Abstract
Human immunodeficiency virus type 1 (HIV-1) transcription is regulated by the viral Tat protein, which relieves a block to elongation by recruiting an elongation factor, P-TEFb, to the viral promoter. Here, we report the discovery of potent Tat inhibitors that utilize a localization signal to target a dominant negative protein to its site of action. Fusing the Tat activation domain to some splicing factors, particularly to the Arg-Ser (RS) domain of U2AF65, creates Tat inhibitors that localize to subnuclear speckles, sites where pre-mRNA processing factors are stored for assembly into transcription complexes. A U2AF65 fusion named T-RS interacts with the nonphosphorylated C-terminal domain of RNA polymerase II (RNAP II) via its RS domain and is loaded into RNAP II holoenzyme complexes. T-RS is recruited efficiently to the HIV-1 promoter in a TAR-independent manner before RNAP II hyperphosphorylation but not to cellular promoters. The “preloading” of T-RS into HIV-1 preinitiation complexes prevents the entry of active Tat molecules, leaving the complexes in an elongation-incompetent state and effectively suppressing HIV-1 replication. The ability to deliver inhibitors to transcription complexes through the use of targeting/localization signals may provide new avenues for designing viral and transcription inhibitors.
Dominant negative proteins typically are nonfunctional variants that form inactive oligomers with a wild-type subunit or otherwise compete for functionally essential protein-protein or protein-nucleic acid interactions (21). Transcription complexes have provided prime targets for dominant-negative inhibition due to the large number of interfaces formed during transcription and the dynamic nature of transcription factor interactions during key steps of complex assembly and disassembly (8, 20). However, inhibition typically requires high levels of expression of the mutant protein to inactivate, at least partially, the wild-type protein activity (13, 17, 21, 44). Dominant negative proteins have been developed as potential human immunodeficiency virus type 1 (HIV-1) therapeutics, including some aimed toward altering viral transcription (19, 38, 48).
In HIV-1, the viral Tat protein is essential for regulating transcription initiation complex assembly (40) and also for recruiting P-TEFb (positive transcription elongation factor b) to a promoter-proximal site on the nascent HIV-1 pre-mRNA (the transactivation response element [TAR]) to assemble elongation-competent, activated transcription complexes (4). Without Tat, RNA polymerase II (RNAP II) complexes are inefficiently converted to the elongating form, which requires phosphorylation of the C-terminal domain (CTD) of the large RNAP II subunit (1, 24). P-TEFb is a heterodimer of cyclin T1 (CycT1) and its associated Cdk9 catalytic subunit and is required by many, but not all, activators for CTD phosphorylation, either at the promoter or during elongation (3, 18, 37). In the case of HIV-1, the Tat activation domain (AD; residues 1 to 48), in the absence of its RNA-binding domain (RBD), functions as a weak dominant negative that is believed to form inactive complexes with P-TEFb (12, 19, 33, 35). Their potential use in therapeutic strategies has been hindered, in part, by their low potency.
The unusual function of Tat as an RNA-binding transcription factor has allowed the development of the Tat hybrid assay, in which the Tat AD fused to a heterologous RBD activates an HIV-1 long-terminal-repeat (LTR) reporter containing a cognate RNA-binding site in place of TAR (26). In developing the Tat hybrid assay to screen libraries for RNA-protein interactions, we discovered a novel class of highly potent dominant negatives, exemplified by fusions with splicing factors, whose potencies appear to be dictated by their efficient recruitment to the HIV-1 promoter. We report that tethering a targeting/localization motif, such as a splicing factor Arg-Ser (RS) domain, to a dominant negative domain strongly enhances inhibitory activity by facilitating the loading of such an inhibitor into HIV-1 transcription complexes. This recruitment-based mechanism effectively co-opts the transcriptional machinery, impairing Tat loading at the promoter, blocking transcription elongation, and inhibiting viral replication.
MATERIALS AND METHODS
Transcriptional activation and inhibition reporter assays.
For fluorescence-activated cell sorter analyses, HeLa cells were transfected with green fluorescent protein (GFP) or DsRed reporter plasmids and appropriate amounts of Tat activator and inhibitor plasmids by using PolyFect (Qiagen). Reporter activity was measured 48 h posttransfection by using a Becton-Dickinson FACSCalibur instrument. Activation (n-fold) was calculated by determining the number of cells in the appropriate quadrant, multiplied by their average fluorescence, relative to the same values calculated for the reporters alone. All LTR reporter plasmids utilize the HIV-1 LTR and various RNA elements in place of TAR (see Table 1) and contain an internal ribosome entry site upstream of the firefly luciferase (FFL) gene to ensure efficient translation irrespective of the 5′ untranslated region sequence used. For FFL reporter assays, transcriptional activities in the linear range were obtained by transfecting HeLa cells with 5 ng of Tat activator and 1 or 5 ng of Tat fusion (referred to as T-fusion) plasmids (see Table 2) (1:0.2 and 1:1 ratios of activator to inhibitor). All transfection mixtures included an appropriate FFL reporter and pCMV-RL (with Renilla luciferase) (Promega) to normalize for transfection efficiency, and activities were measured using a Dual-Glo luciferase assay kit (Promega). Activation assays were performed in triplicate, and data are presented as means ± standard deviations.
TABLE 1.
Reporter nomenclature
| Reporter | Promoter/RNA element | Reporter protein |
|---|---|---|
| HIV-1 LTR-HTAR-FFL | HIV-1 LTR/HIV-1 TAR | FFL |
| HIV-1 LTR-BTAR-FFL | HIV-1 LTR/BIV TAR | FFL |
| HIV-1 LTR-RREIIB-FFL | HIV-1 LTR/HIV-1 RREIIB | FFL |
| HIV-1 LTR-ΔTAR-FFL | HIV-1 LTR/no TAR | FFL |
| HIV-1 LTR-BPS-FFL | HIV-1 LTR/intronic BPS | FFL |
| HIV-1 LTR-BTAR-DsRed | HIV-1 LTR/BIV TAR | DsRed |
| HIV-1 LTR-BPS-GFP | HIV-1 LTR/intronic BPS | GFP |
TABLE 2.
T-fusion nomenclature
| Fusion | Tat portion/fusion partner(s) | Tag(s) used |
|---|---|---|
| T-BIV | HIV-1 Tat AD/BIV Tat RBD | HA |
| T-SF1 | HIV-1 Tat AD/splicing factor 1 | HA |
| T-U2AF65 | HIV-1 Tat AD/U2AF65 | HA, GFP |
| T(K41A)-U2AF65 | HIV-1 Tat AD K41A/U2AF65 | HA, GFP |
| T-U2AF65ΔRS | HIV-1 Tat AD/U2AF65ΔRS domain | GFP |
| Tat-U2AF65 | HIV-1 Tat/U2AF65 | HA |
| T | HIV-1 Tat AD/no fusion | GFP |
| T-Rev | HIV-1 Tat AD/HIV-1 Rev RBD (residues 2-73) | HA, GFP |
| T-NLS | HIV-1 Tat AD/SV40 T-Ag NLS | GFP, HF |
| T-RS | HIV-1 Tat AD/U2AF65 RS domain | HA, GFP, HF |
| T(K41A)-RS | HIV-1 Tat AD K41A/U2AF65 RS domain | HA, GFP |
| T-MS2cp | HIV-1 Tat AD/MS2 coat protein | HA |
| T-U2AF35 | HIV-1 Tat AD/U2AF35 | HA |
| T-RS25-63 | HIV-1 Tat AD/U2AF65 RS dipeptides of residues 25-63 | HA |
| T-RE | HIV-1 Tat AD/U2AF65 RS dipeptides of residues 25-63 mutated to RE | HA |
| T-RG | HIV-1 Tat AD/U2AF65 RS dipeptides of residues 25-63 mutated to RG | HA |
| T-GS | HIV-1 Tat AD/U2AF65 RS dipeptides of residues 25-63 mutated to GS | HA |
| T-BIV-U2AF65 | HIV-1 Tat AD/BIV Tat RBD and U2AF65 |
RNase protection assay.
HeLa cells were transfected with reporter alone or with activator- and inhibitor-expressing plasmids. Total RNA was extracted by use of TRIzol (Invitrogen), and 15 μg of each sample was hybridized with proximal and distal probes corresponding to the HIV-1 LTR promoter and FFL open reading frame regions, respectively. The antisense probes were synthesized from linearized templates using a MaxiScript kit (Ambion), and hybridization was performed with an [α-32P]CTP-labeled probe {in 80% formamide, 40 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], 400 mM NaCl, 1 mM EDTA} incubated at 42°C overnight. RNase digestion was performed for 1.5 h at 30°C (in 10 mM Tris [pH 8.0], 300 mM NaCl, 5 mM EDTA, 11 units/μl of RNase A, 11 units/μl RNase T1), and duplexes were purified. RNAs were separated on a 6% polyacrylamide-8 M urea gel and visualized and quantified by using a Typhoon phosphorimager (Molecular Dynamics). Experiments were performed in duplicate with similar results.
Expression analyses by Western blotting and fluorescence microscopy.
To quantitatively assess relative inhibitor and activator expression levels by Western blotting (see Fig. 1), HeLa cells were cotransfected with 0.3 μg of pEGFPN3 (Clontech) and 1.35 μg of pSV2-HA-tagged T-fusion plasmids in six-well plates. Nuclear extracts were prepared using a NE-PER kit (Pierce), and samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose, and probed with antibodies (see Table S1 in the supplemental material). To examine localization of the GFP-tagged T-fusions (see Fig. 4), HeLa cells were grown to 50% confluence on glass coverslips, transfected with 100 ng of plasmid DNA, fixed in 4% paraformaldehyde in 1× phosphate-buffered saline (PBS) buffer (pH 7.4) at 24 h posttransfection, rinsed twice with PBS, and permeabilized with PBS-0.5% Triton X-100 for 10 min at 4°C. Nonspecific antibody sites were blocked in 1× PBS, 3% goat serum, and 4% bovine serum albumin for 1 h at room temperature, and cells were incubated with primary antibodies for 1 h at room temperature, washed three times with PBS, incubated with appropriate Alexa 488-coupled secondary antibodies (Molecular Probes), and washed three times with PBS. Coverslips were then mounted on DAPI (4′,6-diamidino-2-phenylindole)-containing Vectashield slides (Vector Labs) and examined using an LSM510 confocal microscope (Zeiss).
FIG. 1.
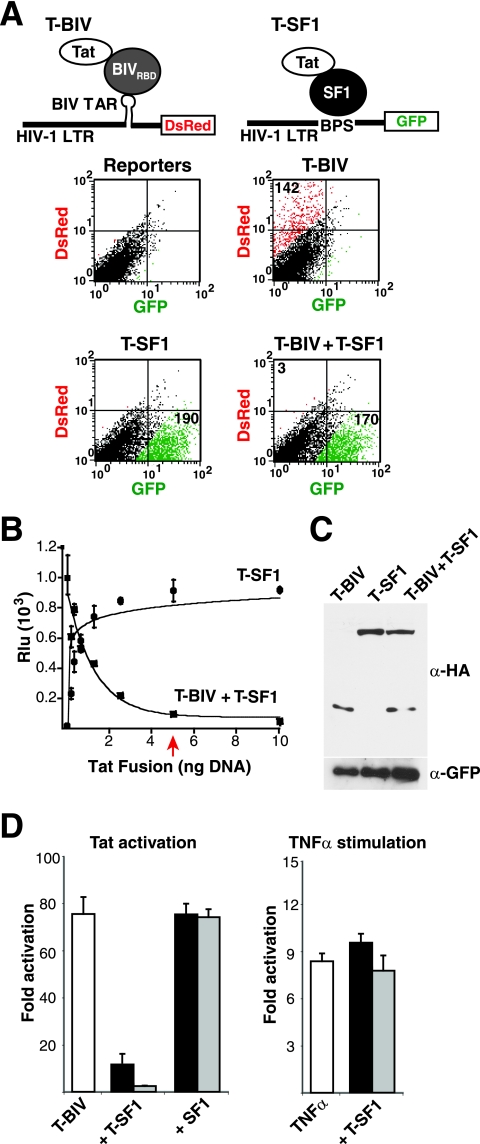
A potent dominant negative Tat inhibitor identified in a reporter assay. (A) The top illustrations represent a schematic for a dual-reporter fluorescence assay in which T-BIV is used to activate an HIV-1 LTR-BTAR-DsRed reporter. The T-SF1 fusion protein is used to activate an HIV-1 LTR-BPS-GFP reporter (Tables 1 and 2). The graphs shows results for HeLa cells cotransfected with both reporters and T-BIV and/or T-SF1 activators as indicated and sorted by flow cytometry. Expression of GFP is shown in green, and expression ofDsRed is shown in red. Numbers in each quadrant represent activation (n-fold). (B) Dose-response curves of T-SF1 activation on an HIV-1 LTR-BPS-FFL reporter and T-SF1-mediated inhibition of T-BIV activity on an HIV-1 LTR-BTAR-FFL reporter. Rlu, relative luciferase units. The arrow indicates stoichiometric DNA concentrations of inhibitor and activator. (C) Western blot of HeLa cells cotransfected with HA-tagged versions of the T-BIV activator and/or the T-SF1 inhibitor along with a GFP expressor to normalize for transfection efficiency. α, anti. (D) The left panel shows results for HeLa cells transiently transfected with the HIV-1 LTR-BTAR-FFL reporter plasmid and different ratios of T-BIV activator to T-SF1 or unfused SF1 plasmids (1:0.2, black bars; 1:1, gray bars). The right panel shows results for HeLa cells transiently transfected with the HIV-1 LTR-BTAR-FFL reporter plasmid and two amounts of T-SF1 plasmid (1 ng, black bars; 5 ng, gray bars), and transcription was stimulated 16 h posttransfection by incubation with TNF-α (10 ng/ml) for 4 h.
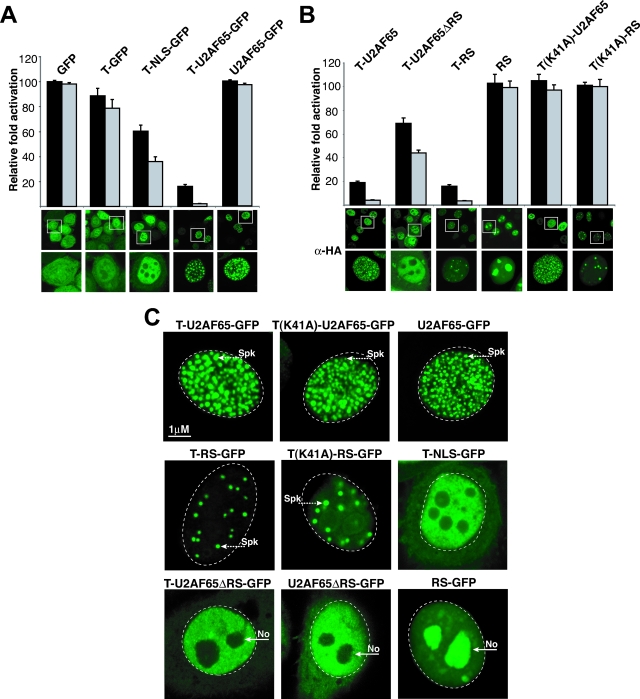
FIG. 4.
Domains that contribute to inhibition and subcellular localization. (A) HeLa cells were transiently cotransfected with an HIV-1 LTR-RREIIB-FFL reporter plasmid, T-Rev activator, and various inhibitors at 1:0.2 (black bars) or 1:1 (gray bars) ratios of activator to inhibitor. Activation levels are plotted as relative levels of activation, normalized to the level of activation without inhibitor (85-fold). Confocal images of each GFP-tagged fusion, including images with magnification of ×3 (lower images, representing the boxed cells shown in the upper images) to highlight the subcellular compartments, are shown below the graph. T-NLS-GFP contains the eight-amino-acid NLS (PPKKKRKV) of simian virus 40 T-Ag (Table 2). (B) Relative activities of T-U2AF65 deletion mutants and Tat AD variants, as determined in experiments whose results are shown in panel A. Corresponding confocal anti-HA (α-HA) immunofluorescence images are shown. T-U2AF65ΔRS-HA has a deletion of the first 90 amino acids of U2AF65, and T-RS contains only residues 2 to 73. (C) Subnuclear localization of active and inactive Tat inhibitors. HeLa cells were transfected with pEGFP-N3 plasmids expressing GFP fused to U2AF65, T-U2AF65 (dominant negative), T(K41A)-U2AF65 (inactive dominant negative), RS (U2AF65 RS domain only), T-RS (active dominant negative), T(K41A)-RS (inactive dominant negative), U2AF65ΔRS, and T-U2AF65ΔRS. Spk indicates the position of nuclear speckles; No indicates nucleoli.
Selection and characterization of HIV-1 LTR reporter-transfected HeLa cell lines.
To generate cell lines with an integrated reporter for chromatin immunoprecipitation (ChIP) analyses, HeLa cells were transfected with an Ssp1-linearized pcDNA3.1 (Invitrogen) template bearing either the HIV-1 LTR-RREIIB (Rev response element, stem-loop IIB)-FFL or HIV-1 LTR-ΔTAR-FFL reporter (Table 1) by use of PolyFect (Qiagen). Clones were selected for more than four weeks in Dulbecco's modified Eagle's medium-10% fetal bovine serum with 750 μg/ml of Geneticin (Gibco). Twenty clones selected from the transfection with HIV-1 LTR-RREIIB-FFL were analyzed for transcription activation with T-Rev (Table 2), and those with reproducible activity levels were selected. From those, a single active clone with the promoter integrated at a single locus was chosen for the ChIP analyses (see Fig. 5). To generate the HIV-1 LTR-ΔTAR-FFL-containing cell line, 12 clones were selected and a clone with a single integration site was chosen based on its response to tumor necrosis factor alpha (TNF-α) (53). The number of integration sites was determined by quantitative PCR (ABI) using genomic DNA extracted with Flexigene (Qiagen) and digested with PacI/EcoRI, which liberates the promoter and amplified with HIV-1 LTR primers (see Table S2 in the supplemental material). Analyses were done using a standard curve derived from the amplification of different amounts of pcDNA-HIV-1 LTR-FFL template and the following formula: (transgene mass/genomic DNA mass) = (transgene bp/haploid genome bp).
FIG. 5.
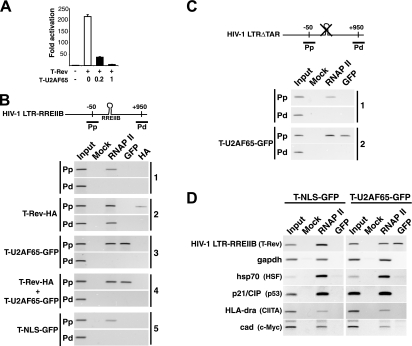
Recruitment of the dominant negative to the HIV-1 promoter via RNAP II. (A) Transcription activation and dose-responsive inhibition of T-Rev in the integrated HIV-1 LTR-RREIIB-FFL reporter-containing cell line used for the ChIP assays, with the molar ratios of the T-U2AF65-GFP inhibitor to T-Rev activator indicated. (B) ChIP from mock-transfected HeLa cells with HIV-1 LTR-RREIIB-FFL (panel 1) or cells transfected with the indicated tagged proteins (panels 2 to 5), using the antibodies shown to monitor occupancy in the Pp and Pd regions. Mock lanes used normal rabbit immunoglobulin G for the immunoprecipitation as a specificity control, and input refers to PCRs from isolated chromatin samples. (C) ChIP from mock-transfected HeLa cells (transfected with HIV-1 LTRΔTAR-FFL; panel 1) or cells transfected with T-U2AF65 (panel 2), using the antibodies shown to monitor occupancy in the Pp and Pd regions. (D) ChIP assays were carried out for the HIV-1 LTR-RREIIB-FFL-transfected HeLa cell line transfected with T-NLS-GFP or T-U2AF65-GFP and using primers to amplify the indicated promoters. Known transcription factors that activate each promoter are indicated in parentheses.
ChIP assay.
The reporter cell lines were transfected with Tat and the indicated T-fusion plasmids (5 μg each) by using calcium phosphate, incubated for 36 h, and washed in PBS. Chromatin was cross-linked with 1% formaldehyde for 5 min at room temperature, and the reaction was stopped by the addition of glycine (150 mM). Cells were washed with PBS and harvested in radioimmunoprecipitation assay (RIPA) buffer (46), and samples were sonicated to generate <500-bp DNA fragments. For immunoprecipitation, 1 mg of protein extract was precleared for 2 h with 40 μl of a 1:1 protein A/G-agarose mixture (Santa Cruz) before the addition of 2 μg of antibody (see Table S1 in the supplemental material). Reaction mixtures were incubated overnight at 4°C in the presence of 40 μl of protein A/G beads preblocked with 1 mg/ml bovine serum albumin and 0.3 mg/ml of salmon sperm DNA. Beads were washed twice with RIPA buffer, four times with ChIP wash buffer (100 mM Tris-HCl [pH 8.5], 500 mM LiCl, 1% [vol/vol] Nonidet P-40, 1% [wt/vol] deoxycholic acid), twice with RIPA buffer, and twice with 1× Tris-EDTA. Immunocomplexes were eluted for 10 min at 65°C with 1% SDS, and cross-linking was reversed by adjusting the solution to 200 mM NaCl and incubating it for 5 h at 65°C. A fraction of the DNA was used as the template in PCRs, which were performed in the exponential range of amplification (25 to 32 cycles, depending on the primer combination and antibody used). To ensure linearity, parallel control PCRs were performed at (−1) cycle with twice the amount of the sample and at (+1) cycle with half the amount of the sample. Amplification products (200 to 250 bp) were electrophoresed in 2% agarose gels and visualized by use of ethidium bromide. PCR primer sequences are shown in Table S2 in the supplemental material. Relative quantification of PCR products was performed with Scion Image1.63 software by using PCR products from known amounts of input DNA as standards.
Protein coimmunoprecipitation and GST pull-down assays.
To examine the association of dominant negative inhibitors with RNAP II (see Fig. 6), HeLa cells were transiently transfected with GFP-tagged T-fusion proteins and protein extracts were prepared with RIPA buffer. Half of the extract was used directly for immunoprecipitation, and the remaining half was treated first with 10 μg of RNase A, which was sufficient to quantitatively digest the RNA from 106 HeLa cells. RNAP II was immunoprecipitated using agarose-conjugated 8WG16 or H14 antibodies overnight at 4°C with mild shaking. Similarly, GFP- and hemagglutinin (HA)-tagged proteins were immunoprecipitated using agarose-conjugated primary antibodies (see Table S1 in the supplemental material). Complexes were dissociated by boiling for 10 min in 2× SDS loading buffer, and proteins were resolved by SDS-PAGE, transferred to nitrocellulose, and analyzed by Western blotting. Interactions between the HA-tagged U2AF65 RS domain (1 μg) and glutathione S-transferase (GST)-CTD (2 to 5 μg) prebound to glutathione-S-Sepharose beads were assessed by incubating both proteins in 500 μl buffer containing 20 mM HEPES (pH 8.0), 100 mM NaCl, and 0.5% NP-40 for 2 h at 4°C, extensively washing the beads, and analyzing them by SDS-PAGE and Western blotting.
FIG. 6.
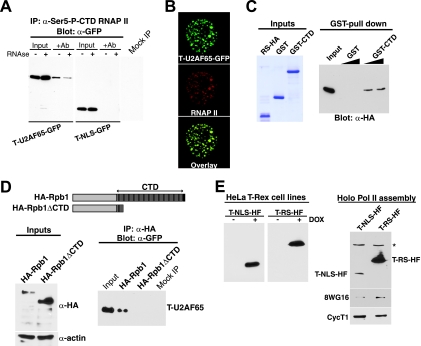
Interactions with the CTD and colocalization with RNAP II. α, anti. (A) GFP-tagged T-U2AF65 and T-NLS proteins were immunoprecipitated from cell extracts and analyzed by Western blotting using the indicated antibodies (Ab). (B) Confocal microscopy of HeLa cells transfected with GFP-tagged T-U2AF65 and immunostained with H14 antibody. (C) In vitro pull-down assays for HA-tagged U2AF65 RS domain with GST-CTD or with GST alone as a control. Inputs show the purified Coomassie blue-stained recombinant proteins used in the pull-down assays. (D) HeLa cells were transfected with the HA-Rpb1 or HA-Rpb1ΔCTD constructs shown (top), and expression levels were assessed by Western blot analysis relative to antiactin antibody (bottom left). Cells were then cotransfected with T-U2AF65-GFP, and extracts were immunoprecipitated (IP) with anti-HA and probed for T-U2AF65-GFP by Western blotting with anti-GFP (bottom right). (E) The left panel shows doxycycline (DOX)-induced expression of HF-tagged Tat AD (T-NLS-HF) and T-RS-HF in stable HeLa T-Rex cell lines analyzed in whole-cell extracts with anti-FLAG. The right panel shows extracts bound and eluted from a GST-TFIIS column that were probed with the antibodies indicated. The asterisk indicates a cross-reacting protein.
Holo-Pol II complex purification and analyses.
A transcription factor IIS (TFIIS) affinity resin was used to purify polymerase II holoenzyme (holo-Pol II) complexes (34, 42). To prepare the resin, Escherichia coli BL21(DE3) cells (Stratagene) expressing GST or GST-TFIIS(N) (a TFIIS N-terminal fragment) (34) were lysed by sonication in Tris-buffered saline buffer containing protease inhibitors and clarified supernatants were bound to a glutathione-agarose resin (Sigma). Holo-Pol II-containing HeLa total extracts were prepared as described previously (34) and further purified by gel filtration on a Sepharose CL-2B column (Amersham).
Selection of inducible T-fusion cell lines.
We constructed T-NLS and T-RS (Table 2) inducible cells lines by cloning the genes into a pcDNAT04 vector (Invitrogen) with a C-terminal His-FLAG (HF) tag and transfecting the plasmids into HeLa T-Rex cells (Invitrogen) that express the tetracycline repressor. A stable cell population was selected in 200 μg/ml zeocin for almost two months, and basal and inducible protein expression were characterized by incubation of cells in Dulbecco's modified Eagle's medium containing 1 μg/ml doxycycline for 16 h and analysis of nuclear extracts by Western blotting with an M2 anti-FLAG antibody (Sigma). The selected populations were used to purify Holo-Pol II complexes (see Fig. 6) as described above.
Dominant negative-expressing SupT1 cells and viral replication kinetics.
Plasmids expressing Tat and/or U2AF65 fusions were constructed in a pBMN retroviral vector (kindly provided by G. Nolan) carrying a simian virus 40 promoter. Plasmids were transfected into øNX packaging cells by using PolyFect (Qiagen), and the retrovirus-containing supernatant was used to transduce human CD4+ SupT1 cells. Populations of stable integrants were selected by growing cells in 2 mg/ml G418 (Invitrogen) for at least 4 weeks. Each stable SupT1 population was infected with an HIV-1 Tat-TAR-dependent or bovine immunodeficiency virus (BIV) Tat-TAR-dependent virus (54) to determine the specificity of inhibition. Supernatant samples were harvested at different intervals following infection, and the amount of viral replication was monitored by p24 antigen expression using enzyme-linked immunosorbent assay (Immuno Diagnostics). Each experiment was performed in duplicate, and mean values for p24 were calculated.
Genomic DNA extraction from SupT1-infected cells and viral genome sequencing.
SupT1-T-U2AF65-, SupT1-T-BIV-U2AF65-, and SupT1-Tat-U2AF65-infected populations (about 1 × 106 cells) were harvested 30 days postinfection and genomic DNA was extracted using Flexigene (Qiagen). DNA was amplified by PCR using Taq polymerase and primer pairs specific to regions of the HIV-1 promoter and surrounding the Tat coding sequence. PCR-amplified DNA was gel purified and cloned into a TOPO vector (Invitrogen). Eight clones from each cell population were sequenced, and sequences were compared to the original HXB2 viral isolate by using the NCBI BLAST algorithm.
Expression levels of Tat domains or fusions from stable SupT1 cell lines.
SupT1 cells (1 × 108) were harvested by centrifugation, resuspended in lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1% NP-40, 0.2 mM EDTA, protease inhibitor cocktail [Roche]), and lysed by sonication on ice using a Branson Sonifier 250 with five 30-s pulses. The lysate (1 ml) was precleared with agarose for 1 h and then incubated with an anti-Tat antibody (MMS-116, 1:100 dilution; Covance) coupled to protein G-agarose (Sigma) for 3 h at 4°C. A parallel control immunoprecipitation was done with an anti-β-actin antibody (Santa Cruz). Samples were washed four times in 1 ml lysis buffer, and proteins were eluted with 0.1 M glycine-HCl (pH 3) and neutralized with 1 M Tris-HCl (pH 10). Half of each sample was electrophoresed on an 8 to 16% gradient gel (Bio-Rad) and probed with the relevant antibody.
RESULTS
A potent dominant negative fusion protein identified in a reporter assay.
Tat can activate transcription through a heterologous RNA site when fused to a corresponding RNA-binding protein (43, 45). We have been developing variants of this Tat hybrid assay to screen for new RNA-binding proteins (26, 47). While modifying the assay to simultaneously monitor two pairs of reporters and T-fusions, we found that certain fusion proteins, particularly those including splicing factors, are potent dominant negatives of Tat activation. Here, we report the exploration of the efficacies of these inhibitors and their mechanisms of action.
To initially calibrate a dual-reporter Tat hybrid assay in a format for fluorescence, we made use of two HIV-1 LTR reporters engineered with corresponding RNA-binding sites (refer to Table 1 for reporter nomenclature) and two previously characterized T-fusions (refer to Table 2 for T-fusion nomenclature). T-BIV, a fusion of the HIV-1 Tat AD and the RBD of BIV Tat, was used to activate an HIV-1 LTR-BIV TAR (BTAR)-DsRed reporter (Fig. 1A), while T-SF1, a fusion of Tat to human splicing factor 1, was used to activate an HIV-1 LTR-branch point sequence (BPS)-GFP reporter (36). When transfected on their own, both T-fusions strongly activated only their cognate RNA reporters (Fig. 1A). Strikingly, activation by T-BIV was strongly inhibited when cotransfected with T-SF1 (3-fold activation), whereas activation by T-SF1 was unaffected (170-fold activation) (Fig. 1A).
Using a more-quantitative luciferase reporter assay, we found that inhibition was remarkably potent, with a stoichiometric amount of T-SF1 plasmid sufficient to almost completely block T-BIV-mediated activation (Fig. 1B). The dose-response curve corresponding to inhibition by T-SF1 mirrors that corresponding to the activation of its cognate BPS reporter (Fig. 1B), demonstrating that T-SF1 is a fully functional activator. The high potency observed by transfection accurately reflects relative protein stoichiometries (Fig. 1C) and requires the fusion of Tat and SF1, as SF1 alone does not inhibit (Fig. 1D) and the Tat AD alone is only a very weak dominant negative (see below). Inhibition is specific to Tat-mediated activation, as T-SF1 does not inhibit TNF-α-induced transcription (Fig. 1D), prompting us to examine the effects on Tat mechanism in more detail.
Specificity of inhibition by T-fusions and RNA-binding requirements.
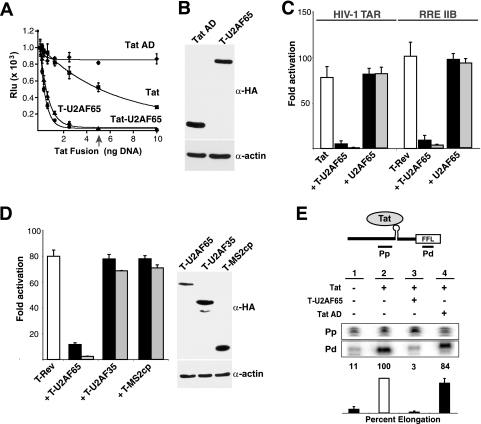
Given that several splicing factors interact with RNAP II- or CTD-associated factors (2, 24, 25, 42), we hypothesized that the SF1 moiety targets the T-fusion to RNAP II. If the targeting hypothesis is correct, then T-fusions with other RNAP II-localized splicing factors, such as U2AF65, might show a similar phenotype. To also assess whether the RBD of Tat contributes to the dominant negative activity, we generated fusions of U2AF65 with full-length Tat (Tat-U2AF65) or the Tat AD alone (T-U2AF65) (Table 2) and measured their effects by using an HIV-1 LTR-BTAR-FFL reporter recognized by the T-BIV activator (Fig. 2A). Indeed, both Tat-U2AF65 and T-U2AF65 inhibited activation more than 10-fold at substoichiometric DNA levels relative to activator (Fig. 2A) and were even more potent than T-SF1, prompting us to focus on the U2AF65 fusions. In contrast, the Tat AD and full-length Tat showed little inhibition, even though the Tat AD and T-U2AF65 are expressed at comparable levels (Fig. 2B), consistent with previous reports of their weak dominant negative activity (7, 12, 19). U2AF65 fusions with full-length Tat and with the AD alone are equally potent, showing that TAR RNA-binding activity is dispensable for inhibition.
FIG. 2.
Tat RNA-binding activity is dispensable for transcription elongation inhibition. α, anti. (A) Dose-response curves indicating inhibition of T-BIV-mediated activation on an HIV-1 LTR-BTAR-FFL reporter by the Tat AD, Tat, T-U2AF65, and Tat-U2AF65 (Table 2). The arrow indicates the position corresponding to stoichiometric DNA concentrations (5 ng) of inhibitor and activator. (B) HeLa cells were cotransfected with HA-tagged versions of the Tat AD and T-U2AF65, and total cell extracts were probed for expression levels with anti-HA antibody or with antiactin to control for protein loading. (C) Activation by HIV-1 Tat and T-Rev (Tat AD fused to Rev) (Table 2) measured in the absence (white bars) or presence (black bars) of T-U2AF65 or unfused U2AF65 by using the HIV-1 LTR-FFL and HIV-1 LTR-RREIIB-FFL reporters indicated. Black bars are 1:0.2 and gray bars are 1:1 ratios of activator to T-U2AF65 or unfused U2AF65. (D) The left panel shows results for activity of T-Rev (white bar) or T-Rev cotransfected with T-U2AF65, T-U2AF35, or T-MS2cp at 1:0.2 (black bars) and 1:1 (gray bars) ratios of T-Rev activator to T-fusion by using the HIV-1 LTR-RREIIB-FFL reporter. The right panel shows results for expression of the T-fusions in nuclear extracts, probed with anti-HA or antiactin to control for protein loading. (E) T-U2AF65 blocks transcription elongation. Cells were transfected with HIV-1 Tat, the Tat AD, or T-U2AF65 as indicated, and RNase protection was performed with a Pp probe directed to the HIV-1 LTR and a Pd probe directed to the FFL open reading frame to quantify transcription rates. The experiment was performed in duplicate with similar results.
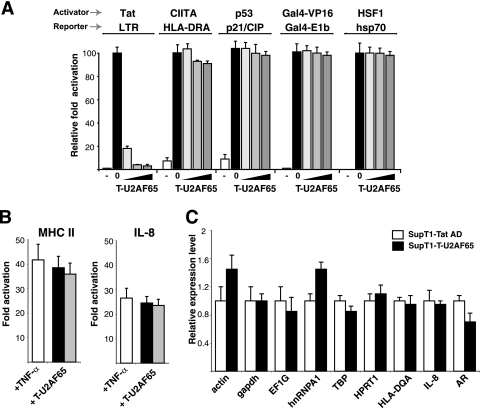
To test whether T-U2AF65 can inhibit activation when Tat is delivered to RNA sites other than BIV TAR, we measured its activity by using a previously described HIV-1 Tat-Rev fusion (T-Rev) (Table 2) and HIV-1 LTR-RREIIB FFL reporter (45), as well as a wild-type HIV-1 Tat and HIV-1 LTR-FFL reporter pair. Activity was in both cases inhibited potently by T-U2AF65 but not by unfused U2AF65 (Fig. 2C), indicating that inhibition does not rely on the nature or affinity of the RNA-protein interaction used to recruit the activator and consistent with the dispensability of TAR-binding activity for inhibition. Since U2AF65 is itself an RNA-binding protein, we asked whether simply appending any RBD would contribute to dominant negative activity. This does not appear to be the case, as T-fusions with U2AF35 (T-U2AF35) or MS2 coat protein (T-MS2cp) (Table 2) are not inhibitors yet are expressed at levels comparable to those of T-U2AF65 (Fig. 2D). Because Tat works primarily at the elongation level, unlike most other activators, we examined the step in transcription affected by the inhibitor. RNase protection assays (Fig. 2E) indicate that T-U2AF65 blocks transcription elongation, but not initiation, at the HIV-1 promoter and that the Tat AD alone is a very weak inhibitor. Specificity of inhibition was confirmed further by the observation that only Tat-mediated activation of the HIV-1 promoter was inhibited and not basal or activated transcription by other activators on their cognate promoters, including the P-TEFb-dependent promoters HLA-DRA (a class II major histocompatibility complex [MHC]) and interleukin-8 (IL-8) (28) (Fig. 3A and B). As well, expression levels from a variety of endogenous promoters were not affected (Fig. 3C).
FIG. 3.
Promoter specificity of the T-U2AF65 fusion. (A) Mammalian cells were transiently transfected with reporter plasmid and different ratios of activator to T-U2AF65 plasmids (from left to right, 1:0, 1:0.2, 1:1, and 1:2). Activities were normalized to activation by HIV-1 Tat alone and to a cotransfected pCMV-RL internal control. To measure heat shock response, endogenous HSF1 was activated 24 h posttransfection by treatment of HeLa cells with 50 μM AsNO2 for 12 h. The activity of transfected p53 was measured in SAOS2 cells. (B) HeLa cells were transiently transfected with the indicated luciferase reporter plasmids (with MHC class II and IL-8) and different ratios of activator to T-U2AF65 plasmids (1:0.2, black bars; 1:1, gray bars). Transcription was stimulated 16 h posttransfection by incubation with TNF-α (10 ng/ml) for 4 h. (C) Relative expression levels of endogenous transcripts in SupT1 cells lines stably expressing the Tat AD or T-U2AF65 using quantitative real-time PCR of total RNA extracted to monitor housekeeping genes (those encoding β-actin [actin], glyceraldehyde-3-phosphate dehydrogenase [gapdh], and eukaryotic translation elongation factor 1 gamma [EF1G]), regulatory factor genes (those encoding heterogeneous nuclear ribonucleoprotein 1 [hnRNPA1], TBP, hypoxanthine phosphoribosyltransferase 1 (HPRT1)]), and P-TEFb regulated genes (those encoding HLA-DQA1 MHC class II [HLA-DQA], IL-8, and androgen receptor [AR]).
Domains required for inhibition and subnuclear localization.
Splicing factors often are localized to subnuclear compartments, where they are recruited to sites of active transcription and splicing (32). If the T-fusions are targeted to RNAP II, like splicing factors are, we would anticipate similar localization patterns. Indeed, T-U2AF65-GFP showed a striking subnuclear speckle pattern, like U2AF65-GFP (Fig. 4A) or U2AF65 itself (14). In contrast, T-fusions to GFP without or with a nuclear localization signal (NLS) (T-GFP or T-NLS-GFP), which had weak dominant negative activity (Fig. 4A), showed very different localization patterns. T-GFP was distributed mostly in the cytoplasm and nucleus, like unfused GFP, whereas >95% of T-NLS-GFP is nuclear, and T-NLS-GFP is absent from the nucleolus (Fig. 4A and C), indicating that nuclear localization is not the major factor contributing to potency.
Previous results implicate splicing factor RS domains in a subnuclear localization (32). To test whether the U2AF65 RS domain is responsible for T-U2AF65 localization and potent inhibition activity, we generated a T-fusion lacking this portion (T-U2AF65ΔRS, which contains U2AF65 residues 91 to 475) and a second with the RS domain alone (T-RS, which contains U2AF65 residues 2 to 73). Of these, T-RS remained a potent inhibitor and showed a speckle pattern, whereas T-U2AF65ΔRS was a weak inhibitor and showed a different nuclear pattern with nucleolar exclusion (Fig. 4B and C). The RS domain alone without the Tat AD or the Tat AD alone is a poor inhibitor and is not localized to speckles. Thus, both the Tat AD and RS domain portions of T-RS are needed for speckle localization and inhibition activity. To confirm the importance of the Tat AD, we generated T-U2AF65 and T-RS mutants with a Lys41-to-Ala substitution (K41A) in the AD that abrogates the P-TEFb interaction and therefore Tat function (15). Both fusions are inactive as inhibitors despite having the same localization patterns as the nonmutant versions, indicating that a functional AD is important (Fig. 4B and C). We hypothesize that the RS domain efficiently delivers the Tat AD into compartments where RNAP II complexes assemble for recruitment to the HIV-1 promoter.
Recruitment of the inhibitor to the HIV-1 promoter.
We next used ChIP assays to test the hypothesis that T-U2AF65 is efficiently targeted to the HIV-1 promoter. To assess complex assembly in the context of integrated chromatin, we generated a stable HeLa cell line carrying a single integrated HIV-1 LTR-RREIIB-FFL reporter. This cell line was activated by T-Rev and inhibited by T-U2AF65 in a dose-responsive manner (Fig. 5A). In the absence of T-Rev activator, RNAP II was detected in the promoter-proximal (Pp) region but not in the promoter-distal (Pd) region (Fig. 5B, panel 1), implying a block to elongation, while RNAP II was seen in both regions following activator expression (panel 2). The level of RNAP II detected in the Pp region increased ∼5-fold in the presence of the activator, consistent with its proposed role in transcription complex assembly (40). The T-Rev activator also was detected in the Pp region (panel 2), but notably, the T-U2AF65-GFP inhibitor showed an even higher level of occupancy (panel 3), confirming that the targeting/localization moiety facilitates recruitment to the HIV-1 promoter. To more directly evaluate competition between the activator and inhibitor, we cotransfected activator and inhibitor plasmids together and analyzed promoter occupancy. We observed a high level of occupancy of the promoter by T-U2AF65-GFP, but not by T-Rev (panel 4), suggesting that the inhibitor is able to displace or efficiently competes with the Tat activator for binding to the viral promoter. Furthermore, the AD alone (T-NLS-GFP) was not detectable at the promoter (panel 5), nor was the T(K41A)-U2AF65 mutant (data not shown), indicating the importance of the modular composition (U2AF65 RS domain and Tat AD) for transcription complex assembly.
Because inhibition does not require TAR and occurs when different RNA sites are used to deliver Tat (Fig. 2), we reasoned that the inhibitor does not use TAR to load into the HIV-1 promoter. To test this hypothesis, we generated a stable HeLa cell line carrying a single integrated copy of an HIV-1 LTR reporter with a deletion of TAR (HIV-1 LTR-ΔTAR-FFL) and examined bound factors by ChIP (Fig. 5C). In the absence of Tat activator, RNAP II is associated with the viral promoter (Fig. 5C, panel 1), and more importantly, T-U2AF65 is recruited to the HIV-1 promoter in a TAR-independent manner (panel 2).
This TAR-independent assembly prompted us to measure T-U2AF65 occupancy at non-HIV-1 promoters by ChIP. We found that of five cellular promoters analyzed, including the P-TEFb-dependent MHC class II and hsp70 promoters, none showed detectable T-U2AF65-GFP (Fig. 5D). Thus, the ChIP experiments support the hypothesis that T-U2AF65 is specifically recruited to the HIV-1 promoter, blocking entry of the Tat activator.
The RS domain of the inhibitor interacts with the CTD of RNAP II.
Because splicing factor RS domains can interact with the RNAP II CTD (10, 32) and because the RS domain of T-U2AF65 is important for inhibition, we tested whether T-U2AF65 interacts with RNAP II complexes by coimmunoprecipitating with the nonphosphorylated (RNAP IIa) form of the CTD. T-U2AF65-GFP formed complexes with RNAP IIa in an RNA-independent manner (Fig. 6A) but interacted only weakly with T-NLS-GFP lacking the U2AF65 moiety (data not shown). Immunofluorescence further confirmed the interaction, showing partial colocalization of T-U2AF65-GFP with RNAP IIa (Fig. 6B and data not shown). Furthermore, a direct interaction was observed using the recombinant CTD and the U2AF65 RS domain (Fig. 6C). To further corroborate the hypothesis that the inhibitor is loaded into transcription complexes via an interaction with the CTD, we cotransfected T-U2AF65-GFP into HeLa cells along with an α-amanitin-resistant Rpb1 mutant containing either a full-length CTD (52 heptad repeats) or a CTD deletion mutant (5 heptad repeats) (11, 16) and coimmunoprecipitated the complexes (Fig. 6D). Both the full-length and deletion-containing Rpb1 forms were expressed, but T-U2AF65 immunoprecipitated only with the full-length protein.
To more precisely determine which RNAP II complexes interact with the inhibitor, we purified polymerase holoenzyme complexes by using a GST-TFIIS affinity column, in which the TFIIS elongation factor selectively binds transcription complexes that have not yet assembled into elongation complexes (23, 34). We generated HeLa cell lines inducibly expressing T-NLS or T-RS (Table 2) and purified holo-Pol II by using the GST-TFIIS column (Fig. 6E). The isolated complexes were purified further by gel filtration and migrated as a single 2- to 4-MDa peak (data not shown). Both the unfused Tat AD (T-NLS) and the T-RS inhibitor were found in holo-Pol II complexes along with nonphosphorylated RNAP II and CycT1 (Fig. 6E), among other expected proteins, such as TATA-box-binding protein (TBP) and Cdk7 (34) (data not shown). T-RS was particularly abundant, presumably due to a combination of RS domain-CTD- and Tat AD-mediated interactions.
Amino acid requirements of the RS domain.
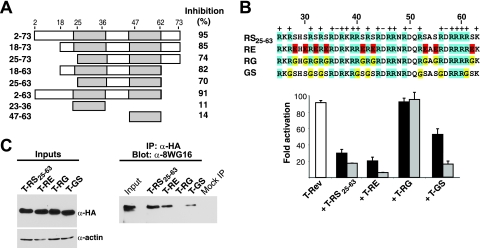
We wished to more precisely pinpoint residues in the U2AF65 RS domain important for inhibitor function and RNAP II interaction. The domain has a negatively charged N terminus (residues 2 to 22) followed by a region containing nine RS dipeptides (residues 25 to 63). Mutants with deletions within the domain from residues 2 to 73 of T-RS showed a progressive loss of inhibitor potency, but a minimal fragment spanning the RS dipeptides (T-RS25-63) still retained significant activity (Fig. 7A). To test the importance of the Arg and Ser residues, we replaced the nine RS dipeptides in T-RS25-63 with Arg-Glu (T-RE), Arg-Gly (T-RG), or Gly-Ser (T-GS). Interestingly, T-RE was as potent as T-RS, whereas T-RG was inactive and T-GS showed intermediate potency (Fig. 7B). To further test the hypothesis that the RS domain-RNAP II CTD interaction is important for inhibition, we sought to determine whether complex formation between the dipeptide mutants and RNAP II correlated with their inhibition activities. Indeed, immunoprecipitating RNAP IIa pulled down substantial amounts of T-RS25-63 and T-RE but no T-RG and an intermediate amount of T-GS (Fig. 7C), correlating well with inhibitor potency.
FIG. 7.
Inhibition activities and RNAP II interactions of RS domain mutants. (A) Various T-RS domain deletion mutants were tested for dominant negative inhibition on an HIV-1 LTR-RREIIB-FFL reporter, and the values shown represent percent inhibition at a 1:1 activator/inhibitor plasmid ratio relative to activation without inhibitor. The data are mean values from three independent experiments. (B) Sequences of RS dipeptide substitutions and their inhibition activities with a T-Rev activator (white bar) and HIV-1 LTR-RREIIB-FFL reporter at 1:0.2 (black bars) and 1:1 (gray bars) ratios of activator to inhibitor. (C) HeLa cells were transfected with HA-tagged versions of the T-RS25-63 domain or mutants (expression levels shown on the left top panel) with loading controls (levels shown on left bottom panel). Amounts of associated RNAP IIa were assessed by immunoprecipitation (IP) with anti-HA (α-HA) and Western blotting with the 8WG16 (α-8WG16) antibody (right panel).
The fact that the U2AF65 RS domain also directly contacts the pre-mRNA during splicing (22, 50) prompted us to evaluate whether RNA binding might contribute to the dominant negative effect. We performed RNA-binding assays with T-RS25-63, T-RE, T-RG, T-GS, and the unfused Tat AD by using U2 snRNA and HIV-1 RRE RNA coupled with beads, and we found that T-RS25-63, T-RE, and T-RG bound the two forms of RNA equally well, T-GS bound 50% less efficiently, and the Tat AD showed no appreciable binding (data not shown). Thus, RNA binding appears to correlate with arginine content, as previously proposed (50), but not with inhibition activity, consistent with the need for the RS domain for CTD targeting.
T-RS inhibits viral replication and generates a chronically replicating cell line.
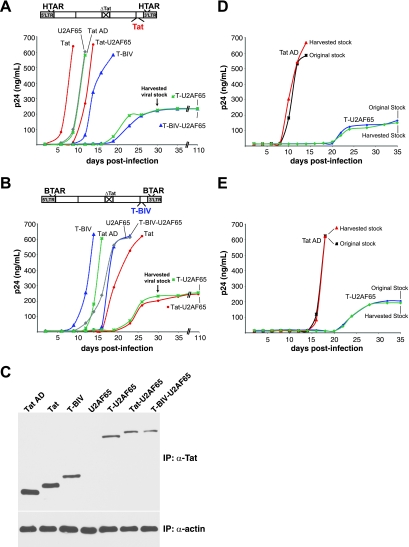
The high potency of the Tat dominant negatives and the requirement of Tat for viral replication suggested that the dominant negatives might be effective viral inhibitors. Thus, we generated SupT1 lymphocyte cell lines stably expressing T-U2AF65, Tat-U2AF65, or T-BIV-U2AF65 dominant negatives or the nonfusion controls Tat AD, Tat, T-BIV, or U2AF65 and monitored viral replication rates by using viruses engineered with either the HIV-1 Tat-TAR or BIV Tat-TAR interaction (54). Only viruses containing a cognate Tat-TAR pair will efficiently replicate, and this allowed us to examine a matched set of viruses with RNA-binding specificity controls in the replication assays.
We observed striking specificity of the dominant negative proteins, as replication was inhibited only in viruses driven by a noncognate RNA-protein interaction. Expression of T-U2AF65, lacking a TAR RBD, markedly suppressed replication of both viruses compared to the Tat AD or U2AF65 controls, with no p24 antigen detectable until 18 to 20 days after infection. Expression of Tat-U2AF65 or T-BIV-U2AF65 inhibited replication of the noncognate virus to an extent similar to that of T-U2AF65 and only slightly inhibited the cognate virus (Fig. 8A and B). Interestingly, expression of Tat or T-BIV activators accelerated replication of the cognate viruses but not the noncognate viruses, suggesting that Tat levels in these viruses may be limiting. The strong levels of inhibition are notable, given that expression levels of the inhibitors are low in these cell lines, as judged by Western blot analysis of samples immunoprecipitated with a Tat antibody (Fig. 8C) and reverse transcription-PCR and reporter assays (data not shown), explaining why virus expression ultimately is seen.
FIG. 8.
Expression of the Tat dominant negative blocks viral replication and generates a latency-like state. SupT1 cells stably expressing the Tat domains or fusion proteins indicated were infected with either HIV-1 Tat-TAR-dependent (A) or BIV Tat-TAR-dependent (B) virus (54), and the kinetics of p24 antigen expression were monitored by enzyme-linked immunosorbent assay. (C) Expression levels of Tat domains or fusions analyzed by immunoprecipitation followed by Western blotting using an anti-Tat (α-Tat) antibody, with an anti-β-actin (α-actin) antibody control. (D) HIV-1 Tat-TAR-dependent viruses emerging from the T-U2AF65 inhibitor-expressing cell lines was harvested at day 30 (arrows) and used to reinfect SupT1 cells expressing Tat AD and T-U2AF65. (E) Reinfection using the BIV Tat-TAR-dependent virus.
Viruses that emerged from the inhibitor cell lines after 18 to 20 days displayed slow replication kinetics and reached a low plateau of p24 expression that remained constant for at least 110 days without producing cytopathic effects. Reinfection experiments with Tat AD- and T-U2AF65-expressing cell lines indicate that the viruses do not acquire resistance mutations during this time period but rather grow poorly under these conditions of dominant negative inhibitor expression and continuously suppress viral replication (Fig. 8D and E).
DISCUSSION
Previously described dominant negative Tat proteins containing the Tat AD alone have shown relatively modest levels of inhibition (6, 19, 35). The extraordinarily potent proteins described here represent a new mechanistic class of transcriptional inhibitor, and a splicing factor (U2AF65 or SF1) or short RS domain acts as a targeting/localization moiety for the tethered Tat AD. The localization function provided by this targeting moiety allows the inhibitor to function at stoichiometric levels, without the need for substantial overexpression typically required by dominant negatives that operate by squelching or other simple competition mechanisms (13, 17, 21). Ptashne and Gann proposed the concept of “regulated localization,” activity and specificity being imposed by simple binding interactions between a locator (transcription factor), the transcriptional machinery, and the DNA (39). Such localization can include subcellular compartmentalization, in which molecular crowding can enhance the assembly of large macromolecular complexes (30, 31), in our case, favoring assembly into holo-Pol II complexes. We further suggest that combining multiple targeting functions within a single polypeptide provides an entropic benefit, allowing T-RS to load efficiently into early transcription complexes through CTD and CycT1 interactions, thereby blocking the entry of wild-type Tat.
It is especially interesting that RNA binding is not required for the delivery of T-RS to the HIV-1 promoter, raising a number of questions about the mechanisms and timing of T-RS and Tat recruitment into HIV-1 transcription complexes and also whether other dominant negative transcription factors can be recruited efficiently to their promoters by a similar cotranscriptional tethering strategy. Preliminary experiments with HSF1, CIITA, and GAL4-VP16 activators show little or no effect of an appended RS domain (data not shown), suggesting that Tat assembly may represent a special case, perhaps related to its role in elongation or dependence on RNA binding. The Tat AD itself associates directly with holo-Pol II complexes (9; this work), perhaps explaining why the U2AF65 RS moiety, known to associate with preinitiation or early transcription complexes (42, 49), enhances T-RS assembly. The proposed targeting function of the RS domain to the CTD is consistent with the observations that U2AF65 is found in early transcription complexes (42, 49) and is recruited to the HIV-1 promoter (5). Our experiments demonstrate a direct interaction between the U2AF65 RS domain and CTD in vitro and a CTD-dependent interaction in vivo (Fig. 6), agreeing with the observation that splicing factors copurify with nonphosphorylated RNAP II (10). Mutagenesis of T-RS indicates that the serines of the RS dipeptides are important for CTD binding and dominant negative activity, whereas the positively charged arginines contribute to nonspecific RNA binding, as previously proposed (50), but have little effect on inhibition (Fig. 7).
Still, the CTD-tethering hypothesis is insufficient to explain the specificity of recruitment to the HIV-1 promoter, given that T-RS, which requires interactions with CycT1 in addition to the CTD, would be expected to bind to and inhibit activation of other P-TEFb-dependent promoters, yet this is not the case (Fig. 3 and 5). Perhaps the transient nature or precise timing of splicing factor-CTD interactions (29), interactions of the Tat AD or P-TEFb with other HIV-1 promoter-specific factors, or the assembly of TBP-associated-factor-less TBP complexes (40) differ among promoters and determine whether stable “ChIPable” complexes can form. Irrespective of the details of assembly, it is clear that mechanisms used to cotranscriptionally load RNA-processing factors at promoters (2, 41) can be co-opted to deliver the dominant negative Tat AD to the HIV-1 promoter and thereby generate an extraordinarily potent inhibitor.
Given the high inhibitor potency, viral replication is substantially inhibited by low-level expression of the dominant negative in stable cell lines, even without optimizing and selecting for lines with high activity. It is interesting that these cells establish a chronic infection without cytopathic effects, reminiscent of other cellular environments that may resemble latent stages of HIV-1 infection (27). The amount of Tat clearly affects viral replication rates (51) and also can drive phenotypic diversity (52), and here, we show that expression of the dominant negative provides another means to alter Tat function. It will be interesting to examine mechanisms by which resistance to the Tat dominant negative might arise and to evaluate its therapeutic potential.
Supplementary Material
Acknowledgments
I.D. dedicates this work to Ileana Cuevas and to the memory of Agustín “Guity” Mairal. We thank R. Andino for critical discussions and K. Yamamoto, A. Johnson, C. Guthrie, V. Calabro, M. Daugherty, and other members of the Frankel laboratory for critical readings of the manuscript. We thank M. Green, R. Tjian, R. Voellmy, R. Kingston, J. Cáceres, A. Kornblitth, W. Wang, J. Greenblatt, and B. M. Peterlin for sharing reagents and advice.
I.D. is a postdoctoral fellow of the Human Frontier Science Program and Fundación Antorchas. J.R.G. was a postdoctoral fellow of the Damon-Runyon Foundation. C.D. was supported by a predoctoral fellowship from the American Foundation for Pharmaceutical Education. This work was supported by NIH grant AI29135 to A.D.F. R.L.N. and A.D.F. are founders of Advanced Genetic Systems, a biotechnology company with an interest in developing antiviral therapeutics.
Footnotes
Published ahead of print on 30 July 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Ahn, S. H., M. Kim, and S. Buratowski. 2004. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell 1367-76. [DOI] [PubMed] [Google Scholar]
- 2.Bentley, D. L. 2005. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 17251-256. [DOI] [PubMed] [Google Scholar]
- 3.Boehm, A. K., A. Saunders, J. Werner, and J. T. Lis. 2003. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol. Cell. Biol. 237628-7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brady, J., and F. Kashanchi. 2005. Tat gets the “green” light on transcription initiation. Retrovirology 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brès, V., N. Gomes, L. Pickle, and K. A. Jones. 2005. A human splicing factor, SKIP, associates with P-TEFb and enhances transcription elongation by HIV-1 Tat. Genes Dev. 191211-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caputo, A., M. P. Grossi, R. Bozzini, C. Rossi, M. Betti, P. C. Marconi, G. Barbanti-Brodano, and P. G. Balboni. 1996. Inhibition of HIV-1 replication and reactivation from latency by tat transdominant negative mutants in the cysteine rich region. Gene Ther. 3235-245. [PubMed] [Google Scholar]
- 7.Carroll, R., B. M. Peterlin, and D. Derse. 1992. Inhibition of human immunodeficiency virus type 1 Tat activity by coexpression of heterologous trans activators. J. Virol. 662000-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cramer, P. 2004. RNA polymerase II structure: from core to functional complexes. Curr. Opin. Genet. Dev. 14218-226. [DOI] [PubMed] [Google Scholar]
- 9.Cujec, T. P., H. Cho, E. Maldonado, J. Meyer, D. Reinberg, and B. M. Peterlin. 1997. The human immunodeficiency virus transactivator Tat interacts with the RNA polymerase II holoenzyme. Mol. Cell. Biol. 171817-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Das, R., J. Yu, Z. Zhang, M. P. Gygi, A. R. Krainer, S. P. Gygi, and R. Reed. 2007. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol. Cell 26867-881. [DOI] [PubMed] [Google Scholar]
- 11.Fong, N., G. Bird, M. Vigneron, and D. L. Bentley. 2003. A 10 residue motif at the C-terminus of the RNA pol II CTD is required for transcription, splicing and 3′ end processing. EMBO J. 224274-4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fraisier, C., D. A. Abraham, M. van Oijen, V. Cunliffe, A. Irvine, R. Craig, and E. A. Dzierzak. 1998. Inhibition of Tat-mediated transactivation and HIV replication with Tat mutant and repressor domain fusion proteins. Gene Ther. 5946-954. [DOI] [PubMed] [Google Scholar]
- 13.Friedman, A. D., S. J. Triezenberg, and S. L. McKnight. 1988. Expression of a truncated viral trans-activator selectively impedes lytic infection by its cognate virus. Nature 335452-454. [DOI] [PubMed] [Google Scholar]
- 14.Gama-Carvalho, M., R. D. Krauss, L. Chiang, J. Valcarcel, M. R. Green, and M. Carmo-Fonseca. 1997. Targeting of U2AF65 to sites of active splicing in the nucleus. J. Cell Biol. 137975-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garber, M. E., P. Wei, V. N. KewalRamani, T. P. Mayall, C. H. Herrmann, A. P. Rice, D. R. Littman, and K. A. Jones. 1998. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 123512-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerber, H. P., M. Hagmann, K. Seipel, O. Georgiev, M. A. West, Y. Litingtung, W. Schaffner, and J. L. Corden. 1995. RNA polymerase II C-terminal domain required for enhancer-driven transcription. Nature 374660-662. [DOI] [PubMed] [Google Scholar]
- 17.Gill, G., and M. Ptashne. 1988. Negative effect of the transcriptional activator GAL4. Nature 334721-724. [DOI] [PubMed] [Google Scholar]
- 18.Gomes, N. P., G. Bjerke, B. Llorente, S. A. Szostek, B. M. Emerson, and J. M. Espinosa. 2006. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 20601-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green, M., M. Ishino, and P. M. Loewenstein. 1989. Mutational analysis of HIV-1 Tat minimal domain peptides: identification of trans-dominant mutants that suppress HIV-LTR-driven gene expression. Cell 58215-223. [DOI] [PubMed] [Google Scholar]
- 20.Hahn, S. 2004. Structure and mechanism of the RNA polymerase II transcription machinery. Nat. Struct. Mol. Biol. 11394-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herskowitz, I. 1987. Functional inactivation of genes by dominant negative mutations. Nature 329219-222. [DOI] [PubMed] [Google Scholar]
- 22.Hertel, K. J., and B. R. Graveley. 2005. RS domains contact the pre-mRNA throughout spliceosome assembly. Trends Biochem. Sci. 30115-118. [DOI] [PubMed] [Google Scholar]
- 23.Kim, B., A. I. Nesvizhskii, P. G. Rani, S. Hahn, R. Aebersold, and J. A. Ranish. 2007. The transcription elongation factor TFIIS is a component of RNA polymerase II preinitiation complexes. Proc. Natl. Acad. Sci. USA 10416068-16073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 142452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kornblihtt, A. R., M. de la Mata, J. P. Fededa, M. J. Munoz, and G. Nogues. 2004. Multiple links between transcription and splicing. RNA 101489-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Landt, S. G., R. Tan, and A. D. Frankel. 2000. Screening RNA-binding libraries using Tat-fusion system in mammalian cells. Methods Enzymol. 318350-363. [DOI] [PubMed] [Google Scholar]
- 27.Lassen, K., Y. Han, Y. Zhou, J. Siliciano, and R. F. Siliciano. 2004. The multifactorial nature of HIV-1 latency. Trends Mol. Med. 10525-531. [DOI] [PubMed] [Google Scholar]
- 28.Luecke, H. F., and K. R. Yamamoto. 2005. The glucocorticoid receptor blocks P-TEFb recruitment by NFkappaB to effect promoter-specific transcriptional repression. Genes Dev. 191116-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mabon, S. A., and T. Misteli. 2005. Differential recruitment of pre-mRNA splicing factors to alternatively spliced transcripts in vivo. PLoS Biol. 3e374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minton, A. P. 2000. Implications of macromolecular crowding for protein assembly. Curr. Opin. Struct. Biol. 1034-39. [DOI] [PubMed] [Google Scholar]
- 31.Misteli, T. 2007. Beyond the sequence: cellular organization of genome function. Cell 128787-800. [DOI] [PubMed] [Google Scholar]
- 32.Misteli, T., and D. L. Spector. 1999. RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol. Cell 3697-705. [DOI] [PubMed] [Google Scholar]
- 33.Orsini, M. J., and C. M. Debouck. 1996. Inhibition of human immunodeficiency virus type 1 and type 2 Tat function by transdominant Tat protein localized to both the nucleus and cytoplasm. J. Virol. 708055-8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan, G., T. Aso, and J. Greenblatt. 1997. Interaction of elongation factors TFIIS and elongin A with a human RNA polymerase II holoenzyme capable of promoter-specific initiation and responsive to transcriptional activators. J. Biol. Chem. 27224563-24571. [DOI] [PubMed] [Google Scholar]
- 35.Pearson, L., J. Garcia, F. Wu, N. Modesti, J. Nelson, and R. Gaynor. 1990. A transdominant tat mutant that inhibits tat-induced gene expression from the human immunodeficiency virus long terminal repeat. Proc. Natl. Acad. Sci. USA 875079-5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peled-Zehavi, H., J. A. Berglund, M. Rosbash, and A. D. Frankel. 2001. Recognition of RNA branch point sequences by the KH domain of splicing factor 1 (mammalian branch point binding protein) in a splicing factor complex. Mol. Cell. Biol. 215232-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peterlin, B. M., and D. H. Price. 2006. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 23297-305. [DOI] [PubMed] [Google Scholar]
- 38.Plavec, I., M. Agarwal, K. E. Ho, M. Pineda, J. Auten, J. Baker, H. Matsuzaki, S. Escaich, M. Bonyhadi, and E. Bohnlein. 1997. High transdominant RevM10 protein levels are required to inhibit HIV-1 replication in cell lines and primary T cells: implication for gene therapy of AIDS. Gene Ther. 4128-139. [DOI] [PubMed] [Google Scholar]
- 39.Ptashne, M., and A. Gann. 1998. Imposing specificity by localization: mechanism and evolvability. Curr. Biol. 8R812-R822. [DOI] [PubMed] [Google Scholar]
- 40.Raha, T., S. W. Cheng, and M. R. Green. 2005. HIV-1 Tat stimulates transcription complex assembly through recruitment of TBP in the absence of TAFs. PLoS Biol. 3e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reed, R., and E. Hurt. 2002. A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell 108523-531. [DOI] [PubMed] [Google Scholar]
- 42.Robert, F., M. Blanchette, O. Maes, B. Chabot, and B. Coulombe. 2002. A human RNA polymerase II-containing complex associated with factors necessary for spliceosome assembly. J. Biol. Chem. 2779302-9306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Selby, M. J., and B. M. Peterlin. 1990. Trans-activation by HIV-1 Tat via a heterologous RNA binding protein. Cell 62769-776. [DOI] [PubMed] [Google Scholar]
- 44.Shamah, S. M., and C. D. Stiles. 1995. Transdominant negative mutations. Methods Enzymol. 254565-576. [DOI] [PubMed] [Google Scholar]
- 45.Southgate, C., M. L. Zapp, and M. R. Green. 1990. Activation of transcription by HIV-1 Tat protein tethered to nascent RNA through another protein. Nature 345640-642. [DOI] [PubMed] [Google Scholar]
- 46.Szak, S. T., D. Mays, and J. A. Pietenpol. 2001. Kinetics of p53 binding to promoter sites in vivo. Mol. Cell. Biol. 213375-3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan, R., and A. D. Frankel. 1998. A novel glutamine-RNA interaction identified by screening libraries in mammalian cells. Proc. Natl. Acad. Sci. USA 954247-4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trono, D., M. B. Feinberg, and D. Baltimore. 1989. HIV-1 Gag mutants can dominantly interfere with the replication of the wild-type virus. Cell 59113-120. [DOI] [PubMed] [Google Scholar]
- 49.Ujvári, A., and D. S. Luse. 2004. Newly initiated RNA encounters a factor involved in splicing immediately upon emerging from within RNA polymerase II. J. Biol. Chem. 27949773-49779. [DOI] [PubMed] [Google Scholar]
- 50.Valcárcel, J., R. K. Gaur, R. Singh, and M. R. Green. 1996. Interaction of U2AF65 RS region with pre-mRNA branch point and promotion of base pairing with U2 snRNA. Science 2731706-1709. [DOI] [PubMed] [Google Scholar]
- 51.Verhoef, K., M. Koper, and B. Berkhout. 1997. Determination of the minimal amount of Tat activity required for human immunodeficiency virus type 1 replication. Virology 237228-236. [DOI] [PubMed] [Google Scholar]
- 52.Weinberger, L. S., J. C. Burnett, J. E. Toettcher, A. P. Arkin, and D. V. Schaffer. 2005. Stochastic gene expression in a lentiviral positive-feedback loop: HIV-1 Tat fluctuations drive phenotypic diversity. Cell 122169-182. [DOI] [PubMed] [Google Scholar]
- 53.Williams, S. A., L. F. Chen, H. Kwon, C. M. Ruiz-Jarabo, E. Verdin, and W. C. Greene. 2006. NF-kappaB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J. 25139-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xie, B., M. A. Wainberg, and A. D. Frankel. 2003. Replication of human immunodeficiency viruses engineered with heterologous Tat-transactivation response element interactions. J. Virol. 771984-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.