Abstract
Mitochondrial gene expression is necessary for proper mitochondrial biogenesis. Genes on the mitochondrial DNA are transcribed by a dedicated mitochondrial RNA polymerase (mtRNAP) that is encoded in the nucleus and imported into mitochondria. In the myxomycete Physarum polycephalum, nucleotides that are not specified by the mitochondrial DNA templates are inserted into some RNAs, a process called RNA editing. This is an essential step in the expression of these RNAs, as the insertion of the nontemplated nucleotides creates open reading frames for the production of proteins from mRNAs or produces required secondary structure in rRNAs and tRNAs. The nontemplated nucleotide is added to the 3′ end of the RNA as the RNA is being synthesized during mitochondrial transcription. Because RNA editing is cotranscriptional, the mtRNAP is implicated in RNA editing as well as transcription. We have cloned the cDNA for the mtRNAP of Physarum and have expressed the mtRNAP in Escherichia coli. We have used in vitro transcription assays based on the Physarum mtRNAP to identify a novel activity associated with the mtRNAP in which non-DNA-templated nucleotides are added to the 3′ end of RNAs. Any of the four ribonucleoside triphosphates (rNTPs) can act as precursors for this process, and this novel activity is observed when only one rNTP is supplied, a condition under which transcription does not occur. The implications of this activity for the mechanism of RNA editing are discussed.
RNA editing is an additional step in gene expression that involves the changing of the RNA sequence relative to the DNA sequence that encodes it (3, 4). RNA editing is a required step in the expression of genes in the mitochondria of Physarum polycephalum. It involves the insertion of nontemplated nucleotides into the RNA (28). In Physarum mitochondrial RNA editing, the nucleotide that is inserted can be any of the four ribonucleotides, and the insertion of ribonucleotides is seen in all three types of RNA, mRNA, tRNA, and rRNA. In mRNAs, the insertion of nucleotides will invariably create an open reading frame from which a protein can be expressed (16, 28), and in tRNAs and rRNAs, it will create the necessary structures required for the proper function of these molecules (1, 27, 32).
Insertional RNA editing can fall into two distinct types based on the mechanism employed: posttranscriptional and cotranscriptional. Posttranscriptional RNA editing involves (i) the release of the RNA from the RNA polymerase (RNAP), (ii) the cleavage of the RNA backbone with an RNA endonuclease, (iii) the addition of a nontemplated nucleotide with a terminal transferase-like activity, and (iv) the ligation of the RNA with an RNA ligase to restore the RNA backbone. This posttranscriptional type of RNA editing is seen in trypanosome mitochondrial mRNAs, in which uridine nucleotides are inserted in or deleted from the RNA relative to the DNA template, creating open reading frames from which proteins can be produced (15, 17).
In contrast, cotranscriptional RNA editing involves the addition of the nontemplated nucleotide to the 3′ end of the RNA as the RNA is being synthesized. The nucleotide can be added either by the RNAP itself or by an RNA-editing activity associated with the RNAP. The insertion of nontemplated nucleotides into the Physarum mitochondrial RNAs has been shown to occur as the RNA is being synthesized by templated transcription, making it a cotranscriptional process (12, 42, 43). This finding implicates the Physarum mitochondrial RNAP (mtRNAP) in the editing of RNAs in the Physarum mitochondria. This mtRNAP is the first to be implicated in an RNA-editing mechanism.
In general, mtRNAPs consist of a single catalytic protein (19, 41). These proteins are evolutionarily conserved, and the mtRNAPs in all eukaryotic organisms with mitochondria, as well as bacteriophage RNAPs, share common features (10, 11, 25, 30, 33). The gene for the mtRNAP is usually located on a nuclear chromosome, and so the mRNA for the mtRNAP is produced in the nucleus and transported to the cytoplasm, where it is translated on cytoplasmic ribosomes (35). mtRNAPs prefer double-stranded DNA templates to single-stranded DNA templates for the initiation of transcription. For example, it has been shown previously that the Saccharomyces cerevisiae mtRNAP will initiate transcription efficiently from a linear, double-stranded piece of DNA in the presence of its promoter and its required transcription factor (MTF1), and the requirement for the transcription factor can be relieved if the DNA is supercoiled (31). In both instances, however, there is a requirement for the promoter to be present and for the promoter to be double stranded. Bacteriophage RNAPs also have the requirement for a double-stranded promoter, although the template strand can be single stranded downstream of the initiation site (20).
While it has been shown previously that RNA editing in Physarum mitochondria is a cotranscriptional process, it is not known how the nontemplated nucleotides are added to the RNA since there are still questions remaining about what enzymatic activity or activities are required for the addition of the nontemplated nucleotide, i.e., the activity of the mtRNAP itself or some other associated activity (6). Additionally, it is unknown what defines the editing site location and the identity of the nucleotide to be inserted (8). Unlike other systems that require RNA editing for RNA maturation, the RNA editing in Physarum mitochondrial gene expression is unique in that all four nucleotides are inserted at RNA-editing sites; i.e., single cytidine and occasionally single uridine insertions are present, as are dinucleotide insertions in specific combinations, although to a lesser extent (7, 32). To date, consensus sequences defining RNA-editing site locations have not been identified and antisense RNAs able to define editing sites have not been detected (39, 44).
In order to elucidate the mechanism by which nucleotides are inserted, Byrne and Gott (6) have used a partially purified mitochondrial transcription elongation complex (mtTEC) to identify the sequence determinants for the RNA-editing location. With this run-on in vitro transcription system, they have shown that sequences in the DNA template that may define editing sites must reside within 15 to 20 bp on either side of the nucleotide insertion site and, therefore, that any primary sequence or secondary structure in the synthesized RNA which is necessary to specify the editing site location, if present, must be within 15 nucleotides upstream of the editing site (6). There is a requirement for all four nucleotides to be supplied in order for transcription to occur in Physarum mitochondrial extracts (12). While limiting the concentration of one of the four nucleotides supplied in vitro decreases the efficiency of transcription and RNA editing, it does not affect the fidelity of such activities (43). Supplying only one nucleotide in the in vitro transcription reaction mixture prevents transcription from occurring in P. polycephalum mtTECs (12). However, it has been shown that in the presence of only one nucleotide, T7 RNAP is capable of adding a nontemplated nucleotide to the 3′ end of RNAs (22).
As an alternative approach to studying the involvement of the mtRNAP in RNA editing, Miller et al. (33) cloned the cDNA of the gene encoding the mtRNAP of Physarum downstream of the gene for the maltose binding protein and expressed it in Escherichia coli as an N-terminal fusion of the maltose binding protein with the mtRNAP. The resulting fusion protein was isolated and used to develop an in vitro transcription assay to analyze the mtRNAP for its ability to transcribe various synthetic and natural templates. Miller et al. (33) have shown that the mtRNAP is able to transcribe various DNA templates. The initiation of transcription is nonspecific, but a DNA template is required for transcription to occur. In contrast to the run-on in vitro transcription system employed by Cheng and Gott (12), this in vitro system produces RNA through nonspecific initiation of transcription and allows non-DNA templates to be supplied to the mtRNAP for in vitro transcription to address how templates affect RNA editing as well as transcription. In addition, because purified, recombinant mtRNAP is used, the mtRNAP can also be systematically altered to observe the effects of those alterations on transcription and RNA editing and partially purified fractions of Physarum mitochondria can be added to determine whether additional factors are necessary for the specific initiation of transcription and/or RNA editing.
In this study, DNA, mixtures of RNA and DNA, and/or RNA alone was used to examine the effect(s) on transcription by the mtRNAP, as well as the potential of the mtRNAP to edit RNA. Using this in vitro transcription system, we have identified a novel, non-DNA-templated activity consisting of the addition of nucleotides to the 3′ end of RNA(s) by the Physarum mtRNAP. The addition of nucleotides can be achieved with only one nucleotide supplied to the in vitro reaction mixture, a condition that does not support transcription (12) but may support RNA editing. The ability to add non-DNA-templated nucleotides to the 3′ end of RNAs can be blocked by the treatment of the RNA with cordycepin. Using a novel technique that we have developed, poly(A)-anchored reverse transcription-PCR (RT-PCR), we were able to confirm the identity of the nucleotide(s) added to the 3′ end of the RNAs in this in vitro system.
MATERIALS AND METHODS
Template preparation.
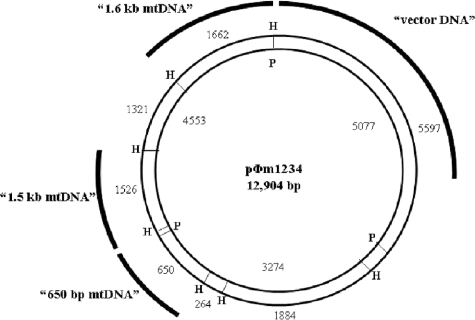
Plasmids containing Physarum mitochondrial DNA (mtDNA) inserts were created from pΦm1234, which contains a 10-kb XbaI fragment of Physarum mtDNA (Fig. 1) (21, 33). p650 is a plasmid containing a 650-bp HindIII/HindIII mtDNA insert which encodes the 5′ end of the large-subunit rRNA. This insert was cloned into the vector pT7/T3α19 (Ambion), which contains promoter recognition sequences for both T7 and T3 bacteriophage RNAPs. Likewise, plasmids p1.5 and p1.6 (Fig. 1) contain HindIII/HindIII mtDNA inserts of 1.5 and 1.6 kb, respectively, cloned in a manner similar to that used for the p650 plasmid.
FIG. 1.
Schematic representation of plasmid pΦm1234. The expected fragment sizes after digestion with HindIII (H; outside of circle) or PstI (P; inside of circle) are shown. Fragments cloned to make plasmids p650, p1.5, and p1.6 (see Materials and Methods) are shown by concentric arcs.
In vitro transcription.
RNAs supplied exogenously to the in vitro transcription reaction mixture for the Physarum mtRNAP activity were produced using T7 RNAP. T7 RNAP was used to produce runoff transcripts from the plasmids p650, p1.5, and p1.6 to create RNAs (650-nucleotide mitochondrial RNA [mtRNA], 1.5-kb mtRNA, 1.6-kb mtRNA, and >1.6-kb mtRNA). All of the T7 runoff transcripts end within Physarum mtDNA sequences, except the >1.6-kb mtRNA transcript, which includes a small portion of vector sequence. RNAs produced by T7 RNAP were treated with DNase I (Promega) to remove DNA from the subsequent transcription reactions. DNase I reactions were performed for 30 min at 37°C. After DNase I treatment, reaction mixtures were subjected to phenol-chloroform extraction to remove the DNase I as well as the T7 RNAP from the in vitro-transcribed RNAs.
In vitro transcription assays based on the mtRNAP were performed by the method of Miller et al. (33). The assays were performed at 37°C for 30 min in transcription buffer consisting of 40 mM Tris-HCl (pH 8.0), 8 mM MgCl2, 1 mM dithiothreitol, 2 mM spermidine, 200 μM (each) ATP, GTP, and CTP, and 20 μM UTP. Single nucleotides were used at concentrations of 200 μM for 3′-end-addition experiments.
Southern blotting and hybridization probes.
Plasmid pΦm1234 was digested with the restriction enzyme HindIII or PstI (Fig. 1), and the digestion products were transferred onto nitrocellulose filters overnight using the method of Southern (38). The filters were cut into strips and prehybridized at 37°C overnight in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 1× Denhardt's buffer, 50 mM sodium phosphate (pH 7.5), 0.1% sodium dodecyl sulfate, and 25 mg of sheared calf thymus DNA/ml (29). Probes for hybridization were the [α-32P]UTP radiolabeled products produced in the in vitro transcription system from Physarum mtRNAP. For each experiment, a master mix containing all reaction components, minus the experimental template to be used, was created. The master mix was aliquoted into tubes containing the RNA, DNA, or a mixture of RNA and DNA, and the reaction was allowed to proceed. After the completion of the reaction, the products were dispensed into hybridization buffer and added to the Southern blots for hybridization. After hybridization overnight at 37°C, filters were washed three times, each time in 2× SSC, 20 mM sodium phosphate (pH 7.5), and 0.1% sodium dodecyl sulfate at room temperature. Hybridization patterns were visualized by exposure to a phosphor screen (Molecular Dynamics) and scanned on a Storm 840 phosphorimager (Amersham Biosciences). Quantification of the phosphorimage was performed using ImageQuant software version 5.2 (Molecular Dynamics).
Sequence analysis of the 3′-end nucleotide addition using poly(A)-anchored RT-PCR.
RNAs treated with the mtRNAP under in vitro conditions for 3′-end addition were subjected to poly(A) polymerase treatment at 37°C for 60 min in buffer consisting of 50 mM Tris-HCl (pH 8.0), 25 mM NaCl, 10 mM MgCl2, 1 mM ATP, and 2.5 mM MnCl2. Subsequent to poly(A) polymerase treatment, the RNAs were reverse transcribed in the presence of an oligo(dT) primer (5′-CGGGATCCATGGT18-3′). Following reverse transcription, products were PCR amplified using a primer corresponding to an internal sequence of the p650 runoff RNA produced by T7 RNAP (5′-GGAATTCCGTTCGCTCACCACTAC-3′). PCR products were sequenced directly using the p650 internal primer or cut with EcoRI and BamHI, subcloned into pUC19, and sequenced (at Macrogen, Inc.) using the M13 forward primer M13F-pUC (−40) (5′-GTTTTCCCAGTCACGAC-3′) (see Fig. 5).
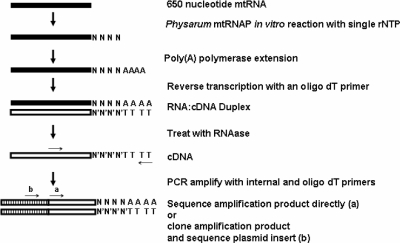
FIG. 5.
Outline of the poly(A)-anchored RT-PCR method. The 650-nucleotide mtRNA was used in the standard Physarum mtRNAP in vitro transcription system in the absence of any DNA template and in the presence of only one ribonucleoside triphosphate (rNTP). After treatment with the recombinant mtRNAP, RNAs were extended with poly(A) polymerase and ATP. Following poly(A) polymerase extension, RNAs were reverse transcribed in the presence of an oligo(dT) primer and reverse transcriptase (see Materials and Methods). After reverse transcription, the cDNAs were PCR amplified with an internal primer and the oligo(dT) primer. PCR products were sequenced directly or digested with EcoRI and BamHI, subcloned into pUC19, and sequenced using the M13F-pUC19 (−40) primer (Macrogen, Inc.).
RESULTS
In vitro transcription by the Physarum mtRNAP.
In order to better understand the transcription activities of the Physarum mtRNAP, we have developed an in vitro transcription system based on the Physarum mtRNAP (33). When the mtRNAP is provided with a double-stranded DNA template, the four ribonucleoside triphosphates, [α-32P]UTP, and a low-salt buffer with MgCl2, radiolabeled UTP is incorporated into RNase-sensitive material; i.e., RNA is synthesized.
Previously, the mtRNAP of Physarum had been characterized for its ability to transcribe various DNA templates (33). Consistent with the report from Matsunaga and Jaehning (31) on the yeast mtRNAP, transcription by the Physarum mtRNAP in vitro from a supercoiled, double-stranded DNA template is more efficient than that from nonsupercoiled DNA (33). Perhaps this effect is due to the absence of a transcription factor and/or an initiation sequence required for the efficient initiation of transcription in the absence of supercoiled DNA. To confirm that the RNA synthesized was directed by the DNA template, the radiolabeled RNA was used as a hybridization probe for Southern blots on which template DNA was immobilized. The hybridization of the probe was specific to the template DNA, showing that RNA synthesis is template directed and that the mtRNAP is a DNA-directed RNAP (33). For example, a 1.5-kb HindIII fragment (Fig. 1) of Physarum mtDNA (coordinates 46777 to 48306) (39) was cloned into the E. coli vector pT7/T3α19 (Ambion). The use of the recombinant plasmid with the 1.5-kb mtDNA insert as a template for RNA synthesis in the mitochondrial in vitro transcription system produced radiolabeled RNA that specifically hybridized to the 1.5-kb HindIII fragment on the Southern blot, as well as to the common vector portion of the plasmid (Fig. 2B, lane 1). This result indicates that the RNA product was specific to its template and that the initiation of transcription likely occurred nonspecifically in both the vector and the mtDNA insert. The RNA produced in the in vitro transcription reaction varied in length from 9 nucleotides to more than 600 nucleotides (data not shown; reference 33), indicating that under the conditions of the assay, the mtRNAP is relatively processive.
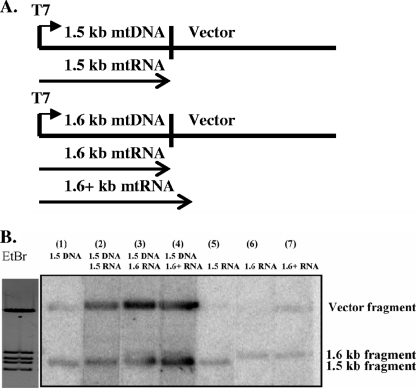
FIG. 2.
In vitro transcription in the presence of DNA and RNAs. (A) Schematic representation of the plasmids used to produce the RNAs added to the Physarum mtRNAP in vitro transcription system. RNAs were produced by T7 RNAP runoff transcription from the T7 RNAP promoter in the p1.5 and p1.6 plasmids to PstI sites (1.5-kb mtRNA and 1.6-kb mtRNA) or to a PvuII site (>1.6-kb [1.6+ kb] mtRNA). Arrows indicate the extent of RNAs relative to the plasmid DNA template. (B) Southern blot of pΦm1234 digested with HindIII and transferred onto nitrocellulose. An ethidium bromide (EtBr) stain of the gel used for the Southern blot is shown on the left. Probes for the Southern blot were radiolabeled products ([α32-P]UTP) from the Physarum mtRNAP in vitro transcription reactions using supercoiled plasmid p1.5 alone (lane 1); supercoiled plasmid p1.5 in the presence of the 1.5-kb mtRNA (lane 2), the 1.6-kb mtRNA (lane 3), or the >1.6-kb mtRNA (lane 4); or each of the RNAs (1.5-, 1.6-, and >1.6-kb RNAs) alone (lanes 5, 6, and 7, respectively).
Addition of non-DNA-templated radiolabeled UTP to the 3′ end of RNAs under standard transcription conditions.
Supplying the in vitro transcription system with templates other than double-stranded DNA allows for the investigation of processes outside of standard transcription. Radiolabeled products from the in vitro transcription reaction were used to probe a Southern blot made from HindIII-digested pΦm1234 (21) (Fig. 1 and 2A) to determine specifically what nucleic acid was being used as a template. When exogenous RNA was added to the transcription reaction, transcription activity from the p1.5 plasmid DNA template was stimulated (Fig. 2B, lanes 2 to 4). That is, the addition of an RNA that was either identical (1.5-kb mtRNA) (Fig. 2B, lane 2), nonidentical (1.6-kb mtRNA) (Fig. 2B, lane 3), or partially identical (>1.6-kb mtRNA) (Fig. 2B, lane 4) to the mtDNA template resulted in an increased amount of radiolabeled material hybridizing to the specific bands in the Southern blot corresponding to the p1.5 DNA template. The increased amount of radiolabel hybridizing to the Southern blot must correspond to increased transcription because all reactions were performed using the same master mix and all blots were hybridized and washed with the same stringency. The stimulation of transcription appears to be independent of the type of RNA added, since identical and nonidentical RNAs stimulated transcription with equal efficiencies (Fig. 2B, lanes 2 to 4).
Additionally, when RNAs that were not identical to the DNA template (i.e., 1.6- and >1.6-kb mtRNAs) were supplied to the in vitro reaction mixture, a unique 1.6-kb band that was specific to the RNA supplied was detected (Fig. 2B, lanes 3 and 4). In reactions lacking the Physarum mtRNAP, neither hybridization to the bands associated with the 1.5-kb mtDNA plasmid (transcription) nor hybridization to the 1.6-kb band (RNA labeling) was observed (data not shown). This result indicates that in addition to the role of the Physarum mtRNAP in transcription, this mtRNAP possesses a unique activity in which ribonucleotides are incorporated into RNA without the use of a DNA template. There are two possible explanations for the hybridization of radiolabeled material to the 1.6-kb mtRNA band: (i) RNA labeling through the addition of a nontemplated nucleotide to one of the ends of the RNA or, less likely, the insertion of a nontemplated nucleotide within the RNA or (ii) RNA-directed RNA transcription using the RNA supplied to the reaction mixture as a template, as has been seen previously with T7 RNAP (2, 5, 9).
To determine whether there is a requirement for a DNA template to be present for the radiolabeled nucleotide to be incorporated into RNA, the RNAs were incubated in the absence of any DNA template but in the presence of the mtRNAP and the four ribonucleoside triphosphates, including the radiolabeled nucleotide. The radiolabeled nucleotide was incorporated into RNA in the absence of a DNA template (Fig. 2B, lanes 5 to 7), and the hybridization signal correlated to the RNA supplied to the reaction mixture. In the absence of any nucleic acid (RNA or DNA), no hybridization was observed (data not shown). Therefore, the Physarum mtRNAP is capable of adding ribonucleotides to RNAs in the absence of any DNA template.
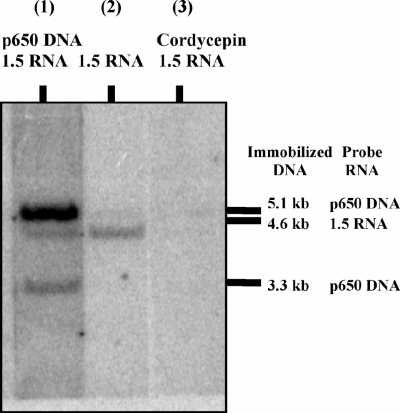
To address the question of whether the addition of the radiolabeled ribonucleotides was occurring at the 3′ end of the RNAs, the 1.5-kb mtRNA was treated with cordycepin and poly(A) polymerase to eliminate the presence of the 3′-OH. Physarum mtRNAP in vitro transcription reactions including (i) the p650 DNA template and the untreated 1.5-kb mtRNA, (ii) only the untreated 1.5-kb mtRNA, or (iii) only the cordycepin-treated 1.5-kb mtRNA were performed, and the labeled nucleic acids produced in the reactions were used as hybridization probes in Southern blots (Fig. 3). Consistent with the data presented in Fig. 2, RNA probes produced in the transcription reactions hybridized to bands corresponding to both the p650 DNA (5.1- and 3.3-kb bands) and the 1.5-kb mtRNA (4.6-kb band) added to the reaction mixture (Fig. 3, lane 1). In the absence of DNA, the untreated 1.5-kb mtRNA probe hybridized only to the band corresponding to the RNA (4.6-kb band) (Fig. 3, lane 2). The 1.5-kb mtRNA probe treated with poly(A) polymerase and cordycepin prior to in vitro transcription was not labeled and so did not expose the 4.6-kb band (Fig. 3, lane 3). This result indicates that the radiolabeled nucleotide was being added exclusively at the 3′ end of the 1.5-kb mtRNA in the absence of any DNA template and rules out the possibility of 5′-end nucleotide addition to the 1.5-kb mtRNA, internal incorporation of nucleotides into the 1.5-kb mtRNA, or the incorporation of nucleotides during de novo RNA synthesis using the 1.5-kb mtRNA as a template.
FIG. 3.
In vitro transcription in the presence of cordycepin-treated RNA. A Southern blot was prepared from an agarose gel in which fragments from a PstI digest of pΦm1234 (Fig. 1) were separated by electrophoresis. The fragments produced from this digest were 5.1, 4.6, and 3.3 kb. RNA produced from Physarum mtRNAP transcription using the plasmid p650 as the template hybridizes with the 5.1- and 3.3-kb bands; the 1.5-kb mtRNA produced from T7 RNAP runoff transcription using the p1.5 plasmid as the template hybridizes with the 4.6-kb band. Probe RNAs for the Southern blot were radiolabeled products ([α32-P]UTP) from the Physarum mtRNAP in vitro transcription assay with nucleic acids added as follows: (i) p650 plasmid in the presence of the 1.5-kb mtRNA (lane 1), (ii) the 1.5-kb mtRNA alone (lane 2), and (iii) the 1.5-kb mtRNA treated with poly(A) polymerase and the chain terminator cordycepin prior to the in vitro transcription assay (lane 3).
The addition of non-DNA-templated nucleotides requires only one ribonucleoside triphosphate to be added and occurs in the absence of transcription.
The activities of transcription and non-DNA-templated addition of nucleotides to RNA can be distinguished based upon whether the in vitro system is supplied with all four ribonucleotides or just one. Cheng and Gott (12) have shown that transcription in Physarum mitochondria requires all four ribonucleotides to be present. The addition of non-DNA-templated ribonucleotides to RNAs occurs when only UTP is added (Fig. 4A, lane 1, and B). The Physarum mtRNAP does not appear to discriminate which nucleotide is added, since all nucleotides that have been tested can be added to the 3′ end of the 650-nucleotide mtRNA (data not shown). When the RNA probe produced by incubating the mtRNA labeled in the presence of the mtRNAP and the four ribonucleotides, including radiolabeled UTP (Fig. 4A, lane 2), was compared with the hybridization signal produced by labeling the RNA in the presence of radiolabeled UTP only (Fig. 4A, lane 1), the intensities of the signals of hybridization to the 650-bp DNA band were essentially equal while hybridization to the vector DNA band was absent in both cases. This result indicates that significant addition of nucleotides to the 3′ end of the RNA occurs in the absence of DNA-templated transcription and that the presence of the additional three cold nucleotides does not prevent the addition of UTP to the 3′ end. That is, the addition of a nucleotide does not prevent or compete with the addition of a further nucleotide, although this result may indicate that nucleotide addition at the 3′ end proceeds more efficiently in the presence of all four ribonucleotide triphosphates than in the presence of a single ribonucleotide triphosphate. Because the in vitro reaction mixtures corresponding to Fig. 4A, lanes 3 and 4, contained a DNA template that promotes transcription, the labeled RNA hybridized to the vector DNA band in these lanes. Consistent with previous data, the addition of nucleotides to the 3′ end of RNAs also occurred in conjunction with DNA-templated transcription by the Physarum mtRNAP (Fig. 4A, lanes 3 and 4). Therefore, the signal corresponding to the 650-bp mtDNA band in lane 3 is somewhat more intense than the signal corresponding to the 650-bp band in lane 4, since the labeled RNA was derived from both de novo transcription and the addition of a radiolabeled nucleotide to the 3′ end of the 650-nucleotide RNA.
FIG. 4.
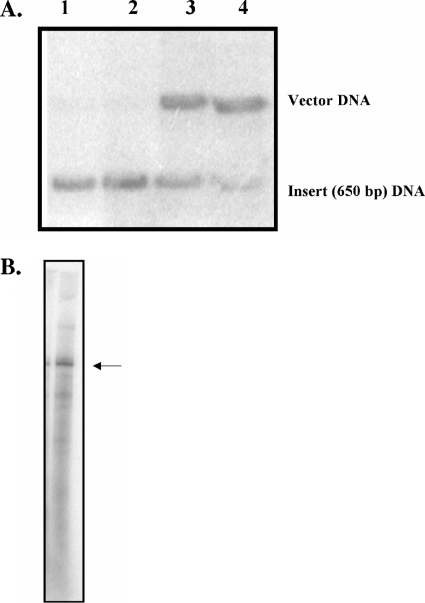
Addition of radiolabeled nucleotide to the 3′ ends of RNAs using a single radiolabeled ribonucleoside triphosphate. (A) A 650-nucleotide RNA produced by T7 RNAP runoff transcription from the p650 plasmid was radiolabeled in vitro using the Physarum mtRNAP in the presence of a single radiolabeled ribonucleoside triphosphate and was hybridized with a Southern blot prepared from an agarose gel containing PstI-digested pΦm1234 DNA (5.1-, 4.6-, and 3.3-kb bands) (Fig. 1). The Southern blot was hybridized with RNA probes produced by the Physarum mtRNAP in in vitro reactions with mixtures containing (i) 650-nucleotide mtRNA in the presence of radiolabeled UTP (lane 1), (ii) 650-nucleotide mtRNA in the presence of radiolabeled UTP with cold 200 μM (each) ATP, CTP, and GTP (lane 2), (iii) 650-nucleotide mtRNA, the p650 mtDNA plasmid, and radiolabeled UTP with cold 200 μM (each) ATP, CTP, and GTP (lane 3), and (iv) the p650 mtDNA plasmid with radiolabeled UTP and cold 200 μM (each) ATP, CTP, and GTP (lane 4). (B) Autoradiograph of a 2% agarose-6 M urea gel of the 650-nucleotide mtRNA treated in the presence of the mtRNAP and radiolabeled UTP. The arrow indicates the full-length transcript as measured by ethidium bromide staining.
To provide direct evidence that the RNA itself was being radiolabeled, the 650-nucleotide mtRNA was incubated with [α32-P]UTP and the mtRNAP and the products were run on a 2% agarose-6 M urea gel (Fig. 4B). When a parallel experiment was performed with T7 RNAP, a similar result was obtained (data not shown), consistent with reports that T7 RNAP is able to add nontemplated nucleotides to the 3′ end of RNAs (22).
Sequence of the nucleotides added to the 3′ end of RNAs.
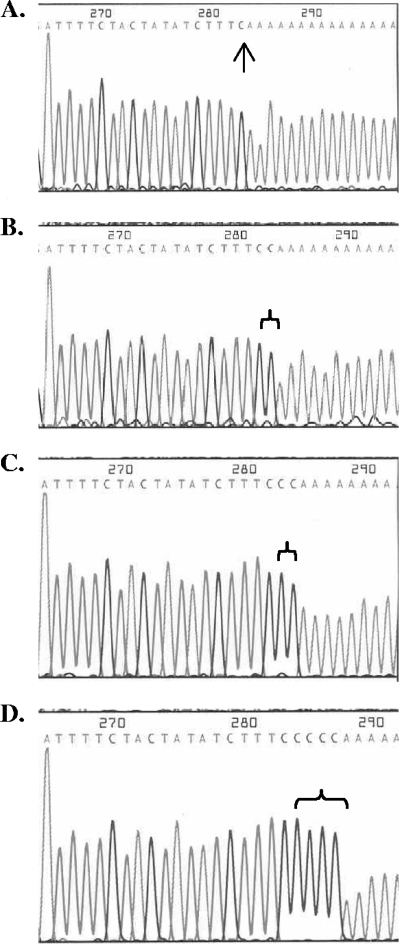
To determine the identities and numbers of nucleotides added to the 3′ end of RNAs, we have developed a technique called poly(A)-anchored RT-PCR in which the 3′ end of RNAs can be sequenced as cDNA amplification products (Fig. 5). When plasmids created by cloning individual PCR amplification products derived from the poly(A)-anchored RT-PCR were sequenced, the sequences showed that in fact, non-DNA-templated nucleotides were added to the 3′ end of RNAs. In the presence of the Physarum mtRNAP and CTP, the addition of C residues to the 3′ end of the RNAs in the in vitro reaction was observed. The sequences of individual clones indicated that from one to several nucleotides were being added (Fig. 6). In control experiments, no nucleotides were added to the RNA if the RNA was not exposed to the mtRNAP or if no nucleotides were added to the in vitro reaction mixture. Of the RNAs that were exposed to mtRNAP in the presence of nucleotides, about half had no nucleotides added (e.g., Fig. 6B), consistent with the low levels of this addition activity. The remainder had one nucleotide added (53%) (e.g., Fig. 6B), two nucleotides added (27%) (e.g., Fig. 6C), or rarely, three (13%) or four (7%) nucleotides added. These data confirm the addition of non-DNA-templated nucleotides at the 3′ end of RNAs, an activity consistent with the type of RNA editing seen in Physarum mitochondria.
FIG. 6.
Sequence analysis of nucleotides added to the 3′ end of RNAs. Plasmid p650 was used as a template for in vitro RNA synthesis using T7 RNAP in runoff transcription to the PstI site. The 650-nucleotide runoff mtRNA was incubated in the standard Physarum mtRNAP in vitro assay in the presence of only 200 μM CTP. The treated RNAs were subjected to the poly(A)-anchored RT-PCR method (Fig. 5) and sequenced directly or cloned into pUC19 and sequenced using the M13F-pUC (−40) primer (see Materials and Methods). Results show no addition of nucleotide (A), one nucleotide added (B), two nucleotides added (C), and four nucleotides added (D). The arrow in panel A indicates the natural 3′ end of the 650-nucleotide mtRNA. Nontemplated nucleotides are indicated by brackets in panels B to D.
DISCUSSION
The Physarum mtRNAP in vitro transcription system (33) allows flexibility in the type of template supplied to the mtRNAP because the mtRNAP is able to initiate transcription de novo. We have discovered and characterized a novel activity associated with the mtRNAP of P. polycephalum in which non-DNA-templated nucleotides are added to the 3′end of RNAs. The addition of the non-DNA-templated nucleotides to the 3′ end of RNAs (i) is not specific to the RNA receiving the addition, (ii) can be produced from any of the four ribonucleotides, and (iii) is observed when only one type of ribonucleotide is provided.
It is not unprecedented for DNA polymerases (DNAPs) and RNAPs to add nontemplated nucleotides. Single-polypeptide mtRNAPs are related to single-polypeptide bacteriophage RNAPs (33), and the prototype bacteriophage RNAP, T7 RNAP, has been shown previously to add nontemplated nucleotides when transcribing a DNA template (22). These RNAPs have structural and primary sequence features similar to those of, and are therefore thought to be related to, DNA- and RNA-directed DNAPs (34), such as the Klenow fragment (single-polypeptide DNA-directed DNAP) (36), Taq DNAP (single-polypeptide DNA-directed DNAP) (14), and human immunodeficiency virus reverse transcriptase (single-polypeptide RNA-directed DNAP) (18, 23, 34). These and other DNAPs have been shown to have nontemplated-nucleotide activity on 3′ blunt ends of DNA (14), an activity for which Taq DNAP is notorious (26). The fourth type of template-directed polymerase, RNA-directed RNAP, has more recently been shown to also be structurally related to the other single-polypeptide polymerases (40), and here again nontemplated nucleotide addition has been documented (37).
Since nontemplated nucleotide addition is inherent to these related polymerases, it is expected that this activity may be present in all or most mtRNAPs. However, to our knowledge, this is the first demonstration of nontemplated nucleotide addition by an mtRNAP. This activity is by definition an RNA-editing activity (3, 4), although it is not identical to the previously characterized RNA-editing activity in Physarum mitochondria in which a specific nucleotide is inserted at a specific site prior to the resumption of transcription (28). This is the first indication that the mtRNAP, and not an associated factor, may be responsible for the addition of nontemplated nucleotides, an activity consistent with the proposed, cotranscriptional mechanism of RNA editing in Physarum mitochondria. While this activity may be the basis of RNA editing in Physarum, it alone cannot explain the unique specificity of editing site location and nucleotide identity in Physarum RNA editing. The acquisition of the specificity of nontemplated nucleotide addition may have constituted the establishment of the RNA-editing activity in the myxomycetes, while the nonspecific activity documented in this paper may have served as an existing preadaptation (24). In this scenario, it is the development of a mechanism to specify where and when to add the nontemplated nucleotide and to specify what nucleotide is to be added to the nascent RNA that is necessary to produce the previously characterized RNA-editing activity in Physarum mitochondria. Characterization of the information that provides this specificity is key to understanding the mechanism of RNA editing in Physarum mitochondria. This specificity may be provided by an additional factor or factors or may have developed within the mtRNAP itself.
While we have demonstrated with this in vitro transcription system based on the Physarum mtRNAP that non-DNA-templated nucleotides can be added to RNAs, it is formally possible that other factors associated with the RNAP in vivo may be necessary to achieve specific RNA editing, perhaps to detect the signal(s) required for the identification of editing sites, as well as the determination of the identity of the nucleotide that is to be inserted at a particular editing site. In our defined in vitro system, these factors, if they exist, must be absent. However, these data support the notion that the mtRNAP is by itself sufficient to add a non-DNA-templated nucleotide(s) to the 3′ end of RNAs, albeit at present, the addition of nucleotides is nonspecific in the sense that any nucleotide can be added to the 3′ end of any RNA.
In this Physarum mtRNAP in vitro transcription system, the identities of the nontemplated nucleotides added to the 3′ end of RNAs are dependent on what ribonucleotide is supplied to the in vitro reaction. The mtRNAP does not appear to discriminate as to what nucleotide is being added, nor does it necessarily appear to discriminate where in the RNA it is positioned as it adds the nontemplated nucleotide. Consistent with the RNA editing seen in Physarum mitochondria, the mtRNAP can add any of the four nucleotides to the 3′ end of RNAs in vitro. Also consistent with the RNA editing seen in Physarum mitochondria is the ability to add the nucleotide to any RNA sequence. The evolutionary analysis of Krishnan et al. (24) has shown that RNA-editing sites can be at essentially any location relative to the primary sequence and secondary structure of an RNA. However, this in vitro system cannot reproduce what is seen in vivo, where only cytidines and uridines can be added as single nucleotides to the edited RNA while adenosine and guanosine must be added as dinucleotides in specific combinations with other nucleotides (7, 44). This in vitro transcription system based on the recombinant mtRNAP will be useful to establish what signals are involved in the identification of specific sites and the determination of nucleotide identities in the mtRNAs, since isolated mitochondrial components can be added to the system to determine their effects on RNA-editing specificity.
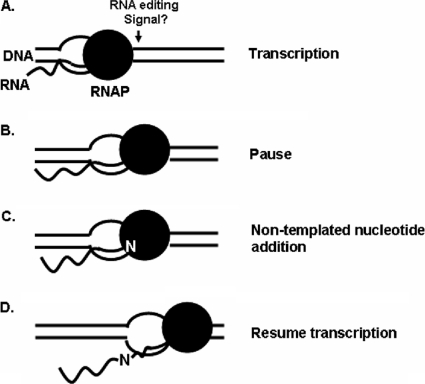
Regardless of what is specifying the editing site location and identity, the mtRNAP is competent to add nucleotides in a non-DNA-templated fashion. It has been shown previously (12) that for RNA editing in Physarum mitochondria, the addition of nontemplated nucleotides occurs in close proximity to the synthesis of RNAs by the Physarum mtRNAP and that the nascent RNA is a substrate for the RNA-editing activity. Furthermore, the editing of RNAs in the mitochondria of Physarum is physically and/or mechanistically linked to the action of transcription (42, 43). It has been shown that the addition of nucleotides to a nascently synthesized RNA in isolated mtTECs requires local features of the template and that transcription elongation and non-DNA-templated addition of nucleotides are separate and competing processes (13). This finding is consistent with a model for cotranscriptional RNA editing in Physarum (Fig. 7) in which some information or signal (Fig. 7A) causes an elongating RNAP to pause in its template-directed synthesis (Fig. 7B), leading to a temporary switch to a non-DNA-templated mode, after which a specific nontemplated nucleotide is added to the 3′ end of the nascent RNA (Fig. 7C), followed by the resumption of transcription (Fig. 7D). The data presented here argue that it is more likely the RNAP itself, and not an associated factor, that adds the nontemplated ribonucleotide, an activity which may constitute a portion of the proposed mechanism of the RNA editing seen in Physarum mitochondria. However, the signal(s) that specifies the editing site location, the identity of the nucleotide to be inserted, and what is recognizing these signals are currently unknown. Our in vitro transcription system based on the Physarum mtRNAP will be useful to examine the basis of the specificity seen in RNA editing in Physarum mitochondria.
FIG. 7.
Model of cotranscriptional RNA editing in Physarum mitochondria. (A) Transcriptional elongation on the DNA template continues until an RNA-editing site signal is encountered. (B) Polymerase pauses and switches from the transcriptional to the nontemplated mode. (C) Nontemplated addition of nucleotide (N). (D) Templated elongation resumes.
Footnotes
Published ahead of print on 23 June 2008.
REFERENCES
- 1.Antes, T., H. Costandy, R. Mahendran, M. R. Spottswood, and D. L. Miller. 1998. Insertional editing of mitochondrial tRNAs of Physarum polycephalum and Didymium nigripes. Mol. Cell. Biol. 187521-7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnaud-Barbe, N., V. Cheynet-Sauvion, G. Oriol, B. Mandrand, and F. Mallet. 1998. Transcription of RNA templates by T7 RNA polymerase. Nucleic Acids Res. 263550-3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benne, R. (ed.) 1993. RNA editing: the alteration of protein coding sequences of RNA. Ellis Horwood, London, United Kingdom.
- 4.Benne, R., J. van den Burg, J. P. J. Brakenhoff, P. Sloof, J. H. van Boom, and M. C. Tromp. 1986. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 46819-826. [DOI] [PubMed] [Google Scholar]
- 5.Biebricher, C. K., and R. Luce. 1996. Template-free generation of RNA species that replicate with bacteriophage T7 RNA polymerase. EMBO J. 153458-3465. [PMC free article] [PubMed] [Google Scholar]
- 6.Byrne, E. M., and J. Gott. 2002. Cotranscriptional editing of Physarum mitochondrial RNA requires local features of the native template. RNA 81174-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byrne, E. M., and J. Gott. 2004. Unexpectedly complex editing patterns at dinucleotide insertion sites in Physarum mitochondria. Mol. Cell. Biol. 247821-7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Byrne, E. M., A. Stout, and J. Gott. 2002. Editing site recognition and nucleotide insertion are separable processes in Physarum mitochondria. EMBO J. 216154-6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cazenave, C., and O. C. Uhlenbeck. 1994. RNA template-directed RNA synthesis by T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 916972-6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cermakian, N., T. M. Ikeda, R. Cedergren, and M. W. Gray. 1996. Sequences homologous to yeast mitochondrial and bacteriophage T3 and T7 RNA polymerases are widespread throughout the eukaryotic lineage. Nucleic Acids Res. 24648-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cermakian, N., T. M. Ikeda, P. Miramontes, B. F. Lang, and M. W. Gray. 1997. On the evolution of the single-subunit RNA polymerases. J. Mol. Evol. 45671-681. [DOI] [PubMed] [Google Scholar]
- 12.Cheng, Y.-W., and J. Gott. 2000. Transcription and RNA editing in a soluble in vitro system from Physarum mitochondria. Nucleic Acids Res. 283695-3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng, Y.-W., L. M. Visomirski-Robic, and J. M. Gott. 2001. Non-templated addition of nucleotides to the 3′ end of nascent RNA during RNA editing in Physarum. EMBO J. 201405-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark, J. M. 1988. Novel non-templated nucleotide addition reactions catalyzed by procaryotic and eucaryotic DNA polymerases. Nucleic Acids Res. 169677-9686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gott, J. 2003. Two distinct roles for terminal uridylyl transerases in RNA editing. Proc. Natl. Acad. Sci. USA 10010583-10584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gott, J., L. M. Visomirski-Robic, and J. L. Hunter. 1993. Substitutional and insertional RNA editing of the cytochrome c oxidase subunit 1 mRNA of Physarum polycephalum. J. Biol. Chem. 26825483-25486. [PubMed] [Google Scholar]
- 17.Hadjuk, S. L., and R. S. Sabatini. 1998. Mitochondrial mRNA editing in kinetoplastid protozoa, p. 377-394. In H. Grosjean and R. Benne (ed.), Modification and editing of RNA. American Society for Microbiology, Washington, DC.
- 18.Jacobo-Molina, A., J. Ding, R. G. Nanni, A. D. Clark, Jr., X. Lu, C. Tantillo, R. L. Williams, G. Kamer, A. L. Ferris, P. Clark, A. Hizi, S. H. Hughes, and E. Arnold. 1993. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 A resolution shows bent DNA. Proc. Natl. Acad. Sci. USA 906320-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaehning, J. A. 1993. Mitochondrial transcription: is a pattern emerging? Mol. Microbiol. 81-4. [DOI] [PubMed] [Google Scholar]
- 20.Jiang, M., M. Rong, C. T. Martin, and W. T. McAllister. 2001. Interrupting the template strand of the T7 promoter facilitates translocation of the DNA during initiation, reducing transcript slippage and the release of abortive products. J. Mol. Biol. 310509-522. [DOI] [PubMed] [Google Scholar]
- 21.Jones, E. P., R. Mahendran, M. R. Spottswood, Y.-C. Yang, and D. L. Miller. 1990. Mitochondrial DNA of Physarum polycephalum: physical mapping, cloning and transcription mapping. Curr. Genet. 17331-337. [DOI] [PubMed] [Google Scholar]
- 22.Kao, C., M. Zheng, and S. Rudisser. 1999. A simple and efficient method to reduce nontemplated nucleotide addition at the 3′ terminus of RNAs transcribed by T7 RNA polymerase. RNA 51268-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kohlstaedt, L., J. Wang, J. Friedman, P. Rice, and T. Steitz. 1992. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science 2561783-1890. [DOI] [PubMed] [Google Scholar]
- 24.Krishnan, U., A. Barsamian, and D. L. Miller. 2007. Evolution of RNA editing sites in the mitochondrial small subunit rRNA of the Myxomycota. Methods Enzymol. 424197-220. [DOI] [PubMed] [Google Scholar]
- 25.Li, J., J. A. Maga, N. Cermakian, R. Cedergren, and J. E. Feagin. 2001. Identification and characterization of a Plasmodium falciparum RNA polymerase gene with similarity to mitochondrial RNA polymerases. Mol. Biochem. Parasitol. 113261-269. [DOI] [PubMed] [Google Scholar]
- 26.Magnuson, V., D. Ally, S. Nylund, Z. Karanjawala, J. Rayman, J. Knapp, A. Lowe, S. Ghosh, and F. Collins. 1996. Substrate nucleotide-determined non-templated addition of adenine by Taq DNA polymerase: implications for PCR-based genotyping and cloning. BioTechniques 21700-709. [DOI] [PubMed] [Google Scholar]
- 27.Mahendran, R., M. R. Spottswood, A. Ghate, M.-L. Ling, K. Jeng, and D. Miller. 1994. Editing of the mitochondrial small subunit rRNA in Physarum polycephalum. EMBO J. 13232-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahendran, R., M. R. Spottswood, and D. L. Miller. 1991. RNA editing by cytidine insertion in mitochondria of Physarum polycephalum. Nature 349434-438. [DOI] [PubMed] [Google Scholar]
- 29.Maniatis, T., E. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 30.Masters, B. S., L. L. Stohl, and D. A. Clayton. 1987. Yeast mitochondrial RNA polymerase is homologous to those encoded by bacteriophages T3 and T7. Cell 5189-99. [DOI] [PubMed] [Google Scholar]
- 31.Matsunaga, M., and J. A. Jaehning. 2004. Intrinsic promoter recognition by a “core” RNA polymerase. J. Biol. Chem. 27944239-44242. [DOI] [PubMed] [Google Scholar]
- 32.Miller, D., R. Mahendran, M. R. Spottswood, M. Ling, S. Wang, N. Yang, and H. Costandy. 1993. RNA editing in mitochondria of Physarum polycephalum, p. 87-103. In R. Benne (ed.), RNA editing: the alteration of protein coding sequences of RNA. Ellis Horwood, London, United Kingdom.
- 33.Miller, M. L., T. J. Antes, F. Qian, and D. L. Miller. 2006. Identification of a putative mitochondrial RNA polymerase from Physarum polycephalum: characterization, expression, purification, and transcription in vitro. Curr. Genet. 49259-271. [DOI] [PubMed] [Google Scholar]
- 34.Moras, D. 1993. Two sisters and their cousin. Nature 364572-573. [DOI] [PubMed] [Google Scholar]
- 35.Neupert, W. 1997. Protein import into mitochondria. Annu. Rev. Biochem. 66863-917. [DOI] [PubMed] [Google Scholar]
- 36.Ollis, D., P. Brick, and T. Steitz. 1985. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. Nature 313765-769. [DOI] [PubMed] [Google Scholar]
- 37.Sivakumaran, K., and C. C. Kao. 1999. Initiation of genomic plus-strand RNA synthesis from DNA and RNA templates by a viral RNA-dependent RNA polymerase. J. Virol. 736415-6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Southern, E. M. 1975. Transfer of DNA fragments to Millipore filters after restriction endonuclease cleavage followed by agarose gel electrophoresis. Mol. Gen. Genet. 98503-517. [Google Scholar]
- 39.Takano, H., T. Abe, R. Sakurai, Y. Moriyama, Y. Miyazawa, H. Nozaki, S. Kawano, N. Sasaki, and T. Kuroiwa. 2001. The complete DNA sequence of the mitochondrial genome of Physarum polycephalum. Mol. Gen. Genet. 264539-545. [DOI] [PubMed] [Google Scholar]
- 40.Thompson, A., and O. Peersen. 2004. Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase. EMBO J. 233462-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tracy, R. L., and D. B. Stern. 1995. Mitochondrial transcription initiation: promoter structures and RNA polymerases. Curr. Genet. 28205-216. [DOI] [PubMed] [Google Scholar]
- 42.Visomirski-Robic, L. M., and J. Gott. 1997. Insertional editing in isolated Physarum mitochondria is linked to RNA synthesis. RNA 3821-837. [PMC free article] [PubMed] [Google Scholar]
- 43.Visomirski-Robic, L. M., and J. Gott. 1997. Insertional editing of nascent mitochondrial RNAs in Physarum. Proc. Natl. Acad. Sci. USA 944324-4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, S. S., R. Mahendran, and D. L. Miller. 1999. Editing of cytochrome b mRNA in Physarum polycephalum. J. Biol. Chem. 2742725-2731. [DOI] [PubMed] [Google Scholar]