Abstract
We previously discovered that microphthalmia transcription factor (MITF) and upstream stimulatory factor 2 (USF2) each forms a complex with its inhibitor histidine triad nucleotide-binding 1 (Hint-1) and with lysyl-tRNA synthetase (LysRS). Moreover, we showed that the dinucleotide diadenosine tetraphosphate (Ap4A), previously shown to be synthesized by LysRS, binds to Hint-1, and as a result the transcription factors are released from their suppression. Thus, transcriptional activity is regulated by Ap4A, suggesting that Ap4A is a second messenger in this context. For Ap4A to be unambiguously established as a second messenger, several criteria have to be fulfilled, including the presence of a metabolizing enzyme. Since several enzymes are able to hydrolize Ap4A, we provided here evidence that the “Nudix” type 2 gene product, Ap4A hydrolase, is responsible for Ap4A degradation following the immunological activation of mast cells. The knockdown of Ap4A hydrolase modulated Ap4A accumulation, resulting in changes in the expression of MITF and USF2 target genes. Moreover, our observations demonstrated that the involvement of Ap4A hydrolase in gene regulation is not a phenomenon exclusive to mast cells but can also be found in cardiac cells activated with the β-agonist isoproterenol. Thus, we have provided concrete evidence establishing Ap4A as a second messenger in the regulation of gene expression.
The dinucleotide diadenosine tetraphosphate (Ap4A) is composed of two adenosine moieties joined in 5′-5′ linkage by a chain of four phosphates. As far as we know, Ap4A is found in all living cells, and ever since its discovery in 1966 (42), it has been proposed to be more than just a “toxic waste” substance (28). This distinctive nucleotide has attracted much research, especially during the early 1980s. Ap4A concentration was shown to increase after exposure of cells to various forms of metabolic stress such as heat, oxidative, nutritional, and DNA damage (13). In addition, increased Ap4A levels have been associated with reduced replication times (28). It has also been suggested that Ap4A acts as an intracellular “alarmone” both in prokaryotes (15) and in eukaryotes (37).
Microphthalmia transcription factor (MITF) is a basic helix-loop-helix leucine zipper (bHLH-Zip) DNA-binding protein and a key transcription factor in mast cells. Many of the MITF-responsive genes are essential for differentiation and survival. These include c-kit (36), mouse mast cell protease (mMCP) 2, 4, 5, 6, and 9 (5, 10, 20, 22), the p75 receptor of nerve growth factor (11), integrin alpha 4 (12), SgIGSF (8), NDST-2 (21), granzyme B, and tryptophan hydroxylase (TPH) (9). Similarly, in osteoclasts and melanocytes MITF regulates the transcription of highly important genes such as the cathepsin K (23) and tyrosinase genes (1), respectively. Our studies have focused on the regulation network of MITF in immunologically activated mast cells for over a decade (18, 26, 27, 31) and in the heart for the past few years (34, 35).
We have shown that both MITF and another bHLH-Zip transcription factor, upstream stimulatory factor 2 (USF2) form a tertiary complex with their inhibitor histidine triad nucleotide-binding 1 (Hint-1) and with lysyl-tRNA synthetase (LysRS) (16, 17). Furthermore, we have shown that Ap4A can bind to Hint-1 and cause its release from MITF and from USF2 (16, 17). Ap4A is known to be produced by certain aminoacyl tRNA synthetases such as LysRS. We have demonstrated that intracellular levels of Ap4A in mast cells are substantially increased following immunoglobulin E-antigen (IgE-Ag) stimulation (16). Thus, our hypothesis is that Ap4A plays a critical role in the regulation of several transcription factors through its ability to control gene expression. These findings led to the proposal that Ap4A acts as a second messenger.
The “Nudix” hydrolase family is a group of enzymes containing the consensus motif GX5EX5[UA]XREX2EEXGU, where X is any amino acid and U is a bulky aliphatic residue (19). In addition to the Nudix signature, the enzymes share another common feature in that they all hydrolyze nucleoside diphosphate linked to another moiety, X. Since these metabolites may be toxic to the cell upon accumulation, as an analogy to the “housekeeping” or maintenance genes, those coding for the Nudix hydrolases were termed “housecleaning” genes by Bessman et al. in 1996 (2).
The Nudix type 2 (nudt2) gene product, Ap4A hydrolase, has been isolated and cloned and its structure has been determined (33). This enzyme hydrolyzes Ap4A into AMP and ATP. Several other enzymes, including members of the ectonucleotide pyrophosphatase/phosphodiesterase family, are also able to hydrolyze Ap4A (38). Until now, evidence for the critical role of Ap4A hydrolase in intracellular Ap4A metabolism following stimuli has been lacking.
Here we present direct evidence that Ap4A hydrolase is indeed responsible for Ap4A degradation in immunologically activated mast cells. We also describe the time course of Ap4A accumulation in this type of cell and find that its levels are rapidly induced by IgE-Ag but return to basal levels within 2 h after activation.
In previous studies, we used cold shock to introduce Ap4A into quiescent cells in order to demonstrate a direct effect of Ap4A on transcriptional regulation and found that Ap4A application resulted in an increased expression of MITF- and USF2-regulated genes. This external administration of Ap4A is considerably different from the physiological situation. Physiological manipulation of Ap4A has not been used before in studies of higher eukaryotes and is of great importance in any attempt to define Ap4A as a second messenger. In the present study, we manipulated intracellular Ap4A levels in a physiological manner by inhibiting Ap4A hydrolase.
Using the short interfering RNA (siRNA) technique, in this study we demonstrated that Ap4A hydrolase plays a critical role in the transcriptional regulation of MITF and USF2. We provide evidence regarding the role of a stimulus-induced Ap4A increase in gene regulation, both in mast cells and in isoproterenol-induced cardiomyocytes.
MATERIALS AND METHODS
Antibodies.
The antibodies anti-Ap4A hydrolase, anti-MITF, and anti-Hint-1 were custom made against the specially designed determinants FKEMKATLQEGHQFLC (Ap4A hydrolase), IIKQEPVLENCSQEL (MITF), and VLGGRQMNWPPG (Hint-1) (Hy Laboratories Ltd., Israel). The antibody against NPP1 (anti-PC-1) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Nucleotide assay.
The nucleotide assay detects the amount of Ap4A present in extracts of mammalian cells. For each determination, cells from one well of a six-well plate were grown to about 80% confluence. The cell layer was washed with warm serum-free medium and was lysed with trichloroacetic acid. Extraction and measurement by luminometry of the nucleotides were performed as described previously (24). Results were normalized by using the Bradford protein assay. It is important to note that this assay will also measure nucleotides other than Ap4A with the form Ap4N, where N is any nucleoside. However, based on previous calculations by Murphy and McLennan (25), the actual Ap4A concentration is roughly 0.75 times the values quoted.
Cell growth.
H9C2 cells were maintained at 37°C in growth medium, which was Dulbecco's modified Eagle's medium supplemented with 2 mM l-glutamine, 2 mM nonessential amino acids, 100 units/ml penicillin, 100 μg/ml streptomycin (Biological Industries, Beit Haemek, Israel), and 10% fetal calf serum (Biological Industries, Beit Haemek, Israel). Rat basophilic leukemia (RBL)-2H3 cells were maintained in RPMI 1640 medium as previously described (29). RBL cells were first sensitized with anti-2,4-dinitrophenol (DNP) IgE monoclonal antibody (SPE-7; Sigma-Aldrich Corp., St. Louis, MO) and then challenged with DNP (Sigma-Aldrich Corp.). IgE antibody was centrifuged (18,000 × g for 5 min) before use to remove aggregates.
Gel electrophoresis and Western blotting.
Proteins were resolved by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis under reducing conditions and were transferred to nitrocellulose membranes. Visualization of reactive proteins was performed by enhanced chemiluminescence.
Immunoprecipitation.
The immunoprecipitation of the specific proteins from RBL and H9C2 cells was carried out as previously described (18).
Real-time quantitative PCR.
Candidate MITF- and USF2-responsive gene transcriptions were measured using real-time quantitative PCR. mRNAs of the target genes were quantified by Sybr green incorporation (Sybr green ROX mix; ABgene). Real-time PCR was performed on a Rotor-Gene sequence detection system (Corbett, Australia). The genes whose mRNA levels were quantified by real-time PCR were rat TPH, GrB, telomerase reverse transcriptase (TERT), myosin light chain 1a (MLC-1a), and β-actin.
siRNA.
Cells were transfected with an siRNA duplex consisting of two complementary 21-nucleotide RNA strands with 3′ dTdT overhangs (Qiagen Inc., California) in order to downregulate Ap4A hydrolase. siRNAs were designed to be complementary to nucleotide sequences found in rat mRNAs of Ap4A hydrolase, and a nonrelevant (NR) nucleotide sequence was used as the control (NR siRNA). The target sequence of the specific siRNAs was CCCGGCGAGAATGACTTAGAA for Ap4A hydrolase. The nonrelevant control sequence was AATTCTCCGAACGTGTCACGT.
Transfection.
Nucleofector technology (Amaxa, Cologne, Germany) was used for transfecting cells. A total of 2 × 106 cells were transfected with 3 μg of the selected siRNA oligonucleotide according to the manufacturer's protocol. Briefly, the cells were resuspended in 100 μl of Nucleofector solution, RNA was added, and the mixture was transferred into an electroporation cuvette. Nucleofector solution was used to stabilize the cells during electroporation, which was performed using the T-20 program.
Statistical analysis.
Either the Friedman test or an analysis of variance was performed when appropriate, with differences being considered significant at a P of <0.05.
RESULTS
Kinetics of Ap4A levels in IgE-Ag-activated mast cells.
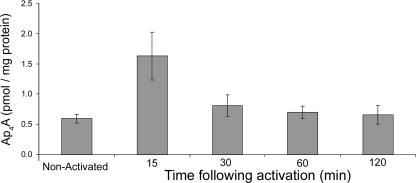
In order to determine the kinetics of intracellular Ap4A accumulation, RBL cells were stimulated for specific times with IgE and Ag. As shown in Fig. 1, Ap4A levels peaked 15 min after stimulation, reaching a concentration of 1.6 pmol/mg protein, followed by a sharp drop and a return to the basal level within 60 min. The concentrations of Ap4A are similar to those published recently for HEK 293 cells under different treatments (4).
FIG. 1.
Kinetics of Ap4A accumulation following the IgE-Ag stimulation of mast cells. Upon stimulation of RBL cells with 100 ng/ml IgE anti-DNP and 100 ng/ml DNP, cells were harvested at specific times, Ap4A level was measured by a luminescence assay as described in Materials and Methods, and results were normalized by a Bradford protein assay. Error bars represent the mean of replicates ± the standard error of the mean. Results for one representative experiment out of four are shown.
Ap4A hydrolase degrades Ap4A in IgE-Ag-activated mast cells.
Ap4A hydrolase was knocked down in RBL cells using the siRNA approach. The siRNA was designed to be complementary to nucleotide sequences found in rat mRNA of the gene encoding nudt2 (Ap4A hydrolase), and NR siRNA was used as the control. Downregulation of Ap4A hydrolase was observed 24 h after transfection (Fig. 2A), and its low level of expression remained for 48 h after transfection as well (data not shown).
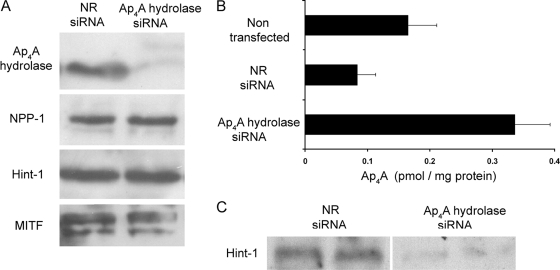
FIG. 2.
Ap4A hydrolase is degrading Ap4A in immunologically activated mast cells. (A) RBL cells were transfected with either NR siRNA or siRNA against the gene encoding Ap4A hydrolase. Twenty-four hours after transfection, Western blot analysis was carried out for cell lysates using antibodies specific for Ap4A hydrolase, NPP-1, Hint-1, and MITF. Results for one representative experiment out of three are shown. (B) RBL cells were transfected with either NR or Ap4A hydrolase siRNA for 24 h. Control and transfected cells were then stimulated by IgE-Ag for 6 h, and Ap4A levels were then determined. The mean and standard error of the mean for three experiments are shown. (C) Coimmunoprecipitation of MITF and Hint-1. MITF was immunoprecipitated from extracts of RBL cells transfected with siRNA against Ap4A hydrolase or NR siRNA. The level of coimmunoprecipitated Hint-1 was determined by Western blot analysis. Results for one representative experiment out of three are shown.
In order to verify the specificity of the siRNA oligonucleotides in the context of our system, we determined the presence of several proteins in lysates taken from Ap4A hydrolase siRNA-treated cells and from NR siRNA-treated cells. As can be seen in Fig. 2A, no significant changes were observed in the levels of Hint-1, MITF, and nucleotide pyrophosphatases/phosphodiesterases 1 (NPP1), another enzyme known to hydrolyze Ap4A (38).
The level of Ap4A in activated NR siRNA-treated cells was similar to that in activated cells that were not transfected (Fig. 2B), whereas a significant intracellular accumulation of Ap4A was observed in activated cells treated with siRNA against Ap4A hydrolase (Fig. 2B). This accumulation of Ap4A was found to be time dependent (data not shown). Thus, these data clearly suggest that Ap4A hydrolase is a major player in the metabolism of Ap4A in activated mast cells.
MITF-Hint dissociation is regulated by Ap4A hydrolase.
Having demonstrated that the elevated Ap4A levels in cells with decreased Ap4A hydrolase protein were maintained even 6 h after activation (Fig. 2B), we examined the association of MITF and Hint-1 in Ap4A hydrolase knockdown RBL cells (Fig. 2C). Endogenous MITF was precipitated by anti-MITF antibody 6 h after the immunological activation of RBL cells, and coimmunoprecipitation of Hint-1 was determined by blotting with anti-Hint-1. As can be seen in Fig. 2C, the accumulated Ap4A in Ap4A hydrolase siRNA-treated cells was found to be sufficient to maintain Hint-1 dissociation from MITF for at least 6 h after activation.
Ap4A hydrolase modulating MITF and USF2 targets gene transcription in mast cells.
To quantitatively assess the direct effect of Ap4A hydrolase on the transcriptional activity of MITF, RBL cells were transfected with a luciferase reporter plasmid containing the mouse mMCP6 promoter (MITF-responsive gene) (22). The transcriptional activity of MITF was determined as the relative luciferase activity in untreated cells or in cells treated either with NR siRNA or with Ap4A hydrolase siRNA (Fig. 3A). A significant increase in activity was detected in cells in which Ap4A hydrolase was depleted.
FIG. 3.
Ap4A hydrolase regulates the transcription of MITF and USF2 target genes in mast cells. (A) RBL cells were transfected with either NR siRNA or siRNA against Ap4A hydrolase. Twenty-four hours later, the cells were transfected with luciferase reporter under the control of the mMCP6 promoter. Cells were then stimulated with 100 ng/ml IgE anti-DNP and 100 ng/ml DNP. The luciferase activity of lysed cells was measured and normalized against the protein concentration. The mean and standard error of the mean for three experiments are shown. (B) RBL cells were transfected with either Ap4A hydrolase siRNA or NR siRNA. Cells were activated with 100 ng/ml IgE anti-DNP and challenged with 100 ng/ml DNP. Twenty-four hours later, cells were lysed and the mRNA quantitations of TPH, c-kit, and TERT were determined by Sybr green incorporation to real-time PCR in RBL cells. Expression levels were normalized to those of the β-actin housekeeping gene. Results are presented relative to NR siRNA-treated cells which were arbitrarily determined as 1. The mean and standard error of the mean for three experiments are shown.
Furthermore, the transcript levels of two of the MITF target genes, encoding TPH and c-kit (9, 36), were measured in immunologically activated RBL cells in which Ap4A hydrolase had been knocked down using the corresponding siRNA. A significant accumulation of TPH mRNA and a moderate increase in c-kit mRNA accumulation was observed in RBL cells, with reduced expression of the Ap4A hydrolase (Fig. 3B). Thus, Ap4A hydrolase is considerably involved in the regulation of MITF transcriptional activity.
USF2 is a bHLH-Zip transcription factor which shares a similar DNA-binding sequence to that of MITF. We have found that USF2, similar to MITF, is negatively regulated by Hint-1 and that Ap4A generated upon IgE-Ag activation acts as a positive regulator of USF2 by dissociating it from its repressor (17). It should be noted that the induction of USF2 transcriptional activity via Ap4A in a mast cell line affected only certain USF2 target genes, such as the gene encoding TERT (17). To determine the direct effect of Ap4A hydrolase on USF2 transcriptional activity, the transcript of TERT was measured after immunological triggering. The introduction of Ap4A hydrolase siRNA specifically increased the transcript level of the gene encoding TERT (Fig. 3B).
Cardiomyocyte stimulation by isoproterenol has a similar effect to mast cell immunological activation.
MITF has been shown to be transcribed in high levels in the heart (6). We have recently demonstrated that MITF plays an essential role in β-adrenergic-induced cardiac hypertrophy (34). Therefore, the role played by Ap4A in MITF regulation was determined in a rat cardiac cell line (H9C2), a cell type that was not derived from the hematopoietic lineage. The cells were exposed for 30 min to the β-agonist isoproterenol, an inducer of cardiac hypertrophy, and then lysed. As shown in Fig. 4A, an increase in the level of Ap4A was observed in cells treated with isoproterenol. The effect of Ap4A elevation on MITF-Hint-1 association/dissociation was determined by immunoprecipitation of MITF followed by immunoblotting with Hint-1 antibody. The increased concentration of Ap4A in stimulated cells correlated with the disassociation of MITF from Hint-1 (Fig. 4B). These data suggest that the role played by Ap4A in the regulation of MITF's association with Hint-1 in IgE-Ag-activated RBL cells could be applied to other cell types.
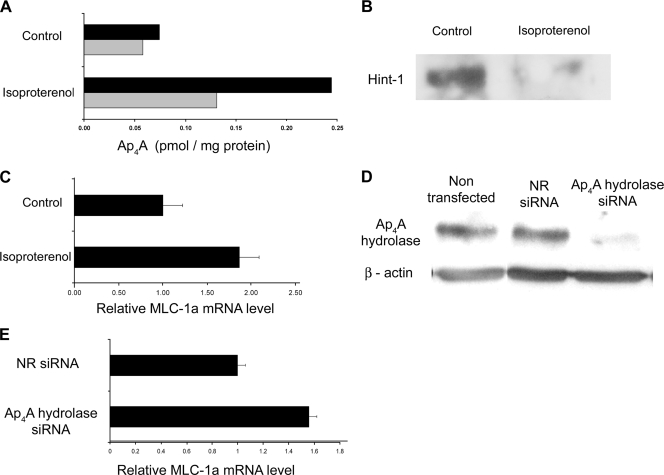
FIG. 4.
Role of Ap4A in the regulation of the MITF target gene, encoding MLC-1a, in activated rat cardiac cells (H9C2). (A) The Ap4A level was measured in H9C2 cells cultured with or without isoproterenol for 30 min. The results of two experiments are shown. (B) Coimmunoprecipitation of MITF and Hint-1. MITF was immunoprecipitated from extracts of H9C2 cells cultured with or without isoproterenol. The level of coimmunoprecipitated Hint-1 was determined by Western blot analysis. Results for one representative experiment out of three are shown. (C) H9C2 cells were cultured with or without isoproterenol. Four hours later, cells were lysed and the mRNA quantitation of MLC-1a was determined by Sybr green incorporation to real-time PCR. Expression levels were normalized to β-actin. The mean and standard error of the mean for three experiments are shown. (D) H9C2 cells were transfected with either NR siRNA or siRNA against Ap4A hydrolase. Forty-eight hours after transfection, Western blot analysis was carried out for cell lysates by using Ap4A hydrolase specific antibody. Results for one representative experiment out of five are shown. (E) H9C2 cells were transfected with either Ap4A hydrolase siRNA or NR siRNA. 24 h after cells were stimulated with isoproterenol they were lysed and the mRNA quantitation of MLC-1a was determined by Sybr green incorporation to real-time PCR in H9C2 cells. Expression levels were normalized to the β-actin housekeeping gene. The result is presented relative to NR siRNA-treated cells, which was arbitrarily determined as 1. The mean and standard error of the mean for three experiments are shown.
Furthermore, recently we identified that the expression of MLC-1a is regulated by the MITF isoform MITF-H in cardiomyocytes (35). To determine whether manipulation of Ap4A level via isoproterenol, which led to dissociation of the MITF-Hint complex, would affect MITF transcriptional activity, we measured the MLC-1a mRNA level. An increase in MLC-1a transcription was observed 4 h after the addition of isoproterenol to H9C2 cells (Fig. 4C).
Ap4A hydrolase is involved in MITF gene regulation in the cardiac cell line.
In order to determine the involvement of Ap4A hydrolase in MITF regulation in H9C2 cells, the enzyme level was depleted using the siRNA technique. As shown in Fig. 4D, the same siRNA sequence used in RBL cells also silenced the hydrolase in the cardiac cell line, as they are both rat cell lines.
The H9C2 cells were treated with either the control NR siRNA or the Ap4A hydrolase siRNA and then stimulated with isoproterenol. Twenty-four hours later, the cells were lysed and the transcript level of MLC-1a was measured. In cells lacking the Ap4A hydrolase, the gene's mRNA level was considerably higher (Fig. 4E), suggesting that Ap4A hydrolase participates in MITF regulation also in other MITF-expressing cells.
DISCUSSION
In this study, we have gathered evidence supporting a possible function of Ap4A as a second messenger in the transcriptional regulation of MITF- and USF2-responsive genes. Several criteria have been established that a molecule has to fulfill in order to be unambiguously assigned as a second messenger (30, 32). These criteria are as follows: (i) the messenger should mimic the effect of the extracellular stimulus when applied intracellularly, (ii) enzymatic machinery should be present to synthesize and metabolize the prospective messenger, (iii) the messenger's intracellular levels should change in response to the extracellular stimulus, (iv) antagonists of the action of the messenger should block the effects of the stimulus whose effects are proposed to be transduced by the messenger, and (v) specific intracellular targets should be present for the candidate messenger. Our previous works confirm that Ap4A fulfills three of these criteria (16, 17). Namely, Ap4A applied intracellularly to mast cells mimics the effect of IgE-Ag stimulation, intracellular Ap4A levels changed in response to extracellular stimuli, and specific intracellular targets were identified for Ap4A. Here, we established Ap4A as a signaling molecule by determining the enzyme directly responsible for its degradation. Furthermore, our observations demonstrated that this novel pathway of gene regulation by Ap4A is broader than expected. We have shown that it is not a phenomenon exclusive to mast cells triggered with IgE and Ag but that it can also be found in heart cells activated with the β-agonist isoproterenol.
The assessment of the role of Ap4A as a possible signal transduction molecule is a complex issue. While this dinucleotide has been postulated to be produced by several aminoacyl tRNA synthetases, LysRS is the major contributor to its production (39). Aminoacyl tRNA synthetases are essential for translation, and therefore any silencing of these proteins can result in unwanted effects within the cell. Thus, manipulation of Ap4A levels through silencing of its degradation offers a unique approach to assess the importance of this nucleotide in any cellular process.
It is clear that degradation of Ap4A is important for any signal transduction pathways in which Ap4A is involved. The main candidate enzyme for degrading stimulus-induced Ap4A is Ap4A hydrolase, even though it is clear that other enzymes, such as NPP1, NPP2, and NPP3, are capable of degrading Ap4A (38).
In Schizosaccharomyces pombe, Aph1 is the known Ap4A hydrolase (7). Disruption of the aph1 gene resulted in substantially increased Ap4A levels in the yeasts without an obvious effect on the morphology of the yeast cells (7). We decided to use a similar approach of downregulation of the proposed Ap4A degrading enzyme by means of the siRNA technique. Depletion of Ap4A hydrolase in mast cells resulted in a substantial increase in Ap4A levels and a delay in the expected return to basal levels several hours after IgE-Ag stimulation (Fig. 2). This is clear evidence that Ap4A hydrolase is indeed the main enzyme responsible for Ap4A degradation following Ap4A induction.
Surprisingly, despite the fact that Ap4A and therefore also Ap4A hydrolase are crucial for some basic functions of cells, no diseases have been reported to be associated with mutations or deletions of the Ap4A hydrolase gene. A possible explanation is that the loss of function of this gene is fatal to the cells. Alternatively, there could be another isoform that has not yet been identified.
The involvement of Hint-1 in the regulation of transcription has been proposed for a few cell systems (14, 29). Our observations demonstrated that Hint-1 is a repressor of MITF (29) and USF2 (17) transcriptional activity, and recently Hint-1 was also found to repress the transcription factor AP-1 (40). Our previous studies suggested that an increase in Ap4A can induce the release of the inhibitory protein Hint-1 from MITF and USF2. A decrease in Hint-1 bound to either of these transcription factors was correlated with an increase in their transcriptional activity. Cellular Ap4A levels were increased in those experiments by the introduction of Ap4A into cells using cold shock. This method of raising cellular Ap4A levels is very different to a more physiologically relevant method, whereby endogenous Ap4A is induced. Thus, in the present study we chose to manipulate Ap4A levels by downregulating Ap4A hydrolase. This was followed by experiments assessing the possible role of Ap4A on the expression of MITF and USF2 target genes. These experiments showed that gene expression is higher in Ap4A hydrolase-depleted cells, providing crucial evidence supporting our previously described model (16, 17) in which induction of Ap4A levels can result in significant increases in the expression of MITF- and USF2-regulated genes. It is important to note that the differences between TPH and c-kit mRNA accumulation can be explained by the observation that several potential binding sites for transcription factors other than MITF, such as AP-2, Sp-1, GATA-1, myb, and Oct-4, were identified in the c-kit promoter (3, 41).
In contrast to MITF, USF2 is ubiquitously expressed in eukaryotic cells. This suggests that the involvement of Ap4A hydrolase in the regulation of bHLH-Zip transcription factors may be universal as opposed to mast cell specific. Indeed, this pathway of gene regulation was also seen in a cardiac cell line. Recently we described the pivotal role of MITF in β-adrenergic-induced cardiac hypertrophy (34). Our hypothesis was that increased intracellular Ap4A levels may also activate MITF in cardiac myocytes. Here we described how isoproterenol, a β-adrenergic agonist, induced an increase in Ap4A levels (Fig. 4). Ap4A hydrolase inhibition in these cells resulted in an increase in the levels of the mRNA of MLC-1a, the cardiac target of MITF (34).
Thus, these data provide further support to the notion that Ap4A is involved in signal transduction regulation of gene transcription. Our finding that Ap4A hydrolase regulates stimuli-induced Ap4A levels in two different biological systems strongly indicates the generality of this mechanism. Further experiments in which Ap4A hydrolase activity is manipulated might resolve the role of this dinucleotide in a variety of biological systems.
In conclusion, data presented in the current article demonstrated the critical role of Ap4A hydrolase in the degradation of Ap4A induced in two different types of cells as described in the proposed model (Fig. 5). These results provide crucial evidence in favor of the hypothesis that Ap4A acts as a second messenger which affects the transcriptional regulation of at least two major transcription factors. The methodology presented in this article should prove highly useful for any further efforts to elucidate the possible role of Ap4A as a second messenger in multiple cellular systems.
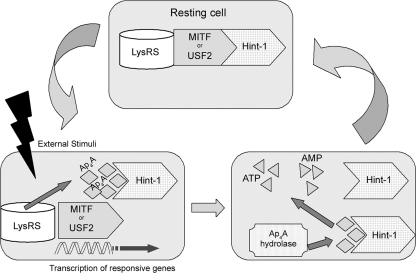
FIG. 5.
Proposed model for the involvement of Ap4A hydrolase in transcription regulation. Upon external stimuli, Ap4A levels are elevated. Ap4A binds to Hint-1, dissociating it from MITF or USF2. With the repression removed, the transcription factors are free to transcribe their responsive genes. Ap4A hydrolase hydrolyzes the accumulated Ap4A into AMP + ATP, returning its level to basal. When Ap4A levels decrease, Hint-1 reassociates with MITF or USF2.
Acknowledgments
This work was supported by the United States-Israel Binational Science Foundation (grant 2003-009 to E. Razin), the Israeli Academy of Science (grant 144/04 to E. Razin), the German-Israel Foundation for Scientific Research and Development (grant I-726-10.2 to E. Razin), and the Legacy Foundation of the Israeli Academy of Science (H. Nechushtan). I. Carmi-Levy is a fellow of the Canadian Friends of Hebrew University.
Footnotes
Published ahead of print on 21 July 2008.
REFERENCES
- 1.Bentley, N. J., T. Eisen, and C. R. Goding. 1994. Melanocyte-specific expression of the human tyrosinase promoter: activation by the microphthalmia gene product and role of the initiator. Mol. Cell. Biol. 147996-8006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bessman, M. J., D. N. Frick, and S. F. O'Handley. 1996. The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J. Biol. Chem. 27125059-25062. [DOI] [PubMed] [Google Scholar]
- 3.Chu, T. Y., and P. Besmer. 1995. Characterization of the promoter of the proto-oncogene c-kit. Proc. Natl. Sci. Counc. Repub. China B 198-18. [PubMed] [Google Scholar]
- 4.Fisher, D. I., and A. G. McLennan. 2008. Correlation of intracellular diadenosine triphosphate (Ap3A) with apoptosis in Fhit-positive HEK293 cells. Cancer Lett. 259186-191. [DOI] [PubMed] [Google Scholar]
- 5.Ge, Y., T. Jippo, Y.-M. Lee, S. Adachi, and Y. Kitamura. 2001. Independent influence of strain difference and mi transcription factor on the expression of mouse mast cell chymases. Am. J. Pathol. 158281-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hodgkinson, C. A., K. J. Moore, A. Nakayama, E. Steingrimsson, N. G. Copeland, N. A. Jenkins, and H. Arnheiter. 1993. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell 74395-404. [DOI] [PubMed] [Google Scholar]
- 7.Ingram, S. W., S. T. Safrany, and L. D. Barnes. 2003. Disruption and overexpression of the Schizosaccharomyces pombe aps1 gene, and effects on growth rate, morphology and intracellular diadenosine 5′,5‴-P1,P5-pentaphosphate and diphosphoinositol polyphosphate concentrations. Biochem. J. 369519-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito, A., T. Jippo, T. Wakayama, E. Morii, Y. Koma, H. Onda, H. Nojima, S. Iseki, and Y. Kitamura. 2003. SgIGSF: a new mast-cell adhesion molecule used for attachment to fibroblasts and transcriptionally regulated by MITF. Blood 1012601-2608. [DOI] [PubMed] [Google Scholar]
- 9.Ito, A., E. Morii, K. Maeyama, T. Jippo, D.-K. Kim, Y.-M. Lee, H. Ogihara, K. Hashimoto, Y. Kitamura, and H. Nojima. 1998. Systematic method to obtain novel genes that are regulated by mi transcription factor: impaired expression of granzyme B and tryptophan hydroxylase in mi/mi cultured mast cells. Blood 913210-3221. [PubMed] [Google Scholar]
- 10.Jippo, T., Y.-M. Lee, Y. Katsu, K. Tsujino, E. Morii, D.-K. Kim, H.-M. Kim, and Y. Kitamura. 1999. Deficient transcription of mouse mast cell protease 4 gene in mutant mice of mi/mi genotype. Blood 931942-1950. [PubMed] [Google Scholar]
- 11.Jippo, T., E. Morii, T. Tsujino, T. Tsujimura, Y.-M. Lee, D.-K. Kim, H. Matsuda, H.-M. Kim, and Y. Kitamura. 1997. Involvement of transcription factor encoded by the mouse mi locus (MITF) in expression of p75 receptor of nerve growth factor in cultured mast cells of mice. Blood 902601-2608. [PubMed] [Google Scholar]
- 12.Kim, S. I., and D. Soll. 1998. Major identity element of glutamine tRNAs from Bacillus subtilis and Escherichia coli in the reaction with B. subtilis glutamyl-tRNA synthetase. Mol. Cells 8459-465. [PubMed] [Google Scholar]
- 13.Kisselev, L. L., J. Justesen, A. D. Wolfson, and L. Y. Frolova. 1998. Diadenosine oligophosphates (Ap(n)A), a novel class of signalling molecules? FEBS Lett. 427157-163. [DOI] [PubMed] [Google Scholar]
- 14.Korsisaari, N., and T. P. Makela. 2000. Interactions of Cdk7 and Kin28 with Hint/PKCI-1 and Hnt1 histidine triad proteins. J. Biol. Chem. 27534837-34840. [DOI] [PubMed] [Google Scholar]
- 15.Lee, P. C., B. R. Bochner, and B. N. Ames. 1983. AppppA, heat-shock stress, and cell oxidation. Proc. Natl. Acad. Sci. USA 807496-7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, Y.-N., H. Nechushtan, N. Figov, and E. Razin. 2004. The function of lysyl-tRNA synthetase and Ap4A as signaling regulators of MITF activity in FcɛRI-activated mast cells. Immunity 20145-151. [DOI] [PubMed] [Google Scholar]
- 17.Lee, Y.-N., and E. Razin. 2005. Nonconventional involvement of LysRS in the molecular mechanism of USF2 transcriptional activity in FcɛRI-activated mast cells. Mol. Cell. Biol. 258904-8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levy, C., H. Nechushtan, and E. Razin. 2002. A new role for the STAT3 inhibitor, PIAS3: a repressor of microphthalmia transcription factor. J. Biol. Chem. 2771962-1966. [DOI] [PubMed] [Google Scholar]
- 19.McLennan, A. G. 2006. The Nudix hydrolase superfamily. Cell. Mol. Life Sci. 63123-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morii, E., T. Jippo, K. Hashimoto, D.-K. Kim, Y.-M. Lee, H. Ogihara, K. Tsujino, H.-M. Kim, and Y. Kitamura. 1997. Abnormal expression of mouse mast cell protease 5 gene in cultured mast cells derived from mutant mi/mi mice. Blood 903057-3066. [PubMed] [Google Scholar]
- 21.Morii, E., H. Ogihara, K. Oboki, C. Sawa, T. Sakuma, S. Nomura, J. D. Esko, H. Handa, and Y. Kitamura. 2001. Inhibitory effect of the mi transcription factor encoded by the mutant mi allele on GA binding protein-mediated transcript expression in mouse mast cells. Blood 973032-3039. [DOI] [PubMed] [Google Scholar]
- 22.Morii, E., T. Tsujimura, T. Jippo, K. Hashimoto, K. Takebayashi, K. Tsujino, S. Nomura, M. Yamamoto, and Y. Kitamura. 1996. Regulation of mouse mast cell protease 6 gene expression by transcription factor encoded by the mi locus. Blood 882488-2494. [PubMed] [Google Scholar]
- 23.Motyckova, G., K. N. Weilbaecher, M. Horstmann, D. J. Rieman, D. Z. Fisher, and D. E. Fisher. 2001. Linking osteopetrosis and pycnodysostosis: regulation of cathepsin K expression by the microphthalmia transcription factor family. Proc. Natl. Acad. Sci. USA 985798-5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy, G. A., D. Halliday, and A. G. McLennan. 2000. The Fhit tumor suppressor protein regulates the intracellular concentration of diadenosine triphosphate but not diadenosine tetraphosphate. Cancer Res. 602342-2344. [PubMed] [Google Scholar]
- 25.Murphy, G. A., and A. G. McLennan. 2004. Synthesis of dinucleoside tetraphosphates in transfected cells by a firefly luciferase reporter gene. Cell. Mol. Life Sci. 61497-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nechushtan, H., and E. Razin. 1998. Deciphering the early-response transcription factor networks in mast cells. Immunol. Today 19441-444. [DOI] [PubMed] [Google Scholar]
- 27.Nechushtan, H., and E. Razin. 2002. The function of MITF and associated proteins in mast cells. Mol. Immunol. 381177. [DOI] [PubMed] [Google Scholar]
- 28.Rapaport, E., and P. C. Zamecnik. 1976. Presence of diadenosine 5′,5‴-P1,P4-tetraphosphate (Ap4A) in mammalian cells in levels varying widely with proliferative activity of the tissue: a possible positive “pleiotypic activator.” Proc. Natl. Acad. Sci. USA 733984-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Razin, E., Z. C. Zhang, H. Nechushtan, S. Frenkel, Y.-N. Lee, R. Arudchandran, and J. Rivera. 1999. Suppression of microphthalmia transcriptional activity by its association with protein kinase C-interacting protein 1 in mast cells. J. Biol. Chem. 27434272-34276. [DOI] [PubMed] [Google Scholar]
- 30.Robison, G. A., R. W. Butcher, and E. W. Sutherland. 1968. Cyclic AMP. Annu. Rev. Biochem. 37149-174. [DOI] [PubMed] [Google Scholar]
- 31.Sonnenblick, A., C. Levy, and E. Razin. 2004. Interplay between MITF, PIAS3, and STAT3 in mast cells and melanocytes. Mol. Cell. Biol. 2410584-10592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sutherland, E. W. 1972. Studies on the mechanism of hormone action. Science 177401-408. [DOI] [PubMed] [Google Scholar]
- 33.Swarbrick, J. D., S. Buyya, D. Gunawardana, K. R. Gayler, A. G. McLennan, and P. R. Gooley. 2005. Structure and substrate-binding mechanism of human Ap4A hydrolase. J. Biol. Chem. 2808471-8481. [DOI] [PubMed] [Google Scholar]
- 34.Tshori, S., D. Gilon, R. Beeri, H. Nechushtan, D. Kaluzhny, E. Pikarsky, and E. Razin. 2006. Transcription factor MITF regulates cardiac growth and hypertrophy. J. Clin. Invest. 1162673-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tshori, S., A. Sonnenblick, N. Yannay-Cohen, G. Kay, H. Nechushtan, and E. Razin. 2007. Microphthalmia transcription factor isoforms in mast cells and the heart. Mol. Cell. Biol. 273911-3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsujimura, T., E. Morii, M. Nozaki, K. Hashimoto, Y. Moriyama, K. Takebayashi, T. Kondo, Y. Kanakura, and Y. Kitamura. 1996. Involvement of transcription factor encoded by the mi locus in the expression of c-kit receptor tyrosine kinase in cultured mast cells of mice. Blood 881225-1233. [PubMed] [Google Scholar]
- 37.Varshavsky, A. 1983. Diadenosine 5′,5‴-P1,P4-tetraphosphate: a pleiotropically acting alarmone? Cell 34711-712. [DOI] [PubMed] [Google Scholar]
- 38.Vollmayer, P., T. Clair, J. W. Goding, K. Sano, J. Servos, and H. Zimmermann. 2003. Hydrolysis of diadenosine polyphosphates by nucleotide pyrophosphatases/phosphodiesterases. Eur. J. Biochem. 2702971-2978. [DOI] [PubMed] [Google Scholar]
- 39.Wahab, S. Z., and D. C. H. Yang. 1985. Synthesis of diadenosine 5′,5‴-P1,P4-tetraphosphate by lysyl-tRNA synthetase and a multienzyme complex of aminoacyl-tRNA synthetases from rat liver. J. Biol. Chem. 2605286-5289. [PubMed] [Google Scholar]
- 40.Wang, L., Y. Zhang, H. Li, Z. Xu, R. M. Santella, and I. B. Weinstein. 2007. Hint1 inhibits growth and activator protein-1 activity in human colon cancer cells. Cancer Res. 674700-4708. [DOI] [PubMed] [Google Scholar]
- 41.Yamamoto, K., A. Tojo, N. Aoki, and M. Shibuya. 1993. Characterization of the promoter region of the human c-kit proto-oncogene. Jpn. J. Cancer Res. 841136-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zamecnik, P. C., M. L. Stephenson, C. M. Janeway, and K. Randerath. 1966. Enzymatic synthesis of diadenosine tetraphosphate and diadenosine triphosphate with a purified lysyl-sRNA synthetase. Biochem. Biophys. Res. Commun. 2491-97. [DOI] [PubMed] [Google Scholar]