Abstract
In HER2-overexpressing mammary epithelial cells, transforming growth factor β (TGF-β) activated phosphatidylinositol-3 kinase (PI3K)/Akt and enhanced survival and migration. Treatment with TGF-β or expression of an activated TGF-β type I receptor (Alk5 with the mutation T204D [Alk5T204D]) induced phosphorylation of TACE/ADAM17 and its translocation to the cell surface, resulting in increased secretion of TGF-α, amphiregulin, and heregulin. In turn, these ligands enhanced the association of p85 with ErbB3 and activated PI3K/Akt. RNA interference of TACE or ErbB3 prevented TGF-β-induced activation of Akt and cell invasiveness. Treatment with TGF-β or expression of Alk5T204D in HER2-overexpressing cells reduced their sensitivity to the HER2 antibody trastuzumab. Inhibition of Alk5, PI3K, TACE, or ErbB3 restored sensitivity to trastuzumab. A gene signature induced by Alk5T204D expression correlated with poor clinical outcomes in patients with invasive breast cancer. These results suggest that by acting on ErbB ligand shedding, an excess of TGF-β may result in (i) conditioning of the tumor microenvironment with growth factors that can engage adjacent stromal and endothelial cells; (ii) potentiation of signaling downstream ErbB receptors, thus contributing to tumor progression and resistance to anti-HER2 therapies; and (iii) poor clinical outcomes in women with breast cancer.
Transforming growth factor β (TGF-β) was originally copurified with TGF-α, a ligand of the epidermal growth factor (EGF) receptor (EGFR), as an activity produced by chemically transformed cells that induced anchorage-independent growth of murine fibroblasts (40, 48). Subsequent studies indicated that signaling by the TGF-β family of cytokines is mediated by a family of transmembrane serine/threonine kinases, namely the type I and type II TGF-β receptors (TβRI and TβRII) (35, 67). Activated TβRI can phosphorylate the transcription factors Smad2 and Smad3, which then associate with Smad4 and translocate to the nucleus, where they regulate gene transcription (36). One of the primary effects of Smad-dependent TGF-β signaling is to limit epithelial proliferation and induce differentiation through a program of cytostatic gene responses (37). Consistent with this tumor-suppressive role of TGF-β, many cancers lose or attenuate TGF-β-mediated antimitogenic action by mutational inactivation of TGF-β receptors or Smads (24, 25, 27, 34, 62, 63). These data suggest that the transforming effects of TGF-β, which led to its discovery, are probably mediated by pathways other than Smads.
Recent studies showed evidence that the phosphatidylinositol-3 kinase (PI3K), extracellular signal-regulated kinase (Erk), c-Jun NH2-terminal kinase, p38 mitogen-activated protein kinase (MAPK), and Rho GTPases are also implicated in the cellular effects of TGF-β (reviewed in references 14 and 61). How TGF-β regulates these non-Smad pathways is not fully understood. Nevertheless, the crucial roles of these pathways in cell survival, motility, and proliferation make them potential mediators of the cancer-promoting effects of TGF-β. In cancer patients, high levels of TGF-β at tumor sites correlate with high histological grade, risk of metastasis, poor response to chemotherapy, and poor patient prognosis. Excess production and/or activation of TGF-β in tumors can accelerate cancer progression by a combination of autocrine and paracrine mechanisms, resulting in enhancement of tumor cell motility and survival, increase in tumor angiogenesis, extracellular matrix production and peritumoral proteases, and the inhibition of immune surveillance mechanisms in the cancer host (reviewed in references 14, 16, and 61).
Synergy between TGF-β and transforming oncogenes has been observed in several animal models of cancer progression. Overexpression of active TGF-β1 or an active mutant of TβRI (Alk5 with the mutation T204D [Alk5T204D]) in the mammary gland of bigenic mice also expressing mouse mammary tumor virus/Neu (ErbB2) accelerates metastases from Neu-induced mammary cancers (41, 42, 54). At the cellular level, exogenous as well as transduced TGF-β confers motility and invasiveness to MCF10A nontransformed human mammary epithelial cells stably expressing HER2 ([ErbB2] MCF10A/HER2 cells) (52, 56). These data suggest that oncogenic signals are permissive for TGF-β-induced signals associated with tumor cell motility and, potentially, metastatic progression. We have previously reported that TGF-β induces the survival and invasion of MCF10A/HER2 cells by activating the Rac1-Pak1 pathway through a PI3K-dependent mechanism (66). In this study we report a novel mechanism by which TGF-β activates PI3K/Akt in mammary cells that overexpress HER2 and describe its potential clinical significance.
MATERIALS AND METHODS
Cell lines, plasmids, and viruses.
MCF10A/HER2 cells were generated and grown as described previously (65). BT474 cells were grown in Iscove's modified Eagle's medium (Cellgro) containing 10% fetal bovine serum (HyClone) in a humidified 5% CO2 incubator at 37°C. SKBR3 cells were grown in Dulbecco's modified Eagle's medium (Cellgro) with 10% fetal bovine serum. Retroviral vectors pBMN-HA-Alk5T204D and pGabe-dnTβRIIK277R were described previously (5). The plasmid encoding the hemagglutinin (HA)-tagged full-length mouse tumor necrosis factor α (TNF-α)-converting enzyme, TACE [HA-TACE(wt); wt is wild type] was described previously (15). Myc-tagged truncated TACE that lacks the cytoplasmic domain (Myc-TACEΔ) was created by replacing the entire intracellular domain of mouse TACE with a Myc tag. Retroviruses expressing HA-Alk5T204D, dominant negative TβRII with the mutation K277R (dnTβRIIK277R), or pBMN vector were produced by transfecting Phoenix-Ampho cells and then utilized to transduce cells, followed by green fluorescent protein selection. Recombinant human TGF-β1, TGF-α, amphiregulin, and heregulin were purchased from R&D Systems. Recombinant human EGF was purchased from Invitrogen. The small-molecule TβRI and TβRII inhibitor LY2109761 was kindly provided by Jonathan Yingling (Eli Lilly Research Laboratories). Trastuzumab was purchased at the Vanderbilt University Medical Center Pharmacy. Pertuzumab was provided by Mark Sliwkowski (Genentech Inc.). Cetuximab was provided by Dan Hicklin (Imclone Systems, Inc.). Neutralizing anti-heregulin and anti-EGF antibodies were purchased from Calbiochem and Chemicon, respectively.
Cell growth, viability, and motility assays.
Coculture cell growth assays were performed using transwell tissue culture inserts with a 0.4-μm-pore-size microporous membrane (Becton Dickinson Labware). Target (test) cells were plated (1.5 × 104) on 12-well plates. Medium-conditioning cells (5 × 103) were plated on the permeable membrane of tissue culture inserts, which were then introduced into the target cell-containing wells. Cocultures were carried out in either growth medium or serum-free medium in the presence or absence of TGF-β. After 72 h, target cells were harvested by trypsinization, and cell number was determined in a Coulter counter. A schematic representation of the coculture experiments is shown in Fig. 3B. For viability assays, cells growing on six-well plates were serum starved for 24, 48, or 72 h and collected for trypan blue staining as described elsewhere (68). Trypan blue-positive cells were counted manually, and their percentages were calculated over the total cell input. Wound closure and transwell assays were performed as described previously (65).
FIG. 3.
TGF-β signaling induces secretion of ErbB ligands. (A) CM was prepared by incubating MCF10A/HER2/Alk5T204D or control cells (C) on 100-mm2 dishes (1 × 106 cells/dish) in serum-free medium for 24 h, followed by filtration through a 0.22-μm-pore-size sterile filter. MCF10A/HER2 or MCF10A/HER2/dnTβRIIK277R cells were serum-starved for 16 h before treatment for 1 h with TGF-β or CM. Cell lysates were subjected to immunoblotting with the indicated antibodies. (B) Coculture cell growth assay was performed as described in Materials and Methods using transwell tissue culture inserts. Cocultures were carried out in growth medium or serum-free medium in the presence or absence of TGF-β (2 ng/ml) for 72 h. Target cells were harvested by trypsinization, and cell number was determined in a Coulter counter. Each data point represents the mean ± standard deviation of three wells. At top is a schematic representation of the coculture system. (C) MCF10A/HER2 cells were labeled with PKH67 green fluorescent cell linker whereas MCF10A/HER2/Alk5T204D cells were labeled with PKH26 red fluorescent cell linker. Labeled cells were immediately seeded in Matrigel for three-dimensional culture in the absence of EGF in the top medium. For single-cell culture, 6 × 103 cells were seeded on day 0 (top). For coculture of different cell types, 3 × 103 cells of each cell type (total of 6 × 103 cells) were seeded on day 0 (bottom). At 12 h after cell seeding, LY2109761 was added to the top medium in the indicated panels. Fluorescent images were captured on day 6. Bar, 100 μm. (D and E) Cells grown on 100-mm2 dishes (1 × 106 cells/dish) were incubated for 24 h in serum-free medium containing cetuximab (10 μg/ml) with or without TGF-β or LY2109761. CM was collected and analyzed for the total amount of TGF-α (D) or amphiregulin (E) by immunoassay as indicated in Materials and Methods. Data are normalized to the number of pg/ml/106 cells/24 h. Each data point represents the mean ± standard deviation of three experiments. (F) Cells were serum starved for 16 h before being incubated in serum-free medium containing TGF-β or LY2109761 for 1 or 16 h, as indicated. Concentrated CM and whole-cell lysates were prepared and subjected to immunoblot analysis using a heregulin antibody. (G) MCF10A/HER2 or BT474 cells were serum starved for 16 h before treatment with TGF-β (2 ng/ml) for 1 h. Neutralizing antibodies against EGF, heregulin, or control IgG was added 1 h prior to TGF-β. Cell lysates were subjected to immunoblot analysis with the indicated antibodies. 10A, MCF10A; Alk5TD, Alk5T204D; Ab, antibody; α, anti; dnRII, dnTβRIIK277R.
Three-dimensional morphogenesis and invasion assay.
Cells were seeded on growth factor-reduced Matrigel (BD Biosciences) or a 1:1 mixture of Matrigel-collagen type I (2 mg/ml collagen; Upstate Biotechnology) in eight-well chamber slides following published protocols (12). TGF-β1 (2 ng/ml) or LY2109761 (0.5 μM) was added into the medium 12 h after cell seeding. In coculture experiments, cells were labeled with PKH67 green fluorescent or PKH26 red fluorescent cell linkers (Sigma) according to the manufacturer's protocol prior to initial seeding.
Cell surface biotinylation, immunoprecipitation, and immunoblot analysis.
Cell surface biotinylation, immunoprecipitation, and immunoblot analysis were performed as described previously (65). Primary antibodies included the following: phospho-Erk1/Erk2 (P-Erk1/2), Erk1/2, Akt phosphorylated at S473 (P-AktS473), Akt, P-Smad2S465/467, P-EGFRY1068, P-HER2Y1248, P-HER3Y1289 (Cell Signaling), EGFR, HER2/ErbB2, heregulin (NeoMarkers), HER3 (Santa Cruz Biotechnology), P-Tyr (4G10), HA, Myc (Vanderbilt Monoclonal Antibody Core), p85 (Upstate), Smad2/3 (BD Biosciences), P-Thr, P-Ser (Zymed), and TACE (Chemicon).
Real-time qPCR analysis.
RNA was isolated using an RNeasy Mini Kit (Qiagen) according to the manufacturer's procedures. After treatment with RQ1 RNase-free DNase (Promega), RNA samples were subjected to first-strand cDNA synthesis using Superscript II reverse transcriptase (Invitrogen) following the manufacturer's protocol. Gene expression was quantified by quantitative PCR (qPCR) using iQ Sybr green Supermix (Bio-Rad) and 50 ng of cDNA per reaction. The sequences of the primer sets used for this analysis are as follows: EGF, 5′-AGCTAACCCATTATGGCAACA-3′ (forward [F]) and 5′-AGTTTTCACTGAGTCAGCTCCAT-3′ (reverse [R]); TGF-α, 5′-GGACAGCACTGCCAGAGA-3′(F) and 5′-CAGGTGATTACAGGCCAAGTAG-3′ (R); heregulin, 5′-TGGCTGACAGCAGGACTAAC-3′ (F) and 5′-CTGGCCTGGATTTCTTC-3′ (R); amphiregulin, 5′-ATATCACATTGGAGTCACTGCCCA-3′ (F) and 5′-GGGTCCATTGTCTTATGATCCAC-3′ (R); heparin binding (HB)-EGF, 5′-GAAAGACTTCCATCTAGTCACAAAGA-3′ (F) and 5′-GGGAGGCCCAATCCTAGA-3′ (R); betacellulin, 5′-TGCCCCAAGCAATACAAGC-3′ (F) and 5′-CGTCTGCTCGGCCACC-3′ (R); epiregulin, 5′-TGCATGCAATTTAAAGTAACTTATTTGACTA-3′ (F) and 5′-ATCTTAAGGTACACAATTATCAAAGCTGA-3′ (R). Human cDNA FLJ22101 fis (GenBank accession number AK025754) was used as a housekeeping gene for normalization of ErbB ligand gene expression. Primer sets for FLJ22101 are as follows: F, 5′-TTCCCTGTGGCACTTGACATT-3′; R, 5′-CTTTTGCCTCTGGCAGTACTCA-3′. An annealing temperature of 57°C was used for all the primers. PCRs were performed in a standard 96-well plate format with a Bio-Rad iQ5 multicolor real-time PCR detection system. For data analysis, the raw threshold cycle (CT) value was first normalized to the housekeeping gene for each sample to get ΔCT. The normalized ΔCT was then calibrated to control cell samples to get ΔΔCT.
TGF-α and amphiregulin immunoassays.
Cells grown on 100-mm2 dishes (1 × 106 cells/dish) were incubated for 24 h in serum-free medium containing TGF-β or LY2109761. Cetuximab (10 μg/ml) was added at the beginning of the incubation to prevent ligand binding to the receptors and internalization. Conditioned medium (CM) was collected, precleared by centrifugation, and analyzed for total amounts of TGF-α and amphiregulin using a Quantikine human TGF-α immunoassay kit (R&D Systems) and DuoSet human amphiregulin kit (R&D Systems), respectively. Plate preparation and assay procedures were performed according to the manufacturer's protocols. Each value was normalized to number of picograms/ml/106 cells/24 h.
RNA interference studies.
Silencer small interfering RNAs (siRNAs) against human TACE were obtained from Qiagen in a FlexiTube format. Four species of siRNAs against different TACE target sequences (TCCCATGAAGAACACGTGTAA, CTGCAGTAAACAATCAATCTA, CAGGATGTAATTGAACGATTT, and AAGAAACAGAGTGCTAATTTA) were mixed in equal amounts to generate a TACE siRNA pool (siTACE). To knock down ErbB3 expression, siRNAs against an ErbB3 target sequence ACCACGGTATCTGGTCATAAA (Qiagen) were used. siRNAs were transfected into cell lines using HiPerFect transfection reagent (Qiagen) according to the manufacturer's procedures. In six-well plate format, a total of 150 ng of siRNAs and 12 μl of HiPerFect reagent were used for each transfection. Mismatched siRNAs were used as negative controls (siCTRL).
In vitro kinase assays.
One milligram of total protein extracted from MCF10A cells transfected by plasmids encoding HA-TACE(wt) or Myc-TACEΔ was immunoprecipitated with HA or Myc antibody. In parallel, 500 μg of total protein extracted from MCF10A/HER2/Alk5T204D cells was precipitated with HA antibody or control immunoglobulin G (IgG). The precipitates were washed twice in NP-40 lysis buffer (20 mM Tris [pH 7.4], 150 mM NaCl, 1% Nonidet P-40, 0.1 mM EDTA, plus protease and phosphatase inhibitors) and twice in kinase buffer (50 mM HEPES [pH 7.5], 10 mM MgCl2, 2 mM MnCl2, 2 mM dithiothreitol, 0.1 mM sodium orthovanadate, and protease inhibitors) and then aliquoted and mixed as indicated in Fig. 4D. Each reaction mixture was brought up to a final volume of 50 μl by adding kinase buffer. The kinase reactions were allowed to proceed for 15 min at 30°C in the presence or absence of ATP (final concentration, 0.1 mM) and/or LY2109761 (1 μM). After the reactions were terminated by adding 5× loading buffer and boiling for 3 min, samples were separated by 7.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by immunoblot analysis.
FIG. 4.
Effect of TGF-β signaling on ErbB ligand mRNAs. (A) MCF10A/HER2 or BT474 cells grown in complete medium were left untreated or were treated with TGF-β for 8 h. Total RNA was extracted and subjected to reverse transcription followed by qPCR as described in Materials and Methods. Data were normalized to untreated cells. Each data point represents the mean ± standard deviation of three experiments. (B) Total RNA extracted from MCF10A/HER2 vector control cells, MCF10A/HER2/Alk5T204D cells, and MCF10A/HER2/Alk5T204D cells pretreated with LY2109761 for 24 h were subjected to quantitative reverse transcription-PCR. Each data point represents the mean ± standard deviation of three experiments. 10A, MCF10A; Alk5TD, Alk5T204D.
Gene expression microarray and breast cancer data set analysis.
Reverse transcription and microarrays were performed by the Vanderbilt Microarray Shared Resource using 30,000 spotted oligonucleotide microarrays printed from the Compugen Human Release, version 2.0, OligoLibrary. The experiments were carried out in triplicates using RNA samples extracted from three independent experiments. The RNAs from BT474 cells were fluorescently tagged with Cy3, and RNAs from BT474 cells expressing Alk5T204D (BT474/Alk5T204D) were tagged with Cy5. Equal amounts of Cy3- and Cy5-labeled RNA samples were hybridized on the arrays. The arrays were scanned using Axon 4000B, and the image analysis was then accomplished using GenePix Pro, version 6.0, reverse transcription. The raw data tables were uploaded to the Vanderbilt Microarray Shared Resource database, and the data were normalized using Lowess for further analysis (11).
The differentially expressed genes were selected by statistical criteria of greater than twofold changes and a P value of <0.01 by a Student t test. The up- and downregulated genes were uploaded to http://www.broad.mit.edu/cmap/for connectivity mapping according to the website instructions. In subsequent analysis, the data were mined by Ingenuity Pathways Analysis ([IPA] Ingenuity Systems [www.ingenuity.com]). The selected differential gene log ratios were submitted to the IPA application. IPA mapped these genes to their corresponding gene objects in the Ingenuity Pathways Knowledge Base. A P value was generated for each mapped network by comparing the number of differential genes that participated in a given network relative to the total number of occurrences of those genes in all the possible network permutations for all the objects in the Ingenuity Pathways Knowledge Base. The network score was assessed for statistical significance, and the generated pathway maps were examined for biological correlation. For comparison of the cell line signature with previously published data sets (10, 28, 58), UniGene identifiers (build no. 184) were assigned to each gene that was present on both Vanderbilt spotted arrays and comparing data set platforms. Genes with identical UniGene identifiers were collapsed by taking the median value within a sample. The data points were quantile normalized across the two platforms. The UniGene identifiers corresponding to the Alk5T204D signature were extracted. The data for each gene were median centered and analyzed with hierarchical clustering (18). Finally, the clustering was visualized by TreeView.
Microarray data accession number.
The raw microarray data for the signature induced by Alk5T204D has been deposited in the Gene Expression Omnibus under accession number GSE11293.
RESULTS
Active TβRI enhances cell survival and motility in HER2-overexpressing cells.
We engineered MCF10A/HER2 cells that stably express a constitutively active mutant of TβRI (Alk5T204D). Compared to control cells, the MCF10A/HER2/Alk5T204D cells showed enhanced survival under serum-free conditions (Fig. 1A). When cultured in growth factor-reduced basement membrane, the control MCF10A/HER2 cells required EGF in the medium to grow whereas the Alk5T204D-expressing cells grew into full-size acini in the absence of added EGF (Fig. 1B). Ligand-independent growth of MCF10A/HER2/Alk5T204D was abolished by LY2109761, a small-molecule inhibitor of type I and type II TGF-β receptor kinases (44, 51) (Fig. 1B). Expression of Alk5T204D also induced cell motility in a wound closure assay (Fig. 1C). Both TGF-β treatment and expression of Alk5T204D resulted in phosphorylation and activation of Smad2, Akt, and Erk1/2; these effects were also abolished by LY2109761 (Fig. 1D). Similar results were observed with BT474 breast cancer cells, which naturally overexpress HER2. These cells harbor a PI3K mutation in the amino-terminal p85 binding domain which does not increase its transforming potential (26). Similarly, expression of Alk5T204D increased cell survival and motility as well as activation of Smad2 in these cells. P-Akt and P-MAPK were upregulated above baseline by expression of Alk5T204D, and these increases were reversed by the addition of LY2109761 (data not shown).
FIG. 1.
Expression of Alk5T204D (Alk5TD) increases cell survival and motility in HER2-overexpressing cells. (A) MCF10A/HER2 cells that stably express Alk5T204D or vector control were grown to 50% confluence on six-well plates (5 × 105 cells/well) and then changed to serum-free medium. After 24, 48, and 72 h, cells were harvested for trypan blue staining. Each data point represents the mean ± standard deviation of three wells. *, P < 0.005. (B) MCF10A/HER2/Alk5T204D or control cells were grown in Matrigel in the absence or presence of EGF. LY2109761 (0.5 μM) was added to the top medium at 12 h after cell seeding. On day 5, acini were photographed and trypsinized, and cell numbers were counted. Each data point represents the mean ± standard deviation of four wells. Bar, 50 μm. *, P < 0.005. (C) Close-to-confluent monolayers of MCF10A/HER2/Alk5T204D or control cells in serum-free medium were wounded with a pipette tip; wound closure was monitored at 0 and 16 h. Bar, 50 μm. Wound width was measured at the same marked position (center of the field) at each time point. Each data point represents the mean ± standard deviation of 10 fields. *, P < 0.05. (D) MCF10A/HER2/Alk5T204D, MCF10A/HER2/dnRIIK277R, or control cells expressing the empty vector were serum starved for 16 h in the absence or presence of LY2109761 (0.5 μM) before treatment with TGF-β (2 ng/ml) or vehicle for 1 h. Cells were next lysed and subjected to immunoblot analyses with the indicated antibodies. (E) MCF10A/HER2 cells were infected with an adenovirus encoding Smad7 or β-galactosidase (β-Gal; multiplicity of infection, 1:5) as described previously (66). At 16 h postinfection, the cells were serum starved for 16 h and treated with TGF-β for 1 h, lysed, and subjected to immunoblot analyses. 10A, MCF10A.
TGF-β increases PI3K activity through activation of ErbB receptors.
Class IA PI3K consists of a p85 regulatory subunit and a p110 catalytic subunit. Tyrosine kinases such as the ErbB, insulin, and insulin-like growth factor I receptors also signal via class IA PI3K. These receptors activate this enzyme by phosphorylating adaptor proteins such as GAB1, GAB2, IRS-1, IRS-2, and ErbB3 (HER3), which have YXXM motifs to engage the SH2 (Src homology 2) domains of p85 (20). As a result, PI3K localizes to the plasma membrane, where it phosphorylates the lipid phosphatidylinositol-4,5-bisphosphate to produce the second messenger, phosphatidylinositol-3,4,5-triphosphate. Because of the association between PI3K and its upstream activator(s), it is possible to identify the activating receptor tyrosine kinase or tyrosine phosphorylated adaptor(s) by coprecipitation with p85. Therefore, p85 was precipitated from MCF10A, MCF10A/HER2, and BT474 cells in the presence and absence of TGF-β. In MCF10A/HER2 and BT474 but not in MCF10A cells treated with TGF-β, a P-Tyr band of ∼200 kDa coprecipitated with the p85 antibody. A previously reported search of proteins containing p85 SH2 binding sites had identified ErbB3 as a likely candidate at this approximate molecular size (19). Indeed, immunoblot analysis confirmed this band to be ErbB3 in both BT474 and MCF10A/HER2 cells(Fig. 2A). These results suggest that in cells with high levels of HER2, TGF-β increases the association of both active HER2 and p85 with ErbB3 which, in turn, activates PI3K/Akt above baseline. Indeed, TGF-β induced phosphorylation of ErbB3, EGFR, and HER2; this was accompanied by activation/phosphorylation of Akt and MAPK (Fig. 2B, first three lanes).
FIG. 2.
TGF-β activates PI3K through ErbB receptors. (A) BT474, MCF10A/HER2 (10A/HER2), or MCF10A cells were serum starved for 16 h before treatment with TGF-β (2 ng/ml) for 1 or 16 h. Cell lysates were precipitated with a p85 antibody; the p85 pull-downs were subjected to P-Tyr (pY), ErbB3, and p85 immunoblot analysis as indicated in Materials and Methods. Whole-cell lysates were subjected to immunoblotting with P-Akt, total Akt, and ErbB3 antibodies. (B) MCF10A/HER2 cells were serum starved for 16 h before treatment with TGF-β for 1 or 8 h. Where indicated, pertuzumab (10 μg/ml) was added to the cells 1 h prior to TGF-β. Cell lysates were precipitated with p85 antibody followed by immunoblot analysis or directly subjected to immunoblotting with the indicated antibodies. (C) BT474 cells were serum starved for 16 h before treatment with TGF-β or vehicle for 1 h. Where indicated, trastuzumab (Tras; 10 μg/ml) or pertuzumab (Pert; 10 μg/ml) was added to the cells 1 h prior to TGF-β. Cell lysates were subjected to immunoblotting with the indicated antibodies. (D) MCF10A/HER2 cells were transfected by siRNA oligonucleotides targeting ErbB3 (siErbB3) or a control sequence (siCTRL). On day 3 after transfection, cells were changed to serum-free medium for 16 h and then treated with TGF-β or vehicle for 1 h. (E) Monolayers of MCF10A/HER2 cells in serum-free medium were wounded with a pipette tip and incubated in serum-free medium containing trastuzumab, pertuzumab, or vehicle in the absence or presence of TGF-β. In the indicated panels, siRNA was transfected into cells 3 days before wounding. Wound closure was monitored at 0 and 16 h. Bar, 50 μm. Wound width was measured as described in the legend of Fig. 1. Each data point represents the mean ± standard deviation of 10 fields. *, P < 0.05. IP, immunoprecipitation.
These effects of TGF-β on ErbB receptors and Akt were not affected by overexpression of Smad7, suggesting that they were independent of Smad function (Fig. 1E). Pertuzumab, an anti-HER2 monoclonal antibody that inhibits HER2 dimerization with ErbB coreceptors and signaling by HER2-containing heterodimers (1, 23), abolished basal and TGF-β-induced association between p85 and ErbB3 as well as the ErbB3 phosphorylation and Akt activation (Fig. 2B). Similar results were observed in BT474 cells, where the HER2 antibodies trastuzumab and pertuzumab blocked both basal and TGF-β-induced P-ErbB3 and P-Akt (Fig. 2C). RNA interference of ErbB3 abolished TGF-β-induced PI3K association with ErbB3, P-ErbB3, and P-Akt (Fig. 2D). In line with this result, both HER2 antibodies as well as knockdown of ErbB3 by RNA interference abolished TGF-β-induced motility of MCF10A/HER2 cells (Fig. 2E).
TGF-β signaling induces secretion of ErbB ligands.
MCF10A/HER2/Alk5T204D cells grew in Matrigel in the absence of exogenous EGF, suggesting that these cells secrete factors that support autocrine cell growth. Therefore, we examined the effect of medium conditioned by Alk5T204D-expressing or control cells on ErbB signaling. When added to MCF10A/HER2 cells, CM from Alk5T204D but not vector control cells induced phosphorylation of ErbB receptors but not Smad2 (Fig. 3A, lanes 1 to 4). CM-induced activation of ErbB receptors was also observed in MCF10A/HER2/dnTβRIIK277R cells that stably express kinase-dead dnTβRIIK277R (17) (Fig. 3A, lanes 5 to 8). In these cells, TGF-β did not activate Smad2, Akt, or Erk1/2 (Fig. 1D).
We next used a coculture system in which MCF10A/HER2 target cells are coincubated with but separated by a 0.4-μm-pore-size filter from the 10A/HER2/Alk5T204D or MCF10A/HER2/vector cells to determine the effect of CM from these two cell lines on MCF10A/HER2 cell growth (diagram in Fig. 3B). After 72 h, MCF10A/HER2 cells grew faster when cocultured with MCF10A/HER2/Alk5T204D than cells cocultured with MCF10A/HER2/vector cells (Fig. 3B). In contrast, addition of TGF-β to the top compartment in the absence of medium-conditioning cells reduced MCF10A/HER2 target cell numbers in the bottom compartment by 40% (Fig. 3B). This suggests that expression of active Alk5 induces the release of growth-promoting factors, which, in turn, stimulate adjacent cells. When MCF10A/HER2/dnTβRIIK277R cells were used as target cells in the bottom compartment, they were refractory to the growth-inhibitory effect of TGF-β. However, they responded to the medium conditioned by MCF10A/HER2/Alk5T204D cells with a 23% increase in cell number (Fig. 3B). We then performed the same coculture experiments under serum-free conditions. Both TGF-β treatment and coculture with Alk5T204D-expressing cells increased survival of MCF10A/HER2 test cells by 35 to 50% (Fig. 3B). TGF-β-enhanced survival was impaired in MCF10A/HER2/dnTβRIIK277R cells. However, CM from Alk5T204D-expressing cells stimulated survival of these cells (Fig. 3B). MCF10A/HER2 cells cocultured with MCF10A/HER2/Alk5T204D cells in Matrigel were able to grow into full-size acini in the absence of exogenous EGF. This effect was blocked by the addition of the TGF-β receptor kinase inhibitor LY2109761 (Fig. 3C).
To determine if TGF-β upregulates expression of ErbB ligands, we used quantitative reverse transcription-PCR. In MCF10A/HER2 cells, treatment with TGF-β did not increase expression of any of the ligands, whereas in BT474 cells TGF-β modestly increased amphiregulin mRNA by 50% (Fig. 4A). Expression of Alk5T204D resulted in an 80% increase of HB-EGF mRNA, which was partially reduced by LY2109761 (Fig. 4B). However, treatment with TGF-β increased protein levels of TGF-α, amphiregulin, and heregulin, as detected by ligand immunoassays and immunoblot analyses (Fig. 3D to F). Consistent with this result, cells expressing Alk5T204D secreted higher levels of ligands, which were modestly inhibited by LY2109761 (Fig. 3D to F). Higher levels of secreted ErbB ligands were also observed in MCF10A cells upon the addition of TGF-β (Fig. 3D to F), suggesting that overexpression of HER2 is not necessary for TGF-β-mediated upregulation of ErbB ligands. Preincubation with a heregulin-neutralizing antibody blocked TGF-β-induced ErbB3 and Akt phosphorylation whereas an EGF-neutralizing antibody or control IgG did not (Fig. 3G).
TGF-β activates ErbB signaling by phosphorylation and translocation of TACE.
The effect of TGF-β on secreted ErbB ligands without an increase in mRNA levels (above) suggested increased shedding of ErbB proligands. All three ErbB ligands induced by TGF-β (Fig. 3) are cleaved by the transmembrane sheddase TACE/ADAM17. Thus, to determine if TACE is required for TGF-β actions, we used RNA interference. Transient transfection of siRNA against human TACE but not control siRNA abolished TGF-β-stimulated ErbB receptor phosphorylation in MCF10A/HER2 cells (Fig. 5A) as well as P-Akt and P-Erk (data not shown). Transfection of a full-length mouse TACE construct [HA-TACE(wt)] but not the truncated TACE that lacks the cytoplasmic domain (Myc-TACEΔ) reconstituted TGF-β-induced ErbB phosphorylation in cells transfected with human TACE siRNA (Fig. 5A). In three-dimensional culture in a Matrigel-collagen mixture, treatment with TGF-β resulted in cell elongation and the formation of acini that invaded the surrounding matrix. These effects were blocked by TACE siRNA and rescued by the full-length mouse TACE but not the truncated version (Fig. 5B), suggesting that the cytoplasmic domain of TACE is necessary for TGF-β-induced ErbB signaling and cell invasiveness.
FIG. 5.
TGF-β modulates TACE phosphorylation and cellular localization. (A) MCF10A/HER2 cells grown on six-well plates were transfected by siRNA oligonucleotides targeting human TACE (siTACE) or a control sequence (siCTRL). To rescue TACE expression, plasmids encoding the full-length mouse TACE [HA-TACE(wt)] or cytoplasmic tail-truncated TACE (Myc-TACEΔ) were cotransfected with siRNA. On day 3 after transfection, cells were changed to serum-free medium and then treated with TGF-β or vehicle for 1 h. Cell lysates were analyzed by immunoblotting using the indicated antibodies. (B) MCF10A/HER2 cells grown on six-well plates were transfected by siTACE or siCTRL with or without TACE-expressing plasmids. At 16 h after transfection, cells were trypsinized and seeded in a 1:1 mixture of Matrigel and collagen as described in Materials and Methods. Where indicated, TGF-β was added to the top medium 12 h after cell seeding. The three-dimensional cultures were photographed on day 5. Acini with an invasive phenotype are indicated by arrows. Bar, 50 μm. (C) MCF10A/HER2 cells were transfected by siTACE or siCTRL, serum starved for 16 h starting from day 3 after transfection, and treated with TGF-β for 1 h. Cell lysates were precipitated with TACE antibody; the TACE pull-downs were subjected to P-Thr, P-Ser, and TACE immunoblot analysis. Positions of the molecular size markers (in kDa) are indicated at the right of each immunoblot. (D) TACE proteins expressed by MCF10A cells transfected by plasmids encoding HA-TACE(wt) or Myc-TACEΔ and HA-Alk5T204D expressed by MCF10A/HER2/Alk5T204D cells were prepared by immunoprecipitation and incubated in the presence or absence of ATP in an in vitro kinase assay described in Materials and Methods. Where indicated, LY2109761 (1 μM) was added to the kinase reaction mixture before the addition of ATP. Reaction products were analyzed by immunoblotting using P-Thr and P-Ser antibodies. Positions of the molecular size markers (in kDa) are indicated. (E) MCF10A/HER2/Alk5T204D cells were incubated in serum-free medium with or without LY2109761 (0.5 μM) for 16 h. Cell lysates were precipitated with an HA antibody (for Alk5T204D) or control IgG; HA pull-downs and whole-cell lysates (WCL) were subjected to immunoblot analysis. Positions of the molecular size markers (in kDa) are indicated. (F) MCF10A/HER2 cells were transfected by siTACE or siCTRL, serum starved for 16 h starting from day 3 after transfection, and treated with TGF-β for 1 h. At the end of the treatment, cells were surface biotinylated as described in Materials and Methods. Cell lysates were incubated with streptavidin beads, and the eluates and whole-cell lysates were subjected to immunoblot analysis with TACE and HER2 antibodies. IP, immunoprecipitation; 10A, MCF10A; Alk5TD, Alk5T204D.
Phosphorylation of the cytoplasmic domain of TACE is required for the shedding of TrkA neurotrophin receptor. Moreover, this phosphorylation results in TACE protein maturation and trafficking from the endoplasmic reticulum to the cell surface (15, 49, 55). Thus, we next examined TACE phosphorylation in cells treated with TGF-β. In MCF10A/HER2 cells, TGF-β increased TACE phosphorylation in both serine and threonine as measured by P-Ser and P-Thr immunoblots of TACE pull-downs (Fig. 5C). To test whether the type I TGF-β receptor can phosphorylate TACE directly, we performed an in vitro kinase assay using HA-tagged Alk5T204D precipitated from MCF10A/HER2/Alk5T204D cells and full-length or truncated TACE precipitated from transfected MCF10A cells. Incubation of TACE with Alk5T204D in the presence of ATP resulted in both Ser and Thr phosphorylation in full-length but not truncated TACE. Phosphorylation at both sites was inhibited by LY2109761 and was not detected in the control reaction when the Alk5T204D kinase was absent (Fig. 5D). We detected only one form of full-length TACE phosphorylated by Alk5T204D; the size of this band suggested it was the 120-kDa proform of TACE. Consistent with previous observations (15), the immunoblot of total TACE [HA-TACE(wt)] in the same figure and in Fig. 5A, C, E, and F suggested that the proform of TACE is more abundant than the mature form; this may explain why only the phosphorylated proform of TACE was detected. In addition, TACE coprecipitated with Alk5T204D in the MCF10A/HER2/Alk5T204D cells; treatment with LY2109761 did not affect this association (Fig. 5E).
Finally, we examined the cell surface localization of TACE by biotinylating MCF10A/HER2 cells at 4°C. TACE localization was detected by streptavidin affinity purification followed by immunoblotting with a TACE antibody. Cell surface biotinylated TACE was markedly increased by the addition of TGF-β whereas the total levels of TACE and of HER2 at the membrane remained unchanged (Fig. 5F).
TGF-β signaling desensitizes BT474 cells to trastuzumab.
Overexpression of ErbB ligands (47) and high PI3K/Akt activity (6, 43) have been associated with resistance to trastuzumab in HER2-overexpressing cells and primary tumors. In the case of increased ErbB ligands, this is the result of increased formation of HER2-containing heterodimers, which are not inhibited by trastuzumab (2). Treatment of BT474 cells with recombinant EGF, TGF-α, amphiregulin, or heregulin reduced trastuzumab-mediated growth inhibition (Fig. 6). Therefore, we speculated that TGF-β might affect the response to trastuzumab through TACE-mediated increased shedding of ErbB ligands. Trastuzumab inhibited BT474 cell growth in a dose-dependent fashion (63% inhibition; 1 μg/ml). In the presence of TGF-β, antibody-mediated inhibition was only 18%. On the other hand, BT474/Alk5T204D cells were inhibited only 22% by a similar dose of trastuzumab. Inhibition of TGF-β signaling with LY2109761 restored the responsiveness to trastuzumab (Fig. 7A). Inhibition of PI3K by LY294002 or knockdown of ErbB3 or TACE by specific siRNA also restored the inhibitory effect of trastuzumab (Fig. 7B). These effects on trastuzumab action were not limited to BT474 cells. TGF-β also increased ErbB/PI3K activation in the HER2-overexpressing breast cancer cells SKBR3 and desensitized them to trastuzumab-mediated growth inhibition (Fig. 8).
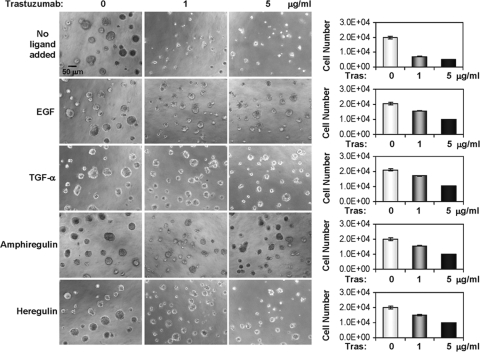
FIG. 6.
Recombinant ErbB ligands induce resistance to trastuzumab in BT474 cells. BT474 cells were plated in Matrigel in eight-well chambers and allowed to grow in the absence or presence of EGF, TGF-α, amphiregulin, and heregulin (20 ng/ml each). Trastuzumab was added to the top medium 12 h after cell seeding. Ligands and inhibitors were replenished every 3 days. Phase-contrast images shown were recorded 9 days after the initial seeding of cells. Bar, 50 μm. For the graphs at right, 9-day acini were trypsinized and total cell number was determined. Each bar graph represents the mean ± standard deviation of four wells. Tras, trastuzumab.
FIG. 7.
TGF-β signaling desensitizes BT474 cells to trastuzumab. (A) BT474 cells that stably express Alk5T204D (Alk5TD) or empty vector were plated in Matrigel in eight-well chambers and allowed to grow in the absence or presence of TGF-β or LY2109761. Trastuzumab was added to the top medium 12 h after cell seeding. TGF-β and inhibitors were replenished every 3 days. Phase-contrast images shown were recorded 9 days after the initial seeding of cells. Bar, 50 μm. For the graphs, 9-day acini were trypsinized, and total cell numbers were determined in a Coulter counter. Each bar graph represents the mean ± standard deviation of four wells. (B) Untransfected or siRNA-transfected BT474 or BT474/Alk5T204D cells were seeded in Matrigel and allowed to grow in the absence or presence of trastuzumab (1 μg/ml). LY294002 (0.5 μM) was added 12 h after cell seeding where indicated. Images shown were recorded 9 days after cell seeding. For the graphs, total cell numbers were determined, and the percentage inhibition by trastuzumab was calculated. Each bar graph represents the mean ± standard deviation of four wells. Tras, trastuzumab; DMSO, dimethyl sulfoxide.
FIG. 8.
TGF-β activates ErbB/PI3K signaling and induces resistance to trastuzumab in SKBR3 cells. (A) SKBR3 cells were serum starved for 16 h before treatment with TGF-β (2 ng/ml) for 1 or 16 h. Cell lysates were precipitated with a p85 antibody; the p85 pull-downs were subjected to ErbB3, EGFR, HER2, and p85 immunoblotting. Whole-cell lysates were subjected to immunoblotting with the indicated antibodies. (B) SKBR3 cells were plated in Matrigel in eight-well chambers and allowed to grow in the absence or presence of TGF-β (2 ng/ml). Trastuzumab was added to the top medium 12 h after cell seeding. TGF-β and trastuzumab were replenished every 3 days. Phase-contrast images shown were recorded 9 days after the initial seeding of cells. Bar, 50 μm. For the graphs, 9-day acini were trypsinized, and total cell numbers were determined. Each bar graph represents the mean ± standard deviation of four wells. Tras, trastuzumab; IP, immunoprecipitation.
Alk5T204D gene expression signature correlates with clinical outcomes in women with breast cancer.
To survey global changes in gene expression due to the constitutive activation of TβRI and to examine these changes in primary human breast cancers, an Alk5T204D signature was determined by selecting genes that were differentially expressed between BT474/Alk5T204D and BT474 control cells. The genes were filtered for expression changes greater than twofold in triplicate arrays with a t test P value of <0.01; 271 unique genes passed the criteria (see Table S1 in the supplemental material). The genes were examined for connectivity using IPA. The most significant molecular and cellular functions were cell signaling, cell growth and proliferation, cell death, cellular movement, and development. The most significant pathways were Wnt/β-catenin, dopamine receptor, cyclic AMP-mediated and TGF-β signaling, and PPARα/RXRα activation. The highest-ranked upregulated gene in log ratio between BT474/Alk5T204D and BT474 controls was TGFBR1, and the highest-ranked downregulated gene was RAPSN. We then used the Alk5T204D expression signature to query the Connectivity Map. This is a collection of genome-wide transcriptional expression data from human cells treated with bioactive small molecules and simple pattern-matching algorithms that together enable the discovery of decisive functional connections between drugs, genes, and diseases through the transitory feature of common gene expression changes (33). Genes whose transcript levels increased or decreased twofold after ectopic Alk5T204D expression were analyzed using the Connectivity Map. Top perturbagens that were associated with a reverse correlation with this gene signature were the PI3K inhibitors LY294002 and wortmannin (data not shown), further suggesting a positive connection between the TGF-β-induced signature and the PI3K pathway.
We next mapped the Alk5T204D signature to a previously published 295-array data set by van de Vijver et al. (58) and Chang et al. (10). There were 90 genes of the 271-gene Alk5T204D signature that demonstrated at least a 1.5-fold change from the median expression in at least two tumors. Even with this partial list of genes, the Alk5T204D signature reflected biological and clinical differences in the 295 tumors. The tumors with a positive correlation with the active TβRI signature were mostly HER2-positive, Basal-like and some luminal B tumors while the tumors with a negative correlation were predominantly luminal A and normal-like tumors (Fig. 9A). Cancers with a positive correlation with the Alk5T204D signature had worse recurrence-free survival (RFS) and overall survival (OS) rates than tumors with a negative correlation (RFS P = 0.05 and OS P = 0.01 by a log rank test) (Fig. 9B). With a Cox proportional hazards model using the signature correlation as a quantitative variable and survival as outcome measures, a higher correlation with the Alk5T204D signature was a significant predictor of poor RFS (P = 0.01) and OS (P = 0.0034).
FIG. 9.
Alk5T204D signature is associated with clinical outcomes in women with breast cancer. (A) Hierarchical clustering of 295 breast tumors (10, 58) using 90 overlapping genes with the 271-gene Alk5T204D (Alk5) signature. The tumors labeled as luminal B, HER2+, and basal-like shared similarities in expression with the Alk5T204D signature while luminal A and normal-like tumors did not. (B) Kaplan-Meier plots for RFS and OS comparing the two groups of tumors with and without a correlation with the Alk5T204D signature. A positive correlation (+) with the Alk5T204D signature was associated with poor survival by a log rank test. −, negative correlation with the Alk5T204D signature. (C) Hierarchical clustering of 22 breast tumors from patients who were treated with navelbine and trastuzumab (28) using 190 overlapping genes with the 271-gene Alk5T204D signature. Cluster 2 showed a positive correlation with the Alk5T204D signature, which indicates the similarities in the gene expression patterns. (D) Box-and-whisker plot of the standard Pearson correlation between the Alk5T204D signature and the clusters determined in panel C.
To explore possible correlation of the Alk5T204D signature with resistance to trastuzumab, we mapped this gene expression signature to an array data set reported by Harris et al. (28) obtained from 22 patients with HER2-overexpressing breast cancer treated with the neoadjuvant trastuzumab and vinorelbine. There were 190 overlapping genes within the 271-gene Alk5T204D signature that changed at least 1.5-fold from the median expression on at least two samples. Hierarchical clustering analysis showed that the tumors in cluster 2 shared similar expression patterns with the Alk5T204D signature demonstrated by the standard Pearson correlation and box-and-whisker plot (Fig. 9C and D). All three patients who achieved a pathological complete response after treatment with trastuzumab were grouped in cluster 1. This result is also in line with the possibility that the Alk5T204D signature reflects resistance to trastuzumab.
DISCUSSION
TGF-β synergizes with the HER2 (ErbB2) signaling network to enhance cancer cell invasiveness and metastatic potential. In this study, we report a mechanism of TGF-β-ErbB receptor cross talk through TβRI (Alk5)-induced phosphorylation and trafficking of TACE to the cell surface, leading to the shedding of TGF-α, amphiregulin, and heregulin. This effect was prevented by dominant negative TβRII or a small-molecule inhibitor of the Alk5 kinase. Simultaneous with upregulation of ErbB ligands, TGF-β increased phosphorylation of ErbB3, Erk, and Akt and cell motility and survival (Fig. 10). Although kinase defective, ErbB3 can be phosphorylated by the EGF receptor (ErbB1) or HER2 (70). The C terminus of ErbB3 contains one NPXY and six YXXM sequence motifs which serve as binding sites for Shc and p85, the regulatory subunit of PI3K (45). Downregulation of ErbB3 or mutagenesis of the PI3K binding sites has been shown to inhibit ErbB receptor-dependent transformation (19, 21, 29, 59).
FIG. 10.
Tumor-promoting function of TGF-β in HER2-overexpressing cancer cells is mediated by TGF-β-driven autocrine and paracrine ErbB ligands. Signals from extracellular TGF-β are transduced into cells that express wild-type TGF-β receptors. Activated TβRI induces a set of Smad-dependent cytostatic gene responses. In addition, TβRI phosphorylates TACE, resulting in its translocation to the cell surface, where TACE cleaves ErbB proligands. ErbB ligands will initiate autocrine and paracrine ErbB signaling in adjacent cells, even if these cells are insensitive to TGF-β due to gene mutations in TβRs and/or Smads.
In HER2-overexpressing breast cancer cells such as BT474 and SKBR3, ErbB3 is constitutively phosphorylated and associated with HER2 (3, 69). Baseline ErbB3 phosphorylation and P-Akt are eliminated by the HER2 tyrosine kinase inhibitor lapatinib (21, 47), suggesting that HER2 phosphorylates ErbB3 which, in turn, binds p85 and activates PI3K and its downstream kinase Akt. Treatment with TGF-β markedly increased the basal association of p85 with P-ErbB3 (Fig. 10). Both basal and ligand-induced p85-ErbB3 coupling and P-ErbB3 were abrogated by the HER2 antibody pertuzumab, implying that its association with the HER2 kinase is required for ErbB3 phosphorylation. Further, exogenous TGF-β or stable expression of an active type I TGF-β receptor (Alk5T204D) upregulated P-Akt and P-Erk and induced cell invasiveness. Finally, RNA interference of TACE or ErbB3 or the HER2 antibodies trastuzumab or pertuzumab abrogated TGF-β-induced cell motility and P-Akt, suggesting that TACE, HER2, and ErbB3 are all necessary for TGF-β action in HER2-overexpressing mammary tumor cells.
The upregulation of ErbB ligands by TGF-β did not require the overexpression of HER2 as it was also observed in untransfected MCF10A cells (Fig. 3D to F). We propose that the overexpression of HER2 in MCF10A/HER2 cells provides a signal amplification mechanism to ligand-activated ErbB coreceptors which changes the phenotypic response to TGF-β and permits its tumor promoter effect. Similar effects of TGF-β on the upregulation of ErbB ligands in some cases with a simultaneous activation of PI3K/Akt, Erk, or p38 have been reported in endothelial cells, fetal rat hepatocytes, hepatoma cells, and lung fibroblasts (9, 13, 30, 57, 60). However, the mechanism(s) of TGF-β-induced activation of PI3K/Akt was unclear in these studies. We previously reported that in cells expressing low levels of HER2, TGF-β activates PI3K through a mechanism initially involving the association of TGF-β receptors with p85 (71). In these cells, expression of ectopic Smad7 prevented TGF-β-induced PI3K/Akt whereas it did not do so in the HER2-overexpressing cells used herein (data not shown). The mechanism(s) of TGF-β-mediated activation of PI3K/Akt in cells with low HER2 levels requires additional investigation. In cells with HER2 overexpression treated with TGF-β or stably expressing Alk5T204D, the upregulation of ErbB receptor ligands and high levels of the HER2 tyrosine kinase cooperate to engage ErbB3 which, in turn, couples to the PI3K pathway.
TACE also mediates the cleavage and solubilization of several other transmembrane proteins, including the precursor for TNF-α, l-selectin, the p75TNFR receptor, and the β-amyloid precursor protein (39). It has been reported that the cytoplasmic domain of TACE is not required for its function to shed TNF, p75TNFR, and interleukin 1 receptor type II upon phorbol ester stimulation (46). However, it has also been shown that Erk mediates phosphorylation of TACE in Thr735, and phosphorylation of TACE in threonine and serine residues in the cytoplasmic domain can be induced by extracellular signals such as phorbol-12-myristate-13-acetate, EGF, and gastrin-releasing peptide (8, 15, 22, 39, 72). In the studies shown here, a form of TACE in which the intracellular domain had been deleted did not rescue TACE activity in cells in which endogenous TACE had been knocked down, suggesting that the intracellular tail of TACE is required for TGF-β-induced responses. Additional evidenced suggested that TGF-β increases TACE levels at the plasma membrane and induces Ser/Thr phosphorylation of its cytoplasmic tail (Fig. 5).
Cleavage of ErbB ligands is also regulated by other sheddases. HB-EGF, a ligand for EGFR and ErbB4, is cleaved by ADAM9 (31). In mouse embryonic fibroblasts, ADAM10 is the main sheddase of EGF and betacelluin (50). In addition to the results shown here, other data suggest a role for ADAMs on TGF-β signaling. In a recent report, ADAM12 was shown to associate with TβRII and facilitate its trafficking to the endosome as well as Smad phosphorylation and transcriptional activity (4). Whether TGF-β also affects other targets of TACE and whether other ADAM family members are regulated by TGF-β need to be further investigated. Nevertheless, targeting an enzyme with multiple substrates would be an effective mechanism by which TGF-β can modulate its tumor promoting activity. Recent studies in lung cancer and breast cancer cells indicated that TACE-mediated ligand cleavage may serve as a therapeutic target (32, 73). In the study herein, RNA interference of TACE reversed the invasive three-dimensional growth induced by TGF-β (Fig. 5B), thus linking TACE to the oncogenic actions of TGF-β. This result has important therapeutic implications. Given the dual effects of TGF-β as an oncogene or a tumor suppressor, it is difficult to define whether and in which cancers this factor should be targeted with a therapeutic intent. Clearly, dissection of the mechanisms responsible for both actions should help strategies that neutralize the prooncogenic responses to TGF-β, leaving its tumor suppressor function intact. The interplay between TGF-β and TACE in breast cancers that bear amplified HER2 facilitates the oncogenic actions of TGF-β, thus implying that targeting TACE may be of therapeutic benefit in this disease, especially if combined with HER2 antagonists.
The PI3K/Akt survival pathway has been identified as a preferential mechanism of resistance to ErbB receptor inhibitors. For example, amplification of the PI3K pathway as a result of loss or low levels of the phosphatase PTEN and/or PIK3CA gene mutations in HER2-overexpressing breast cancers is associated with a lower response to trastuzumab (6, 43). EGFR tyrosine kinase inhibitors (TKIs) are ineffective in high-grade gliomas that lack PTEN (38). Overexpression of constitutively active Akt renders HER2-overexpressing breast cancer cells insensitive to trastuzumab (69). Restoration of PTEN expression into PTEN mutant cancer cells sensitizes them to EGFR antagonists (7, 53), and downregulation of PTEN using short hairpin RNAs dampens the apoptotic effect of EGFR TKIs against receptor-dependent tumor cells (53, 64).
Consistent with these data, addition of TGF-β or stable expression of active Alk5 enhanced PI3K/Akt signaling in HER2-overexpressing cells and increased the 50% inhibitory concentration of trastuzumab (Fig. 7 and 8). Inhibition of PI3K with LY294002 or knockdown of TACE or ErbB3 with siRNA oligonucleotides restored the full inhibitory effect of trastuzumab, suggesting that increased cleavage of ErbB ligands and subsequent activation of ErbB3 and PI3K/Akt attenuated the response to trastuzumab (Fig. 6). A role of ErbB3 in resistance to anti-ErbB drugs is not unprecedented. Indeed, downregulation of ErbB3 signaling is required for EGFR TKIs to induce an antitumor effect in lung cancer cells (19). Lung cancer cells with acquired resistance to EGFR TKIs have been shown to acquire amplification of the MET oncogene. In drug-resistant cells, MET phosphorylates and activates ErbB3, thus maintaining PI3K/Akt activation in the presence of an EGFR TKI. In this study, expression of ErbB3 short hairpin RNAs inhibited Akt and reversed drug resistance (21).
Finally, mammary tumors with a gene expression profile that correlated with the Alk5T204D signature had a worse RFS and OS than tumors without such correlation. When the active Alk5 gene expression signature was examined in 22 tumors from patients who were treated with trastuzumab and vinorelbine, three patients who achieved pathologically complete responses did not share similarities in their expression patterns with the signature. While these data support a correlation between the Alk5 signature and the poor prognosis and lack of clinical response to trastuzumab, they do not establish a direct link between the TGF-β-TACE axis and these clinical parameters. Clinical trials with inhibitors of TGF-β, TACE, ErbB3, and PI3K will be required to establish this link and uncover the therapeutic value of targeting this axis in cancer progression.
Supplementary Material
Acknowledgments
This work was supported by NCI 1K99 CA125892 (S.E.W.), American Cancer Society-Institutional Research Grant IRG-58-009-49 (S.E.W.), grant BFU2006-01813/BMC from the Ministry of Science and Education of Spain (A.P.), NCI R01 CA62212 (C.L.A.), R01 CA80195 (C.L.A.), NIH R01 DE017982 (C.H.C.), the Damon Runyon Clinical Investigator Award CI-28-05 (C.H.C.), Breast Cancer Specialized Program of Research Excellence) P50 CA98131, and Vanderbilt-Ingram Comprehensive Cancer Center support grant P30 CA68485.
We thank Harold Moses, Brent Rexer, Brian Bierie, Nikki Cheng (Vanderbilt), and Ben Turk (Yale University) for valuable comments and the Vanderbilt Microarray Shared Resource for expert assistance.
Footnotes
Published ahead of print on 14 July 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adams, C. W., D. E. Allison, K. Flagella, L. Presta, J. Clarke, N. Dybdal, K. McKeever, and M. X. Sliwkowski. 2006. Humanization of a recombinant monoclonal antibody to produce a therapeutic HER dimerization inhibitor, pertuzumab. Cancer Immunol. Immunother 55717-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agus, D. B., R. W. Akita, W. D. Fox, G. D. Lewis, B. Higgins, P. I. Pisacane, J. A. Lofgren, C. Tindell, D. P. Evans, K. Maiese, H. I. Scher, and M. X. Sliwkowski. 2002. Targeting ligand-activated ErbB2 signaling inhibits breast and prostate tumor growth. Cancer Cell 2127-137. [DOI] [PubMed] [Google Scholar]
- 3.Alimandi, M., A. Romano, M. C. Curia, R. Muraro, P. Fedi, S. A. Aaronson, P. P. Di Fiore, and M. H. Kraus. 1995. Cooperative signaling of ErbB3 and ErbB2 in neoplastic transformation and human mammary carcinomas. Oncogene 101813-1821. [PubMed] [Google Scholar]
- 4.Atfi, A., E. Dumont, F. Colland, D. Bonnier, A. L′Helgoualc'h, C. Prunier, N. Ferrand, B. Clement, U. M. Wewer, and N. Theret. 2007. The disintegrin and metalloproteinase ADAM12 contributes to TGF-β signaling through interaction with the type II receptor. J. Cell Biol. 178201-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bakin, A. V., C. Rinehart, A. K. Tomlinson, and C. L. Arteaga. 2002. p38 mitogen-activated protein kinase is required for TGFβ-mediated fibroblastic transdifferentiation and cell migration. J. Cell Sci. 1153193-3206. [DOI] [PubMed] [Google Scholar]
- 6.Berns, K., H. M. Horlings, B. T. Hennessy, M. Madiredjo, E. M. Hijmans, K. Beelen, S. C. Linn, A. M. Gonzalez-Angulo, K. Stemke-Hale, M. Hauptmann, R. L. Beijersbergen, G. B. Mills, M. J. van de Vijver, and R. Bernards. 2007. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell 12395-402. [DOI] [PubMed] [Google Scholar]
- 7.Bianco, R., I. Shin, C. A. Ritter, F. M. Yakes, A. Basso, N. Rosen, J. Tsurutani, P. A. Dennis, G. B. Mills, and C. L. Arteaga. 2003. Loss of PTEN/MMAC1/TEP in EGF receptor-expressing tumor cells counteracts the antitumor action of EGFR tyrosine kinase inhibitors. Oncogene 222812-2822. [DOI] [PubMed] [Google Scholar]
- 8.Black, R. A., C. T. Rauch, C. J. Kozlosky, J. J. Peschon, J. L. Slack, M. F. Wolfson, B. J. Castner, K. L. Stocking, P. Reddy, S. Srinivasan, N. Nelson, N. Boiani, K. A. Schooley, M. Gerhart, R. Davis, J. N. Fitzner, R. S. Johnson, R. J. Paxton, C. J. March, and D. P. Cerretti. 1997. A metalloproteinase disintegrin that releases tumour-necrosis factor-alpha from cells. Nature 385729-733. [DOI] [PubMed] [Google Scholar]
- 9.Caja, L., C. Ortiz, E. Bertran, M. M. Murillo, M. J. Miro-Obradors, E. Palacios, and I. Fabregat. 2007. Differential intracellular signalling induced by TGF-β in rat adult hepatocytes and hepatoma cells: implications in liver carcinogenesis. Cell Signal 19683-694. [DOI] [PubMed] [Google Scholar]
- 10.Chang, H. Y., D. S. Nuyten, J. B. Sneddon, T. Hastie, R. Tibshirani, T. Sorlie, H. Dai, Y. D. He, L. J. van't Veer, H. Bartelink, M. van de Rijn, P. O. Brown, and M. J. van de Vijver. 2005. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc. Natl. Acad. Sci. USA 1023738-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cleveland, W. S. 1981. LOWESS: A program for smoothing scatterplots by robust locally weighted regression. Am. Stat. 3554. [Google Scholar]
- 12.Debnath, J., S. K. Muthuswamy, and J. S. Brugge. 2003. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 30256-268. [DOI] [PubMed] [Google Scholar]
- 13.Del Castillo, G., M. M. Murillo, A. Alvarez-Barrientos, E. Bertran, M. Fernandez, A. Sanchez, and I. Fabregat. 2006. Autocrine production of TGF-beta confers resistance to apoptosis after an epithelial-mesenchymal transition process in hepatocytes: role of EGF receptor ligands. Exp. Cell Res. 3122860-2871. [DOI] [PubMed] [Google Scholar]
- 14.Derynck, R., R. J. Akhurst, and A. Balmain. 2001. TGF-beta signaling in tumor suppression and cancer progression. Nat. Genet. 29117-129. [DOI] [PubMed] [Google Scholar]
- 15.Diaz-Rodriguez, E., J. C. Montero, A. Esparis-Ogando, L. Yuste, and A. Pandiella. 2002. Extracellular signal-regulated kinase phosphorylates tumor necrosis factor alpha-converting enzyme at threonine 735: a potential role in regulated shedding. Mol. Biol. Cell 132031-2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dumont, N., and C. L. Arteaga. 2003. Targeting the TGF beta signaling network in human neoplasia. Cancer Cell 3531-536. [DOI] [PubMed] [Google Scholar]
- 17.Dumont, N., A. V. Bakin, and C. L. Arteaga. 2003. Autocrine transforming growth factor-beta signaling mediates Smad-independent motility in human cancer cells. J. Biol. Chem. 2783275-3285. [DOI] [PubMed] [Google Scholar]
- 18.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 9514863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engelman, J. A., P. A. Janne, C. Mermel, J. Pearlberg, T. Mukohara, C. Fleet, K. Cichowski, B. E. Johnson, and L. C. Cantley. 2005. ErbB-3 mediates phosphoinositide 3-kinase activity in gefitinib-sensitive non-small cell lung cancer cell lines. Proc. Natl. Acad. Sci. USA 1023788-3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engelman, J. A., J. Luo, and L. C. Cantley. 2006. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat. Rev. Genet. 7606-619. [DOI] [PubMed] [Google Scholar]
- 21.Engelman, J. A., K. Zejnullahu, T. Mitsudomi, Y. Song, C. Hyland, J. O. Park, N. Lindeman, C. M. Gale, X. Zhao, J. Christensen, T. Kosaka, A. J. Holmes, A. M. Rogers, F. Cappuzzo, T. Mok, C. Lee, B. E. Johnson, L. C. Cantley, and P. A. Janne. 2007. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 3161039-1043. [DOI] [PubMed] [Google Scholar]
- 22.Fan, H., C. W. Turck, and R. Derynck. 2003. Characterization of growth factor-induced serine phosphorylation of tumor necrosis factor-alpha converting enzyme and of an alternatively translated polypeptide. J. Biol. Chem. 27818617-18627. [DOI] [PubMed] [Google Scholar]
- 23.Franklin, M. C., K. D. Carey, F. F. Vajdos, D. J. Leahy, A. M. de Vos, and M. X. Sliwkowski. 2004. Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex. Cancer Cell 5317-328. [DOI] [PubMed] [Google Scholar]
- 24.Gobbi, H., W. D. Dupont, J. F. Simpson, W. D. Plummer, Jr., P. A. Schuyler, S. J. Olson, C. L. Arteaga, and D. L. Page. 1999. Transforming growth factor-β and breast cancer risk in women with mammary epithelial hyperplasia. J. Natl. Cancer Inst. 912096-2101. [DOI] [PubMed] [Google Scholar]
- 25.Goggins, M., M. Shekher, K. Turnacioglu, C. J. Yeo, R. H. Hruban, and S. E. Kern. 1998. Genetic alterations of the transforming growth factor beta receptor genes in pancreatic and biliary adenocarcinomas. Cancer Res. 585329-5332. [PubMed] [Google Scholar]
- 26.Gymnopoulos, M., M. A. Elsliger, and P. K. Vogt. 2007. Rare cancer-specific mutations in PIK3CA show gain of function. Proc. Natl. Acad. Sci. USA 1045569-5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hahn, S. A., M. Schutte, A. T. Hoque, C. A. Moskaluk, L. T. da Costa, E. Rozenblum, C. L. Weinstein, A. Fischer, C. J. Yeo, R. H. Hruban, and S. E. Kern. 1996. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science 271350-353. [DOI] [PubMed] [Google Scholar]
- 28.Harris, L. N., F. You, S. J. Schnitt, A. Witkiewicz, X. Lu, D. Sgroi, P. D. Ryan, S. E. Come, H. J. Burstein, B. A. Lesnikoski, M. Kamma, P. N. Friedman, R. Gelman, J. D. Iglehart, and E. P. Winer. 2007. Predictors of resistance to preoperative trastuzumab and vinorelbine for HER2-positive early breast cancer. Clin. Cancer Res. 131198-1207. [DOI] [PubMed] [Google Scholar]
- 29.Holbro, T., R. R. Beerli, F. Maurer, M. Koziczak, C. F. Barbas III, and N. E. Hynes. 2003. The ErbB2/ErbB3 heterodimer functions as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumor cell proliferation. Proc. Natl. Acad. Sci. USA 1008933-8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horowitz, J. C., D. Y. Lee, M. Waghray, V. G. Keshamouni, P. E. Thomas, H. Zhang, Z. Cui, and V. J. Thannickal. 2004. Activation of the prosurvival phosphatidylinositol 3-kinase/AKT pathway by transforming growth factor-β1 in mesenchymal cells is mediated by p38 MAPK-dependent induction of an autocrine growth factor. J. Biol. Chem. 2791359-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Izumi, Y., M. Hirata, H. Hasuwa, R. Iwamoto, T. Umata, K. Miyado, Y. Tamai, T. Kurisaki, A. Sehara-Fujisawa, S. Ohno, and E. Mekada. 1998. A metalloprotease-disintegrin, MDC9/meltrin-γ/ADAM9 and PKCδare involved in TPA-induced ectodomain shedding of membrane-anchored heparin-binding EGF-like growth factor. EMBO J. 177260-7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kenny, P. A., and M. J. Bissell. 2007. Targeting TACE-dependent EGFR ligand shedding in breast cancer. J. Clin. Investig. 117337-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lamb, J., E. D. Crawford, D. Peck, J. W. Modell, I. C. Blat, M. J. Wrobel, J. Lerner, J. P. Brunet, A. Subramanian, K. N. Ross, M. Reich, H. Hieronymus, G. Wei, S. A. Armstrong, S. J. Haggarty, P. A. Clemons, R. Wei, S. A. Carr, E. S. Lander, and T. R. Golub. 2006. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 3131929-1935. [DOI] [PubMed] [Google Scholar]
- 34.Markowitz, S., J. Wang, L. Myeroff, R. Parsons, L. Sun, J. Lutterbaugh, R. S. Fan, E. Zborowska, K. W. Kinzler, B. Vogelstein, et al. 1995. Inactivation of the type II TGF-β receptor in colon cancer cells with microsatellite instability. Science 2681336-1338. [DOI] [PubMed] [Google Scholar]
- 35.Massague, J. 1998. TGF-β signal transduction. Annu. Rev. Biochem. 67753-791. [DOI] [PubMed] [Google Scholar]
- 36.Massague, J., and Y. G. Chen. 2000. Controlling TGF-β signaling. Genes Dev. 14627-644. [PubMed] [Google Scholar]
- 37.Massague, J., and R. R. Gomis. 2006. The logic of TGFβ signaling. FEBS Lett. 5802811-2820. [DOI] [PubMed] [Google Scholar]
- 38.Mellinghoff, I. K., M. Y. Wang, I. Vivanco, D. A. Haas-Kogan, S. Zhu, E. Q. Dia, K. V. Lu, K. Yoshimoto, J. H. Huang, D. J. Chute, B. L. Riggs, S. Horvath, L. M. Liau, W. K. Cavenee, P. N. Rao, R. Beroukhim, T. C. Peck, J. C. Lee, W. R. Sellers, D. Stokoe, M. Prados, T. F. Cloughesy, C. L. Sawyers, and P. S. Mischel. 2005. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N. Engl. J. Med. 3532012-2024. [DOI] [PubMed] [Google Scholar]
- 39.Montero, J. C., L. Yuste, E. Diaz-Rodriguez, A. Esparis-Ogando, and A. Pandiella. 2002. Mitogen-activated protein kinase-dependent and -independent routes control shedding of transmembrane growth factors through multiple secretases. Biochem. J. 363211-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moses, H. L., E. L. Branum, J. A. Proper, and R. A. Robinson. 1981. Transforming growth factor production by chemically transformed cells. Cancer Res. 412842-2848. [PubMed] [Google Scholar]
- 41.Muraoka-Cook, R. S., I. Shin, J. Y. Yi, E. Easterly, M. H. Barcellos-Hoff, J. M. Yingling, R. Zent, and C. L. Arteaga. 2006. Activated type I TGFβ receptor kinase enhances the survival of mammary epithelial cells and accelerates tumor progression. Oncogene 253408-3423. [DOI] [PubMed] [Google Scholar]
- 42.Muraoka, R. S., Y. Koh, L. R. Roebuck, M. E. Sanders, D. Brantley-Sieders, A. E. Gorska, H. L. Moses, and C. L. Arteaga. 2003. Increased malignancy of Neu-induced mammary tumors overexpressing active transforming growth factor β1. Mol. Cell. Biol. 238691-8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagata, Y., K. H. Lan, X. Zhou, M. Tan, F. J. Esteva, A. A. Sahin, K. S. Klos, P. Li, B. P. Monia, N. T. Nguyen, G. N. Hortobagyi, M. C. Hung, and D. Yu. 2004. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell 6117-127. [DOI] [PubMed] [Google Scholar]
- 44.Peng, S. B., L. Yan, X. Xia, S. A. Watkins, H. B. Brooks, D. Beight, D. K. Herron, M. L. Jones, J. W. Lampe, W. T. McMillen, N. Mort, J. S. Sawyer, and J. M. Yingling. 2005. Kinetic characterization of novel pyrazole TGF-β receptor I kinase inhibitors and their blockade of the epithelial-mesenchymal transition. Biochemistry 442293-2304. [DOI] [PubMed] [Google Scholar]
- 45.Prigent, S. A., and W. J. Gullick. 1994. Identification of c-erbB-3 binding sites for phosphatidylinositol 3′-kinase and SHC using an EGF receptor/c-erbB-3 chimera. EMBO J. 132831-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reddy, P., J. L. Slack, R. Davis, D. P. Cerretti, C. J. Kozlosky, R. A. Blanton, D. Shows, J. J. Peschon, and R. A. Black. 2000. Functional analysis of the domain structure of tumor necrosis factor-α converting enzyme. J. Biol. Chem. 27514608-14614. [DOI] [PubMed] [Google Scholar]
- 47.Ritter, C. A., M. Perez-Torres, C. Rinehart, M. Guix, T. Dugger, J. A. Engelman, and C. L. Arteaga. 2007. Human breast cancer cells selected for resistance to trastuzumab in vivo overexpress epidermal growth factor receptor and ErbB ligands and remain dependent on the ErbB receptor network. Clin. Cancer Res. 134909-4919. [DOI] [PubMed] [Google Scholar]
- 48.Roberts, A. B., M. A. Anzano, L. C. Lamb, J. M. Smith, and M. B. Sporn. 1981. New class of transforming growth factors potentiated by epidermal growth factor: isolation from non-neoplastic tissues. Proc. Natl. Acad. Sci. USA 785339-5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rousseau, S., M. Papoutsopoulou, A. Symons, D. Cook, J. M. Lucocq, A. R. Prescott, A. O'Garra, S. C. Ley, and P. Cohen. 2008. TPL2-mediated activation of ERK1 and ERK2 regulates the processing of pre-TNFα in LPS-stimulated macrophages. J. Cell Sci. 121149-154. [DOI] [PubMed] [Google Scholar]
- 50.Sahin, U., G. Weskamp, K. Kelly, H. M. Zhou, S. Higashiyama, J. Peschon, D. Hartmann, P. Saftig, and C. P. Blobel. 2004. Distinct roles for ADAM10 and ADAM17 in ectodomain shedding of six EGFR ligands. J. Cell Biol. 164769-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sawyer, J. S., D. W. Beight, K. S. Britt, B. D. Anderson, R. M. Campbell, T. Goodson, Jr., D. K. Herron, H. Y. Li, W. T. McMillen, N. Mort, S. Parsons, E. C. Smith, J. R. Wagner, L. Yan, F. Zhang, and J. M. Yingling. 2004. Synthesis and activity of new aryl- and heteroaryl-substituted 5,6-dihydro-4H-pyrrolo[1,2-b]pyrazole inhibitors of the transforming growth factor-β type I receptor kinase domain. Bioorg. Med. Chem. Lett. 143581-3584. [DOI] [PubMed] [Google Scholar]
- 52.Seton-Rogers, S. E., Y. Lu, L. M. Hines, M. Koundinya, J. LaBaer, S. K. Muthuswamy, and J. S. Brugge. 2004. Cooperation of the ErbB2 receptor and transforming growth factor β in induction of migration and invasion in mammary epithelial cells. Proc. Natl. Acad. Sci. USA 1011257-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.She, Q. B., D. Solit, A. Basso, and M. M. Moasser. 2003. Resistance to gefitinib in PTEN-null HER-overexpressing tumor cells can be overcome through restoration of PTEN function or pharmacologic modulation of constitutive phosphatidylinositol 3′-kinase/Akt pathway signaling. Clin. Cancer Res. 94340-4346. [PubMed] [Google Scholar]
- 54.Siegel, P. M., W. Shu, R. D. Cardiff, W. J. Muller, and J. Massague. 2003. Transforming growth factor β signaling impairs Neu-induced mammary tumorigenesis while promoting pulmonary metastasis. Proc. Natl. Acad. Sci. USA 1008430-8435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Soond, S. M., B. Everson, D. W. Riches, and G. Murphy. 2005. ERK-mediated phosphorylation of Thr735 in TNFα-converting enzyme and its potential role in TACE protein trafficking. J. Cell Sci. 1182371-2380. [DOI] [PubMed] [Google Scholar]
- 56.Ueda, Y., S. Wang, N. Dumont, J. Y. Yi, Y. Koh, and C. L. Arteaga. 2004. Overexpression of HER2 (ErbB2) in human breast epithelial cells unmasks transforming growth factor β-induced cell motility. J. Biol. Chem. 27924505-24513. [DOI] [PubMed] [Google Scholar]
- 57.Valdes, F., M. M. Murillo, A. M. Valverde, B. Herrera, A. Sanchez, M. Benito, M. Fernandez, and I. Fabregat. 2004. Transforming growth factor-β activates both pro-apoptotic and survival signals in fetal rat hepatocytes. Exp. Cell Res. 292209-218. [DOI] [PubMed] [Google Scholar]
- 58.van de Vijver, M. J., Y. D. He, L. J. van't Veer, H. Dai, A. A. Hart, D. W. Voskuil, G. J. Schreiber, J. L. Peterse, C. Roberts, M. J. Marton, M. Parrish, D. Atsma, A. Witteveen, A. Glas, L. Delahaye, T. van der Velde, H. Bartelink, S. Rodenhuis, E. T. Rutgers, S. H. Friend, and R. Bernards. 2002. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 3471999-2009. [DOI] [PubMed] [Google Scholar]
- 59.Vijapurkar, U., M. S. Kim, and J. G. Koland. 2003. Roles of mitogen-activated protein kinase and phosphoinositide 3′-kinase in ErbB2/ErbB3 coreceptor-mediated heregulin signaling. Exp. Cell Res. 284291-302. [DOI] [PubMed] [Google Scholar]
- 60.Vinals, F., and J. Pouyssegur. 2001. Transforming growth factor β1 (TGF-β1) promotes endothelial cell survival during in vitro angiogenesis via an autocrine mechanism implicating TGF-α signaling. Mol. Cell. Biol. 217218-7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wakefield, L. M., and A. B. Roberts. 2002. TGF-β signaling: positive and negative effects on tumorigenesis. Curr. Opin. Genet. Dev. 1222-29. [DOI] [PubMed] [Google Scholar]
- 62.Wang, D., T. Kanuma, H. Mizunuma, F. Takama, Y. Ibuki, N. Wake, A. Mogi, Y. Shitara, and S. Takenoshita. 2000. Analysis of specific gene mutations in the transforming growth factor-β signal transduction pathway in human ovarian cancer. Cancer Res. 604507-4512. [PubMed] [Google Scholar]
- 63.Wang, J., L. Sun, L. Myeroff, X. Wang, L. E. Gentry, J. Yang, J. Liang, E. Zborowska, S. Markowitz, J. K. Willson, et al. 1995. Demonstration that mutation of the type II transforming growth factor β receptor inactivates its tumor suppressor activity in replication error-positive colon carcinoma cells. J. Biol. Chem. 27022044-22049. [DOI] [PubMed] [Google Scholar]
- 64.Wang, M. Y., K. V. Lu, S. Zhu, E. Q. Dia, I. Vivanco, G. M. Shackleford, W. K. Cavenee, I. K. Mellinghoff, T. F. Cloughesy, C. L. Sawyers, and P. S. Mischel. 2006. Mammalian target of rapamycin inhibition promotes response to epidermal growth factor receptor kinase inhibitors in PTEN-deficient and PTEN-intact glioblastoma cells. Cancer Res. 667864-7869. [DOI] [PubMed] [Google Scholar]
- 65.Wang, S. E., A. Narasanna, M. Perez-Torres, B. Xiang, F. Y. Wu, S. Yang, G. Carpenter, A. F. Gazdar, S. K. Muthuswamy, and C. L. Arteaga. 2006. HER2 kinase domain mutation results in constitutive phosphorylation and activation of HER2 and EGFR and resistance to EGFR tyrosine kinase inhibitors. Cancer Cell 1025-38. [DOI] [PubMed] [Google Scholar]
- 66.Wang, S. E., I. Shin, F. Y. Wu, D. B. Friedman, and C. L. Arteaga. 2006. HER2/Neu (ErbB2) signaling to Rac1-Pak1 is temporally and spatially modulated by transforming growth factor β. Cancer Res. 669591-9600. [DOI] [PubMed] [Google Scholar]
- 67.Wrana, J. L., L. Attisano, R. Wieser, F. Ventura, and J. Massague. 1994. Mechanism of activation of the TGF-β receptor. Nature 370341-347. [DOI] [PubMed] [Google Scholar]
- 68.Wu, F. Y., S. E. Wang, M. E. Sanders, I. Shin, F. Rojo, J. Baselga, and C. L. Arteaga. 2006. Reduction of cytosolic p27(Kip1) inhibits cancer cell motility, survival, and tumorigenicity. Cancer Res. 662162-2172. [DOI] [PubMed] [Google Scholar]
- 69.Yakes, F. M., W. Chinratanalab, C. A. Ritter, W. King, S. Seelig, and C. L. Arteaga. 2002. Herceptin-induced inhibition of phosphatidylinositol-3 kinase and Akt Is required for antibody-mediated effects on p27, cyclin D1, and antitumor action. Cancer Res. 624132-4141. [PubMed] [Google Scholar]
- 70.Yarden, Y., and M. X. Sliwkowski. 2001. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2127-137. [DOI] [PubMed] [Google Scholar]
- 71.Yi, J. Y., I. Shin, and C. L. Arteaga. 2005. Type I transforming growth factor beta receptor binds to and activates phosphatidylinositol 3-kinase. J. Biol. Chem. 28010870-10876. [DOI] [PubMed] [Google Scholar]
- 72.Zhang, Q., S. M. Thomas, V. W. Lui, S. Xi, J. M. Siegfried, H. Fan, T. E. Smithgall, G. B. Mills, and J. R. Grandis. 2006. Phosphorylation of TNF-α converting enzyme by gastrin-releasing peptide induces amphiregulin release and EGF receptor activation. Proc. Natl. Acad. Sci. USA 1036901-6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou, B. B., M. Peyton, B. He, C. Liu, L. Girard, E. Caudler, Y. Lo, F. Baribaud, I. Mikami, N. Reguart, G. Yang, Y. Li, W. Yao, K. Vaddi, A. F. Gazdar, S. M. Friedman, D. M. Jablons, R. C. Newton, J. S. Fridman, J. D. Minna, and P. A. Scherle. 2006. Targeting ADAM-mediated ligand cleavage to inhibit HER3 and EGFR pathways in non-small cell lung cancer. Cancer Cell 1039-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.