Abstract
The activation of NF-κB by T-cell receptor (TCR) signaling is critical for T-cell activation during the adaptive immune response. CARD11 is a multidomain adapter that is required for TCR signaling to the IκB kinase (IKK) complex. During TCR signaling, the region in CARD11 between the coiled-coil and PDZ domains is phosphorylated by protein kinase Cθ (PKCθ) in a required step in NF-κB activation. In this report, we demonstrate that this region functions as an inhibitory domain (ID) that controls the association of CARD11 with multiple signaling cofactors, including Bcl10, TRAF6, TAK1, IKKγ, and caspase-8, through an interaction that requires both the caspase recruitment domain (CARD) and the coiled-coil domain. Consistent with the ID-mediated control of their association, we demonstrate that TRAF6 and caspase-8 associate with CARD11 in T cells in a signal-inducible manner. Using an RNA interference rescue assay, we demonstrate that the CARD, linker 1, coiled-coil, linker 3, SH3, linker 4, and GUK domains are each required for TCR signaling to NF-κB downstream of ID neutralization. Requirements for the CARD, linker 1, and coiled-coil domains in signaling are consistent with their roles in the association of CARD11 with Bcl10, TRAF6, TAK1, caspase-8, and IKKγ. Using Bcl10- and MALT1-deficient cells, we show that CARD11 can recruit signaling cofactors independently of one another in a signal-inducible manner.
The NF-κB family of transcription factors plays important pleiotropic roles in the regulation of cellular activation, proliferation, and survival. The precise regulation of NF-κB activity is critical for several biological processes including innate and adaptive immunity (22, 54), learning and memory (34), epidermal development and homeostasis (2), bone formation and metabolism (65), and embryonic development (19). In most normal cells, NF-κB is inactive but is poised for rapid posttranslational activation by a diverse array of stimuli such as bacterial and viral products, proinflammatory cytokines, antigens recognized by lymphocytes, DNA-damaging agents, and UV irradiation.
A remarkable aspect of NF-κB regulation is that each of these stimuli can activate the IκB kinase complex (IKK complex) (50). An emerging theme is that a ligand-receptor pair that may be specialized to a particular biological context will signal to the IKK complex through adapter molecules that can receive stimulus-specific molecular signals and transmit that information to widely expressed signaling cofactors that are of general use. The IKK complex activates NF-κB by canonical and noncanonical pathways (50), both of which link IKK kinase activity to the phosphorylation and degradative processing of inhibitory proteins that keep NF-κB inactive in the unstimulated state.
NF-κB is one of the key transcription factors that are induced when a T lymphocyte is activated by antigenic stimulation in the adaptive immune response (51). Following the engagement of antigen by the T-cell receptor (TCR) complex and concomitant triggering of costimulatory receptors like CD28, a cascade of signaling results in the activation of the IKK complex. A cadre of TCR-proximal molecules transduces signals from the TCR complex to elicit activation of protein kinase Cθ (PKCθ), a kinase that is required in this pathway (60) and that is recruited to the central portion of the supramolecular activation cluster or immunological synapse (36, 37). TCR-proximal molecules that function between the TCR and PKCθ and have been shown to be required for NF-κB activation include the adapters SLP-76 (23) and SAP (5), the tyrosine kinases Fyn (5) and ZAP-70 (23), the Vav1 GTP/GDP exchange factor (9), phospholipase Cγ1 (11), and the serine/threonine kinases PDK1 (27) and IRAK4 (61). The requirements for these molecules in the pathway can be bypassed by treating T cells with phorbol ester and calcium ionophore (phorbol myristate acetate [PMA] and ionomycin), which are thought to activate PKCθ directly.
The role of PKCθ in TCR-mediated NF-κB activation appears to be to regulate the signaling function of CARD11 (also known as CARMA1 or BIMP3). CARD11 is a multidomain signaling scaffold that contains a caspase recruitment domain (CARD) in addition to coiled-coil, PDZ, Src homology 3 (SH3), and GUK domains (3, 18). CARD11 is expressed in a cell-type-restricted manner (3, 18). The presence of PDZ, SH3, and GUK domains is a signature feature of the membrane-associated guanylate kinase family of proteins, which function in diverse organisms and tissues to localize and assemble clusters of signaling proteins for efficient signaling (13). Several studies have established that CARD11 is required for TCR signaling to NF-κB upstream of IKK complex activation (15, 17, 21, 25, 41, 44, 68).
During TCR signaling, CARD11 is phosphorylated on serine residues 564, 577, and 657 in a region between the coiled-coil and PDZ domains (32, 55). Serine residues 564 and 657 have been shown to be substrates for PKCθ in vitro (32, 55). It has been proposed that the signal-induced phosphorylation of CARD11 somehow converts the protein into an active state that can induce and coordinate signaling to the IKK complex. Molecules that have been implicated in IKK activation downstream of CARD11 include the Bcl10 adapter molecule (31, 47), the MALT1 paracaspase (7, 46, 48), the TRAF2 and TRAF6 E3 ubiquitin ligases (59), the TAK1 kinase (29, 49, 53, 59, 66), and caspase-8 (58), all of which are widely expressed.
Several studies have proposed mechanisms for how the IKK complex is activated in this pathway. The K63-linked ubiquitination of IKKγ has been implicated in this process, mediated either by MALT1 (73) or TRAF6 (59) in a manner promoted by Bcl10 oligomerization. In addition, TCR-mediated IKK activation has also been shown to correlate with the TAK1-mediated phosphorylation of IKKβ (52, 59) and the TRAF6-mediated ubiquitination of MALT1 (43). The proteolytic activity of caspase-8 has also been demonstrated to be required for IKK activation in this pathway, but its relevant substrate has not been reported (58).
CARD11 activity during TCR signaling correlates with the relocalization of Bcl10 and the IKK complex to the T-cell membrane, to the central portion of the supramolecular activation cluster, and to lipid rafts (15, 20, 32, 55). While Bcl10, MALT1, and IKKγ have previously been shown to associate with CARD11 during TCR signaling (17, 56, 67, 69), the precise mechanisms by which CARD11 interprets upstream signals from the TCR and coordinates the activities of cofactors in IKK activation are poorly understood.
In this study we report that the inhibitory domain (ID) that functions to keep CARD11 inactive in the absence of TCR signaling targets both the CARD and the coiled-coil domain of CARD11 and can regulate the association of CARD11 with several signaling cofactors. We investigate which domains in CARD11 function downstream of PKCθ-mediated conversion of the protein to an active scaffolding molecule, and we ascribe recruitment functions to three N-terminal domains in the association of Bcl10, TRAF6, TAK1, caspase-8, and IKKγ. Consistent with the ID-mediated control of the association of CARD11 with TRAF6 and caspase-8, we demonstrate that signaling induces the association of TRAF6 and caspase-8 with CARD11 in T cells. Finally, we demonstrate that the ID-regulated association of TAK1, TRAF6, and IKKγ with CARD11 can occur in Bcl10-deficient and MALT1-deficient cells, indicating that CARD11 can recruit signaling cofactors independently of one another in a signal-inducible manner.
MATERIALS AND METHODS
Generation of Jurkat T cells and HEK293T cells with stable expression of shRNA.
Self-inactivating lentiviral constructs based upon the vector FUGW (30) were constructed to express either sihCARD11-2 (44) or siMUT (44) short hairpin RNAs (shRNAs) downstream of the human H1 RNA promoter, and the puromycin resistance gene downstream of the cytomegalovirus (CMV) enhancer/chicken β-actin promoter fusion as indicated in Fig. 1A. In the lentivirus encoding the siMUT hairpin, the H1 RNA promoter and hairpin are in the opposite orientation to the configuration depicted in Fig. 1A. To package lentiviruses, HEK293T cells were plated at 2.5 × 106 per 10-cm plate 24 h prior to calcium phosphate-mediated transfection with 3.75 μg of pCL-Ampho (40), 3.75 μg of pMDL/pRRE (14), 3.75 μg of RSV-Rev (14), and 3.75 μg of lentiviral construct encoding either no hairpin, the siCARD11-2 hairpin, or the siMUT hairpin. The medium was changed 24 h after transfection, and at 48 h after transfection viral supernatant was supplemented with polybrene (8 μg/ml) and added to Jurkat T-cell cultures. Forty-eight hours after the addition of viral supernatant, Jurkat T cells were resuspended in fresh medium containing puromycin at 0.5 μg/ml and selected for 2 weeks. Puromycin-resistant Jurkat pools were maintained in medium containing 0.5 μg/ml puromycin. Knockdown of CARD11 was assessed by Western analysis of total cell lysates using antiserum raised against murine CARD11 (gift of D. Baltimore) and antibodies to IKKα (sc-7606; Santa Cruz Biotechnology).
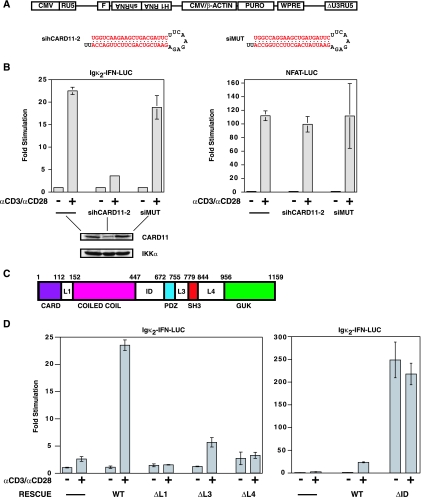
FIG. 1.
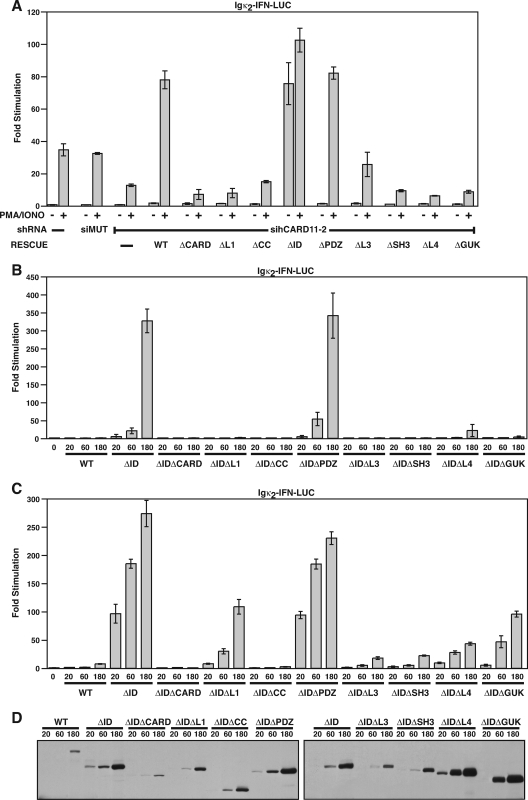
The linker domains in CARD11 are required for regulated TCR signaling to NF-κB. (A) Schematic of lentiviral constructs. Following integration, the human H1 RNA promoter drives expression of an shRNA, and the CMV enhancer/chicken β-actin promoter fusion drives expression of the puromycin resistance gene (PURO). The virus contains a flap sequence (F) for increased nuclear translocation efficiency and a Woodchuck hepatitis virus posttranscriptional regulatory element (WPRE) for enhanced expression. The sequences of the sihCARD11-2 and siMUT shRNAs are depicted. (B) Jurkat T-cell pools infected with viruses expressing no hairpin (-), the sihCARD11-2 hairpin, or the siMUT hairpin were transfected with 200 ng of pCSK-LacZ and 2,800 ng of either Igκ2-IFN-LUC or NFAT-LUC and stimulated with anti-CD3/anti-CD28 cross-linking as indicated. The lower panels are Western blots of total cell extracts of the infected pools probed with antibodies to CARD11 or IKKα. (C) Schematic of the domain structure of CARD11. The numbers indicate the amino acid position according to Pomerantz et al. (44). (D) The Jurkat T-cell pool stably expressing the sihCARD11-2 shRNA was transfected with 200 ng of pCSK-LacZ and 1,800 ng of Igκ2-IFN-LUC in the absence or presence of 200 ng of the indicated CARD11 variant expression construct. Cells were stimulated with anti-CD3/anti-CD28 cross-linking as indicated. The bars represent the mean values of triplicate samples, and error bars indicate the standard deviation. WT, wild type; α, anti.
The KD-GFP, KD-Bcl10, and KD-MALT1 HEK293T (KD, knockdown) cell lines were infected with lentiviruses based on pLKO.1 (35) and expressing shRNAs targeting the following sequences: for green fluorescent protein (GFP), 5′-AAGGCTACGTCCAGGAGCGCA-3′; for Bcl10, 5′-GTTGAATCTATTCGGCGAGAA-3′; for MALT1, 5′-CCTCACTACCAGTGGTTCAAA-3′. To package these lentiviruses, HEK293T cells were plated at 9 × 104 per well in a 24-well plate 24 h prior to calcium phosphate-mediated transfection with 50 ng of pCMV-VSVG (39), 100 ng of pMDL/pRRE, 35 ng of RSV-Rev, and 200 ng of lentiviral construct. At 48 h after transfection viral supernatant was supplemented with polybrene (8 μg/ml) and added to fresh HEK293T cell cultures. Twenty-four hours after the addition of viral supernatant, HEK293T were selected for 2 weeks in the presence of puromycin at 1 μg/ml. Knockdown was assessed by Western analysis of total cell lysates using antibodies to Bcl10 (sc-5273; Santa Cruz Biotechnology) and MALT1 (1664-1; Epitomics). The KD-GFP, KD-Bcl10, and KD-MALT1 Jurkat T-cell lines were infected by the identical pLKO.1-based lentiviruses and selected and maintained in medium containing 0.5 μg/ml puromycin.
Reporter assays of HEK293T cells.
HEK293T cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 U/ml each of penicillin and streptomycin, and 2 mM glutamine in humidified 5% CO2 at 37°C. One day prior to infection, 9 × 104 cells were plated in 0.5 ml of medium in each well of a 24-well plate. On the day of transfection, 376 ng of DNA was transfected by the calcium phosphate method, including 20 ng of the reporter Igκ2-IFN-LUC (44), 6 ng of pCSK-LacZ (8), and up to 350 ng of the expression construct to be tested. In each experiment, each sample was supplemented with empty parental expression vector (pcDNA3; Invitrogen) to keep the total amount of expression vector constant. The medium was changed 20 to 24 h following transfection, and 40 to 44 h following transfection cells were lysed in 100 μl of reporter lysis buffer (Promega) at room temperature. Cells were scraped and spun at 16,000 × g at room temperature for 10 min to pellet debris. Twenty microliters of lysate was used to measure luciferase activity using a luciferase assay system (Promega) and a luminometer (Perkin Elmer TR717) integrating for 10 s after a 2-s delay, according to the manufacturer's instructions. β-Galactosidase (β-Gal) activity was determined using 10 μl of lysate and a chemiluminescent β-Gal reporter gene assay (Roche) according to the manufacturer's instructions. Relative stimulation was calculated for each sample by dividing the luciferase activity, normalized to β-Gal activity, by that observed in the sample containing only empty expression vector.
Reporter assays of Jurkat T cells.
Jurkat T cells were grown in RPMI medium supplemented with 10% fetal bovine serum, 100 U/ml each of penicillin and streptomycin, 2 mM glutamine, and 50 μM β-mercaptoethanol in humidified 5% CO2 at 37°C. On the day of transfection, 5 × 105 cells were plated in 2 ml in six-well plates prior to incubation with DNA-Fugene 6 (Roche) or DNA-Transit LT1 (Mirus) complexes, according to the manufacturers' instructions, using a total of 3 μg of DNA and 9 μl of Fugene 6 or LT1. Assays included either the Igκ2-IFN-LUC reporter for NF-κB or the NFAT-LUC reporter for NFAT (Stratagene). In all assays 200 ng of pCSK-LacZ was included and used as a transfection and extract recovery control. At 40 to 44 h posttransfection, cells were resuspended in 1 ml of medium with or without 1 μg/ml each of anti-human CD3 (BD Pharmingen 555329), anti-human CD28 (BD Pharmingen 555725), and anti-mouse immunoglobulin G1 (BD Pharmingen; 553440) antibodies or 50 ng/ml PMA (Sigma) plus 1 μM ionomycin (Sigma) and incubated for 4 to 6 h. Following stimulation, cells were lysed in 150 μl of reporter lysis buffer (Promega) for 10 min at room temperature. Debris was removed as described above, and luciferase and β-Gal activities were measured using 50 and 25 μl of lysate, respectively. In each experiment, each sample was supplemented with empty parental expression vector (pcDNA3 or pEBB) to keep the total amount of expression vector constant. Stimulation was calculated as described above.
Immunoprecipitations.
HEK293T cells were transfected by the calcium phosphate method as described above except that 1 day prior to transfection, 5 × 105 HEK293T cells were plated in each well of a six-well plate, and a total of 2 μg of DNA was transfected. The medium was changed 20 to 24 h posttransfection, and the cells were harvested 44 to 48 h posttransfection. Cell lysates were made in 440 μl of immunoprecipitation lysis buffer (150 mM NaCl, 1 mM EDTA, 10% glycerol, 1% Igepal, 50 mM HEPES, pH 7.9, and a protease inhibitor cocktail[Sigma P8340]). Debris was removed by centrifugation at 16,000 × g at 4°C for 10 min. The supernatant was precleared twice with a 7-μl bed volume of protein G-Sepharose 4 Fast Flow (GE Healthcare) for 1 h at 4°C with rotation. Ten percent of the precleared lysate was saved for analysis by Western blotting, and the rest was incubated with 1 μg of anti-FLAG antibody (Sigma F7425) for 90 to 120 min at 4°C with rotation. A 7-μl bed volume of protein G-Sepharose that had been blocked with 1% insulin (Sigma I9278) was added and incubated for 60 to 90 min at 4°C with rotation. The resulting immunocomplex was washed with immunoprecipitation lysis buffer four times for 5 min at 4°C with rotation. The samples were boiled in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer, resolved on 10% SDS-PAGE gels, transferred to polyvinylidene difluoride membrane, and analyzed by Western blotting using anti-myc (sc-40; Santa Cruz Biotechnology) and anti-FLAG (M2; Eastman Kodak IB13026) antibodies.
For the experiments shown in Fig. 11 and 12, immunoprecipitations were performed in the same manner as described above, except that immunocomplexes were eluted from protein G-Sepharose beads with two 5-min incubations at room temperature with 50 μl of 100 mM glycine, pH 3.0. Eluates were neutralized with 10 μl of 3 M Tris, pH 8.8, added to SDS loading buffer, boiled, and resolved on 10% acrylamide SDS-PAGE gels. These elution conditions were used in order to eliminate a cross-reacting protein G band at 25 to 30 kDa. The samples were analyzed by Western blotting using anti-myc (Santa Cruz sc-40), anti-FLAG (Eastman Kodak IB13026), anti-Bcl10 (Santa Cruz sc-5273), and anti-MALT1 (Epitomics 1664-1) antibodies.
FIG. 11.
Effect of Bcl10 and MALT1 knockdown on the association of the ΔID with TAK1 and TRAF6 in HEK293T cells. The KD-GFP, KD-Bcl10, and KD-MALT1 cell lines were transfected with expression vectors for the wild-type CARD11 or the ΔID in the absence (−) or presence (+) of expression vectors for FLAG-tagged TAK1 (A) or TRAF6 (B). Anti-FLAG immunoprecipitations were performed as described in Materials and Methods. Where indicated, transfections included 100 ng of expression vector for wild-type CARD11 and 200 ng of expression vector for either TAK1 or TRAF6. To achieve approximately equivalent expression of the ΔID variant in each cell line, 200 ng of ΔID expression vector was used in the KD-GFP line, and 400 ng was used in the KD-Bcl10 and KD-MALT1 lines. The panels show the contents of the immunoprecipitate and lysate input developed with the indicated primary antibodies. In the control lane (C), the equivalent of 5% of the lysate input from the KD-GFP sample was resolved so as to provide a positive control for the Western blot (WB) analysis of the immunoprecipitates for the presence of Bcl10 and MALT1. WT, wild type; α, anti; IP, immunoprecipitation.
FIG. 12.
Effect of Bcl10 and MALT1 knockdown on the association of the ΔID with IKKγ and caspase-8 C360S in HEK293T cells. The KD-GFP, KD-Bcl10, and KD-MALT1 cell lines were transfected with expression vectors for the wild-type CARD11 or the ΔID in the absence (−) or presence (+) of expression vectors for FLAG-tagged IKKγ or caspase-8. Anti-FLAG immunoprecipitations were performed as described in Materials and Methods. In panel A, transfections included 800 ng of expression vector for IKKγ where indicated. To achieve approximately equivalent expression of wild-type and ΔID CARD11 variants in each cell line, the following amounts were used, from left to right (in ng): 100 WT, 200 ΔID, 100 WT, and 200 ΔID (for the KD-GFP cells); 200 WT, 400 ΔID, 100 WT, and 400 ΔID (for KD-Bcl10 cells); 200 WT, 400 ΔID, 100 WT, and 400 ΔID (for KD-MALT1 cells). In panel B, where indicated, transfections included 100 ng of expression vector for wild-type CARD11 and 1,000 ng of expression vector for caspase-8. To achieve approximately equivalent expression of the ΔID variant in each cell line, 200 ng of ΔID expression vector was used in the KD-GFP line, and 400 ng was used in the KD-Bcl10 and KD-MALT1 lines. The panels show the contents of the immunoprecipitate and lysate input developed with the indicated primary antibodies. In the control lane (C), the equivalent of 5% of the lysate input from the KD-GFP sample was resolved so as to provide a positive control for the Western blot (WB) analysis of the immunoprecipitates for the presence of Bcl10 and MALT1. WT, wild type; α, anti; IP, immunoprecipitation.
For immunoprecipitations of endogenous CARD11, TRAF6, IKKα, and caspase-8 proteins, 1 × 108 to 2 × 108 Jurkat T cells per sample were stimulated with 50 ng/ml PMA and 1 μM ionomycin for 3, 5, 6, 9, 12, 15, or 30 min in a volume of 20 ml and then placed on ice for 10 min. Cells were then pelleted at 500 × g for 5 min and lysed on ice for 10 min in 1.5 ml of immunoprecipitation lysis buffer containing 2.5 mM sodium orthovanadate, 10 mM sodium fluoride, and 10 mM β-glycerophosphate. Debris was pelleted by centrifugation at 16,000 × g at 4°C for 10 min. The supernatant was precleared with a 7-μl bed volume of protein G-Sepharose for 1 h at 4°C with rotation. Two to five percent of the precleared lysate was saved for analysis by Western blotting, and the remaining fraction was incubated with 2 μg of either anti-TRAF6 (Santa Cruz sc-8409) or anti-caspase-8 (Santa Cruz sc-6136) antibody overnight at 4°C with rotation or with 2 μg of anti-IKKα (Santa Cruz sc-7606) antibody for 2.5 h at 4°C with rotation. A 7-μl bed volume of protein G-Sepharose that had been blocked with 1% insulin (Sigma I9278) was added and incubated for 60 to 90 min at 4°C with rotation. The resulting immunocomplex was washed with immunoprecipitation lysis buffer four times for 5 min at 4°C with rotation. The samples were boiled in SDS-PAGE loading buffer, resolved on 10% acrylamide SDS-PAGE gels, transferred to polyvinylidene difluoride membrane, and analyzed by Western blotting using anti-CARD11 (Prosci 3189) and anti-caspase-8 (Santa Cruz sc-7890), anti-TRAF6 (Santa Cruz sc-7221), or anti-IKKα (Santa Cruz sc-7218) antibodies.
Glutathione-agarose precipitations.
Glutathione-agarose precipitations were done as described above for immunoprecipitations, except that after the supernatant was precleared with protein G-Sepharose, 10% of the lysate was saved for analysis by Western blotting, and the rest was incubated with a 7-μl bed volume of glutathione-agarose (Molecular Probes) for 90 to 120 min at 4°C with rotation. The resulting complex was washed with immunoprecipitation lysis buffer four times for 5 min with rotation at 4°C. The samples were analyzed as described above by Western blotting using anti-myc and anti-glutathione S-transferase (GST) antibodies (sc-40 and sc-459, respectively; Santa Cruz Biotechnology).
IκBα degradation assays.
The IκBα degradation assays were done as described previously (44) using anti-IκBα (Santa Cruz sc-371) antibody.
Expression constructs.
To generate pcCARD11, a DNA fragment encoding residues 2 to 1159 of murine CARD11 (44) was cloned into pcDNA3 to make an in-frame fusion to an N-terminal c-myc epitope (EQKLISEEDL). All CARD11 deletion constructs were constructed in pcDNA3 using standard molecular biology techniques and verified by sequencing. Each retains the N-terminal myc epitope. The ΔCARD expression construct (pcΔCARD) deletes residues 2 to 115. The ΔL1 construct (pcΔL1) deletes residues 116 to 149, the ΔCC construct (pcΔCC) deletes residues 152 to 447, the ΔID construct (pcΔID) deletes residues 441 to 671, the ΔPDZ construct (pcΔPDZ) deletes residues 669 to 752, the ΔL3 construct (pcΔL3) deletes residues 759 to 775, the ΔSH3 construct (pcΔSH3) deletes residues 779 to 844, the ΔL4 construct (pcΔL4) deletes residues 848 to 952, and the ΔGUK construct (pcΔGUK) deletes residues 956 to 1159. The double deletion constructs pcΔIDΔCARD, pcΔIDΔL1, pcΔIDΔCC, pcΔIDΔPDZ, pcΔIDΔL3, pcΔIDΔSH3, pcΔIDΔL4, and pcΔIDΔGUK contain identical simultaneous deletions in the indicated domains. To generate the pcHA-ID-GST expression vector, a DNA fragment encoding the ID of CARD11, corresponding to residues 441 to 671, was cloned into pcDNA3 in frame with an N-terminal hemagglutinin (HA)-tag (YPYDVPDYA), and a C-terminal GST derived from pGEX-6P-1 (GE Healthcare). To make the pEBB-HA-ID-GST expression vector, a fragment encoding the HA-ID-GST fusion was cloned into pEBB, which drives expression from an EF1α promoter. pEBG is a derivative of pEBB that expresses GST. Expression vectors for FLAG-tagged proteins were kind gifts from G. Nuñez (Bcl10 in pcDNA3), D. Goeddel (TRAF6 in pRK), T. Kawai and S. Akira (TAK1 in pFLAG-CMV4), and M. Boldin and D. Baltimore (IKKγ and caspase-8 C360S in pcDNA3).
RESULTS
The linker domains in CARD11 are required for regulated TCR signaling to NF-κB.
To examine which domains in CARD11 are required for TCR signaling to NF-κB, we established a pool of Jurkat T cells that stably expresses an shRNA previously shown to knock down expression of human CARD11 (44). We infected Jurkat T cells with the lentiviral construct shown in Fig. 1A and selected for infectants in the presence of puromycin. Stable expression of the sihCARD11-2 hairpin resulted in an approximate 70% reduction in the steady-state level of CARD11 protein, while control cells expressing a mutant hairpin or no hairpin displayed wild-type CARD11 levels (Fig. 1B left, lower panel). As expected, CARD11-deficient Jurkat T cells displayed defects in TCR-mediated activation of NF-κB, as judged by the relative induction of the NF-κB-responsive reporter Igκ2-IFN-LUC elicited by anti-CD3/anti-CD28 antibodies (Fig. 1B, left). In contrast, CARD11-deficient Jurkat T cells displayed no defect in the ability of anti-CD3/anti-CD28 cross-linking to activate the NFAT-responsive reporter NFAT-LUC (Fig. 1B, right).
Since the sihCARD11-2 hairpin targets the human CARD11 mRNA in a region that contains nucleotides that differ from the murine CARD11 mRNA, we could rescue CARD11 knockdown by cotransfection of the wild-type murine CARD11, as described previously (44). Using this RNAi rescue assay, we tested a panel of CARD11 deletion mutants for the ability to mediate signaling between the TCR complex and NF-κB. We have previously reported that deletion of the CARD, coiled-coil, SH3, and GUK domains of CARD11 abrogated the ability of CARD11 to signal to NF-κB, while deletion of the PDZ domain had no effect (44) (Fig. 1C depicts the domain structure of CARD11). As shown in Fig. 1D (left), the deletion of several other regions of the protein also abrogated CARD11 signaling, including the linker between the CARD and coiled-coil domains (L1), the linker between the PDZ and SH3 domains (L3), and the linker between the SH3 and GUK domains (L4). These three deletion mutants failed to restore CARD11 activity over a wide titration range. Western analyses of transfected cells indicated that the expression constructs for CARD11 and these deletion variants result in comparable levels of protein expression per nanogram of DNA transfected (data not shown).
Interestingly, deletion of the linker between the coiled-coil and PDZ domains resulted in a variant of CARD11 that had a greatly enhanced ability to signal to NF-κB (Fig. 1D, right). We therefore termed this region the ID and the hyperactive variant the ΔID. Recently, Sommer et al. (55) and Matsumoto et al. (32) have demonstrated that during TCR signaling CARD11 is phosphorylated in this ID at residues 564, 577, and 657 in a signal-dependent manner. Residues 564 and 657 can be phosphorylated by PKCθ in vitro. The kinase responsible for phosphorylation of serine 577 has not been reported. The phosphorylation of this ID region appears to be required for TCR-induced NF-κB activation and the CARD11-dependent recruitment of Bcl10 and the IKK complex to the membrane and to lipid rafts (32, 55). Sommer et al. (55) have provided evidence in favor of a model in which the signal-induced phosphorylation of the ID reverses its inhibitory effect on CARD11 signaling.
In agreement with the studies of Sommer et al. (55), we observed that the ΔID behaved as a constitutively active CARD11 that had already received the activating signal from the TCR complex that is mediated in part by PKCθ (see Fig. S1A in the supplemental material). The ΔID constitutively activated NF-κB in both Jurkat T cells and in human embryonic kidney cells (HEK293T), indicating that all of the required components downstream of CARD11 in the signaling pathway to NF-κB are present in both lymphoid and nonlymphoid cells (see Fig. S1A and B in the supplemental material). CARD11 may thus be viewed as a cell-type-restricted adapter protein that functions to couple cell-type-specific upstream signaling events to widely expressed downstream machinery.
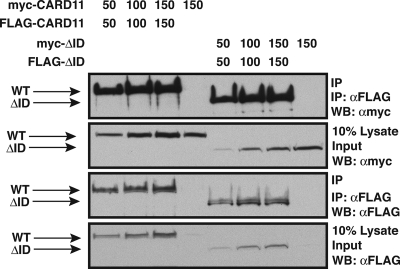
We found that an ID-GST fusion protein could inhibit the ΔID in trans and could associate with the ΔID (see Fig. S1C and D in the supplemental material). These results suggested that the ID inhibits CARD11 by direct interaction with other portions of the CARD11 protein and that, upon signaling, the ID would disengage the other portions of CARD11 and allow the signal-induced recruitment of other signaling cofactors.
The ID targets the CARD and the coiled-coil domain.
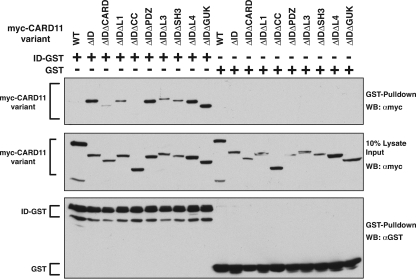
We next determined which domains in CARD11 were required for the interaction with the ID. A panel of double-deletion variants was assayed for the ability to associate with the ID-GST fusion when coexpressed in HEK293T cells. As shown in Fig. 2, the deletion of the coiled-coil domain had the largest deleterious effect on the binding of the ID-GST fusion to the ΔID. The deletion of the CARD also reduced the level of ID-GST binding, while deletions of the other regions of CARD11 did not have significant effects. No interaction was observed with the GST control. Consistent with this analysis, an N-terminal construct containing the CARD and coiled-coil regions of CARD11 could associate with the ΔID, but the isolated CARD or the coiled-coil domain could not (data not shown). These results indicate that the ID binds both the CARD and the coiled-coil domain and that the interaction with both domains is required for highest affinity binding.
FIG. 2.
Determinants of ID association with the CARD11 ΔID variant. (A) HEK293T cells were transfected with expression vectors for the indicated CARD11 variants and 150 ng of expression vector for either the ID-GST fusion (pEBB-ID-GST) or GST (pEBG). Glutathione-agarose precipitations were performed as described in Materials and Methods. To achieve approximately equivalent expression of each CARD11 variant, the following amounts of each expression vector were used (in ng): wild type (WT), 50; ΔID, 50; ΔIDΔCARD, 1,000; ΔIDΔL1, 150; ΔIDΔCC, 75; ΔIDΔPDZ, 100; ΔIDΔL3, 150; ΔIDΔSH3, 150; ΔIDΔL4, 100; ΔIDΔGUK, 100. Western blotting (WB) with the indicated antibodies was performed to develop the contents of the precipitate (top and bottom panels) and the lysate input (middle panel). α, anti.
The CARD11 domains that function in NF-κB activation downstream of ID neutralization.
Our results and those of a previous study (44) indicated that the CARD, L1, coiled-coil, L3, SH3, L4, and GUK domains are each required for NF-κB activation by TCR signaling. Each CARD11 domain that is required for TCR-mediated NF-κB activation could, in principle, act at several levels of the pathway. As part of the scaffolding function of CARD11, a domain may coordinate and be required for signaling events upstream of PKCθ activation. For example, the GUK domain of CARD11 is required for association with the PDK1 kinase, which has been reported to phosphorylate and activate PKCθ in this pathway (27). It is possible that other domains of CARD11 function to coordinate events upstream of PKCθ activation. Alternatively, a domain in CARD11 might be required for the derepressive action of PKCθ on CARD11. For example, a domain in CARD11 might mediate a protein-protein interaction that is required for the efficient association of active PKCθ with CARD11 to enable the phosphorylation of the ID and the resulting derepression of CARD11. Finally, a domain in CARD11 might be required for events downstream of ID domain phosphorylation and neutralization to coordinate or activate proteins that act on the IKK complex.
If a domain were required exclusively for events that activate PKCθ kinase activity, we expected that it would be required for TCR signaling to NF-κB but not be required for NF-κB activation by PMA/ionomycin treatment, which can bypass the requirements for upstream molecules, presumably by activating PKCθ directly. Therefore, we used the RNAi rescue assay described in Fig. 1 to test whether each domain that was required for NF-κB activation by anti-CD3/anti-CD28 cross-linking was also required for PMA-ionomycin-mediated activation. As shown in Fig. 3A, the CARD, L1, coiled-coil, L3, SH3, L4, and GUK domains were required for optimal activation of the Igκ2-IFN-LUC reporter by PMA/ionomycin treatment in Jurkat T cells, indicating that these domains are required for events downstream of PKCθ activation.
FIG. 3.
CARD11 domain requirements for NF-κB activation by PMA/ionomycin treatment and by the CARD11 ΔID variant. (A) Jurkat T cell pools stably expressing either no hairpin (−), the siMUT shRNA, or the sihCARD11-2 shRNA were transfected with 200 ng of pCSK-LacZ and 1,800 ng of Igκ2-IFN-LUC in the absence or presence of 200 ng of the indicated CARD11 variant expression construct. Cells were stimulated with PMA and ionomycin as indicated. (B) Wild-type Jurkat T cells were transfected with 200 ng of pCSK-LacZ and 2,500 ng of Igκ2-IFN-LUC in the presence of the indicated amounts (in ng) of expression vectors for the indicated CARD11 variants. (C) HEK293T cells were transfected with 6 ng of pCSK-LacZ and 20 ng of Igκ2-IFN-LUC in the presence of the indicated amounts (in ng) of expression vectors for the indicated CARD11 variants. (D) The lysates from samples in panel C were probed by Western blotting using anti-myc primary antibody to indicate the relative expression level of each variant. The bars represent the mean values of triplicate samples, and error bars indicate the standard deviations. WT, wild type.
If a domain were required exclusively for the action of activated PKCθ on CARD11, for PKCθ-mediated neutralization of the ID, then the domain should be required for TCR- and PMA/ionomycin-mediated reporter stimulation but not for reporter stimulation by the ΔID, which does not require PKCθ action or ID neutralization for activity. We therefore deleted each domain in the context of the ΔID and tested the effect of the deletion on the ability of the protein to activate the Igκ2-IFN-LUC reporter in Jurkat T cells (Fig. 3B) and HEK293T cells (Fig. 3C). Interestingly, each domain tested, with the exception of the PDZ domain, was required for maximal ΔID activity in both lymphoid and nonlymphoid cell types. These results indicate that the CARD, L1, coiled-coil, L3, SH3, L4, and GUK domains each function, directly or indirectly, in CARD11 signaling to NF-κB downstream of ID neutralization.
The ID regulates the association of CARD11 with Bcl10, TRAF6, TAK1, IKKγ, and caspase-8.
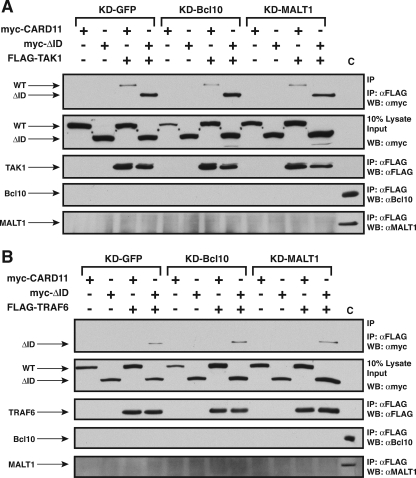
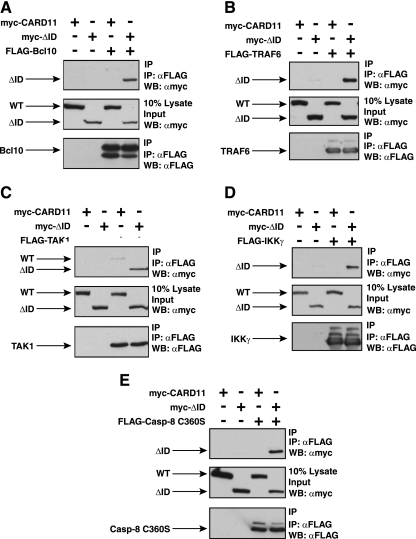
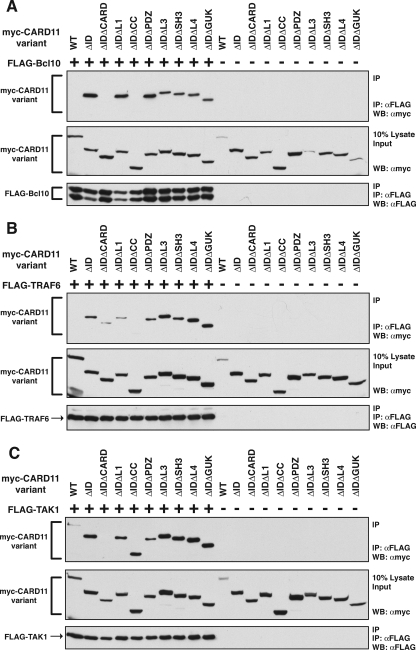
Several widely expressed signaling proteins have been implicated in TCR-mediated activation of the IKK complex, including Bcl10, MALT1, TRAF2, TRAF6, caspase-8, and TAK1. During antigen receptor signaling, CARD11 has been shown to associate with Bcl10 (17, 69), TAK1 (53), and the IKKγ subunit of the IKK complex (56). We suspected that the ID might regulate the ability of CARD11 to associate with one or more of these signaling cofactors. Indeed, Sommer et al. (55) have demonstrated that the ID can regulate the association of CARD11 with Bcl10. We also observed an enhanced ability of the CARD11 ΔID variant to associate with Bcl10 compared to wild-type CARD11. We coexpressed myc-tagged ΔID and wild-type CARD11 with FLAG-tagged Bcl10 and assayed association by coimmunoprecipitation using anti-FLAG antibodies. As shown in Fig. 4A, we readily detected the CARD11 ΔID variant in the immunoprecipitate while wild-type CARD11 was barely detected.
FIG. 4.
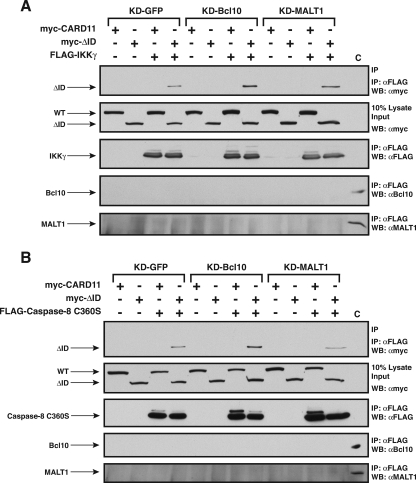
The ID regulates the ability of CARD11 to associate with Bcl10, TRAF6, TAK1, IKKγ, and caspase-8. HEK293T cells were transfected with 50 to 150 ng of the expression vector for wild-type CARD11 or the ΔID in the absence and presence of vectors encoding FLAG-tagged versions of Bcl10 (100 ng), TRAF6 (200 ng), TAK1 (200 ng), IKKγ (1,000 ng), or caspase-8 C360S (1,000 ng), as indicated. Anti-FLAG immunoprecipitations were performed as described in Materials and Methods. In each panel, Western blotting (WB) with the indicated antibodies (at right) was performed to develop the contents of the immunoprecipitate (top and bottom) and the lysate input (middle). WT, wild type; α, anti; IP, immunoprecipitation.
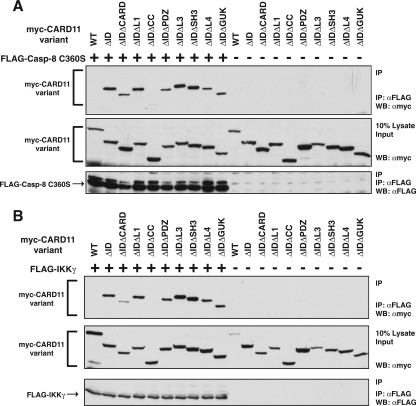
Interestingly, we found that the ΔID also had an enhanced ability, compared to wild-type CARD11, to associate with TRAF6 (Fig. 4B), TAK1 (Fig. 4C), IKKγ (Fig. 4D), and caspase-8 (Fig. 4E) using the same immunoprecipitation assay. TRAF6, IKKγ, and caspase-8 appeared to associate only with the ΔID, indicating a strong ability of the ID to regulate the association of CARD11 with these proteins. TAK1 appeared to be somewhat less susceptible to ID regulation for CARD11 association since there was less of a difference between the wild type and the ΔID in parallel immunoprecipitations. For these experiments we used the catalytically inactive C360S variant of caspase-8 so as not to cause apoptosis in the transfected cells. In similar experiments, we did not detect a reproducible association between the ΔID and either TRAF2 or MALT1 (data not shown). These results indicate that the ID regulates the ability of CARD11 to associate with multiple signaling cofactors.
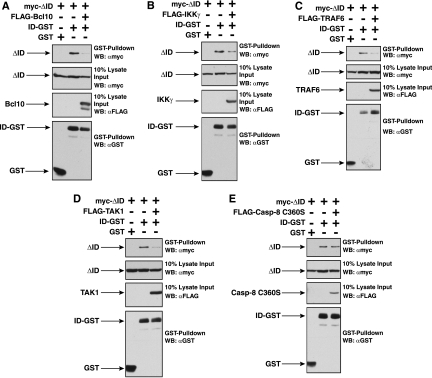
To test whether the ID directly competes for binding of these cofactors to CARD11, we assessed in glutathione-agarose precipitations whether the interaction between the ID-GST fusion and the ΔID would be affected by coexpression of any of these cofactors. As shown in Fig. 5, Bcl10, IKKγ, TRAF6, and TAK1 did reduce the interaction between ID and the ΔID, consistent with a model in which the ID sterically interferes with the binding of these factors to CARD11. Caspase-8 did not reduce the ID-ΔID interaction; this might suggest that caspase-8 associates with CARD11 through a bridging protein that is endogenous to 293T cells. Alternatively, the affinity of caspase-8 for the ΔID or its expression level might be too low to achieve competition with the ID under the conditions of this experiment. Consistent with mutually exclusive binding of the ID or these cofactors with the ΔID, we did not detect any of the FLAG-tagged cofactors in the glutathione-agarose precipitates containing the ID-GST (data not shown).
FIG. 5.
Bcl10, TRAF6, TAK1, and IKKγ compete with the ID in trans for binding to the ΔID variant. HEK293T cells were transfected with expression vectors for the ΔID and either the ID-GST fusion or GST in the absence and presence of vectors encoding Bcl10 (A), IKKγ (B), TRAF6 (C), TAK1 (D), or caspase-8 C360S (E). Glutathione-agarose precipitations were performed as described in Materials and Methods. To achieve approximately equivalent expression of the ΔID and the ID-GST in the absence and presence of each signaling cofactor, the following amount of each expression vector was used (in ng): ΔID, 50 to 100; ID-GST, 100 to 300; GST, 50; Bcl10, 150; IKKγ, 300; TRAF6, 300; TAK1, 100; or caspase-8 C360S, 300. The panels show the contents of the precipitate and lysate input, developed with anti-myc, anti-FLAG, or anti-GST primary antibody, as indicated. α, anti; WB, Western blotting.
Inducible association of CARD11 with caspase-8 and TRAF6 in T cells.
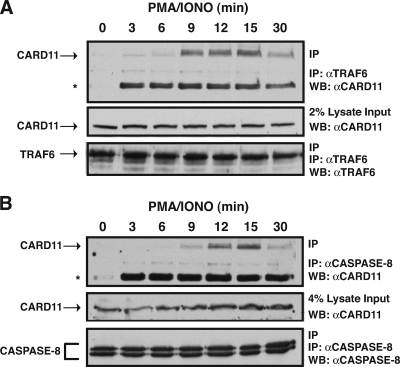
While it has previously been demonstrated that CARD11 can associate with the IKK complex, Bcl10, and TAK1 in a signal-inducible manner during antigen receptor signaling in lymphocytes, such an interaction has not been reported for caspase-8 and TRAF6. The ability of the ID to regulate CARD11 association with caspase-8 and TRAF6 predicts that, in T cells, the neutralization of the ID during TCR signaling should result in the inducible association of these proteins with CARD11. To test this prediction, we treated Jurkat T cells with a time course of PMA/ionomycin, performed immunoprecipitations from extracts using either anti-TRAF6 or anti-caspase-8 antibody, and probed for the presence of CARD11. As shown in Fig. 6, CARD11 associated with these proteins in a signal-inducible manner.
FIG. 6.
Signal-induced association of CARD11 with TRAF6 and caspase-8 in T cells. Jurkat T cells were stimulated with a time course of PMA/ionomycin as indicated. Anti-TRAF6 or anti-caspase-8 immunoprecipitations were performed as described in Materials and Methods. In each panel, Western blotting (WB) with the indicated antibodies (at right) was performed to develop the contents of the immunoprecipitate (top and bottom) and the lysate input (middle). The asterisk indicates an undefined inducible band in the immunoprecipitate that is reactive to the anti-CARD11 primary antibody. α, anti; IP, immunoprecipitation.
The ID does not determine whether CARD11 can oligomerize.
Tanner et al. (62) have reported that the coiled-coil domain of CARD11 mediates oligomerization of the protein. Since the ID interacted with the coiled-coil domain, we tested whether the ID could regulate the ability of CARD11 to oligomerize. When myc-tagged and FLAG-tagged wild-type CARD11 were coexpressed in HEK293T cells, the proteins readily associated, as revealed in the coimmunoprecipitation experiment shown in Fig. 7. The hyperactive ΔID variant displayed no further enhanced oligomerization at comparable protein concentrations, indicating that the ID and signals that regulate the interaction between the ID and the coiled-coil domain do not significantly regulate the oligomerization status of CARD11.
FIG. 7.
The ID does not determine whether CARD11 can self-associate. HEK293T cells were transfected with the indicated amounts (in ng) of expression vectors for wild-type CARD11 or the ΔID variant as either myc-tagged or FLAG-tagged fusions. Anti-FLAG immunoprecipitations were performed as described in Materials and Methods. The panels show the contents of the immunoprecipitate and the lysate input, developed with either anti-myc or anti-FLAG primary antibody, as indicated. WT, wild type; α, anti; IP, immunoprecipitation; WB, Western blotting.
Determinants of ID-regulated association of signaling cofactors with CARD11.
The enhanced ability of the ΔID to associate with Bcl10, TRAF6, TAK1, caspase-8, and IKKγ afforded the opportunity to ascribe recruitment functions to the domains revealed in Fig. 3 to be required for signaling downstream of ID neutralization. We used the immunoprecipitation assay and a panel of double deletion variants of CARD11 to determine which domains of the ΔID are required for the association of Bcl10, TRAF6, TAK1, caspase-8 C360S, and IKKγ. As shown in Fig. 8A, the deletion of either the CARD or coiled-coil domain in the context of the ΔID abrogated the association of Bcl10, indicating that both the CARD and the coiled-coil domain of CARD11 are required for high-affinity association with Bcl10. None of the other domain deletions appeared to affect the association of Bcl10 with the ΔID.
FIG. 8.
Determinants of the association of the ΔID with Bcl10, TRAF6, and TAK1. (A) HEK293T cells were transfected with expression vectors for the indicated CARD11 variants in the absence (−) or presence (+) of the expression vector for FLAG-tagged Bcl10. Anti-FLAG immunoprecipitations were performed as described in Materials and Methods. To achieve approximately equivalent expression of each CARD11 variant, the following amount of each expression vector was used (in ng): wild type (WT), 100; ΔID, 100; ΔIDΔCARD, 1,000; ΔIDΔL1, 100; ΔIDΔCC, 100; ΔIDΔPDZ, 150; ΔIDΔL3, 125; ΔIDΔSH3, 150; ΔIDΔL4, 50; ΔIDΔGUK, 50. To achieve approximately equivalent expression of Bcl10 in each sample, 100 ng of Bcl10 expression vector was used in each sample except that 500 ng was used in the ΔIDΔCARD sample and 300 ng was used in the ΔIDΔPDZ sample. Western blotting (WB) with the indicated antibodies (at right) was performed to develop the contents of the immunoprecipitate (top and bottom panels) and the lysate input (middle panel). (B) Anti-FLAG immunoprecipitations were performed as described in panel A except that 200 ng of the expression vector for FLAG-tagged TRAF6 was used in all indicated samples (+). The following amount (in ng) of expression vector for each CARD11 variant was used: WT, 200; ΔID, 200; ΔIDΔCARD, 2,000; ΔIDΔL1, 200; ΔIDΔCC, 90; ΔIDΔPDZ, 300; ΔIDΔL3, 250; ΔIDΔSH3, 300; ΔIDΔL4, 100; ΔIDΔGUK, 100. (C) Anti-FLAG immunoprecipitations were performed as described in panel A except that 200 ng of the expression vector for FLAG-tagged TAK1 was used in all indicated samples (+). The following amount (in ng) of expression vector for each CARD11 variant was used: WT, 100; ΔID, 100; ΔIDΔCARD, 1,000; ΔIDΔL1, 100; ΔIDΔCC, 100; ΔIDΔPDZ, 150; ΔIDΔL3, 175; ΔIDΔSH3, 150; ΔIDΔL4, 50; ΔIDΔGUK, 50. α, anti; IP, immunoprecipitation.
In the assay for TRAF6 association with the ΔID, the deletion of the coiled-coil domain had the largest deleterious effect (Fig. 8B). In addition, the deletion of the CARD and the L1 domain also reduced the ability of TRAF6 to associate with the ΔID.
Interestingly, TAK1 association with the ΔID was affected in this assay only by deletion of the CARD and not by deletion of any other domain (Fig. 8C). Caspase-8 C360S association with the ΔID was abrogated by deletion of the coiled-coil domain and partially affected by deletion of the CARD (Fig. 9A). IKKγ showed a pattern similar to that of caspase-8; its association with the ΔID was eliminated by deletion of the coiled-coil domain and reduced to a lesser extent by deletion of the CARD (Fig. 9B).
FIG. 9.
Determinants of the association of the ΔID with caspase-8 C360S and IKKγ. (A) Anti-FLAG immunoprecipitations were performed as described in the legend of Fig. 8A except that 1,000 ng of the expression vector for FLAG-tagged caspase-8 C360S was used in all indicated samples (+). The following amount (in ng) of expression vector for each CARD11 variant was used: wild type (WT), 100; ΔID, 100; ΔIDΔCARD, 1,000; ΔIDΔL1, 100; ΔIDΔCC, 50; ΔIDΔPDZ, 150; ΔIDΔL3, 125; ΔIDΔSH3, 150; ΔIDΔL4, 50; ΔIDΔGUK, 50. (B) Anti-FLAG immunoprecipitations were performed as described in the legend of Fig. 8A except that 800 ng of the expression vector for FLAG-tagged IKKγ was used in all indicated samples (+). The following amount (in ng) of expression vector for each CARD11 variant was used: WT, 120; ΔID, 120; ΔIDΔCARD, 1,200; ΔIDΔL1, 120; ΔIDΔCC, 120; ΔIDΔPDZ, 180; ΔIDΔL3, 210; ΔIDΔSH3, 180; ΔIDΔL4, 60; ΔIDΔGUK, 60. α, anti; IP, immunoprecipitation; WB, Western blotting.
Although these proteins in general showed different relative domain dependencies for their association with the hyperactive CARD11 ΔID variant, each required either the CARD or the coiled-coil domain or both for association. These are the same domains that appeared from our analysis in Fig. 2 to be targeted for binding by the ID.
Bcl10 is not required for the association of TAK1, TRAF6, caspase-8, or IKKγ with activated CARD11 in HEK293T cells.
Recent models of CARD11 action in antigen receptor signaling have proposed that the binding of Bcl10 and MALT1 to CARD11 is required for the recruitment of other signaling cofactors into an active signaling complex. In these models, a Bcl10-MALT1 complex binds directly to CARD11 during signaling and then bridges an interaction between CARD11 and other factors that directly bind to either Bcl10 or Bcl10-bound MALT1, such as TRAF6, caspase-8, and the IKK complex (22, 43, 45, 63). However, the domain requirements for the association of cofactors with the ΔID suggested that several proteins may associate with CARD11 in a Bcl10-independent manner. The coiled-coil domain deletion had a greater effect on Bcl10 association than on TAK1 association (Fig. 8), and the CARD deletion had a greater effect on Bcl10 association than on TRAF6, caspase-8 C360S, or IKKγ association with the ΔID (Fig. 8 and 9).
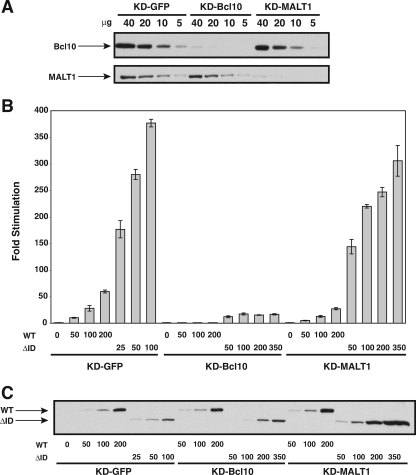
We therefore tested whether the Bcl10 that is endogenously expressed in HEK293T cells was required for the observed association between the hyperactive ΔID and TAK1, TRAF6, caspase-8 C360S, or IKKγ. We infected HEK293T cells with a lentivirus that stably expresses an shRNA designed to knock down human Bcl10 by RNAi (see Materials and Methods for experimental details). As shown in Fig. 10A, expression of this hairpin resulted in approximately 90% knockdown of endogenous Bcl10 expression in this cell line (KD-Bcl10) compared to a control line infected with a lentivirus expressing a hairpin designed to knock down GFP (KD-GFP). As expected, this reduction in Bcl10 expression severely impaired the ability of the ΔID or wild-type CARD11 to activate the Igκ2-IFN-LUC reporter when expressed at levels of protein expression comparable to that in the KD-GFP control line (Fig. 10B and C). We then determined whether this reduction in Bcl10 levels, which so drastically impaired ΔID signaling to NF-κB, would affect the ability of the ΔID to bind to other signaling cofactors. Surprisingly, there was no significant difference in the ability of the ΔID to interact with TAK1 (Fig. 11A), TRAF6 (Fig. 11B), IKKγ (Fig. 12A), or caspase-8 C360S (Fig. 12B) in the Bcl10-deficient line compared to interaction observed in control KD-GFP line. These experiments were conducted using subsaturating levels of the interacting proteins in order to maximize the detection of any effect of Bcl10 deficiency. Consistent with a Bcl10-independent association of these proteins with the ΔID, we did not detect Bcl10 in the immunoprecipitated complexes that contained the ΔID and either TAK1, caspase-8 C360S, TRAF6, or IKKγ in either the KD-GFP or KD-Bcl10 lines (Fig. 11 and 12). These data indicate that the hyperactive ΔID, which corresponds to CARD11 that has been activated by signal-induced ID neutralization, has an enhanced ability to associate with these cofactors that does not depend on Bcl10.
FIG. 10.
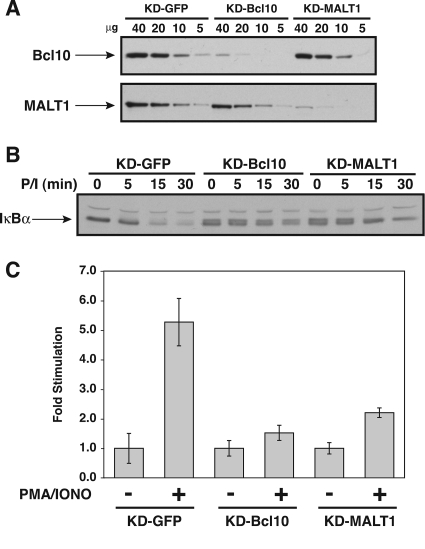
Stable knockdown of Bcl10 and MALT1 in HEK293T cell lines. HEK293T cells were infected with lentiviruses expressing shRNAs that target either Bcl10, MALT1, or GFP as a control, as described in Materials and Methods, to establish the KD-GFP, KD-Bcl10, and KD-MALT1 cell lines. (A) The indicated amounts (in μg) of stably infected cell lysates were assayed by Western blotting for Bcl10 and MALT1 protein levels using anti-Bcl10 and anti-MALT1 primary antibodies. (B) The KD-GFP, KD-Bcl10, and KD-MALT1 cell lines were transfected with 6 ng of pCSK-LacZ, 20 ng of Igκ2-IFN-LUC, and the indicated amounts (in ng) of expression vectors for myc-tagged wild-type CARD11 or the ΔID variant. Stimulation was determined as described in Materials and Methods. (C) The lysates from samples in panel B were probed by Western blotting using anti-myc primary antibody to indicate the relative expression level of each variant. The bars represent the mean values of triplicate samples, and error bars indicate the standard deviations. WT, wild type.
We also determined whether the MALT1 that is endogenously expressed in HEK293T cells was required for the observed association between the hyperactive ΔID and TAK1, TRAF6, caspase-8 C360S, or IKKγ. HEK293T cells were infected with a lentivirus expressing a hairpin that targets the human MALT1 mRNA (see Materials and Methods for experimental details). As shown in Fig. 10A, infection with this virus resulted in a cell line, KD-MALT1, that demonstrated a ∼87% reduction in MALT1 protein levels. Interestingly, this reduction in MALT1 expression resulted in only a mild, approximately twofold reduction in the ability of wild-type CARD11 or the ΔID to activate the Igκ2-IFN-LUC reporter (Fig. 10B) at equivalent levels of protein expression in the KD-GFP and KD-MALT1 lines (Fig. 10C). This result indicates either that the residual levels of MALT1 (∼13% of wild-type levels) are largely sufficient to conduct CARD11-mediated signaling to NF-κB or that MALT1 is not strictly required in this context for CARD11-mediated signaling to NF-κB. Interestingly, Ferch et al. (16) have recently shown that in B cells MALT1 is dispensable for CARD11-mediated IKK activation and for the activation of RelA-containing NF-κB complexes. In contrast, MALT1 is required for B-cell receptor-mediated activation of c-Rel-containing NF-κB heterodimers. It is possible that HEK293T cells rely mainly on RelA-containing heterodimers for NF-κB activity.
As shown in Fig. 11 and 12, the ∼87% reduction in MALT1 protein levels did not affect the ability of the ΔID to interact with TAK1, TRAF6, caspase-8 C360S, or IKKγ. Additionally, we did not detect MALT1 in the immunoprecipitated complexes that contained the ΔID and either TAK1, TRAF6, caspase-8 C360S, or IKKγ in either the KD-GFP, KD-Bcl10, or KD-MALT1 lines. These data indicate that an eightfold reduction in MALT1 levels does not significantly affect the ability of the ΔID to interact with these proteins under these conditions.
Differential dependence on Bcl10 and MALT1 for the signal-induced association of CARD11 with TRAF6, the IKK complex, and caspase-8 in T cells.
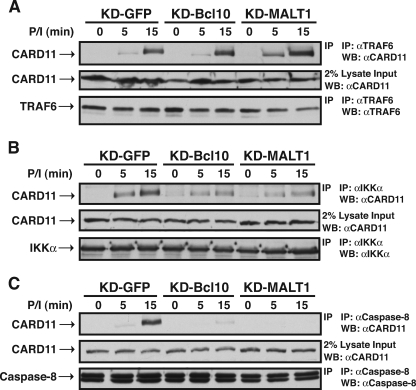
We then determined whether Bcl10 and MALT1 were required for the signal-induced association of cofactors with CARD11 in T cells. We infected Jurkat T cells with the same lentiviruses used in the experiment shown in Fig. 10 and obtained Jurkat lines that exhibited ∼83% reduced expression of Bcl10 (KD-Bcl10) and MALT1 (KD-MALT1) (Fig. 13A). Each of these lines was defective in PMA/ionomycin-induced activation of NF-κB, as judged by IκBα degradation (Fig. 13B) and Igκ2-IFN-LUC reporter stimulation (Fig. 13C), indicating that this level of knockdown of either Bcl10 or MALT1 in Jurkat T cells impaired CARD11-dependent signaling. As shown in Fig. 14A, despite the 83% reduction in Bcl10 or MALT1, there was no defect in the PMA/ionomycin-induced association between CARD11 and TRAF6 in the KD-Bcl10 or KD-MALT1 Jurkat T cell lines, as indicated by the coimmunoprecipitation of CARD11 with TRAF6 using anti-TRAF6 antibodies. This result confirmed that, at endogenous protein concentrations in T cells, neither Bcl10 nor MALT1 is required for the signal-induced association of CARD11 and TRAF6.
FIG. 13.
Stable knockdown of Bcl10 and MALT1 in Jurkat T cell lines. Jurkat T cells were infected with lentiviruses expressing shRNAs that target either Bcl10, MALT1, or GFP as a control, as described in Materials and Methods, to establish the KD-GFP, KD-Bcl10, and KD-MALT1 cell lines. (A) The indicated amounts (in μg) of stably infected cell lysates were assayed by Western blotting for Bcl10 and MALT1 protein levels using anti-Bcl10 and anti-MALT1 primary antibodies. (B) The KD-GFP, KD-Bcl10, and KD-MALT1 cell lines were stimulated with PMA (50 ng/ml) and ionomycin (1 μM) (P/I) for the indicated times. Lysates were assayed by Western blotting for IκBα degradation using anti-IκBα primary antibody. (C) The KD-GFP, KD-Bcl10, and KD-MALT1 cell lines were transfected with 200 ng of pCSK-LacZ, 1,500 ng of Igκ2-IFN-LUC, and 1,300 ng of pBSKII+ and stimulated with 50 ng/ml PMA and 1 μM ionomycin for 1 h. Stimulation was determined as described in Materials and Methods. The bars represent the mean values of triplicate samples, and error bars indicate the standard deviations.
FIG. 14.
Effect of Bcl10 and MALT1 knockdown on the inducible association of CARD11 with cofactors in Jurkat T cells. The KD-GFP, KD-Bcl10, and KD-MALT1 cell lines were stimulated with a time course of PMA-ionomycin (P/I) as indicated. Anti-TRAF6 (A), anti-IKKα (B), and anti-caspase-8 (C) immunoprecipitations were performed as described in Materials and Methods. In each panel, Western blotting (WB) with the indicated antibodies (at right) was performed to develop the contents of the immunoprecipitate (top and bottom) and the lysate input (middle). α, anti; IP, immunoprecipitation.
We assessed the association of the IKK complex with CARD11 in these lines by immunoprecipitation with anti-IKKα antibodies. As shown in Fig. 14B, the signal-induced association of the IKK complex with CARD11 was only partially reduced in the KD-Bcl10 and KD-MALT1 lines, indicating that the recruitment of the IKK complex to CARD11 is largely independent of Bcl10 and MALT1.
In sharp contrast, the inducible coimmunoprecipitation of CARD11 with caspase-8 was severely affected by the knockdown of either Bcl10 or MALT1 (Fig. 14C), indicating that both Bcl10 and MALT1 are required for caspase-8 to stably associate with CARD11 during signaling. These results indicate that at endogenous levels of expression in T cells, during the signal-induced conversion of CARD11 to an active scaffold, CARD11 can recruit cofactors independently of one another in a manner that differentially depends on Bcl10 and MALT1.
DISCUSSION
CARD11 is a critical adapter molecule that functions to couple the recognition of antigen and costimulatory signals at the surface of the T cell to the induction of NF-κB activity that is required for T-cell activation and proliferation in the adaptive immune response. As a signaling adapter, CARD11 must contain determinants that allow it to receive pathway-specific upstream signals and transmit the information to non-pathway-specific molecules that are used widely for the activation of the IKK complex and NF-κB.
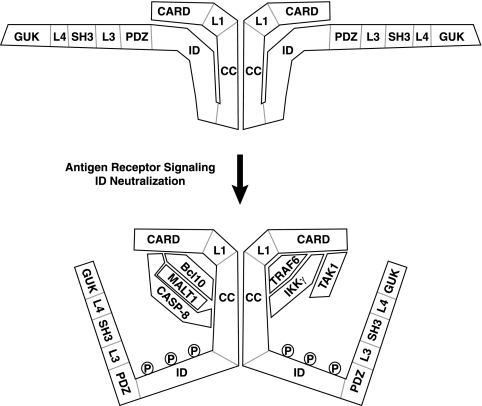
Our results illuminate how CARD11 is converted from an inactive state to an active signaling scaffold so that upstream TCR signaling is translated into the assembly of an active signaling complex (Fig. 15). Prior to signaling, the ID interacts with the CARD and the coiled-coil domain to prevent the recruitment of signaling cofactors to CARD11. The interaction of the ID with the CARD and the coiled-coil domain may occur within the same CARD11 molecule or between two or more oligomerized molecules of CARD11. Signaling downstream of the TCR results in the neutralization of the activity of the ID, which requires the phosphorylation of the ID on serine residues 564 and 657 by PKCθ and on serine 577 by an undetermined kinase (32, 55). Once the ID is neutralized, the CARD, L1, and coiled-coil domains present surfaces for the recruitment of Bcl10, TAK1, TRAF6, caspase-8, and IKKγ. MALT1 is presumably recruited through its interaction with Bcl10, and caspase-8 stably associates during signaling in a Bcl10- and MALT1-dependent manner. These factors then function to promote the activation of the IKK complex and NF-κB, which other investigators have shown to involve the TRAF6-mediated ubiquitination of MALT1 (43) and IKKγ (59), the MALT1-mediated ubiquitination of IKKγ (73), the ubiquitination of Bcl10 by an undetermined ligase (71), the TAK1-mediated phosphorylation of IKKβ (52, 59), and the proteolytic activity of caspase-8 on an undefined substrate (58).
FIG. 15.
Model of the signal-induced conversion of CARD11 to an active signaling scaffold. Prior to TCR engagement, the ID of CARD11 interacts with the CARD and the coiled-coil (CC) domain to prevent the association of signaling cofactors. Although it is depicted here as an intramolecular interaction in the inhibited state, it is also possible that the ID interacts with the CARD and the coiled-coil domain of another CARD11 molecule in the oligomer. TCR signaling leads to the neutralization of ID activity, in part through the phosphorylation of the ID by PKCθ. Once the ID is neutralized, signaling cofactors can be recruited to the N-terminal portion of CARD11. The model indicates which domains of CARD11 are required for the association with the cofactors analyzed. TAK1 requires only the CARD, while Bcl10 requires the CARD and coiled-coil domain. TRAF6 requires the CARD and the L1 and coiled-coil domains, while caspase-8 and IKKγ each require the CARD and the coiled-coil domain. MALT1 is presumably recruited through interactions with Bcl10. Caspase-8 (CASP-8) requires both Bcl10 and MALT1 to associate with CARD11 during signaling. The association of TAK1, TRAF6, and IKKγ with activated CARD11 can occur in a Bcl10-independent and MALT1-independent manner. The ID does not appear to influence whether CARD11 oligomerizes, suggesting that oligomerization occurs both before and after signaling through a region of the coiled-coil domain that is not targeted by the ID. The depicted interactions between CARD11 and signaling cofactors may be direct or may require other proteins to bridge the associations.
It is important to note that our association studies do not distinguish between direct and indirect association between CARD11 and these cofactors since the experiments were performed using cell lysates. Other proteins that are present in HEK293T and Jurkat T cells may bridge the interactions we studied.
The determinants in CARD11 that control its association with Bcl10, TAK1, TRAF6, IKKγ, and caspase-8 lie within the CARD-L1-coiled-coil region (Fig. 15). The ΔID hyperactive variant, which corresponds to activated CARD11 in which the ID has been neutralized, required both the CARD and the coiled-coil domain to associate with Bcl10. The TAK1 association required only the CARD, while the TRAF6 association required the CARD, L1, and coiled-coil domains. The association of IKKγ and caspase-8 required the coiled-coil domain and, to a lesser extent, the CARD. Bcl10, TAK1, and IKKγ have previously been shown to associate with CARD11 in an inducible manner during antigen receptor signaling (17, 53, 56, 69). Our observation that the ΔID had an enhanced ability to associate with TRAF6 and caspase-8 compared to wild-type CARD11 predicted that both of these proteins would also associate with CARD11 in T cells in a signal-dependent manner. This was indeed the case (Fig. 6). Caspase-8 and TRAF6 have been shown to interact, and this interaction may be important for signaling events that occur subsequent to their recruitment to CARD11 (4).
Although the ID regulates the association of multiple cofactors with CARD11, it does not appear to regulate CARD11 oligomerization. Both wild-type and ΔID variants of CARD11 appeared to have equivalent capacities to self-associate (Fig. 7). We have shown that the coiled-coil domain is targeted by the ID, and Tanner et al. have demonstrated that the coiled-coil domain is required for CARD11 oligomerization (62). This indicates that CARD11 oligomerization is likely mediated by surfaces of the coiled-coil domain that are not engaged by the ID and is not influenced by signals that neutralize activity of the ID.
Recent models have proposed that during TCR signaling, Bcl10 is recruited to CARD11 and functions to recruit other signaling cofactors into a CARD11-containing complex so that IKK complex activation may proceed (43, 45, 51, 52, 63). Our data in HEK293T cells indicate that once the ID is neutralized, CARD11 has an intrinsic enhanced affinity for TAK1, TRAF6, caspase-8, and IKKγ and can recruit these molecules in a Bcl10-independent manner (Fig. 10 to 12).
In Jurkat T cells, the signal-induced recruitment of TRAF6 and the IKK complex to CARD11 were largely Bcl10 and MALT1 independent (Fig. 13 and 14), consistent with a Bcl10- and MALT1-independent ability of CARD11 to associate with these cofactors once the ID is neutralized by upstream signaling. In contrast, the inducible association of caspase-8 with CARD11 in T cells exhibited a much greater dependence on Bcl10 and MALT1 than the other factors examined (Fig. 14). This observation was surprising since the ΔID could associate with caspase-8 C360S in HEK293T cells in a manner that was not affected by the ∼90% reduction in Bcl10 or MALT1 levels. The difference might be explained by the fact that we assayed association in HEK293T cells using the catalytically inactive C360S variant of caspase-8. Caspase-8 is known to undergo conformational changes after the activation of its proteolytic activity (6). Since caspase-8 is activated during TCR signaling, it is likely that the associations we detected in Jurkat T cells involved catalytically active caspase-8. It is possible that the inactive conformation of caspase-8 can be recruited to CARD11 during signaling in a Bcl10-independent manner and then convert to the active conformation which stably associates with CARD11 through Bcl10 and MALT1. We were not able to reproducibly assay the signal-induced recruitment of TAK1 to CARD11 in Jurkat T cells with the available anti-TAK1 antibodies. However, the CARD11-TAK1 association was not as inhibited by the ID in cis as were the other cofactor associations, and TAK1 required only the CARD for association with the ΔID, while the other factors required other domains. These data strongly suggest that TAK1 is recruited to CARD11 in a manner that is independent of the other factors we studied.
The association of Bcl10, TAK1, TRAF6, IKKγ, and caspase-8 with the CARD and with the L1 and coiled-coil domains explains why these domains of CARD11 are required after the inhibitory activity of the ID is neutralized by upstream signals. We also demonstrated that each of the L3, SH3, L4, and GUK domains of CARD11 is also necessary for signaling after ID neutralization. Each of these domains is required for PMA/ionomycin-mediated signaling to NF-κB in T cells, and each is required for constitutive signaling by the CARD11 ΔID (Fig. 3).
The L3, SH3, L4, and GUK domains may mediate the recruitment of other factors required for IKK complex activation, may target CARD11 and associated factors to a cellular locale that is required for signaling, or may promote conformational states of the CARD and the coiled-coil domain that are required to bind cofactors that we did not study. All of these domains are required for the activity of the ΔID in both lymphoid and nonlymphoid cells, indicating that they function in concert with factors that are not specific to the antigen-receptor signaling pathway and do not depend on the specific spatial architecture of the immunological synapse. The SH3 domain has been shown to be involved in membrane recruitment of CARD11 (67), and it is likely that membrane recruitment of CARD11 is required for PKCθ to act on the ID. However, it is not clear whether SH3-mediated membrane association or a distinct function of the SH3 domain is required for events downstream of PKCθ action. The PDZ-L3-SH3 region of CARD10 (also called CARMA3) has been demonstrated in a yeast two-hybrid assay to present a binding surface for IKKγ (56); however, our results suggest that these domains in CARD11 are not required for IKKγ association, which requires the coiled-coil domain and CARD instead. The CARD11 GUK domain has been shown to bind PDK1 (27), a kinase that functions in this pathway in PKCθ activation and in recruitment of CARD11 to lipid rafts in T cells. Since we demonstrate a role for the GUK domain subsequent to ID neutralization, other GUK-binding proteins are likely to be involved in this pathway. Finally, the adhesion and degranulation-promoting adapter protein (ADAP) has been shown to interact with the C-terminal half of CARD11 containing the PDZ, L3, SH3, L4, and GUK domains (33). Since ADAP is required for TCR signaling to NF-κB, this protein may explain the requirement for one or more of these CARD11 domains in this pathway in T cells. However, since ADAP is expressed in a tissue-restricted fashion and since L3, SH3, L4, and GUK domains function in both lymphoid and nonlymphoid cells, other proteins are likely to interact with these domains during CARD11-mediated signaling.
The integrative signaling function of CARD11 that is required for normal antigen receptor signaling is also likely to play a critical role in the progression of some types of cancer. Lymphomas of mucosa-associated lymphoid tissue (MALT lymphoma) are associated with chromosomal translocations that lead to the overexpression of Bcl10 or MALT1 or to the expression of a hyperactive API2-MALT1 fusion protein (1, 12, 24, 31, 38, 57, 64, 70, 72). It is possible that in MALT lymphoma the association of these molecules with CARD11 is required for their dysregulated signaling to NF-κB, which is thought to contribute to lymphoma progression. The activated-B-cell-like subtype of diffuse large B-cell lymphoma displays aberrant activation of the IKK complex and NF-κB in a manner that is required for proliferation and survival (10, 26). Recently, CARD11 has been shown to be a required upstream signaling molecule in this lymphoma (42), and missense mutations in CARD11 have been found in ∼10% of activated-B-cell-like diffuse large B-cell lymphoma tumor biopsies (28). Intriguingly, several of these missense mutations increase the signaling ability of CARD11 and map to the coiled-coil domain (28). Based upon our studies, we suggest that oncogenic coiled-coil mutations could lead to enhanced signaling activity, most likely by disrupting the inhibitory interaction that we have described between the ID and the coiled-coil domain. Alternatively, one or more of these mutations could enhance the affinity of CARD11 for Bcl10, IKKγ, TRAF6, or caspase-8. Each of the surfaces that CARD11 uses to coordinate signaling to NF-κB may provide a target for therapeutic amelioration of lymphomas that depend on CARD11 signaling. Importantly, since CARD11 appears to be critical in a limited number of NF-κB-inducing pathways, pharmacological agents that target the scaffolding function of CARD11 might not perturb the important biological effects of the diverse array of physiological stimuli that regulate NF-κB in other contexts.
Supplementary Material
Acknowledgments
We thank V. Schowinsky for excellent technical assistance; M. Boldin, D. Baltimore, G. Nuñez, T. Kawai, S. Akira, and D. Goeddel for reagents; R. Lamason, S. Lew, V. Schowinsky, and J. McLellan for critical reading of the manuscript; and M. Meffert for helpful advice and discussions.
This work was supported by grant RSG-06-172-01-LIB from the American Cancer Society and funds from the Johns Hopkins Institute for Cell Engineering. J.L.P. is a recipient of a Kimmel Scholar Award from the Sidney Kimmel Foundation for Cancer Research and a Rita Allen Foundation Scholar.
Footnotes
Published ahead of print on 14 July 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Akagi, T., M. Motegi, A. Tamura, R. Suzuki, Y. Hosokawa, H. Suzuki, H. Ota, S. Nakamura, Y. Morishima, M. Taniwaki, and M. Seto. 1999. A novel gene, MALT1 at 18q21, is involved in t(11;18) (q21;q21) found in low-grade B-cell lymphoma of mucosa-associated lymphoid tissue. Oncogene 185785-5794. [DOI] [PubMed] [Google Scholar]
- 2.Bell, S., K. Degitz, M. Quirling, N. Jilg, S. Page, and K. Brand. 2003. Involvement of NF-κB signalling in skin physiology and disease. Cell Signal. 151-7. [DOI] [PubMed] [Google Scholar]
- 3.Bertin, J., L. Wang, Y. Guo, M. D. Jacobson, J. L. Poyet, S. M. Srinivasula, S. Merriam, P. S. DiStefano, and E. S. Alnemri. 2001. CARD11 and CARD14 are novel caspase recruitment domain (CARD)/membrane-associated guanylate kinase (MAGUK) family members that interact with BCL10 and activate NF-κB. J. Biol. Chem. 27611877-11882. [DOI] [PubMed] [Google Scholar]
- 4.Bidere, N., A. L. Snow, K. Sakai, L. Zheng, and M. J. Lenardo. 2006. Caspase-8 regulation by direct interaction with TRAF6 in T cell receptor-induced NF-κB activation. Curr. Biol. 161666-1671. [DOI] [PubMed] [Google Scholar]
- 5.Cannons, J. L., L. J. Yu, B. Hill, L. A. Mijares, D. Dombroski, K. E. Nichols, A. Antonellis, G. A. Koretzky, K. Gardner, and P. L. Schwartzberg. 2004. SAP regulates TH2 differentiation and PKC-θ-mediated activation of NF-κB1. Immunity 21693-706. [DOI] [PubMed] [Google Scholar]
- 6.Chang, D. W., Z. Xing, V. L. Capacio, M. E. Peter, and X. Yang. 2003. Interdimer processing mechanism of procaspase-8 activation. EMBO J. 224132-4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Che, T., Y. You, D. Wang, M. J. Tanner, V. M. Dixit, and X. Lin. 2004. MALT1/paracaspase is a signaling component downstream of CARMA1 and mediates T cell receptor-induced NF-κB activation. J. Biol. Chem. 27915870-15876. [DOI] [PubMed] [Google Scholar]
- 8.Condie, B. G., A. H. Brivanlou, and R. M. Harland. 1990. Most of the homeobox-containing Xhox 36 transcripts in early Xenopus embryos cannot encode a homeodomain protein. Mol. Cell. Biol. 103376-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costello, P. S., A. E. Walters, P. J. Mee, M. Turner, L. F. Reynolds, A. Prisco, N. Sarner, R. Zamoyska, and V. L. Tybulewicz. 1999. The Rho-family GTP exchange factor Vav is a critical transducer of T cell receptor signals to the calcium, ERK, and NF-κB pathways. Proc. Natl. Acad. Sci. USA 963035-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis, R. E., K. D. Brown, U. Siebenlist, and L. M. Staudt. 2001. Constitutive nuclear factor κB activity is required for survival of activated B cell-like diffuse large B cell lymphoma cells. J. Exp. Med. 1941861-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dienz, O., A. Moller, A. Strecker, N. Stephan, P. H. Krammer, W. Droge, and M. L. Schmitz. 2003. Src homology 2 domain-containing leukocyte phosphoprotein of 76 kDa and phospholipase Cγ1 are required for NF-κB activation and lipid raft recruitment of protein kinase Cθ induced by T cell costimulation. J. Immunol. 170365-372. [DOI] [PubMed] [Google Scholar]
- 12.Dierlamm, J., M. Baens, I. Wlodarska, M. Stefanova-Ouzounova, J. M. Hernandez, D. K. Hossfeld, C. De Wolf-Peeters, A. Hagemeijer, H. Van den Berghe, and P. Marynen. 1999. The apoptosis inhibitor gene API2 and a novel 18q gene, MLT, are recurrently rearranged in the t(11;18)(q21;q21) associated with mucosa-associated lymphoid tissue lymphomas. Blood 933601-3609. [PubMed] [Google Scholar]
- 13.Dimitratos, S. D., D. F. Woods, D. G. Stathakis, and P. J. Bryant. 1999. Signaling pathways are focused at specialized regions of the plasma membrane by scaffolding proteins of the MAGUK family. Bioessays 21912-921. [DOI] [PubMed] [Google Scholar]
- 14.Dull, T., R. Zufferey, M. Kelly, R. J. Mandel, M. Nguyen, D. Trono, and L. Naldini. 1998. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 728463-8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egawa, T., B. Albrecht, B. Favier, M. J. Sunshine, K. Mirchandani, W. O'Brien, M. Thome, and D. R. Littman. 2003. Requirement for CARMA1 in antigen receptor-induced NF-κB activation and lymphocyte proliferation. Curr. Biol. 131252-1258. [DOI] [PubMed] [Google Scholar]
- 16.Ferch, U., C. M. zum Buschenfelde, A. Gewies, E. Wegener, S. Rauser, C. Peschel, D. Krappmann, and J. Ruland. 2007. MALT1 directs B cell receptor-induced canonical nuclear factor-κB signaling selectively to the c-Rel subunit. Nat. Immunol. 8984-991. [DOI] [PubMed] [Google Scholar]
- 17.Gaide, O., B. Favier, D. F. Legler, D. Bonnet, B. Brissoni, S. Valitutti, C. Bron, J. Tschopp, and M. Thome. 2002. CARMA1 is a critical lipid raft-associated regulator of TCR-induced NF-κB activation. Nat. Immunol. 3836-843. [DOI] [PubMed] [Google Scholar]
- 18.Gaide, O., F. Martinon, O. Micheau, D. Bonnet, M. Thome, and J. Tschopp. 2001. Carma1, a CARD-containing binding partner of Bcl10, induces Bcl10 phosphorylation and NF-κB activation. FEBS Lett. 496121-127. [DOI] [PubMed] [Google Scholar]
- 19.Gerondakis, S., R. Grumont, R. Gugasyan, L. Wong, I. Isomura, W. Ho, and A. Banerjee. 2006. Unravelling the complexities of the NF-κB signalling pathway using mouse knockout and transgenic models. Oncogene 256781-6799. [DOI] [PubMed] [Google Scholar]
- 20.Hara, H., C. Bakal, T. Wada, D. Bouchard, R. Rottapel, T. Saito, and J. M. Penninger. 2004. The molecular adapter Carma1 controls entry of IκB kinase into the central immune synapse. J. Exp. Med. 2001167-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hara, H., T. Wada, C. Bakal, I. Kozieradzki, S. Suzuki, N. Suzuki, M. Nghiem, E. K. Griffiths, C. Krawczyk, B. Bauer, F. D'Acquisto, S. Ghosh, W. C. Yeh, G. Baier, R. Rottapel, and J. M. Penninger. 2003. The MAGUK family protein CARD11 is essential for lymphocyte activation. Immunity 18763-775. [DOI] [PubMed] [Google Scholar]
- 22.Hayden, M. S., A. P. West, and S. Ghosh. 2006. NF-κB and the immune response. Oncogene 256758-6780. [DOI] [PubMed] [Google Scholar]
- 23.Herndon, T. M., X. C. Shan, G. C. Tsokos, and R. L. Wange. 2001. ZAP-70 and SLP-76 regulate protein kinase C-θ and NF-κB activation in response to engagement of CD3 and CD28. J. Immunol. 1665654-5664. [DOI] [PubMed] [Google Scholar]
- 24.Isaacson, P. G., and M. Q. Du. 2004. MALT lymphoma: from morphology to molecules. Nat. Rev. Cancer 4644-653. [DOI] [PubMed] [Google Scholar]
- 25.Jun, J. E., L. E. Wilson, C. G. Vinuesa, S. Lesage, M. Blery, L. A. Miosge, M. C. Cook, E. M. Kucharska, H. Hara, J. M. Penninger, H. Domashenz, N. A. Hong, R. J. Glynne, K. A. Nelms, and C. C. Goodnow. 2003. Identifying the MAGUK protein Carma-1 as a central regulator of humoral immune responses and atopy by genome-wide mouse mutagenesis. Immunity 18751-762. [DOI] [PubMed] [Google Scholar]
- 26.Lam, L. T., R. E. Davis, J. Pierce, M. Hepperle, Y. Xu, M. Hottelet, Y. Nong, D. Wen, J. Adams, L. Dang, and L. M. Staudt. 2005. Small molecule inhibitors of IκB kinase are selectively toxic for subgroups of diffuse large B-cell lymphoma defined by gene expression profiling. Clin. Cancer Res. 1128-40. [PubMed] [Google Scholar]
- 27.Lee, K. Y., F. D'Acquisto, M. S. Hayden, J. H. Shim, and S. Ghosh. 2005. PDK1 nucleates T cell receptor-induced signaling complex for NF-κB activation. Science 308114-118. [DOI] [PubMed] [Google Scholar]
- 28.Lenz, G., R. E. Davis, V. N. Ngo, L. Lam, T. C. George, G. W. Wright, S. S. Dave, H. Zhao, W. Xu, A. Rosenwald, G. Ott, H. K. Muller-Hermelink, R. D. Gascoyne, J. M. Connors, L. M. Rimsza, E. Campo, E. S. Jaffe, J. Delabie, E. B. Smeland, R. I. Fisher, W. C. Chan, and L. M. Staudt. 2008. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science 3191676-1679. [DOI] [PubMed] [Google Scholar]
- 29.Liu, H. H., M. Xie, M. D. Schneider, and Z. J. Chen. 2006. Essential role of TAK1 in thymocyte development and activation. Proc. Natl. Acad. Sci. USA 10311677-11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lois, C., E. J. Hong, S. Pease, E. J. Brown, and D. Baltimore. 2002. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science 295868-872. [DOI] [PubMed] [Google Scholar]
- 31.Lucas, P. C., M. Yonezumi, N. Inohara, L. M. McAllister-Lucas, M. E. Abazeed, F. F. Chen, S. Yamaoka, M. Seto, and G. Nunez. 2001. Bcl10 and MALT1, independent targets of chromosomal translocation in malt lymphoma, cooperate in a novel NF-κB signaling pathway. J. Biol. Chem. 27619012-19019. [DOI] [PubMed] [Google Scholar]
- 32.Matsumoto, R., D. Wang, M. Blonska, H. Li, M. Kobayashi, B. Pappu, Y. Chen, and X. Lin. 2005. Phosphorylation of CARMA1 plays a critical role in T cell receptor-mediated NF-κB activation. Immunity 23575-585. [DOI] [PubMed] [Google Scholar]
- 33.Medeiros, R. B., B. J. Burbach, K. L. Mueller, R. Srivastava, J. J. Moon, S. Highfill, E. J. Peterson, and Y. Shimizu. 2007. Regulation of NF-κB in T cells via association of the adapter proteins ADAP and CARMA1. Science 316754-758. [DOI] [PubMed] [Google Scholar]
- 34.Meffert, M. K., and D. Baltimore. 2005. Physiological functions for brain NF-κB. Trends Neurosci. 2837-43. [DOI] [PubMed] [Google Scholar]
- 35.Moffat, J., D. A. Grueneberg, X. Yang, S. Y. Kim, A. M. Kloepfer, G. Hinkle, B. Piqani, T. M. Eisenhaure, B. Luo, J. K. Grenier, A. E. Carpenter, S. Y. Foo, S. A. Stewart, B. R. Stockwell, N. Hacohen, W. C. Hahn, E. S. Lander, D. M. Sabatini, and D. E. Root. 2006. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 1241283-1298. [DOI] [PubMed] [Google Scholar]
- 36.Monks, C. R., B. A. Freiberg, H. Kupfer, N. Sciaky, and A. Kupfer. 1998. Three-dimensional segregation of supramolecular activation clusters in T cells. Nature 39582-86. [DOI] [PubMed] [Google Scholar]
- 37.Monks, C. R., H. Kupfer, I. Tamir, A. Barlow, and A. Kupfer. 1997. Selective modulation of protein kinase C-θ during T-cell activation. Nature 38583-86. [DOI] [PubMed] [Google Scholar]
- 38.Morgan, J. A., Y. Yin, A. D. Borowsky, F. Kuo, N. Nourmand, J. I. Koontz, C. Reynolds, L. Soreng, C. A. Griffin, F. Graeme-Cook, N. L. Harris, D. Weisenburger, G. S. Pinkus, J. A. Fletcher, and J. Sklar. 1999. Breakpoints of the t(11;18)(q21;q21) in mucosa-associated lymphoid tissue (MALT) lymphoma lie within or near the previously undescribed gene MALT1 in chromosome 18. Cancer Res. 596205-6213. [PubMed] [Google Scholar]
- 39.Naldini, L., U. Blomer, F. H. Gage, D. Trono, and I. M. Verma. 1996. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc. Natl. Acad. Sci. USA 9311382-11388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Naviaux, R. K., E. Costanzi, M. Haas, and I. M. Verma. 1996. The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses. J. Virol. 705701-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Newton, K., and V. M. Dixit. 2003. Mice lacking the CARD of CARMA1 exhibit defective B lymphocyte development and impaired proliferation of their B and T lymphocytes. Curr. Biol. 131247-1251. [DOI] [PubMed] [Google Scholar]
- 42.Ngo, V. N., R. E. Davis, L. Lamy, X. Yu, H. Zhao, G. Lenz, L. T. Lam, S. Dave, L. Yang, J. Powell, and L. M. Staudt. 2006. A loss-of-function RNA interference screen for molecular targets in cancer. Nature 441106-110. [DOI] [PubMed] [Google Scholar]
- 43.Oeckinghaus, A., E. Wegener, V. Welteke, U. Ferch, S. C. Arslan, J. Ruland, C. Scheidereit, and D. Krappmann. 2007. Malt1 ubiquitination triggers NF-κB signaling upon T-cell activation. EMBO J. 264634-4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pomerantz, J. L., E. M. Denny, and D. Baltimore. 2002. CARD11 mediates factor-specific activation of NF-κB by the T cell receptor complex. EMBO J. 215184-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rawlings, D. J., K. Sommer, and M. E. Moreno-Garcia. 2006. The CARMA1 signalosome links the signalling machinery of adaptive and innate immunity in lymphocytes. Nat. Rev. Immunol. 6799-812. [DOI] [PubMed] [Google Scholar]
- 46.Ruefli-Brasse, A. A., D. M. French, and V. M. Dixit. 2003. Regulation of NF-κB-dependent lymphocyte activation and development by paracaspase. Science 3021581-1584. [DOI] [PubMed] [Google Scholar]
- 47.Ruland, J., G. S. Duncan, A. Elia, I. del Barco Barrantes, L. Nguyen, S. Plyte, D. G. Millar, D. Bouchard, A. Wakeham, P. S. Ohashi, and T. W. Mak. 2001. Bcl10 is a positive regulator of antigen receptor-induced activation of NF-κB and neural tube closure. Cell 10433-42. [DOI] [PubMed] [Google Scholar]
- 48.Ruland, J., G. S. Duncan, A. Wakeham, and T. W. Mak. 2003. Differential requirement for Malt1 in T and B cell antigen receptor signaling. Immunity 19749-758. [DOI] [PubMed] [Google Scholar]
- 49.Sato, S., H. Sanjo, T. Tsujimura, J. Ninomiya-Tsuji, M. Yamamoto, T. Kawai, O. Takeuchi, and S. Akira. 2006. TAK1 is indispensable for development of T cells and prevention of colitis by the generation of regulatory T cells. Int. Immunol. 181405-1411. [DOI] [PubMed] [Google Scholar]
- 50.Scheidereit, C. 2006. IκB kinase complexes: gateways to NF-κB activation and transcription. Oncogene 256685-6705. [DOI] [PubMed] [Google Scholar]
- 51.Schulze-Luehrmann, J., and S. Ghosh. 2006. Antigen-receptor signaling to nuclear factor kappa B. Immunity 25701-715. [DOI] [PubMed] [Google Scholar]
- 52.Shambharkar, P. B., M. Blonska, B. P. Pappu, H. Li, Y. You, H. Sakurai, B. G. Darnay, H. Hara, J. Penninger, and X. Lin. 2007. Phosphorylation and ubiquitination of the IκB kinase complex by two distinct signaling pathways. EMBO J. 261794-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shinohara, H., T. Yasuda, Y. Aiba, H. Sanjo, M. Hamadate, H. Watarai, H. Sakurai, and T. Kurosaki. 2005. PKC β regulates BCR-mediated IKK activation by facilitating the interaction between TAK1 and CARMA1. J. Exp. Med. 2021423-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Siebenlist, U., K. Brown, and E. Claudio. 2005. Control of lymphocyte development by nuclear factor-κB. Nat. Rev. Immunol. 5435-445. [DOI] [PubMed] [Google Scholar]
- 55.Sommer, K., B. Guo, J. L. Pomerantz, A. D. Bandaranayake, M. E. Moreno-Garcia, Y. L. Ovechkina, and D. J. Rawlings. 2005. Phosphorylation of the CARMA1 linker controls NF-κB activation. Immunity 23561-574. [DOI] [PubMed] [Google Scholar]
- 56.Stilo, R., D. Liguoro, B. Di Jeso, S. Formisano, E. Consiglio, A. Leonardi, and P. Vito. 2004. Physical and functional interaction of CARMA1 and CARMA3 with Iκ kinase γ-NFκB essential modulator. J. Biol. Chem. 27934323-34331. [DOI] [PubMed] [Google Scholar]
- 57.Streubel, B., A. Lamprecht, J. Dierlamm, L. Cerroni, M. Stolte, G. Ott, M. Raderer, and A. Chott. 2003. t(14;18)(q32;q21) involving IGH and MALT1 is a frequent chromosomal aberration in MALT lymphoma. Blood 1012335-2339. [DOI] [PubMed] [Google Scholar]
- 58.Su, H., N. Bidere, L. Zheng, A. Cubre, K. Sakai, J. Dale, L. Salmena, R. Hakem, S. Straus, and M. Lenardo. 2005. Requirement for caspase-8 in NF-κB activation by antigen receptor. Science 3071465-1468. [DOI] [PubMed] [Google Scholar]
- 59.Sun, L., L. Deng, C. K. Ea, Z. P. Xia, and Z. J. Chen. 2004. The TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MALT1 in T lymphocytes. Mol. Cell 14289-301. [DOI] [PubMed] [Google Scholar]
- 60.Sun, Z., C. W. Arendt, W. Ellmeier, E. M. Schaeffer, M. J. Sunshine, L. Gandhi, J. Annes, D. Petrzilka, A. Kupfer, P. L. Schwartzberg, and D. R. Littman. 2000. PKC-theta is required for TCR-induced NF-κB activation in mature but not immature T lymphocytes. Nature 404402-407. [DOI] [PubMed] [Google Scholar]
- 61.Suzuki, N., S. Suzuki, D. G. Millar, M. Unno, H. Hara, T. Calzascia, S. Yamasaki, T. Yokosuka, N. J. Chen, A. R. Elford, J. Suzuki, A. Takeuchi, C. Mirtsos, D. Bouchard, P. S. Ohashi, W. C. Yeh, and T. Saito. 2006. A critical role for the innate immune signaling molecule IRAK-4 in T cell activation. Science 3111927-1932. [DOI] [PubMed] [Google Scholar]
- 62.Tanner, M. J., W. Hanel, S. L. Gaffen, and X. Lin. 2007. CARMA1 coiled-coil domain is involved in the oligomerization and subcellular localization of CARMA1 and is required for T cell receptor-induced NF-κB activation. J. Biol. Chem. 28217141-17147. [DOI] [PubMed] [Google Scholar]
- 63.Thome, M., and R. Weil. 2007. Post-translational modifications regulate distinct functions of CARMA1 and BCL10. Trends Immunol. 28281-288. [DOI] [PubMed] [Google Scholar]
- 64.Uren, A. G., K. O'Rourke, L. A. Aravind, M. T. Pisabarro, S. Seshagiri, E. V. Koonin, and V. M. Dixit. 2000. Identification of paracaspases and metacaspases: two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol. Cell 6961-967. [DOI] [PubMed] [Google Scholar]
- 65.Wada, T., T. Nakashima, N. Hiroshi, and J. M. Penninger. 2006. RANKL-RANK signaling in osteoclastogenesis and bone disease. Trends Mol. Med. 1217-25. [DOI] [PubMed] [Google Scholar]
- 66.Wan, Y. Y., H. Chi, M. Xie, M. D. Schneider, and R. A. Flavell. 2006. The kinase TAK1 integrates antigen and cytokine receptor signaling for T cell development, survival and function. Nat. Immunol. 7851-858. [DOI] [PubMed] [Google Scholar]
- 67.Wang, D., R. Matsumoto, Y. You, T. Che, X. Y. Lin, S. L. Gaffen, and X. Lin. 2004. CD3/CD28 costimulation-induced NF-κB activation is mediated by recruitment of protein kinase C-θ, Bcl10, and IκB kinase β to the immunological synapse through CARMA1. Mol. Cell. Biol. 24164-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang, D., Y. You, S. M. Case, L. M. McAllister-Lucas, L. Wang, P. S. DiStefano, G. Nunez, J. Bertin, and X. Lin. 2002. A requirement for CARMA1 in TCR-induced NF-κB activation. Nat. Immunol. 3830-835. [DOI] [PubMed] [Google Scholar]
- 69.Wegener, E., A. Oeckinghaus, N. Papadopoulou, L. Lavitas, M. Schmidt-Supprian, U. Ferch, T. W. Mak, J. Ruland, V. Heissmeyer, and D. Krappmann. 2006. Essential role for IκB kinase β in remodeling Carma1-Bcl10-Malt1 complexes upon T cell activation. Mol. Cell 2313-23. [DOI] [PubMed] [Google Scholar]
- 70.Willis, T. G., D. M. Jadayel, M. Q. Du, H. Peng, A. R. Perry, M. Abdul-Rauf, H. Price, L. Karran, O. Majekodunmi, I. Wlodarska, L. Pan, T. Crook, R. Hamoudi, P. G. Isaacson, and M. J. Dyer. 1999. Bcl10 is involved in t(1;14)(p22;q32) of MALT B cell lymphoma and mutated in multiple tumor types. Cell 9635-45. [DOI] [PubMed] [Google Scholar]
- 71.Wu, C. J., and J. D. Ashwell. 2008. NEMO recognition of ubiquitinated Bcl10 is required for T cell receptor-mediated NF-κB activation. Proc. Natl. Acad. Sci. USA 1053023-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang, Q., R. Siebert, M. Yan, B. Hinzmann, X. Cui, L. Xue, K. M. Rakestraw, C. W. Naeve, G. Beckmann, D. D. Weisenburger, W. G. Sanger, H. Nowotny, M. Vesely, E. Callet-Bauchu, G. Salles, V. M. Dixit, A. Rosenthal, B. Schlegelberger, and S. W. Morris. 1999. Inactivating mutations and overexpression of BCL10, a caspase recruitment domain-containing gene, in MALT lymphoma with t(1;14)(p22;q32). Nat. Genet. 2263-68. [DOI] [PubMed] [Google Scholar]
- 73.Zhou, H., I. Wertz, K. O'Rourke, M. Ultsch, S. Seshagiri, M. Eby, W. Xiao, and V. M. Dixit. 2004. Bcl10 activates the NF-κB pathway through ubiquitination of NEMO. Nature 427167-171. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.