Abstract
While early steps of gene expression, such as transcription preinitiation, are known to often be rate limiting and to be regulated by such stimuli as steroid hormones, the potential impact of downstream steps, including splicing, on the mRNA production rate is unknown. In this work, we studied the effects of the transcriptional stimulus estradiol on cyclin D1, PS2, and c-fos gene expression by measuring the levels of RNA polymerase II on the DNA templates, the levels of nascent transcripts associated with RNA polymerase II, and the levels of unspliced, partially spliced, and fully spliced RNAs. We demonstrated that the efficiency of cotranscriptional splicing of the first intron was higher in the case of cyclin D1 than with PS2 and potentiated the cyclin D1 mRNA production rate. The mechanism involved in cotranscriptional splicing depended on the level of serine 5 phosphorylation of RNA polymerase II at the gene 5′ end and on the recruitment of CBP80, one of the two subunits of the cap binding complex, which stimulates splicing of the promoter-proximal intron. Our data indicate that mRNA production from a subset of estradiol-stimulated genes, such as cyclin D1, could occur in a very efficient “assembly line.” In contrast, we demonstrated for the first time that despite a strong transcriptional activation of the PS2 gene, the production of mRNA is not optimized owing to inefficient cotranscriptional RNA processing.
Gene expression plays a key role in stimulus-dependent regulation of cellular metabolism and fate. Gene expression is a multistep process starting in the nucleus with the synthesis of premessenger RNAs (pre-mRNAs) and with RNA processing (including 5′- and 3′-end processing and splicing). The mature mRNAs are then exported to the cytosol, where they are translated. Many stimuli, such as steroid hormones, affect the cellular levels of various mRNAs by essentially modulating the transcriptional activities of their target genes. Indeed, steroid hormones (e.g., estrogens) bind to intracellular receptors, which act as ligand-dependent transcription factors and belong to the nuclear receptor superfamily (for reviews, see references 19 and 34). When activated by ligands, nuclear receptors bind to their target gene promoters and serve as platforms for the subsequent recruitment of transcriptional coregulators (for a recent review, see reference 33). With few exceptions (1, 26, 49), most of the efforts to understand the effects of steroid hormones on mRNA production by their target genes have been made by studying their impact on early steps of the transcriptional process. In this context, a large set of transcriptional coregulators has been shown to play a key role in transcription preinitiation by modulating the chromatin structure of the DNA templates and by recruiting RNA polymerase II (Pol II) (33).
However, the transitions between preinitiation, initiation, and transcription elongation can also be rate-limiting steps in various models (8, 43, 44). These transitions involve specific phosphorylations of the carboxy-terminal domain (CTD) of the large subunit of Pol II. The Pol II CTD is composed of 52 repeats of a conserved heptapeptide motif (YSPTSPS) that is subject to phosphorylation at serine 5 (Ser5) and serine 2 (Ser2) (39, 44). While unphosphorylated forms of Pol II are loaded on gene promoters, Ser5 and Ser2 phosphorylation must occur to permit transcription initiation and elongation, respectively (39, 44). In addition, although only a few studies have investigated this possibility, the processing of a subset of RNAs can be rate limiting under certain situations, as recently shown for yeast (41).
In this context, it is now widely accepted that transcription and RNA processing are connected. In particular, it has been shown that the Pol II CTD interacts with splicing factors and could be a landing platform favoring the interaction of these splicing factors with the nascent RNA (6, 14, 27, 36, 42). It has also been proposed that the coupling between transcription and splicing could enhance splicing efficiency (13, 18, 20). However, this is still a matter of debate (30). Importantly, although some reports have indicated that the splicing of a subset of pre-mRNAs occurs during transcription (29, 32, 47), cotranscriptional splicing is not mandatory (46, 47). Finally, despite some exceptions (4, 32), most studies on the coupling of transcription to splicing in metazoans have been done in vitro or using transfected minigenes and have not been done in the context of endogenous gene transcriptional activation by stimuli. Therefore, more studies are required to better understand the extent and potential physiological relevance of the coupling between transcription and splicing.
To test whether steps downstream of transcription preinitiation, particularly splicing, can influence the mRNA production rate in response to estrogens, we performed a time course analysis of the impact of estradiol on the expression levels of CCND1 (cyclin D1), PS2 (trefoil factor 1), and c-fos, which are three well-known estrogen target genes, by measuring successively Pol II levels and phosphorylation status on the DNA templates by using the chromatin immunoprecipitation (ChIP) assay, levels of unspliced and partially spliced nascent transcripts associated with Pol II by using the RNA-ChIP assay (23), and levels of the mature (i.e., fully spliced) RNA products. We demonstrated that the efficiency of splicing of the first intron during transcription was gene specific and potentiated the mRNA production rate. The efficiency of cotranscriptional splicing depended on the Pol II Ser5 phosphorylation level at the gene 5′ end and on the recruitment of CBP80, one of the two subunits of the cap binding complex involved in the recruitment of the U1 snRNP. Our data indicate that mRNA production from a subset of estradiol-stimulated genes, such as that for cyclin D1, occurs in a very efficient “assembly line,” while in the case of the PS2 gene, the mRNA production is not optimized because of inefficient cotranscriptional splicing.
MATERIALS AND METHODS
Cell culture, treatment, and antibodies.
MCF-7 cells were grown in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum at 37°C under 5% CO2. Before estradiol (E2) treatment, 2 × 106 cells were plated per 10-cm dish and were kept for 72 h in red phenol-free Dulbecco's modified Eagle medium supplemented with 2% charcoal-treated fetal bovine serum. Cells were treated with a final concentration of 10−8 M E2 for given time periods and/or with DRB (5,6-dichloro-β-d-ribofuranosyl benzimidazole), H8, and H7 (Sigma) at a final concentration of 25 μM for 1 h. Forty micrograms of proteins were used for Western blot analysis using PS2 (sc-28925), cyclin D1 (DCS6), and c-fos (sc-7202). Small interfering RNA (siRNA) targeting estrogen receptor alpha (ERα) from Dharmacon and CBP80 (35) was transfected using RNAiMax, following the manufacturer's instructions (Invitrogen).
RNA preparation and RT-PCR.
Nuclear and cytosolic fractions were prepared as previously described (3). RNAs were prepared using Trizol, and 1 μl of Glycoblue (Ambion) was added before RNA precipitation. Each RNA preparation was treated with DNase I (DNA-free; Ambion). Reverse transcription (RT) was performed with 0.1 to 1 μg of total RNA using Superscript II (Invitrogen) and random primers. The RT reactions were diluted to contain in fine 2.5 ng/μl of initial RNA, except where indicated in Fig. S1 posted at http://www.fast-db.com/SupplementalMaterialBittencourtMCB2231-07Vol28No18.pdf, and 2.5 μl of the diluted reverse transcriptase was used in the quantitative PCR (qPCR) mixtures. qPCR was performed using Master Sybr green I (Roche) on a Roche LightCycler Primers, PCR conditions, and absolute quantification by qPCR are described in Fig. S1 posted at the URL mentioned above. All qPCR results were standardized by measuring 18S RNA.
Immunoselection of RNAs with anti-Cap monoclonal antibody (H20) was performed as previously described (22).
RNA and chromatin immunoprecipitation.
A detailed protocol of the RNA-ChIP assay is provided in Fig. S3 posted at the URL mentioned above. Nuclear extracts from one dish of cells were always prepared in parallel to measure the nuclear levels of the different RNA molecules, and this was considered the input. The ChIP assay was performed as for the RNA-ChIP assay except that reverse cross-linking was done at 65°C overnight, and DNA was purified from the supernatant by using the QIAquick PCR purification kit (Qiagen). In addition to the CTD4H8 antibody (Upstate), we also used the 8WG16, H14, and H5 antibodies (ascites fluid from Covence) and control antibodies (mouse immunoglobulin G [IgG] or IgM). Immunoprecipitate (IP) (2.5 μl) or input (diluted 1:3) was used in 10-μl qPCR mixtures. A fraction of input was used to standardize the values obtained. The relative proportions of coimmunoprecipitated DNA or RNA fragments were determined on the basis of the threshold cycle (CT) for each PCR product. The data sets were normalized to input values (percent input; 2CT(input) − CT(IP) × 100). The effect of E2 was calculated by dividing the normalized value (percent input) obtained in the presence of E2 by the normalized value (percent input) obtained in the control experiments. The CBP80 ChIP was performed as previously described (32). Primers and qPCR conditions are described in Fig. S1 posted at the URL mentioned above.
Standard errors of the means (SEM) shown in figures are the standard deviations for the samples divided by the square root of the sample size. For results shown in each figure, at least three independent experiments were performed, ensuring statistical independence of the values in the sample.
RESULTS
Differential effects of estradiol on the expression levels of PS2 and cyclin D1 pre-mRNAs and mRNAs.
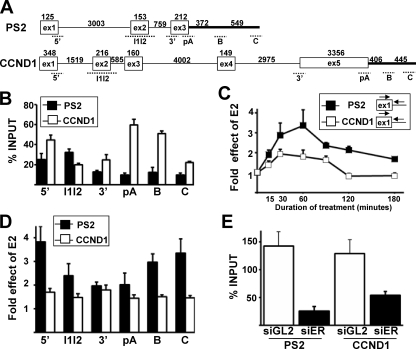
The cyclin D1 (CCND1) and trefoil factor 1 (PS2) genes are two well-characterized genes that are transcriptionally stimulated by E2 in MCF-7 cells (9, 16, 21). To investigate the effect of E2 on the CCND1 and PS2 gene transcriptional activity, the pattern of Pol II on these genes was analyzed using the CTD4H8 antibody, which recognizes both phosphorylated and unphosphorylated forms of Pol II (8), with the ChIP assay. The Pol II pattern on CCND1 and PS2 genes was analyzed by qPCR using several sets of primers that cover the entire length of the genes (Fig. 1A). More Pol II was detected on the CCND1 gene than on the PS2 gene in the absence of E2, as measured by normalizing the ChIP values to the input values (percent input) (Fig. 1B). Interestingly, a strong level of Pol II around the CCND1 gene polyadenylation (pA) site was detected, and this could be due to the presence of a transcriptional enhancer that has been characterized within this region (16).
FIG. 1.
E2 treatment induces a stronger increase in the Pol II levels on the PS2 gene than on the CCND1 gene. (A) Exon-intron structure of the PS2 and CCND1 genes. Exons are depicted as boxes, and introns and the 3′-flanking gene region are depicted as horizontal lines. The sizes of introns and exons are given in base pairs. Regions amplified by PCR are represented by dotted lines. (B) qPCR analysis of the Pol II levels on the PS2 (black) or CCND1 (white) gene at various locations in the absence of E2. The ChIP values are expressed as percentages of input. (C) qPCR analysis of the E2-mediated induction of Pol II levels on the PS2 (black) or CCND1 (white) gene 5′ ends measured by ChIP at given times (minutes) following the onset of E2 treatment. The values at baseline (0 h) were assigned 1. (D) Effect of E2 treatment for 1 h on the Pol II levels on the PS2 (black) or CCND1 (white) gene at various locations. The values at baseline (0 h) were assigned 1. (E) qPCR analysis of the Pol II levels at the PS2 and CCND1 gene 5′ ends in the presence of E2 for 1 h when cells were transfected with a control siRNA (siGL2, white) or an siRNA targeting ERα (siER, black). The ChIP values are expressed as percentages of input. Experiments were performed at least three times. Data are represented as means ± SEM.
A time course analysis of E2 treatment was then performed, and the percent input values obtained in the presence of E2 were divided by the percent input values obtained in the control experiments to measure the effect of E2 on the gene Pol II level. This time course analysis showed that E2 increased Pol II levels on both genes within 15 min (Fig. 1C). The effect of E2 was maximal between 30 and 60 min of treatment on both genes but lasted longer in the case of PS2 (Fig. 1C). In addition, the effect of E2 on Pol II levels after 1 h of treatment was stronger all along the PS2 gene than with the CCND1 gene (Fig. 1D). A marked high induction of the Pol II level downstream of the PS2 gene pA site was observed (positions B and C on Fig. 1D). This marked increase in the Pol II level downstream of the PS2 pA site in the presence of E2 might be related to 3′-end RNA processing (see Fig. 6). Altogether, these data indicated that E2 treatment induced a stronger increase in the Pol II levels on the PS2 gene than on the CCND1 gene. Finally, when cells were transfected with an siRNA targeting the ERα, the level of Pol II induced by a treatment with E2 for 1 h strongly decreased on both genes (Fig. 1E). These expected results indicated that E2-stimulated Pol II recruitment depended on ERα (9, 21, 40, 48).
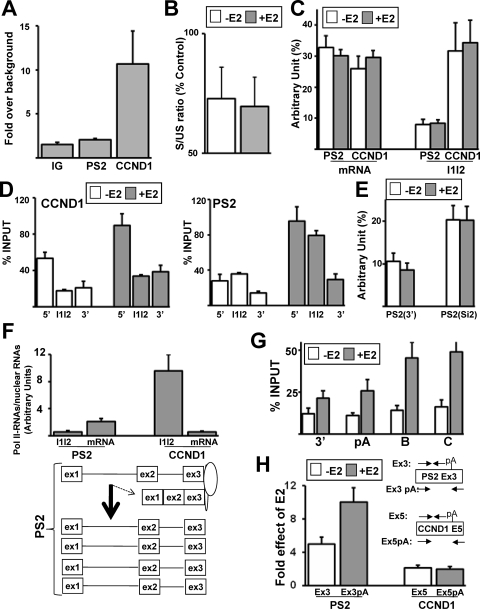
FIG. 6.
Differential cotranscriptional recruitment of CBP80 on PS2 and CCND1 genes. (A) qPCR analysis of the recruitment of CBP80 on the PS2 and CCND1 genes and in an intergenic region (IG) measured by ChIP in the presence of E2 for 1 h. (B) RT-qPCR analysis of the effect of an siRNA against CBP80 on the spliced-to-unspliced ratio both in the presence and in the absence of E2. Results are expressed as percentages of control values (transfection of the siGL2 control siRNA). (C) Nuclear RNAs were prepared from cells treated or not with E2 for 1 h. The levels of PS2 and CCND1 mRNAs or pre-mRNAs immunoprecipitated with anti-Cap antibody were measured and compared to their respective levels measured in the input (percent, arbitrary unit). (D) qPCR analysis of the Pol II levels on different regions of the PS2 gene in the absence (white) or presence (gray) of E2 for 1 h. The ChIP values are expressed as percentages of input. (E) Same as panel C, using different primer sets: PS2(3′) (see Fig. 1A) and PS2(Si2) (see Fig. 4D). (F) Proportion of Pol II-associated RNAs. MCF-7 cells were treated with E2 for 1 h and were used to perform the RNA-ChIP assay or to prepare nuclear extracts. PS2 and CCND1 pre-mRNAs (I1I2) and mRNAs were measured by RT-qPCR in the RNA-ChIP assay using the same fraction of immunoprecipitated RNAs and in the nuclear extracts using the same amount of nuclear RNAs. The value obtained in the RNA-ChIP assay was divided by the value obtained for nuclear extract. (G) Levels of Pol II (calculated as a percentage of input) determined by ChIP and qPCR analysis on different regions (3′, pA, B, and C, as shown in Fig. 1A) of PS2 genes in the absence (white) or presence (gray) of E2 for 1 h. (H) RT-qPCR analysis of E2 effects on the levels of PS2 and CCND1 RNAs in nuclear extracts using primers upstream of or flanking the RNA pA/cleavage site as indicated.
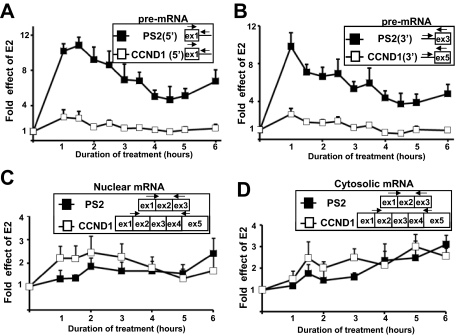
When measuring, using RT-qPCR, the PS2 and CCND1 pre-mRNA levels prepared from the nuclei of E2-treated cells, we next observed that E2 induced a stronger increase in the PS2 pre-mRNA level than in the CCND1 pre-mRNA level (Fig. 2A). After 1 h of treatment, where the effect of E2 was maximal, PS2 pre-mRNA levels increased by 10 times while CCND1 pre-mRNAs levels increased only by 2 to 3 times (Fig. 2A). In addition, the effect of E2 on the PS2 pre-mRNA levels lasted longer (Fig. 2A). Similar results were obtained with primers at the 3′ end of the transcripts (Fig. 2B), demonstrating that E2-stimulated Pol II synthesized full-length PS2 and CCND1 pre-mRNA molecules. The stronger effect of E2 on PS2 than on CCND1 pre-mRNA levels was consistent with the stronger E2-mediated increase in Pol II levels on the PS2 gene than on the CCND1 gene (Fig. 1C and D), although the differential effect of E2 on both genes was more pronounced when compared at the pre-mRNA levels.
FIG. 2.
Differential effects of E2 on the CCND1 and PS2 pre-mRNA and mRNA levels. (A) RT-qPCR analysis of E2 effects on the PS2 (black) or CCND1 (white) pre-mRNA levels in nuclear extracts using primers at the 5′ ends of the RNAs at given times (hours) following the onset of E2 treatment. (B) Same experimental conditions as for panel A, but 3′ regions of PS2 and D1 were analyzed. (C) RT-qPCR analysis of E2 effects on the PS2 (black) or CCND1 (white) mRNA levels in nuclear extracts at given times (hours) following the onset of E2 treatment. The amplification of fully spliced mRNAs was done using a forward primer overlapping the exon 1-exon 2 junction and a reverse primer in the last exon. (D) RT-qPCR analysis of effects of E2 on the PS2 (black) or CCND1 (white) mRNA levels in cytoplasmic (C) extracts at given times (hours) following the onset of E2 treatment. Each value was standardized by 18S RNA measurement, and the values at baseline (0 h) for each product were assigned 1. Experiments were performed three times. Data are represented as means ± SEM.
However, E2 treatment did not induce a stronger increase in the nuclear PS2 mRNA levels than in those of CCND1, as shown by RT-qPCR using primers that amplify the fully spliced mRNAs (Fig. 2C). The unexpected lesser increase in the nuclear level of the PS2 mRNA was not due to a faster export to the cytosol. Indeed, the cytosolic level of the PS2 mRNAs increased moderately and less rapidly than the cytosolic level of the CCND1 mRNA (Fig. 2D). In addition, the ratio between the nuclear and cytosolic PS2 mRNA concentrations was not affected by E2 treatment, and E2 treatment for 3 to 12 h induced similar and constant increases in the levels of cytosolic and nuclear PS2 mRNA (data not shown). This demonstrated that the nuclear and cytosolic levels of PS2 mRNAs increased with similar kinetics. Finally, the small increase in the PS2 nuclear mRNA level was not due to an excessively high steady-state level, because the nuclear PS2 pre-mRNAs represented a significant proportion of all nuclear PS2 RNAs (see Fig. S2 posted at http://www.fast-db.com/SupplementalMaterialBittencourtMCB2231-07Vol28No18.pdf). In conclusion, E2 treatment resulted in a stronger transcriptional activation of the PS2 gene than of the CCND1 gene but resulted in a lesser increase in PS2 mRNA levels than in CCND1 mRNA levels. This pointed out a possible lower rate of PS2 RNA processing than for CCND1.
Inefficient cotranscriptional splicing of PS2 intron 1.
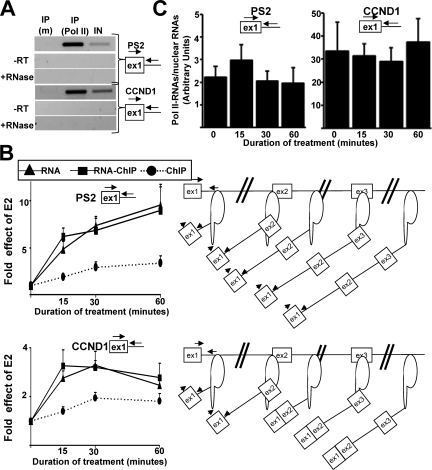
To further investigate the fate of the PS2 pre-mRNAs made in response to E2, we used an assay that has been referred to as RNA-ChIP (23) and that measures the levels of RNAs associated with Pol II. As detailed in Materials and Methods (see also Fig. S3 at the URL mentioned above), this assay is similar to the ChIP assay, but coimmunoprecipitated RNAs are purified instead of coimmunoprecipitated DNAs. Therefore, this assay measures the levels of RNAs associated with Pol II by using Pol II antibodies. As shown on Fig. 3A, the RNA-ChIP assay allowed specific immunoprecipitation of RNAs associated with Pol II using the CTD4H8 antibody (Fig. 3A). PCR signals were not due to potential DNA contaminations, because no PCR signals were detected in the absence of an RT reaction or when RNase was added before the RT reaction.
FIG. 3.
A smaller proportion of PS2 pre-mRNAs than CCND1 pre-mRNAs were associated with Pol II. (A) RT-PCR analysis of RNA immunoprecipitated by RNA-ChIP with either the CTD4H8 antibody against Pol II [IP (Pol II)] or a control mouse IgG antibody [IP (m)]. IN represents the input fraction. Samples treated with RNase before RT (+RNase) or where Superscript was not added to the RT reaction (−RT) did not show any PCR signal. (B) qPCR analysis of E2 effects on the Pol II levels for the PS2 and CCND1 genes (ChIP, black circles), pre-mRNA levels in nuclear extracts (RNA, black triangles), and Pol II-associated pre-mRNA levels (RNA-ChIP, black boxes) at given times (min) following the onset of E2 treatment. The controls for each assay are described in Materials and Methods. Experiments were performed three times. The values at baseline (0 h) for each product were assigned 1. (C) Proportion of Pol II-associated RNAs. MCF-7 cells were treated with E2 for different times (min) and were used to perform the RNA-ChIP assay or to prepare nuclear extracts. PS2 and CCND1 pre-mRNAs were measured by RT-qPCR in the RNA-ChIP assay using the same fraction of immunoprecipitated RNAs and in the nuclear extracts using the same amount of nuclear RNAs. The value obtained in the RNA-ChIP assay was divided by the value obtained in the nuclear extract. Experiments were performed at least three times. Data are represented as means ± SEM.
Because there was a peak in the PS2 pre-mRNA levels after E2 treatment for 1 h (Fig. 2A), we performed a time course analysis at 15, 30, and 60 min of E2 treatment and we compared the results obtained by the RNA-ChIP assays and by the ChIP assays. In addition, in the same set of experiments, we measured the nuclear level of the RNAs from which at least a proportion (if not all) is associated with Pol II. We observed that for both CCND1 and PS2, the increase in the pre-mRNA levels was similar in nuclear extracts and Pol II immunoprecipitation (Fig. 3B, RNA versus RNA-ChIP). Our previous observation that the PS2 pre-mRNA level increased more than the CCND1 pre-mRNA level after 1 h of E2 treatment (Fig. 2A) was confirmed by this analysis and was extended to very short durations of treatment (Fig. 3B, compare the PS2 and CCND1 panels). Interestingly, while the curves obtained from the ChIP and RNA-ChIP assays converged after 60 min of E2 treatment in the case of CCND1 (Fig. 3B, lower panel, RNA-ChIP versus ChIP), these curves diverged over time in the case of PS2 (Fig. 3B, upper panel, RNA-ChIP versus ChIP). This discrepancy pointed out a possible difference between CCND1 and PS2 intron 1 splicing.
Sonication, which is performed during both the ChIP and the RNA-ChIP protocols, results in DNA but not RNA fragmentation (see Fig. S4 posted at the URL mentioned above) (37). Therefore, only Pol II localized at the gene 5′ end permits immunoprecipitation of the DNA 5′ end of genes, whereas the pre-mRNA 5′ end can be immunoprecipitated by Pol II molecules located within the gene. In this context, one expects during transcriptional activation that although a small proportion of the Pol II molecules reside on the gene 5′ end, increasing amounts of Pol II distributed all along the gene permit immunoprecipitation of nascent RNAs having the first exon/intron; this may explain the discrepancy between the ChIP and RNA-ChIP curves obtained in the case of PS2 (Fig. 3B, upper panels). However, because the primers we used were at the boundary between exon 1 and intron 1, the discrepancy between RNA versus DNA 5′-end enrichment in the Pol II immunoprecipitation may also depend on the efficiency of cotranscriptional splicing. Indeed, a rapid splicing of intron 1 during transcription would result in a decrease of the PCR signal obtained from the RNAs. Therefore, only Pol II localized close to the gene 5′ end would permit immunoprecipitation of RNA bearing intron 1 in the case where splicing occurs during transcription. Consequently, in the case of CCND1 (Fig. 3B, lower panels), the convergence of the RNA-ChIP and ChIP curves after 30 min of E2 treatment could be explained by cotranscriptional splicing of intron 1 starting 15 to 30 min after the initial rise of pre-mRNA synthesis.
In addition, one can anticipate that a smaller proportion of unspliced pre-mRNAs would be associated with Pol II in the case where cotranscriptional splicing was inefficient. Indeed, unspliced pre-mRNA could, at least transiently, accumulate in the nucleus after transcription. We observed that the proportion of pre-mRNAs associated with Pol II was 15 times smaller in the case of PS2 than in that of CCND1 (Fig. 3C). Altogether, these results showed that compared to CCND1, increasing amounts of Pol II-associated PS2 RNAs contained intron 1 over time (Fig. 3B) and a larger proportion of nuclear PS2 pre-mRNAs were not associated with Pol II (Fig. 3C). This result raised the possibility that a larger proportion of PS2 RNAs containing intron 1 was released from Pol II and strengthened our previous conclusion that cotranscriptional splicing of PS2 intron 1 was less efficient than cotranscriptional splicing of CCND1 intron 1.
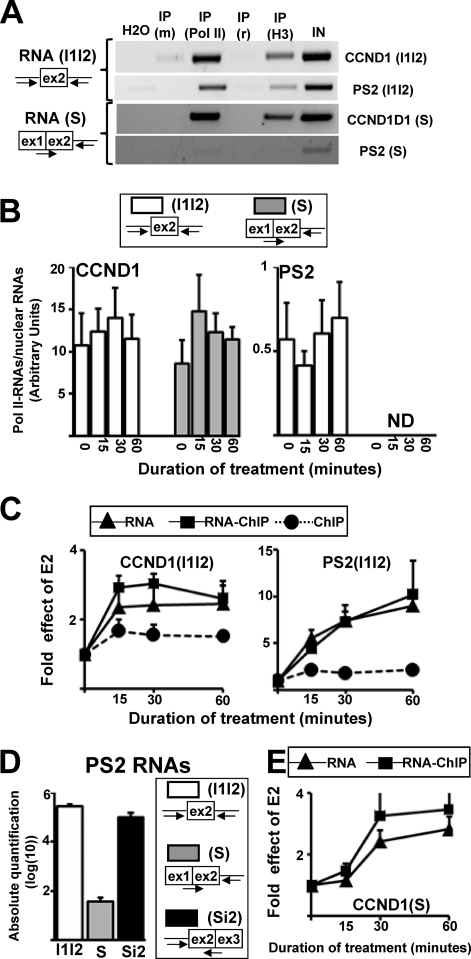
To further test this possibility, we next measured the levels of the partially spliced transcripts [CCND1(S) and PS2(S)], which corresponded to RNAs in which intron 1 was spliced but that still contained intron 2 (Fig. 4A). These RNA molecules were cloned and sequenced, and then we verified that the primers used to quantify the partially spliced RNAs did not amplify the unspliced RNAs (see Fig. S5 posted at the URL mentioned above). We also measured the levels of the unspliced CCND1(I1I2) and PS2(I1I2) RNAs, which contain both introns 1 and 2 (Fig. 4A). Using the RNA-ChIP assay described above, we first tested whether CCND1 and PS2 RNA splicing occurred in the close vicinity of the DNA template while transcripts were still associated with Pol II. To this end, we used antibodies against either Pol II or histone H3. As shown on Fig. 4A, unspliced CCND1(I1I2) and PS2(I1I2) RNAs, as well as the spliced CCND1(S) RNAs, were specifically immunoprecipitated using antibody against Pol II or H3. Meanwhile, we detected a very low level of the spliced PS2(S) RNAs even in the nuclear extracts, as further detailed below.
FIG. 4.
Inefficient cotranscriptional splicing of PS2 intron 1. (A) RT-PCR analysis of CCND1 and PS2 unspliced (I1I2) or partially spliced (S) RNAs immunoprecipitated by RNA-ChIP with control antibodies against mouse or rabbit Ig [IP (m) and IP (r), respectively] or with antibodies against Pol II [IP (Pol II)] or histone 3 [IP (H3)]. The input (IN) was obtained from purified nuclear RNAs. (B) Proportion of Pol II-associated RNAs. MCF-7 cells were treated with E2 for different times (min) and were used to perform the RNA-ChIP assay or to prepare nuclear extracts. Immunoprecipitated or nuclearly purified RNAs were used to quantify the PS2 or CCND1 (I1I2) and (S) transcripts. At each specific time, the value obtained in the RNA-ChIP assay was divided by the value obtained for the nuclear extracts. ND, not determined. (C) qPCR analysis of E2-mediated induction of Pol II levels on the CCND1 and PS2 genes (ChIP; black circles), pre-mRNA levels in nuclear extracts (RNA; black triangles), and Pol II-associated pre-mRNA levels (RNA-ChIP; black boxes) at given times (min) following the onset of E2 treatment. The values at baseline (0 h) for each product were assigned 1. Data are represented as means ± SEM. (D) Absolute quantification of PS2 unspliced (I1I2), partially spliced (S), and Si2 RNA (see Fig. S5 posted at http://www.fast-db.com/SupplementalMaterialBittencourtMCB2231-07Vol28No18.pdf). (E) qPCR analysis of E2-mediated induction of partially spliced CCND1(S) in nuclear extracts (RNA, black triangles) or associated with Pol II (RNA-ChIP, black boxes), as described for panel C.
We then quantified by RT-qPCR the levels of the unspliced and spliced RNAs that were associated with Pol II. Several major differences were observed between CCND1 and PS2. First, as already shown in Fig. 3C, the proportion of unspliced PS2 RNAs associated with Pol II was about 20 times smaller than the proportion of unspliced CCND1 RNAs associated with Pol II [comparing CCND1(I1I2) and PS2(I1I2)] (Fig. 4B). Second, while the unspliced and spliced CCND1 RNAs were enriched in a similar manner in the Pol II immunoprecipitation [Fig. 4B, left panel, compare CCND1(I1I2) and CCND1(S)], we could not quantify the PS2(S) RNAs associated with Pol II due to their very low level (Fig. 4B, right panel; also see below). Finally, as shown in Fig. 4C, the RNA-ChIP and ChIP curves diverged after the onset of E2 treatment in the case of PS2, which demonstrated that nascent PS2 RNAs were not spliced [compare RNA-ChIP and ChIP in the right panel, PS2(I1I2); Fig. 4C], in contrast to what we observed in the case of CCND1 (Fig. 4C). This observation was in agreement with our previous observations using primers at the RNA and DNA 5′ end (Fig. 3B). Altogether, these results demonstrated that a large proportion of nascent PS2 RNAs that were still associated with Pol II were not spliced, compared to CCND1 RNAs; because a large proportion of unspliced PS2 RNAs were not associated with Pol II, we concluded that cotranscriptional splicing of PS2 intron 1 was not efficient.
Moreover, the level of the spliced PS2(S) RNA was very low, as already mentioned (Fig. 4A and B). Because the order of intron removal is governed by preferential binding of splicing factors rather than being a sequential numerical order (25, 47), we also measured the levels of the RNAs labeled PS2(Si2), which contain intron 1 but not intron 2. By performing absolute quantification of the PS2(S), PS2(Si2), and PS2(I1I2) RNAs (see Fig. S5 posted at http://www.fast-db.com/SupplementalMaterialBittencourtMCB2231-07Vol28No18.pdf), we observed that the nuclear concentration of the PS2(S) RNA was more than 200 times lower than the concentration of the PS2(I1I2) and PS2(Si2) RNAs (Fig. 4D). Altogether these results demonstrated that cotranscriptional splicing of PS2 intron 1 was not as efficient as cotranscriptional splicing of CCND1 intron 1 and that PS2 intron 2 (which is a small intron, as shown in Fig. 1A) was more frequently removed before intron 1.
However, the PS2(Si2) RNAs were also poorly enriched in the Pol II immunoprecipitation (not shown). This raises the possibility that the partially spliced PS2 RNA molecules having intron 2 but not intron 1 spliced may be released from Pol II either before intron 1 splicing or before degradation in the nucleoplasm (see below). In contrast to the case with PS2, CCND1 intron 1 splicing occurred while the RNAs were still associated with Pol II (Fig. 3B and 4C). While the maximal level of the spliced RNA occurred at between 30 and 60 min of E2 treatment (Fig. 4E), the maximal level of the unspliced RNA occurred at between 15 and 30 min (Fig. 4C). This short time lag was consistent with our previous observations showing that cotranscriptional splicing of intron 1 could start 15 to 30 min after the initial rise of pre-mRNA synthesis (Fig. 3B).
Ser5 phosphorylation of the Pol II CTD was involved in cotranscriptional splicing of CCND1 intron 1.
To investigate the mechanism by which CCND1 but not PS2 splicing occurs during transcription, we next analyzed the status of Pol II phosphorylation on both the CCND1 and PS2 genes, because it has been reported that Pol II phosphorylation plays an important role in communication between the transcriptional and splicing machineries (see the introduction). In addition to the CTD4H8 antibody, we used the 8WG16, H14, and H5 antibodies, which preferentially recognize the unphosphorylated, Ser5-phosphorylated (Ser5P), and Ser2-phosphorylated (Ser2P) epitopes, respectively (10, 39). However, the level of Ser2P epitopes was very low on both genes, and we did not observe significant differences between the PS2 and CCND1 genes (data not shown).
As already shown (Fig. 1), E2 treatment resulted in a stronger increase in the total Pol II level on the PS2 gene than on the CCND1 gene, as measured by normalizing the ChIP values to the input values (percent input, CTD4H8; Fig. 5A). A similar level of Pol II was detected on the CCND1 and PS2 gene 5′ ends in the presence of E2 (Fig. 5A). However, while the Pol II level dropped on CCND1 exon 2, similar levels of Pol II were observed on PS2 exons 1 and 2 both in the presence and in the absence of E2 (compare 5′ and I1I2 in Fig. 5A). This was not an artifact of the ChIP assay, because PS2 intron 1 is about twice the size of CCND1 intron 1 (Fig. 1A), which decreases the risk of contamination by 5′-end-bound Pol II. Instead, this discrepancy might reflect differential dynamics of Pol II on the PS2 and CCND1 genes (see below).
FIG. 5.
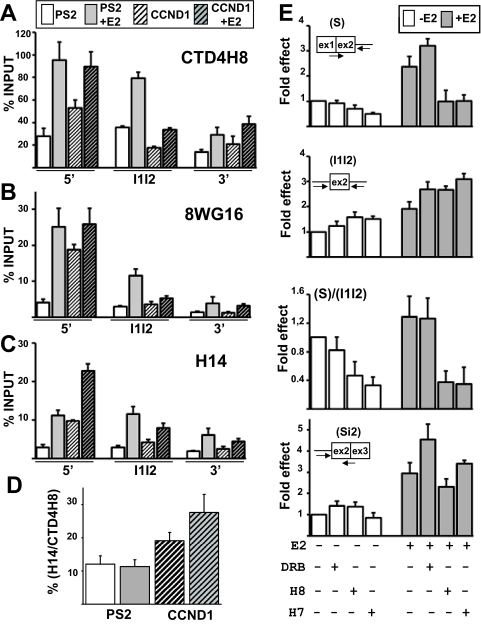
Pol II phosphorylation status for CCND1 and PS2 genes. (A to C) Levels of Pol II (calculated as percentage of input) determined by ChIP and qPCR analysis on different regions (5′, I1I2, and 3′, as shown in Fig. 1A) of PS2 (not hatched) and CCND1 (hatched) genes in the absence (white) or in the presence (gray) of E2 for 1 h. Antibodies against Pol II used in ChIP were CTD4H8 (A), 8WG16 (B), and H14 (C). (D) The values obtained with the H14 antibody (Ser5-phosphorylated Pol II) were expressed as a percentage of the values obtained with the CTD4H8 antibody (phosphorylated and unphosphorylated Pol II) in the absence (white) or in the presence (gray) of E2. (E) RT-qPCR analysis of the levels of CCND1(S), CCND1(I1I2), and CCND1(Si2) RNAs in nuclear extracts prepared from cells treated with E2 for 1 h or left untreated and with or without DRB, H8, and H7. The values obtained in controls (without treatment) for each product were assigned 1. Experiments were performed three times. Data are represented as means ± SEM.
As expected, the number of unphosphorylated epitopes strongly decreased from the gene 5′ to 3′ ends (Fig. 5B), because transcribing Pol II is known to be phosphorylated (10, 39). While the level of unphosphorylated epitopes slightly increased after 1 h of E2 treatment on the CCND1 gene 5′ end (Fig. 5B), it strongly increased on the PS2 gene 5′ end (by about 7 times; Fig. 5B). This result suggested that E2 had a stronger impact on transcription preinitiation in the case of PS2 than in the case of CCND1.
Remarkably, while the levels of total or unphosphorylated Pol II forms were similar on the CCND1 and PS2 gene 5′ ends in the presence of E2 (Fig. 5A and B), the levels of Ser5P epitopes were higher on the CCND1 gene than on the PS2 gene 5′ end (by more than two times [Fig. 5C]). After results were normalized to the total Pol II level, there were between two and three times more Ser5P epitopes on the CCND1 gene 5′end than on the PS2 gene 5′ end both in the absence and in the presence of E2 (Fig. 5D).
To test whether the higher level of Ser5P epitopes on the CCND1 gene 5′ end plays a role in splicing, we next investigated the impact of inhibitors of Ser5 phosphorylation. We used the H7 and H8 molecules, which preferentially inhibit cdk7, which is involved in Ser5 phosphorylation, rather than cdk9, which is involved in Ser2 phosphorylation (7, 15). As a control, we used DRB, which preferentially inhibits cdk9 rather than cdk7 (7, 15). As expected, DRB, which inhibits transcription elongation, induced a decrease in the 3′- to 5′-end ratio at the level of the pre-mRNAs (data not shown).
As shown in Fig. 5E, H7 and H8 molecules decreased the level of the spliced CCND1(S) RNAs, particularly in the presence of E2, and the spliced-to-unspliced (S/I1I2) ratio decreased by two to four times. DRB had no effect on the spliced-to-unspliced (S/I1I2) ratio (Fig. 5E), and the effect of H7 and H8 molecules was specific to CCND1 intron 1 splicing, because minor effects on the levels of the CCND1(Si2) RNAs that contain intron 1 but not intron 2 were observed (Fig. 5E, lower panel). Altogether, these results suggested that Ser5P level plays an important role in the splicing of CCND1 intron 1. In addition, because it has been shown that H7 and H8 molecules inhibit cotranscriptional RNA splicing rather than posttranscriptional splicing (7), these results strengthen our conclusion that CCND1 intron 1 splicing occurred during transcription.
Differential cotranscriptional recruitment of CBP80 on PS2 and CCND1 genes.
In addition to the CTD of Pol II, several reports have indicated that the cap binding complex (CBC) that binds to the RNA 5′-end cap structure plays an important role in the splicing of the first intron by recruiting the U1 snRNP, thereby enhancing the recognition of the promoter or cap-proximal 5′ splice site (31, 36). Therefore, we performed a ChIP assay as recently described (32) by using an antibody against CBP80, which is one of the two subunits of the CBC (31, 36). Remarkably, CBP80 was specifically enriched on the CCDN1 gene 5′ end compared to the PS2 gene 5′ end and to an intergenic region (Fig. 6A). To test whether CBC was necessary for the splicing of CCND1 intron 1, we next investigated the impact of an siRNA against CBP80 (35) on CCND1 splicing. As expected, the depletion of CBP80 reduced the efficiency of the removal of the first CCND1 intron, since it decreased the spliced-to-unspliced ratio by about 30%, both in the presence and in the absence of E2 (Fig. 6B). Therefore, the results obtained with the CCND1 gene are in agreement with a model where the recruitment of CBC favors the splicing of the first intron (31, 36).
In addition, the higher level of CPB80 enrichment on the CCND1 gene correlated with a higher proportion of CCND1 than PS2 pre-mRNAs being capped. Indeed, after the immunoselection of RNAs with anti-Cap monoclonal antibody, as already described (22), we performed RT-qPCR using primer sets flanking exon 2 in the first and second introns (I1I2) of CCND1 or PS2. We next compared the amount of RNAs coimmunoprecipitated with the anti-Cap antibody to the amount of RNAs present in the input (nuclear extract). We measured that the proportion of CCND1 pre-mRNAs containing introns 1 and 2 was more than three times higher than the proportion of PS2 pre-mRNAs containing introns 1 and 2 in the anti-Cap immunoprecipitation (I1I2; Fig. 6C). As a control, there was the same proportion of mature PS2 and CCND1 mRNAs that was immunoprecipitated with the anti-Cap antibody (“mRNA,” Fig. 6C). This result was expected, because decapped mRNAs are degraded (5).
Remarkably, we also noted that cotranscriptional splicing of CCND1 intron 1 correlated with a decrease in the Pol II level between exon 1 and exon 2, both in the presence and in the absence of E2 (Fig. 6D, left panel). Meanwhile, PS2 intron 1 was not efficiently spliced during transcription, and the level of Pol II remained constant between exon 1 and exon 2 on the PS2 gene (Fig. 6D, right panel). However, the Pol II level decreased between exon 2 and exon 3 on the PS2 gene. This observation was interesting because PS2 intron 2 was more frequently spliced before intron 1 (Fig. 4D). In addition, we noted that the proportion of partially spliced PS2(Si2) RNAs (Fig. 4D) that were capped was ∼2 times higher than the proportion of unspliced PS2 RNAs (Fig. 6E). Altogether, these results raised the possibility that at least a fraction of PS2 RNAs could be spliced during transcription.
As already mentioned, a large proportion of unspliced PS2 RNAs were not associated with Pol II compared to results for CCND1 RNAs (Fig. 3C and 4B). This result could arise if splicing occurred after transcription or if a large proportion of PS2 unspliced RNAs were released from Pol II before their degradation in the nucleoplasm. In the latter case, fully spliced PS2 mRNAs may come from a small proportion of nascent PS2 RNAs being spliced during transcription (with a larger proportion of neosynthesized unspliced PS2 RNAs being released from Pol II before degradation in the nucleoplasm). As a consequence, a larger proportion of the PS2 mRNAs was expected to be detected in the Pol II immunoprecipitation than would be the case for unspliced PS2 RNAs. Remarkably, we detected a larger proportion (∼4 times) of PS2 mRNAs than of unspliced (I1I2) PS2 pre-mRNAs in the Pol II immunoprecipitation (Fig. 6F). This result demonstrated that although cotranscriptional splicing was poorly efficient in the case of PS2 (leading to the release of unspliced PS2 RNAs from Pol II), a small fraction of nascent PS2 RNAs could be spliced when they were still associated with Pol II. The splicing of at least a small proportion of PS2 RNAs in the vicinity of the PS2 gene was in agreement with a recent report indicating an increase in spliced products close to the PS2 gene in response to E2 (38).
In contrast, when looking at CCND1 RNAs, we observed opposite results. There was a larger proportion (∼17 times) of unspliced RNAs (I1I2) than mature mRNA in the Pol II immunoprecipitation (Fig. 6F). This result was in agreement with our previous findings, because there was a small fraction of pre-mRNA being released from Pol II since cotranscriptional splicing was efficient.
We also noted that the fraction of mRNAs present in the Pol II immunoprecipitation was about four times higher in the case of PS2 than in that of CCND1 (Fig. 6F). By analyzing the Pol II pattern at the gene 3′ ends and downstream of the pA sites (Fig. 1A), we noted that transcriptional activation of the PS2 gene resulted in a marked increase in the Pol II level downstream of the PS2 pA site (positions B and C on Fig. 1D). Compared to the basal level, E2 treatment resulted in a marked increase in the Pol II level downstream of the PS2 pA site, as measured by normalizing the ChIP values to the input values (Fig. 6G, % input). This accumulation of Pol II downstream of the PS2 pA site in the presence of E2, which we did not observe for the CCND1 gene (not shown), was likely to be due to increased pausing of Pol II, which is associated with a possible delay in the initiation of 3′-end RNA processing, as recently reported (24).
To further test this possibility, we used a set of primers within the last exon but downstream of the pA site and a set of primers flanking the pA/RNA cleavage site (Fig. 6H). We noted that E2 treatment resulted in a stronger increase in the level of the RNA molecules when primers downstream of the pA/RNA cleavage site were used in the case of PS2 but not in the case of CCND1 (Fig. 6H). Altogether, these results suggested that in the case of PS2, the 3′-end RNA processing that is associated with mRNA release from the site of transcription (11, 36) may require more time when PS2 gene transcription is activated. This result may explain why there was a larger proportion of PS2 than of CCND1 mRNAs in the Pol II immunoprecipitation (Fig. 6F).
Estradiol selectively induced Ser5 phosphorylation of the Pol II CTD in a gene-specific manner.
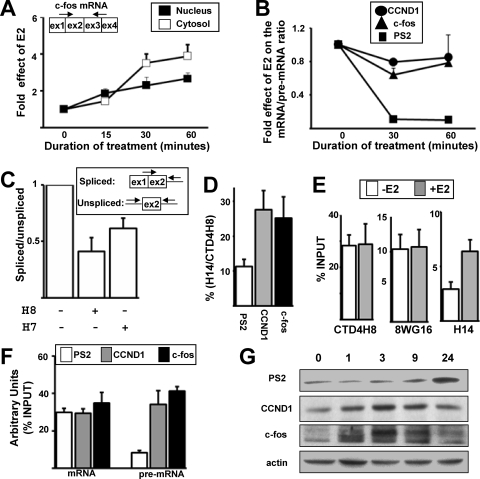
To further analyze the role of Pol II Ser5 phosphorylation, we looked for a third E2-stimulated gene. We selected the c-fos gene because it has been reported that c-fos splicing occurred during transcription upon induction with a calcium ionophore (32). As shown in Fig. 7A, the levels of fully spliced c-fos mRNA rapidly increased in response to E2 in both the nucleus and the cytosol.
FIG. 7.
Pol II Ser5 phosphorylation can be selectively stimulated by E2 in a gene-specific manner. (A) RT-qPCR analysis of effects of E2 on the fully spliced c-fos mRNA in nuclear (black) or cytoplasmic (white) extracts at given times (min) following the onset of E2 treatment. (B) PS2, CCND1, and c-fos mRNAs and pre-mRNAs were quantified by RT-qPCR using purified nuclear RNAs. The values obtained with the mRNAs were divided by the values obtained with the pre-mRNAs. The value of the ratio at baseline (0 h) for each product was assigned 1. (C) The c-fos spliced and unspliced RNAs were quantified by RT-qPCR using nuclear RNAs from cells treated with E2 in the presence or not of H8 or H7 molecules. The spliced-to-unspliced ratio obtained in the control experiment (without H7 or H8) was assigned 1. (D) Proportion of Ser5P epitopes on the PS2, CCND1, and c-fos gene 5′ ends in the presence of E2 for 1 h. The value obtained in the ChIP assay using the H14 antibody was divided by the value obtained with the CTD4H8 antibody. (E) Pol II levels calculated as a percentage of input on the c-fos gene 5′ end, determined by ChIP in the absence (white) or in the presence (gray) of E2 for 1 h. Experiments were performed at least three times. Data are represented as means ± SEM. (F) Nuclear RNAs were prepared from cells treated with E2 for 1 h. The levels of PS2, CCND1, and c-fos mRNAs or pre-mRNAs immunoprecipitated with anti-Cap antibody were measured and compared to their respective levels measured in the input. (G) Western blot analysis of PS2, CCND1, c-fos, and actin proteins after treatment with E2 for different times (hours).
When we compared the effects of E2 on the nuclear mRNA-to-pre-mRNA ratio, we observed very similar curves for CCND1 and c-fos (Fig. 7B). In contrast, E2 treatment strongly decreased the nuclear PS2 mRNA/pre-mRNA ratio (Fig. 7B). Therefore, these results suggested that E2 had similar effects on c-fos and CCND1 RNA synthesis/maturation, which differed from its effects on PS2.
To test whether Pol II Ser5 phosphorylation also plays a role in the splicing of c-fos pre-mRNA produced in response to E2, cells were treated with H7 and H8 molecules before E2 was added. As in the case of CCND1 (Fig. 5E), the H7 or H8 molecules decreased the spliced-to-unspliced ratio in the case of c-fos (Fig. 7C). In addition, the percentage of Ser5P epitopes in the presence of E2 on the c-fos gene 5′ end was similar to that on the CCND1 gene 5′ end and was higher than that on the PS2 gene 5′ end (Fig. 7D). Remarkably, the level of Ser5P epitopes (H14) selectively increased at the c-fos gene 5′ end in response to E2, because no effect was observed with the CTD4H8 and 8WG16 antibodies (Fig. 7E). Altogether, these results demonstrated that E2 had differential effects on the Pol II status depending on the target genes. While E2 induced a selective increase in the Ser5P epitope level on the c-fos gene (Fig. 7E), it increased both phosphorylated and unphosphorylated Pol II forms on the CCND1 and PS2 genes (Fig. 5). The Ser5 phosphorylated form increased more than the other Pol II forms in the case of CCND1, while the unphosphorylated form increased more than the other Pol II forms in the case of PS2 (Fig. 5). Therefore, E2 stimulated the transcription of three different target genes by different mechanisms, and Pol II Ser5 phosphorylation associated with cotranscriptional splicing was playing an unexpected critical role in the mRNA production by a subset of E2 target genes. Furthermore, we also observed that, similar to the case with CCND1, the proportion of c-fos pre-mRNAs being capped was higher than that in the case of PS2 (Fig. 7F).
Finally, we analyzed the PS2, CCND1, and c-fos protein levels after treating the cells with E2 for different periods of time. As previously reported (40, 48), the PS2 protein level increased much later than the CCND1 and c-fos protein levels, which were induced rapidly and transiently by E2 (Fig. 7G). Altogether, our data suggested that the efficiency of cotranscriptional RNA processing may play a critical role for gene products that are tightly regulated. This might be particularly relevant to gene products that mediate the rapid induction of cell cycle progression by estradiol in breast cells (9, 21, 40).
DISCUSSION
Although several studies have investigated the coupling between transcription and splicing, this was generally not performed in the context of transcriptional activation of endogenous genes. In addition, many studies investigated the transcriptional mechanisms by which such stimuli as estrogens impact regulation of gene expression without taking RNA splicing into account. To fill in these gaps, we investigated the impact of E2 on the expression levels of endogenous genes by examining the Pol II levels and phosphorylation status on the DNA templates (ChIP assay), the levels of unspliced and partially spliced RNAs associated with Pol II (RNA-ChIP assay), and the nuclear levels of unspliced, partially spliced, and fully spliced RNAs.
The ChIP assay indicated that E2 induced a globally stronger induction of Pol II on the PS2 gene than on the CCND1 gene (Fig. 1 and 5), and this observation was consistent with the stronger E2-mediated induction of PS2 pre-mRNA levels than of CCND1 pre-mRNA levels (Fig. 2 and 3). However, this was in sharp contrast with the lesser effect of E2 on PS2 mRNA levels than on CCND1 mRNA levels (Fig. 2C and D). The effects of E2 on the PS2 and CCND1 mRNA levels were consistent with previous reports (9, 21). Using the RNA-ChIP assay, which permits immunoprecipitation of RNAs associated with Pol II, we demonstrated that increasing amounts of Pol II-associated PS2 RNAs contained PS2 intron 1 during E2 treatment, which contrasts with what we observed in the case of CCND1 (Fig. 3B and 4C). This observation raised the possibility that a larger proportion of PS2 RNAs associated with Pol II were not spliced, in contrast with CCND1 RNAs. Supporting this hypothesis, a larger proportion of PS2 than CCND1 pre-mRNAs were not associated with Pol II (Fig. 3C and 4B).
We next demonstrated that the mechanism by which cotranscriptional splicing of intron 1 was more efficient in the case of CCND1 than in that of PS2 was dependent on the Pol II status on the DNA templates. Indeed, Pol II, which plays a critical role in the communication between the transcriptional and splicing machineries (see the introduction), had very different patterns in a comparison of the PS2 and CCND1 genes. In particular, a higher proportion of the Pol II Ser5-phosphorylated form was observed at the CCND1 gene 5′ end than at that of PS2 (Fig. 5D). The inhibition of Ser5 phosphorylation by H7 and H8 molecules (but not inhibition of Pol II Ser2 phosphorylation by DRB) decreased specifically the levels of the spliced CCND1(S) RNAs (Fig. 5E), which demonstrates a direct link between Pol II Ser5 phosphorylation and splicing of the first intron. Because H7 and H8 molecules alter cotranscriptional but not posttranscriptional splicing (7), these observations strengthen our conclusion that CCND1 intron 1 was spliced during transcription.
In addition to a higher level of Ser5 phosphorylation of the Pol II CTD (Fig. 4D), we detected a higher level of CBP80 (one of the two subunits of CBC) at the CCND1 gene 5′ end than at the PS2 gene 5′ end (Fig. 6A); this correlated with a higher proportion of CCND1 pre-mRNAs being capped than was the case for PS2 pre-mRNAs (Fig. 6C). Importantly, it has been shown that CBC plays a critical role in the recognition of the cap-proximal 5′ splice site (31, 36). Supporting the notion that CBC plays an important role in cotranscriptional splicing of CCND1 intron 1, the depletion of CBP80 resulted in a decrease in the spliced-to-unspliced ratio (Fig. 6B). Therefore, in the case of CCND1, a high level of Ser5 phosphorylation of the Pol II CTD and the recruitment of CBC (Fig. 4D and 6A) facilitated the recognition and splicing of the first intron (Fig. 5E and 6B). Interestingly, the recruitment of CBP80 and the splicing of the first intron may in turn permit Pol II to elongate more efficiently.
Indeed, we noted that Pol II levels dropped between exon 1 and exon 2 on the CCND1 gene [compare CCND1(5′) and CCND1(I1I2) in Fig. 6D] but not on the PS2 gene [compare PS2(5′) and PS2(I1I2) in Fig. 6D]. This was not an artifact of the ChIP assay, because PS2 intron 1 is about twice the size of D1 intron 1, which decreases the risk of contamination by 5′-end-bound Pol II (Fig. 1A). Therefore, in the case of CCND1, where cotranscriptional splicing of intron 1 was efficient, there was a change in the pattern of Pol II between exon 1 and exon 2, in sharp contrast with PS2, where cotranscriptional splicing of intron 1 was inefficient. One explanation for the pattern of Pol II on the CCND1 gene could be that CBC helps the recruitment of the U1 snRNP. This may not only enhance splicing, but the recruitment of the U1 snRNA may also stimulate Pol II transcription as has been recently shown (12, 28). Such a mechanism would also explain why a similar level of Pol II was detected between exon 1 and exon 2 on the PS2 gene (Fig. 6D), since CBP80 was not recruited and cotranscriptional splicing of PS2 intron 1 was inefficient.
In this context, we noted that PS2 intron 2 was more frequently spliced before intron 1 (Fig. 4D), and this correlated with an increase in the proportion of capped RNAs (Fig. 6E) and also with a decrease in the Pol II level between exon 2 and exon 3 on the PS2 gene (Fig. 6D). These data indicated a strong correlation between Pol II dynamics and splicing and suggested that at least a portion of PS2 RNAs may be spliced during transcription. Supporting a model where only a small fraction of PS2 RNA molecules could be processed during transcription and give rise to mature PS2 mRNAs, we detected a larger proportion of PS2 mRNAs than PS2 pre-mRNAs in the Pol II immunoprecipitation (Fig. 6F). This result is in agreement with a recent report showing that spliced PS2 RNAs can be detected at the gene site (38) and suggested that a large proportion of unspliced PS2 RNAs could be released from Pol II (Fig. 3C, 4B, and 6F).
The large fraction of PS2 mRNA in the Pol II immunoprecipitation was also consistent with the delay in PS2 3′-end RNA processing when transcription was activated (Fig. 6G and H). Importantly, increasing evidence supports a coupling between RNA 3′-end processing and mRNA release that also involves Pol II CTD (11, 36). Therefore, the delay in 3′-end RNA processing may result in the transient accumulation of PS2 mRNA before release from the transcriptional site. The low rate of PS2 3′-end processing might be due either to the PS2 pA site being not optimal or to the small size of the last PS2 exon being only 212 nucleotides (Fig. 1A). Alternatively, the lower efficiency of PS2 3′-end RNA processing might be due to the absence of the recruitment of CBC on PS2 RNAs, because it has been shown that CBC may help the recruitment of 3′-end processing factors (17).
Noteworthily, inefficient cotranscriptional splicing of PS2 intron 1 did not result from E2 treatment. Indeed, the partially spliced PS2(S) RNA level was low even in the absence of E2 (Fig. 4D). In addition, the proportion of unspliced PS2 RNAs associated with Pol II was not affected by E2 treatment (Fig. 3C and 4B). Finally, compared to results for CCND1, the amount of Pol II Ser5P epitopes on the PS2 gene 5′ end was low both in the presence and in the absence of E2 (Fig. 5D). Therefore, the inefficient cotranscriptional splicing of PS2 intron 1 may be intrinsic to the PS2 gene, which was revealed only by E2 treatment. Similarly, the efficient cotranscriptional splicing of CCND1 intron 1 may be independent of the E2 signaling pathway.
In this context, our data interestingly indicated that E2 stimulated transcription by different mechanisms depending on the gene context. E2 strongly increased the unphosphorylated form of Pol II on the PS2 gene (Fig. 5B), which suggested a major effect on the transcriptional preinitiation step on this gene. Meanwhile, although the unphosphorylated form of Pol II increased only slightly on the CCND1 gene 5′ end after 1 h of E2 treatment (Fig. 5B), our data showed that all the transcriptional steps were similarly enhanced in the case of CCND1. Indeed, almost all the parameters reflecting CCND1 gene activity increased about twofold in response to E2 (Fig. 1, 2, and 5). This observation suggests that E2 treatment may “simply” increase a process already efficient in the absence of E2. Finally, our results of a selective increase in the Ser5P epitope level in response to E2 on the c-fos gene 5′ end (Fig. 7E) demonstrated that E2 treatment resulted in the specific improvement of steps downstream of preinitiation in the case of the c-fos gene. Therefore, the action of E2 strongly depends on the “intrinsic” proprieties of its target genes.
Our data clearly indicated that the effect of E2 on the mRNA levels produced by its target genes depends not only on its transcriptional effects but also on the efficiency of the processing of the nascent pre-mRNAs. In particular, a high level of Pol II Ser5 phosphorylation increased the efficiency of mRNA production owing to the coupling between transcription and splicing. In other words, only a subset of E2-regulated mRNAs might be produced in a very efficient way in what has been referred to as the “mRNA factory” (6). Because the PS2 protein level increased later than the CCND1 and c-fos protein levels (Fig. 7G), our data collectively suggest that the efficiency of cotranscriptional RNA processing might play a critical role for gene products whose expression level is tightly regulated. This might be particularly relevant for genes whose products play a role in cell cycle progression. In this context, we must underline that this study was performed using the MCF-7 breast cancer cell line, whose growth depends on estrogens. The cells were maintained for several days in medium containing stripped serum, which results in inhibition of MCF-7 cell growth. The MCF-7 growth inhibition is reversed by addition of E2, resulting in rapid production of cell cycle activators, such as CCND1 and c-fos (9, 21). The efficient maturation of the CCND1 and c-fos gene products in response to E2 may be a key component of the rapid mitogenic response of MCF-7 cells to the estrogen signaling pathway. Because a recent transcriptomic analysis with yeast demonstrated that modulation of splicing efficiency plays a critical role in the rapid regulation of the expression levels of a subset of genes in response to stress (41), these observations support a model in which RNA processing efficiency might be a major level of regulation during the cellular response to growth conditions.
In conclusion, our data add an important piece of evidence for considering the splicing process in the studies of the effects of transcriptional stimuli on gene expression levels. We and others have shown that transcriptional stimuli impact the ratio of the alternative splicing variants produced by their target genes (2). This is critical in the gene expression process, because alternative splicing can result in the production of protein isoforms having different and even opposite biological activities (45). In this report, we now have shown that the levels of mRNAs produced could not be explained only by the impact of transcriptional stimuli on their target gene transcriptional activity: although E2 had stronger effects on PS2 than on CCND1 gene transcriptional activity, PS2 RNA processing was not as efficient as CCND1 RNA processing. Importantly, the estrogen signaling pathway is a major pharmacological target in several diseases, and the study of the regulation of the PS2 mRNA expression level is one of the most popular models used to study the impact of E2 and E2-related pharmaceutical molecules on gene transcriptional activity. Because our data showed that the production of the PS2 mRNA was limited by splicing, strong effects on PS2 transcriptional activity may not result in significant effects at the mRNA level. More generally, the study of gene expression regulation by transcriptional stimuli by considering steps all along the “assembly line” of mRNA production is opening the opportunity to identify new gene-selective rate-limiting steps and therefore to guide the development of pharmaceutical molecules with increased efficiency and selectivity.
Acknowledgments
We are grateful to R. Lührmann and E. Izaurralde for providing the H20 and CBP80 antibodies. We thank B. O'Malley, O. Bensaude, and K. Neugebauer for their critical reading of the manuscript.
This work was supported by the INSERM AVENIR program, Agence Nationale de la Recherche, and by the EC (6th PCRD, NoE EURASNET). D.B. was supported by the Ile-de-France Council and by ARC, M.D. by INSERM, G.S. by Chancellerie des Universités de Paris, J.B. by the French Ministry of Education, and L.G. by INCa.
We declare that no competing interests exist.
Footnotes
Published ahead of print on 21 July 2008.
REFERENCES
- 1.Aiyar, S. E., J. L. Sun, A. L. Blair, C. A. Moskaluk, Y. Z. Lu, Q. N. Ye, Y. Yamaguchi, A. Mukherjee, D. M. Ren, H. Handa, and R. Li. 2004. Attenuation of estrogen receptor alpha-mediated transcription through estrogen-stimulated recruitment of a negative elongation factor. Genes Dev. 182134-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auboeuf, D., A. Honig, S. M. Berget, and B. W. O'Malley. 2002. Coordinate regulation of transcription and splicing by steroid receptor coregulators. Science 298416-419. [DOI] [PubMed] [Google Scholar]
- 3.Barbier, J., M. Dutertre, D. Bittencourt, G. Sanchez, L. Gratadou, P. de la Grange, and D. Auboeuf. 2007. Regulation of H-ras splice variant expression by cross talk between the p53 and nonsense-mediated mRNA decay pathways. Mol. Cell. Biol. 277315-7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batsche, E., M. Yaniv, and C. Muchardt. 2006. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat. Struct. Mol. Biol. 1322-29. [DOI] [PubMed] [Google Scholar]
- 5.Beelman, C. A., and R. Parker. 1995. Degradation of mRNA in eukaryotes. Cell 81179-183. [DOI] [PubMed] [Google Scholar]
- 6.Bentley, D. 2002. The mRNA assembly line: transcription and processing machines in the same factory. Curr. Opin. Cell Biol. 14336-342. [DOI] [PubMed] [Google Scholar]
- 7.Bird, G., D. A. Zorio, and D. L. Bentley. 2004. RNA polymerase II carboxy-terminal domain phosphorylation is required for cotranscriptional pre-mRNA splicing and 3′-end formation. Mol. Cell. Biol. 248963-8969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brodsky, A. S., C. A. Meyer, I. A. Swinburne, G. Hall, B. J. Keenan, X. S. Liu, E. A. Fox, and P. A. Silver. 2005. Genomic mapping of RNA polymerase II reveals sites of co-transcriptional regulation in human cells. Genome Biol. 6R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cascio, S., V. Bartella, C. Garofalo, A. Russo, A. Giordano, and E. Surmacz. 2007. Insulin-like growth factor 1 differentially regulates estrogen receptor-dependent transcription at estrogen response element and AP-1 sites in breast cancer cells. J. Biol. Chem. 2823498-3506. [DOI] [PubMed] [Google Scholar]
- 10.Cheng, C., and P. A. Sharp. 2003. RNA polymerase II accumulation in the promoter-proximal region of the dihydrofolate reductase and gamma-actin genes. Mol. Cell. Biol. 231961-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Custodio, N., M. Carmo-Fonseca, F. Geraghty, H. S. Pereira, F. Grosveld, and M. Antoniou. 1999. Inefficient processing impairs release of RNA from the site of transcription. EMBO J. 182855-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Damgaard, C. K., S. Kahns, S. Lykke-Andersen, A. L. Nielsen, T. H. Jensen, and J. Kjems. 2008. A 5′ splice site enhances the recruitment of basal transcription initiation factors in vivo. Mol. Cell 29271-278. [DOI] [PubMed] [Google Scholar]
- 13.Das, R., K. Dufu, B. Romney, M. Feldt, M. Elenko, and R. Reed. 2006. Functional coupling of RNAP II transcription to spliceosome assembly. Genes Dev. 201100-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das, R., J. Yu, Z. Zhang, M. P. Gygi, A. R. Krainer, S. P. Gygi, and R. Reed. 2007. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol. Cell 26867-881. [DOI] [PubMed] [Google Scholar]
- 15.Dubois, M. F., V. T. Nguyen, S. Bellier, and O. Bensaude. 1994. Inhibitors of transcription such as 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole and isoquinoline sulfonamide derivatives (H-8 and H-7) promote dephosphorylation of the carboxyl-terminal domain of RNA polymerase II largest subunit. J. Biol. Chem. 26913331-13336. [PubMed] [Google Scholar]
- 16.Eeckhoute, J., J. S. Carroll, T. R. Geistlinger, M. I. Torres-Arzayus, and M. Brown. 2006. A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes Dev. 202513-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flaherty, S. M., P. Fortes, E. Izaurralde, I. W. Mattaj, and G. M. Gilmartin. 1997. Participation of the nuclear cap binding complex in pre-mRNA 3′ processing. Proc. Natl. Acad. Sci. USA 9411893-11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosh, S., and M. A. Garcia-Blanco. 2000. Coupled in vitro synthesis and splicing of RNA polymerase II transcripts. RNA 61325-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heldring, N., A. Pike, S. Andersson, J. Matthews, G. Cheng, J. Hartman, M. Tujague, A. Strom, E. Treuter, M. Warner, and J. A. Gustafsson. 2007. Estrogen receptors: how do they signal and what are their targets. Physiol. Rev. 87905-931. [DOI] [PubMed] [Google Scholar]
- 20.Hicks, M. J., C. R. Yang, M. V. Kotlajich, and K. J. Hertel. 2006. Linking splicing to Pol II transcription stabilizes pre-mRNAs and influences splicing patterns. PLoS Biol. 4e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikeda, K., S. Ogawa, T. Tsukui, K. Horie-Inoue, Y. Ouchi, S. Kato, M. Muramatsu, and S. Inoue. 2004. Protein phosphatase 5 is a negative regulator of estrogen receptor-mediated transcription. Mol. Endocrinol. 181131-1143. [DOI] [PubMed] [Google Scholar]
- 22.Kabrane-Lazizi, Y., X. J. Meng, R. H. Purcell, and S. U. Emerson. 1999. Evidence that the genomic RNA of hepatitis E virus is capped. J. Virol. 738848-8850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaneko, S., and J. L. Manley. 2005. The mammalian RNA polymerase II C-terminal domain interacts with RNA to suppress transcription-coupled 3′ end formation. Mol. Cell 2091-103. [DOI] [PubMed] [Google Scholar]
- 24.Kaneko, S., O. Rozenblatt-Rosen, M. Meyerson, and J. L. Manley. 2007. The multifunctional protein p54nrb/PSF recruits the exonuclease XRN2 to facilitate pre-mRNA 3′ processing and transcription termination. Genes Dev. 211779-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kessler, O., Y. Jiang, and L. A. Chasin. 1993. Order of intron removal during splicing of endogenous adenine phosphoribosyltransferase and dihydrofolate reductase pre-mRNA. Mol. Cell. Biol. 136211-6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kininis, M., B. S. Chen, A. G. Diehl, G. D. Isaacs, T. Zhang, A. C. Siepel, A. G. Clark, and W. L. Kraus. 2007. Genomic analyses of transcription factor binding, histone acetylation, and gene expression reveal mechanistically distinct classes of estrogen-regulated promoters. Mol. Cell. Biol. 275090-5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 142452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwek, K. Y., S. Murphy, A. Furger, B. Thomas, W. O'Gorman, H. Kimura, N. J. Proudfoot, and A. Akoulitchev. 2002. U1 snRNA associates with TFIIH and regulates transcriptional initiation. Nat. Struct. Biol. 9800-805. [DOI] [PubMed] [Google Scholar]
- 29.Lacadie, S. A., D. F. Tardiff, S. Kadener, and M. Rosbash. 2006. In vivo commitment to yeast cotranscriptional splicing is sensitive to transcription elongation mutants. Genes Dev. 202055-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lazarev, D., and J. L. Manley. 2007. Concurrent splicing and transcription are not sufficient to enhance splicing efficiency. RNA 131546-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis, J. D., E. Izaurralde, A. Jarmolowski, C. McGuigan, and I. W. Mattaj. 1996. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′ splice site. Genes Dev. 101683-1698. [DOI] [PubMed] [Google Scholar]
- 32.Listerman, I., A. K. Sapra, and K. M. Neugebauer. 2006. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat. Struct. Mol. Biol. 13815-822. [DOI] [PubMed] [Google Scholar]
- 33.Lonard, D. M., and B. W. O'Malley. 2006. The expanding cosmos of nuclear receptor coactivators. Cell 125411-414. [DOI] [PubMed] [Google Scholar]
- 34.Mangelsdorf, D. J., C. Thummel, M. Beato, P. Herrlich, G. Schutz, K. Umesono, B. Blumberg, P. Kastner, M. Mark, P. Chambon, et al. 1995. The nuclear receptor superfamily: the second decade. Cell 83835-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Narita, T., T. M. Yung, J. Yamamoto, Y. Tsuboi, H. Tanabe, K. Tanaka, Y. Yamaguchi, and H. Handa. 2007. NELF interacts with CBC and participates in 3′ end processing of replication-dependent histone mRNAs. Mol. Cell 26349-365. [DOI] [PubMed] [Google Scholar]
- 36.Neugebauer, K. M. 2002. On the importance of being co-transcriptional. J. Cell Sci. 1153865-3871. [DOI] [PubMed] [Google Scholar]
- 37.Niranjanakumari, S., E. Lasda, R. Brazas, and M. A. Garcia-Blanco. 2002. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods 26182-190. [DOI] [PubMed] [Google Scholar]
- 38.Nunez, E., Y. S. Kwon, K. R. Hutt, Q. Hu, M. D. Cardamone, K. A. Ohgi, I. Garcia-Bassets, D. W. Rose, C. K. Glass, M. G. Rosenfeld, and X. D. Fu. 2008. Nuclear receptor-enhanced transcription requires motor- and LSD1-dependent gene networking in interchromatin granules. Cell 132996-1010. [DOI] [PubMed] [Google Scholar]
- 39.Palancade, B., and O. Bensaude. 2003. Investigating RNA polymerase II carboxyl-terminal domain (CTD) phosphorylation. Eur. J. Biochem. 2703859-3870. [DOI] [PubMed] [Google Scholar]
- 40.Planas-Silva, M. D., J. L. Donaher, and R. A. Weinberg. 1999. Functional activity of ectopically expressed estrogen receptor is not sufficient for estrogen-mediated cyclin D1 expression. Cancer Res. 594788-4792. [PubMed] [Google Scholar]
- 41.Pleiss, J. A., G. B. Whitworth, M. Bergkessel, and C. Guthrie. 2007. Rapid, transcript-specific changes in splicing in response to environmental stress. Mol. Cell 27928-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robert, F., M. Blanchette, O. Maes, B. Chabot, and B. Coulombe. 2002. A human RNA polymerase II-containing complex associated with factors necessary for spliceosome assembly. J. Biol. Chem. 2779302-9306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ryser, S., T. Fujita, S. Tortola, I. Piuz, and W. Schlegel. 2007. The rate of c-fos transcription in vivo is continuously regulated at the level of elongation by dynamic stimulus-coupled recruitment of positive transcription elongation factor b. J. Biol. Chem. 2825075-5084. [DOI] [PubMed] [Google Scholar]
- 44.Sims, R. J., III, S. S. Mandal, and D. Reinberg. 2004. Recent highlights of RNA-polymerase-II-mediated transcription. Curr. Opin. Cell Biol. 16263-271. [DOI] [PubMed] [Google Scholar]
- 45.Stamm, S., S. Ben-Ari, I. Rafalska, Y. Tang, Z. Zhang, D. Toiber, T. A. Thanaraj, and H. Soreq. 2005. Function of alternative splicing. Gene 3441-20. [DOI] [PubMed] [Google Scholar]
- 46.Tardiff, D. F., S. A. Lacadie, and M. Rosbash. 2006. A genome-wide analysis indicates that yeast pre-mRNA splicing is predominantly posttranscriptional. Mol. Cell 24917-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wetterberg, I., G. Bauren, and L. Wieslander. 1996. The intranuclear site of excision of each intron in Balbiani ring 3 pre-mRNA is influenced by the time remaining to transcription termination and different excision efficiencies for the various introns. RNA 2641-651. [PMC free article] [PubMed] [Google Scholar]
- 48.Zacharewski, T. R., K. L. Bondy, P. McDonell, and Z. F. Wu. 1994. Antiestrogenic effect of 2,3,7,8-tetrachlorodibenzo-p-dioxin on 17 beta-estradiol-induced pS2 expression. Cancer Res. 542707-2713. [PubMed] [Google Scholar]
- 49.Zhang, H., L. Sun, J. Liang, W. Yu, Y. Zhang, Y. Wang, Y. Chen, R. Li, X. Sun, and Y. Shang. 2006. The catalytic subunit of the proteasome is engaged in the entire process of estrogen receptor-regulated transcription. EMBO J. 254223-4233. [DOI] [PMC free article] [PubMed] [Google Scholar]