assay was performed with a biotin-(TTAGGG)6 oligonucleotide instead of the C-rich oligonucleotide and the telomeric signal was probed with 32P-(TTAGGG)4, no telomeric signal was detected in the pulldown (data not shown).
Finally, we applied the last nucleosome assay to a setting in which telomeres become fused and are therefore expected to lose their 3′ overhang. For this control, we used Cre-mediated deletion of TRF2 from DNA ligase IV-proficient cells, focusing on a late time point when ∼30% of chromosome ends have undergone NHEJ (8). TRF2 deletion from these cells resulted in a 20 to 40% reduction in the percentage of total telomeric signal pulled down in the last nucleosome assay compared to the percentage for the control (Fig. 4A and B). The difference between Cre-infected cells and the controls became less pronounced with increasing MNase concentration (Fig. 4B), suggesting that the optimal range for interpreting results from the last nucleosome assay was at MNase concentrations of less than 80 U/ml.
The terminal nucleosome is not evicted following depletion of TRF2.
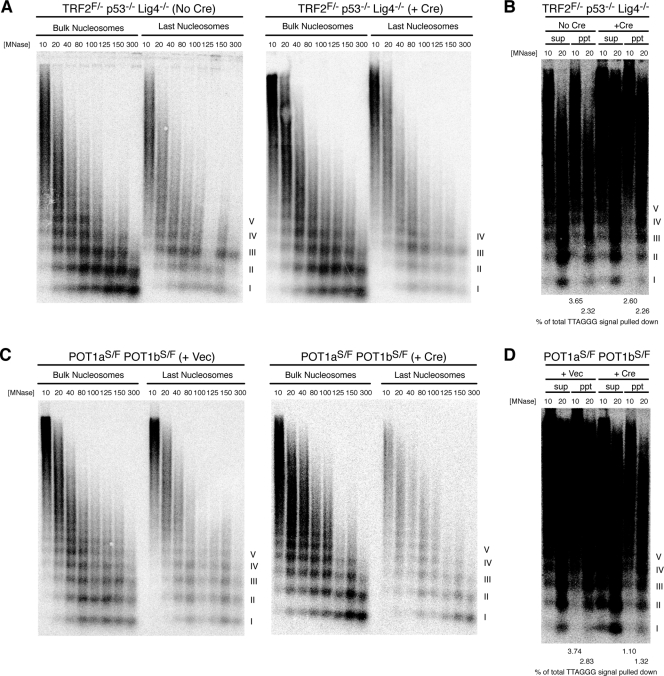
Since the 3′ overhang remains intact following deletion of TRF2 in cells deficient for DNA ligase IV (8), the assay for the last nucleosome could be used to examine whether terminal nucleosomes are evicted following telomere deprotection in this setting. Thus, we isolated the DNA fragments associated with the last nucleosome in TRF2FLOX/− p53−/− Lig4−/− MEFs infected with Cre. Whereas degradation of the overhang would decrease the yield of the terminal DNA, thereby reducing the signal intensity of the ladder representing the last nucleosomes, we observed no change in the signal intensity of last nucleosomal DNA from cells infected with Cre compared to that of uninfected cells (Fig. 5A). Upon longer exposure, the signal due to the last nucleosome appeared similar with and without TRF2 and quantification indicated a comparable percentage of total telomeric signal pulled down by the assay in both cases (Fig. 5B). Furthermore, no significant shift in the last nucleosome was observed, further supporting the absence of nucleosome eviction from telomere ends lacking TRF2.
FIG. 5.
No eviction of terminal nucleosomes following TRF2 or POT1 deletion. (A and C) MNase sensitivity of bulk and last telomeric nucleosomes assessed by the last nucleosome assay in TRF2FLOX/− p53−/− Lig4−/− cells not infected with Cre (left) or at 90 h following Cre-mediated TRF2 deletion (right) (A) and POT1aS/FLOX POT1bS/FLOX cells infected with pWzl vector (left) or pWzl-Cre (right) at 5 days postselection with hygromycin (C). MNase concentrations in U/ml are shown. (B and D) Results of longer exposure of the telomere blots for the last nucleosome assay in cells with TRF2 and POT1 deleted, respectively, with 4% of total supernatant and 100% of total pulldown loaded onto the gel. Quantitation of the percentage of total TTAGGG signal pulled down is provided below the blot. Percentage of total TTAGGG signal pulled down was calculated with the formula pulldown signal/[pulldown signal + (25 × supernatant signal)]. Roman numerals represent oligonucleosomes formed by partial digestion. F, FLOX; S, STOP; +, present; −, absent; Vec, vector; sup, supernatant; ppt, pulldown.
We further assayed the MNase sensitivity of the last nucleosome following telomere deprotection in the TRF2ts system. After 6 h of incubation at 37°C, there was no change in the MNase digestion pattern in the last nucleosome compared to that in cells maintained at the permissive temperature (see Fig. S4 in the supplemental material). The relative telomeric signal pulled down by the assay also remained roughly similar before and after the removal of TRF2.
POT1 deletion leads to increased MNase susceptibility of the overhang, but no overt nucleosome eviction.
We next determined whether the DNA damage response associated with POT1 deficiency involves nucleosome eviction from the chromosome end. Although POT1 double-knockout (DKO) cells show a two- to threefold excess of single-stranded overhang signal, the amount of single-stranded telomeric signal in the fragments recovered after MNase digestion was comparable to that from control cells following the same treatment (see Fig. S2B in the supplemental material). This suggested that MNase digestion of POT1 DKO nuclei led to some degradation of the excess single-stranded DNA, although the last nucleosome assay could still be used to detect chromatin organization at the telomere ends with intact overhangs. We used the last nucleosome assay to compare the MNase digestion pattern of the last nucleosomes in Cre-infected POT1aSTOP/FLOX POT1bSTOP/FLOX cells to that in vector-infected control cells. First, we found that the percentage of total telomeric signal pulled down in POT1 DKO cells was significantly less than that in control cells, corroborating the data showing that some of the overhangs were degraded (Fig. 5D). However, POT1 deletion did not significantly alter the MNase pattern of the detectable last nucleosomes (Fig. 5C). Partial digestion products representing 6 or 7 nucleosomes from the chromosome end were seen in cells with and without POT1a and -b. Furthermore, deletion of either POT1a or POT1b alone also did not affect the MNase digestion pattern of the terminal nucleosomes (see Fig. S5A and C in the supplemental material). In addition, deletion of POT1b led to no change in the fraction of last nucleosomal fragments pulled down in the assay (see Fig. S5D in the supplemental material). On the other hand, deletion of POT1a alone resulted in a reduction in the percentage of total telomeric signal pulled down, suggesting some degradation of the single-stranded overhang by MNase (see Fig. S5B in the supplemental material).
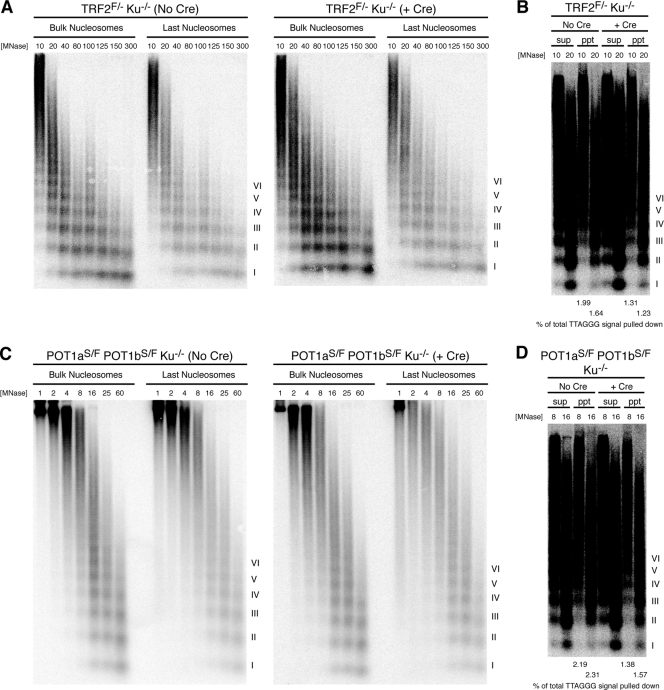
TRF2 deletion in Ku-deficient cells does not affect nucleosome organization, while deletion of POT1 and Ku leads to increased MNase susceptibility of the overhang.
Since nucleosome displacement appears particularly important in DSB repair by HR (10, 39, 40), we further explored the possibility that nucleosome eviction occurs in the absence of factors that protect telomeres from HR. Recombination between sister telomeres is frequent in cells that lack both Ku70 and TRF2 (9), and increased T-SCEs have been observed in cells triply deficient in Ku70 and both POT1a and POT1b (Palm, Hockemeyer, and de Lange, unpublished). Therefore, we examined the MNase digestion pattern of bulk telomeric chromatin, as well as that of the last nucleosomes, in cells deficient in Ku70 and either TRF2 or POT1a and -b. Ku70 deficiency alone led to a typical MNase digestion pattern of bulk and last nucleosomes (Fig. 6A and C, left panels). Further removal of TRF2 did not alter the MNase digestion pattern of either the bulk telomeric chromatin or terminal nucleosomes (Fig. 6A, right panel). Furthermore, there was no change in the signal intensity of the MNase digestion ladder for the last nucleosomes, indicating no degradation of the overhang (Fig. 6B). On the other hand, deletion of POT1a and -b in Ku-deficient cells did not affect the MNase digestion pattern of telomeric nucleosomes but reduced the percentage of total telomeric signal that could be isolated with the last nucleosome assay, suggesting an increased susceptibility of the overhang to MNase (Fig. 6C and D). The results seen in the absence of POT1 and Ku appeared similar to those observed with POT1 deletion alone.
FIG. 6.
No eviction of terminal nucleosomes in cells doubly deficient for Ku70 and TRF2 or POT1a and -b. (A and C) MNase sensitivity of bulk and last telomeric nucleosomes in TRF2FLOX/− Ku−/− cells without Cre infection (left) or at 96 h after Cre-mediated TRF2 deletion (right) (A) and POT1aSTOP/FLOX POT1bSTOP/FLOX Ku−/− cells not infected with Cre (left) or at 96 h after Cre infection (right) (C). (B and D) Results of longer exposure of the telomere blots for the last nucleosome assay in TRF2/Ku-deficient and POT1/Ku-deficient cells, respectively, with 4% of total supernatant and 100% of total pulldown loaded onto the gel. Quantitation of the percentage of total TTAGGG signal pulled down is provided below the blot. MNase concentrations in U/ml are shown. Roman numerals represent oligonucleosomes formed by partial digestion. F, FLOX; S, STOP; sup, supernatant; ppt, pulldown.
DISCUSSION
Here, we used conventional MNase assays and a newly developed assay specific to the terminal part of the telomere to probe the nucleosomal organization at chromosome ends after loss of telomere protection. The novel assay specifically detects fragments containing a G-rich telomeric overhang after the recovery of DNA from MNase-digested nuclei. We found that MNase treatment left the single-stranded overhang largely intact in wild-type and TRF2-deficient cells but led to some degradation following deletion of POT1a or POT1a and -b. The absence of the NHEJ factor Ku70 did not affect the protection of the overhang from MNase. As MNase rapidly degrades the single-stranded telomeric DNA in naked DNA, the results suggest that POT1 proteins, in particular POT1a, block MNase from digesting the telomeric overhang in chromatin. Nevertheless, even in POT1a-deficient cells, DNA fragments recovered after MNase digestion retained a significant fraction of the overhang DNA, suggesting an alternative mode of protection, potentially involving other single-strand binding proteins, such as replication protein A, or the strand invasion of the overhang into duplex telomeric DNA as it occurs in the t-loop.
Our results revealed that the organization of terminal nucleosomes in wild-type cells resembles that of bulk telomeric chromatin. The results of previous work have shown that short telomeres yield a more diffuse MNase pattern than long telomeres, which led to the proposal that telomere ends may exhibit an unusual chromatin organization that would appear more prominently in short telomeres (38). However, this previous study did not examine the terminal telomeric chromatin directly. Alternative explanations for the altered chromatin of short telomeres may be a difference in shelterin loading or a change in the histone modification state in shorter versus longer telomeres (3, 14, 15).
The results of our studies on cells with deprotected telomeres suggest that a number of DNA damage signaling and repair pathways can occur at damaged telomeres without detectable changes in the organization of the bulk telomeric chromatin or overt nucleosome eviction. The absence of any observable change in the sensitivity of telomeric chromatin to MNase following the conditional deletion of shelterin components demonstrates that damaged telomeres retain their nucleosomal organization while eliciting an ongoing DNA damage signal. Specifically, since no overt chromatin remodeling occurs at either bulk or terminal telomere nucleosomes at time points when TIFs have previously been observed (8, 19), the maintenance of the DNA damage signal at dysfunctional telomeres, as evident from the phosphorylation of H2AX and the recruitment of 53BP1, does not appear to require persistent nucleosome eviction.
Importantly, our work contributes to an understanding of the mechanisms by which the sensing of DNA damage and subsequent activation of ATM signaling occur at dysfunctional telomeres. The Mre11/Rad50/Nbs1 complex has been implicated as a DNA damage sensor that induces ATM activity (30), but the molecular event sensed by the Mre11 complex remains undefined. It has been suggested that chromatin changes directly activate ATM, based on the results of studies in which ATM activation occurred in the presence of chromatin relaxation without DSBs (1). In contrast, the absence of changes in telomeric chromatin following the deletion of TRF2 excludes the requirement of nucleosome eviction for DNA damage sensing and signaling by ATM at damaged telomeres.
Our results further suggest that signaling through the ATR pathway can also occur without overt nucleosome eviction, since cells lacking POT1a and POT1b, which accumulate ATR-dependent TIFs, maintain their nucleosomal organization at the telomeres. In POT1a and -b DKO and POT1a knockout cells, MNase degraded some, but not all, of the single-stranded overhang and hindered the isolation of the last nucleosomal DNA. Nonetheless, the assay retrieved a fraction of ends, whose chromatin organization appeared unchanged following POT1 deletion. In cases where MNase significantly degrades the single-stranded overhang, it remains a possibility that our assay detects a subset of terminal nucleosomes that maintain protection and are not involved in the DNA damage signaling or repair processes.
Finally, our results suggest that NHEJ and HR at damaged telomeres do not require nucleosome eviction, since the telomeric chromatin did not show obvious changes in settings that make telomeres highly susceptible to these repair pathways. It is interesting that no chromatin rearrangements were observed under conditions where T-SCEs occur, since nucleosome displacement has been posited to be particularly important for HR (10, 39, 41).
What accounts for the finding that damaged telomeres do not appear to experience nucleosome eviction, whereas DSBs elsewhere in the genome undergo extensive chromatin remodeling? One explanation may be that the kinetics of DNA damage signaling and repair processes at damaged telomeres differ from those found in other models of the response to DSBs. In our system, distinct steps in signaling by ATM or ATR and in repair by NHEJ or HR may indeed involve nucleosome displacement at damaged telomeres, but these events may occur transiently and/or take place too early to be detected by our methods. The results of our studies using the TRF2ts allele, which allows us to assay chromatin organization within a shorter time frame, argue against the likelihood of this explanation, since we demonstrate the absence of nucleosome eviction as early as 3 to 6 h after the removal of TRF2. Alternatively, nucleosome rearrangement may not be required at chromosome ends lacking TRF2 or POT1 because intrinsic properties of telomeric chromatin render it more accessible to DNA damage factors. For instance, the shelterin component TRF1, which can bind telomeric DNA within a nucleosomal context, has been shown to modulate nucleosome structure (13). It is also interesting to note that nucleosomes assembled on telomeric sequences under physiologic conditions appear intrinsically more mobile than those assembled on average DNA (33) and that the telomeric core particles appear highly sensitive to nucleases. Perhaps these unusual features of telomeric nucleosomes obviate the need for chromatin remodeling, permitting access to DNA repair and signaling factors in the native telomeric chromatin state. Such a model would be consistent with the view that numerous DNA repair factors are present at telomeres, where they presumably act to promote the protective function of telomeres.
The finding that telomeric chromatin does not undergo remodeling in the presence of a persistent and robust DNA damage signal has implications for how shelterin protects the telomere. It has previously been proposed that shelterin may create an alternative nucleosomal organization that prevents the end from being recognized as a DSB. Our findings argue against this possibility, as there appears to be no difference between the organization of telomeric chromatin in protected and deprotected states. Nonetheless, our results do not preclude contributions of chromatin remodeling and histone modifications to shelterin function in other ways that remain to be elucidated.
Supplementary Material
Acknowledgments
P.W. thanks Nadya Dimitrova, Dirk Hockemeyer, and Agnel Sfeir for guidance with protocols and Akimitsu Konishi for the TRF2ts cells. T.D.L. thanks Dirk Hockemeyer and Eros Lazzerini Denchi for instruction in Cre-mediated gene deletion and Jill Donigian for instruction in immunoblotting.
P.W. was supported by NIH MSTP grant GM07739 to the Weill Cornell/RU/MSKCC Tri-Institutional M.D.-Ph.D. program. This work was supported by an NIH Director's pioneer award.
Footnotes
Published ahead of print on 14 July 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bakkenist, C. J., and M. B. Kastan. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421499-506. [DOI] [PubMed] [Google Scholar]
- 2.Baumann, P., and T. R. Cech. 2001. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 2921171-1175. [DOI] [PubMed] [Google Scholar]
- 3.Benetti, R., M. Garcia-Cao, and M. A. Blasco. 2007. Telomere length regulates the epigenetic status of mammalian telomeres and subtelomeres. Nat. Genet. 39243-250. [DOI] [PubMed] [Google Scholar]
- 4.Berkovich, E., R. J. J. Monnat, and M. B. Kastan. 2007. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat. Cell Biol. 9683-690. [DOI] [PubMed] [Google Scholar]
- 5.Bilaud, T., C. Brun, K. Ancelin, C. E. Koering, T. Laroche, and E. Gilson. 1997. Telomeric localization of TRF2, a novel human telobox protein. Nat. Genet. 17236-239. [DOI] [PubMed] [Google Scholar]
- 6.Broccoli, D., A. Smogorzewska, L. Chong, and T. de Lange. 1997. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 17231-235. [DOI] [PubMed] [Google Scholar]
- 7.Budarf, M. L., and E. H. Blackburn. 1986. Chromatin structure of the telomeric region and 3′-nontranscribed spacer of Tetrahymena ribosomal RNA genes. J. Biol. Chem. 261363-369. [PubMed] [Google Scholar]
- 8.Celli, G., and T. de Lange. 2005. DNA processing not required for ATM-mediated telomere damage response after TRF2 deletion. Nat. Cell Biol. 7712-718. [DOI] [PubMed] [Google Scholar]
- 9.Celli, G. B., E. Lazzerini Denchi, and T. de Lange. 2006. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat. Cell Biol. 8885-890. [DOI] [PubMed] [Google Scholar]
- 10.Chai, B., J. Huang, B. R. Cairns, and B. C. Laurent. 2005. Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev. 191656-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chai, W., Q. Du, J. W. Shay, and W. E. Wright. 2006. Human telomeres have different overhang sizes at leading versus lagging strands. Mol. Cell 21427-435. [DOI] [PubMed] [Google Scholar]
- 12.de Lange, T. 2005. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 192100-2110. [DOI] [PubMed] [Google Scholar]
- 13.Galati, A., L. Rossetti, S. Pisano, L. Chapman, D. Rhodes, M. Savino, and S. Cacchione. 2006. The human telomeric protein TRF1 specifically recognizes nucleosomal binding sites and alters nucleosome structure. J. Mol. Biol. 360377-385. [DOI] [PubMed] [Google Scholar]
- 14.Garcia-Cao, M., R. O'Sullivan, A. H. Peters, T. Jenuwein, and M. A. Blasco. 2004. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat. Genet. 3694-99. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalo, S., I. Jaco, M. F. Fraga, T. Chen, E. Li, M. Esteller, and M. A. Blasco. 2006. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat. Cell Biol. 8416-424. [DOI] [PubMed] [Google Scholar]
- 16.Gottschling, D. E., and T. R. Cech. 1984. Chromatin structure of the molecular ends of Oxytricha macronuclear DNA: phased nucleosomes and a telomeric complex. Cell 38501-510. [DOI] [PubMed] [Google Scholar]
- 17.Griffith, J. D., L. Comeau, S. Rosenfield, R. M. Stansel, A. Bianchi, H. Moss, and T. de Lange. 1999. Mammalian telomeres end in a large duplex loop. Cell 97503-514. [DOI] [PubMed] [Google Scholar]
- 18.Hanish, J. P., J. L. Yanowitz, and T. de Lange. 1994. Stringent sequence requirements for the formation of human telomeres. Proc. Natl. Acad. Sci. USA 918861-8865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hockemeyer, D., J. P. Daniels, H. Takai, and T. de Lange. 2006. Recent expansion of the telomeric complex in rodents: two distinct POT1 proteins protect mouse telomeres. Cell 12663-77. [DOI] [PubMed] [Google Scholar]
- 20.Hockemeyer, D., W. Palm, T. Else, J. P. Daniels, K. K. Takai, J. Z. Ye, C. E. Keegan, T. de Lange, and G. D. Hammer. 2007. Telomere protection by mammalian POT1 requires interaction with TPP1. Nat. Struct. Mol. Biol. 14754-761. [DOI] [PubMed] [Google Scholar]
- 21.Hockemeyer, D., W. Palm, R. C. Wang, S. S. Couto, and T. de Lange. 2008. Engineered telomere degradation models dyskeratosis congenita. Genes Dev. 221773-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kacian, D. L., and S. Spiegelman. 1974. Use of micrococcal nuclease to monitor hybridization reactions with DNA. Analytical Biochem. 58534-540. [DOI] [PubMed] [Google Scholar]
- 23.Konishi, A., and T. de Lange. 2008. Cell cycle control of telomere protection and NHEJ revealed by a ts mutation in the DNA-binding domain of TRF2. Genes Dev. 221221-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kruhlak, M. J., A. Celeste, G. Dellaire, O. Fernandez-Capetillo, W. G. Muller, J. G. McNally, D. P. Bazett-Jones, and A. Nussenzweig. 2006. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J. Cell Biol. 172823-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lazzerini Denchi, E., and T. de Lange. 2007. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 4481068-1071. [DOI] [PubMed] [Google Scholar]
- 26.Lejnine, S., V. L. Makarov, and J. P. Langmore. 1995. Conserved nucleoprotein structure at the ends of vertebrate and invertebrate chromosomes. Proc. Natl. Acad. Sci. USA 922393-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makarov, V. L., Y. Hirose, and J. P. Langmore. 1997. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell 88657-666. [DOI] [PubMed] [Google Scholar]
- 28.Makarov, V. L., S. Lejnine, J. Bedoyan, and J. P. Langmore. 1993. Nucleosomal organization of telomere-specific chromatin in rat. Cell 73775-787. [DOI] [PubMed] [Google Scholar]
- 29.McElligott, R., and R. J. Wellinger. 1997. The terminal DNA structure of mammalian chromosomes. EMBO J. 163705-3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morales, M., J. W. Theunissen, C. F. Kim, R. Kitagawa, M. B. Kastan, and J. H. Petrini. 2005. The Rad50S allele promotes ATM-dependent DNA damage responses and suppresses ATM deficiency: implications for the Mre11 complex as a DNA damage sensor. Genes Dev. 193043-3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morrison, A. J., J. Highland, N. J. Krogan, A. Arbel-Eden, J. F. Greenblatt, J. E. Haber, and X. Shen. 2004. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 119767-775. [DOI] [PubMed] [Google Scholar]
- 32.Nikitina, T., and C. L. Woodcock. 2004. Closed chromatin loops at the ends of chromosomes. J. Cell Biol. 166161-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pisano, S., E. Marchioni, A. Galati, R. Mechelli, M. Savino, and S. Cacchione. 2007. Telomeric nucleosomes are intrinsically mobile. J. Mol. Biol. 3691153-1162. [DOI] [PubMed] [Google Scholar]
- 34.Shim, E. Y., S. J. Hong, J. H. Oum, Y. Yanez, Y. Zhang, and S. E. Lee. 2007. RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin. Mol. Cell. Biol. 271602-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shim, E. Y., J. L. Ma, J. H. Oum, Y. Yanez, and S. E. Lee. 2005. The yeast chromatin remodeler RSC complex facilitates end joining repair of DNA double-strand breaks. Mol. Cell. Biol. 253934-3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smogorzewska, A., J. Karlseder, H. Holtgreve-Grez, A. Jauch, and T. de Lange. 2002. DNA ligase IV-dependent NHEJ of deprotected mammalian telomeres in G1 and G2. Curr. Biol. 121635. [DOI] [PubMed] [Google Scholar]
- 37.Takai, H., A. Smogorzewska, and T. de Lange. 2003. DNA damage foci at dysfunctional telomeres. Curr. Biol. 131549-1556. [DOI] [PubMed] [Google Scholar]
- 38.Tommerup, H., A. Dousmanis, and T. de Lange. 1994. Unusual chromatin in human telomeres. Mol. Cell. Biol. 145777-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsukuda, T., A. B. Fleming, J. A. Nickoloff, and M. A. Osley. 2005. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature 438379-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Attikum, H., O. Fritsch, and S. M. Gasser. 2007. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J. 264113-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Attikum, H., O. Fritsch, B. Hohn, and S. M. Gasser. 2004. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119777-788. [DOI] [PubMed] [Google Scholar]
- 42.van Steensel, B., A. Smogorzewska, and T. de Lange. 1998. TRF2 protects human telomeres from end-to-end fusions. Cell 92401-413. [DOI] [PubMed] [Google Scholar]
- 43.Wright, J. H., D. E. Gottschling, and V. A. Zakian. 1992. Saccharomyces telomeres assume a non-nucleosomal chromatin structure. Genes Dev. 6197-210. [DOI] [PubMed] [Google Scholar]
- 44.Wright, W. E., V. M. Tesmer, K. E. Huffman, S. D. Levene, and J. W. Shay. 1997. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 112801-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu, L., A. S. Multani, H. He, W. Cosme-Blanco, Y. Deng, J. M. Deng, O. Bachilo, S. Pathak, H. Tahara, S. M. Bailey, Y. Deng, R. R. Behringer, and S. Chang. 2006. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell 12649-62. [DOI] [PubMed] [Google Scholar]
- 46.Zhao, Y., H. Hoshiyama, J. W. Shay, and W. E. Wright. 2008. Quantitative telomeric overhang determination using a double-strand specific nuclease. Nucleic Acids Res. 36e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.