Abstract
Neonatal injection of recombinant adeno-associated virus serotype 8 (rAAV8) vectors results in widespread transduction in multiple organs and therefore holds promise in neonatal gene therapy. On the other hand, insertional mutagenesis causing liver cancer has been implicated in rAAV-mediated neonatal gene transfer. Here, to better understand rAAV integration in neonatal livers, we investigated the frequency and spectrum of genomic integration of rAAV8 vectors in the liver following intraperitoneal injection of 2.0 × 1011 vector genomes at birth. This dose was sufficient to transduce a majority of hepatocytes in the neonatal period. In the first approach, we injected mice with a β-galactosidase-expressing vector at birth and quantified rAAV integration events by taking advantage of liver regeneration in a chronic hepatitis animal model and following partial hepatectomy. In the second approach, we performed a new, quantitative rAAV vector genome rescue assay by which we identified rAAV integration sites and quantified integrations. As a result, we find that at least ∼0.05% of hepatocytes contained rAAV integration, while the average copy number of integrated double-stranded vector genome per cell in the liver was ∼0.2, suggesting concatemer integration. Twenty-three of 34 integrations (68%) occurred in genes, but none of them were near the mir-341 locus, the common rAAV integration site found in mouse hepatocellular carcinoma. Thus, rAAV8 vector integration occurs preferentially in genes at a frequency of 1 in approximately 103 hepatocytes when a majority of hepatocytes are once transduced in the neonatal period. Further studies are warranted to elucidate the relationship between vector dose and integration frequency or spectrum.
Adeno-associated virus (AAV) is a small, nonpathogenic, nonenveloped, replication-defective virus containing a single-stranded DNA genome of approximately 5 kb. Recombinant AAV (rAAV) devoid of all the virally encoded genes is among the most promising gene delivery vectors due mainly to the ability to transduce various tissues and cell types with high efficiency by in vivo gene delivery approaches. Recently, newly identified alternative AAV serotype vectors that surpass traditional serotype 1 to 6 vectors in many aspects have become available (11, 12). Among them, AAV serotype 2 inverted terminal repeat (ITR)-containing vectors pseudotyped with AAV serotype 8 capsid (rAAV8) (12) and those with AAV serotype 9 capsid (rAAV9) (11) have gained attention due to their exceptionally high in vivo transduction efficiencies and their ability to cross various types of cellular barriers including vascular endothelial cell barriers (2, 18, 45). These robust rAAV serotype vectors can transduce various organs via the bloodstream and have offered a systemic approach to deliver genes to target organs via peripheral routes (2, 14, 18, 32, 40, 45).
With the advent of such advances in rAAV vectors, the systemic administration of the robust serotype rAAV vectors into fetuses or neonates has emerged as a new, promising approach to treat severe genetic diseases that are difficult to treat once pathological changes emerge and progress after birth. Previous studies with rAAV2 and other conventional serotype vectors have demonstrated a proof of principle for the this approach in metabolic diseases such as lysosomal storage disease (7, 10, 17) and glycogen storage diseases (23, 41). During the last couple of years, a series of mouse studies has shown that a single intravascular or intraperitoneal injection of rAAV8 or rAAV9 vector at a dose of 1.0 × 1011 to 3.5 × 1011 vector genomes (vg)/mouse into neonates results in the persistent and widespread global transduction of heart and skeletal muscles throughout the body (2, 14, 40, 45). This observation has opened up a new avenue to treat or even cure systemic degenerative muscular diseases such as Duchenne muscular dystrophy.
Such advances in practical applications of rAAV vectors to early therapeutic interventions have brought tremendous hope to patients with severe early-onset genetic diseases and their families. However, knowledge about the biology of rAAV vectors administered into fetuses or neonates is currently very limited. To better understand the therapeutic efficacy, to develop optimal therapeutic regimens, to further improve the current systems, and to establish the safety and risk profiles of this approach, it is very important to substantially understand the vector biology in neonates. A study has shown an increased incidence of liver tumorigenesis in both wild-type mice and a glycogen storage disease mouse model treated with rAAV at birth (8). In addition, rAAV-mediated insertional mutagenesis has been implicated in four cases of liver cancers that developed in mice injected with rAAV at birth (8). Although it has often been reported that prenatal or neonatal injection with rAAV vectors, even with rAAV8 or rAAV9 vectors, poorly transduces the liver (1-3, 22), it does not necessarily indicate that the liver could be spared from transduction. Poor liver transduction has proven to be attributed to the rapid degradation of vector genomes in growing liver in fetuses and neonates (6, 45). In fact, a substantial amount of rAAV vectors could be taken up by hepatocytes following vector administration, resulting in transient high levels of liver transduction only in the neonatal period (6). Importantly, no matter what organ is the target for gene transfer and no matter via what route the rAAV vector is administered, considerable vector dissemination to fetal and neonatal livers is essentially unavoidable in any of the currently available prenatal and neonatal approaches using robust serotype vectors such as rAAV8.
In the present study, we investigated the frequency and spectrum of the genomic integration of rAAV vectors in the liver following rAAV8 injection into newborn mice to better understand rAAV integration in neonatal livers. An accurate quantification of rAAV integration frequency in the liver has been challenging, although a series of studies has demonstrated that the frequency should be low (27). The rAAV integration site spectrum in neonatal livers has never been investigated. To achieve the goal of the study, we took two approaches. In the first approach, to facilitate quantitative assays, we substantially eliminated extrachromosomal vector genomes from rAAV8-transduced hepatocytes by exploiting both continuous liver regeneration in a chronic hepatitis mouse model and transient liver regeneration following a partial hepatectomy. In the second approach, we established and used a new method to quantify rAAV integrations that does not require any cell manipulations based on a quantitative rAAV vector genome rescue assay. As a result, we show that rAAV integration occurs preferentially in genic regions at a frequency of at least 1 in 2,000 hepatocytes when a majority of hepatocytes are transduced once with rAAV8 in the neonatal period. They appear to be in the form of large concatemers, and only a portion of rAAV integration events results in persistent transgene expression in the liver. Assuming that the number of hepatocytes in human babies is a ∼1010 cell range and if the integration frequency and spectrum are to be the same in humans, at least 5 million rAAV integrations would occur preferentially in genes in developing hepatocytes in human liver, which might be a considerable threat to the genome in newborns.
MATERIALS AND METHODS
rAAV vector production.
rAAV vectors AAV8-CMV-lacZ, AAV8-EF1α-nlslacZ, AAV9-LSP (liver-specific promoter)-hF.IX, AAV8-CMV-hF.IX, and AAV8-ISceI.AO3 were produced based on plasmids pAAV-CMV-lacZ (32), pAAV-EF1α-nlslacZ (36), pAAV-hF.IX16 (37), pAAV-CMV-hF.IX (33), and pAAV-ISceI.AO3 (GenBank accession number EU022316) (19), respectively. Each rAAV vector genome was packaged into corresponding AAV capsids by the triple-plasmid transfection method, and recombinant vector particles were purified by two cycles of cesium chloride gradient ultracentrifugation followed by dialysis as previously described (32). Vector titers were determined by a quantitative dot blot assay.
Animal studies.
All animal experiments were performed according to the guidelines for animal care at Stanford University and the University of Pittsburgh. C57BL/6J mice were purchased from the Jackson Laboratory. Hepatitis B virus large-envelope polypeptide transgenic mice (5) [C57BL/6J-TgN(Alb1HBV)44Bri] were obtained from the Scripps Research Institute, and the colony was established. Wild-type female C57BL/6J mice from the colony and 44Bri female mice heterozygous for the transgene were used for the study in which we injected mice with vectors AAV8-EF1α-nlslacZ and/or AAV9-LSP-hF.IX. Newborn male mice from the 44Bri colony were also injected with vector AAV-EF1α-nlslacZ at birth but were not used for the present study. They were used for a separate long-term study. Genotypes were determined by PCR according to the method recommended by the Jackson Laboratory at 3 to 4 weeks of age. Wild-type male and female C57BL/6J neonates from a breeding pair of wild-type C57BL/6J mice were injected with AAV8-CMV-lacZ or AAV8-ISceI.AO3 in the preliminary study or the study aimed at identifying rAAV integration sites, respectively.
For intraperitoneal injection of the rAAV vector into neonates, neonatal mice at 24 to 48 h of age were held with a hand, and 20 μl of rAAV vector preparation in phosphate-buffered saline (PBS) with 5% sorbitol and, subsequently, 10 μl of air were injected with a 30-gauge needle attached to a Hamilton syringe. Methods for tail vein injection and two-thirds partial hepatectomy were performed as described previously (34, 39). In the partial hepatectomy experiments, mice were sacrificed 6 weeks after hepatectomy for analyses. Blood samples were collected from the retro-orbital plexus under anesthesia with isoflurane.
Blood analyses.
Serum alanine aminotransferase (s-ALT) levels and plasma hF.IX antigen levels were determined as previously described (32).
Anti-β-galactosidase antibody titers in mouse sera were quantified by an enzyme-linked immunosorbent assay (ELISA). Briefly, a microtiter plate (Maxisorp; Nunc, Rochester, NY) was coated with Escherichia coli β-galactosidase (200 ng/ml; Roche, Indianapolis, IN) at 37°C for 1 h, washed three times with PBS-0.05% Tween 20, and then blocked with PBS-5% nonfat dry milk-0.05% Tween 20 at room temperature for 1 h. Mouse serum samples and ELISA standard (anti-E. coli β-galactosidase antibody; Roche) diluted with PBS-5% nonfat dry milk-0.05% Tween 20 were applied to each well and incubated at 37°C for 1 h. The plate was washed five times with PBS-0.05% Tween 20, and absorbed anti-β-galactosidase antibodies in mouse sera were detected with horseradish peroxidase-conjugated sheep anti-mouse immunoglobulin G antibody (diluted 1:1,000 with PBS-5% nonfat dry milk) and O-phenylenediamine dihydrochloride (Sigma, St. Louis, MO). The average antibody titer obtained from 10 naïve mouse sera was 60 ± 19 pg/ml (mean ± standard deviation [SD]). We considered the sample to be antibody positive when the titers were higher than the mean plus 2 SD (97 pg/ml).
The presence or absence of neutralizing anti-AAV8 capsid antibody in mouse sera was assessed as previously described, with modifications (15). Fifteen microliters of mouse sera was incubated with 6.0 × 109 vg of AAV8-CMV-hF.IX at 37°C for 1 h and then added to 293 cells seeded on 24-well plates together with a first-generation adenovirus type 5 vector (a gift from Anja Ehrhardt) at a multiplicity of infection of 10. After an hour of incubation at 37°C, the culture medium was replaced with fresh medium. The amount of hF.IX protein secreted into the medium between 24 and 48 h postinfection was determined by hF.IX ELISA as described above. We considered the sample to be antibody positive when hF.IX expression was inhibited by over 90% compared to that in naïve mouse sera. All the serum samples were heat inactivated by incubation at 56°C for 30 min.
Histological analysis.
Pieces of the left lobe, median lobe, and two right lobes were embedded in Tissue-Tek Optimal Cutting Temperature compound (Sakura Finetek USA, Inc., Torrance, CA) and frozen on dry ice. Ten-micrometer-thick sections were then prepared and mounted onto each microscope slide. For the mice that underwent a two-thirds partial hepatectomy, which removed the left and median lobes, the two right lobes and two caudate lobes that had undergone compensatory regeneration were analyzed. The procedures for 5-bromo-4-chloro-3-indolylphosphate (X-Gal) staining of liver sections were performed as described previously (32), and liver sections were counterstained with light hematoxylin.
Methods for determination of liver transduction efficiency and frequency of hepatocyte cluster formation.
To determine transduction efficiency in liver transduced with vector AAV8-EF1α-nlslacZ expressing β-galactosidase, each set of liver lobes on a histology section was analyzed for the total number of X-Gal-positive hepatocyte nuclei, the total number of clusters of X-Gal-positive hepatocytes, and the total number of hepatocyte nuclei in each hepatocyte cluster. The total number of hepatocyte nuclei in the area of liver sections that we analyzed on a microscope slide was determined based on the average density of hepatocyte nuclei (the number of hepatocyte nuclei per mm2). Liver transduction efficiency was determined as follows: (total number of X-Gal-positive hepatocyte nuclei in an analyzed area)/(total number of hepatocyte nuclei in the same analyzed area) × 100%.
The frequency of hepatocyte cluster formation in the liver was determined in the following manner. First, we defined a cluster as a focus comprising three or more hepatocyte nuclei on the histology sections. The three-dimensional frequency of hepatocyte cluster formation in the liver is not necessarily the same as the frequency of hepatocyte cluster formation in a two-dimensional area of a histology section. To make calculation simple, all hepatocyte clusters were considered to be spheres. The diameter (R) of the spheres can be calculated by the following equation: R(n) (distance units [d.u.]) = (4n/π)0.5, where R is a function of “n,” “n” represents the number of hepatocyte nuclei in a cluster, and 1 d.u. equals to the average distance between neighboring hepatocyte nuclei in liver sections. The average distances were determined to be 34 μm and 41 μm in wild-type and 44Bri mice, respectively, and longer than the thickness of the liver sections. The volume of a hepatocyte cluster sphere (V) was calculated by the equation V(n) = 4π[R(n)/2]3/3 = (4n1.5)/(3π0.5). Assuming that hepatocyte cluster spheres are distributed randomly in the liver, the probability, P(N), that a hepatocyte cluster comprising “N” hepatocytes found in a liver section has originated from a hepatocyte in the same liver section as follows:
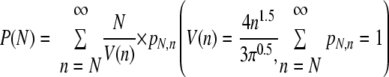 |
where pN,n is the probability that the diameter of three-dimensional hepatocyte cluster sphere is R(n) d.u. when the observed diameter of the two-dimensional hepatocyte cluster in a liver section on a slide is R(N). pN,n was calculated based on histograms of the sizes of hepatocyte clusters observed in our study. The table for the pN,n values will be provided upon request. Finally, the frequency of hepatocyte cluster formation in the liver was determined as follows:
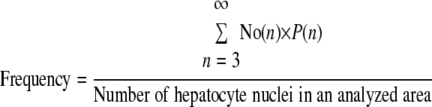 |
where No(n) is the number of total hepatocyte clusters that have “n” hepatocyte nuclei in a liver section on a slide.
Molecular analyses of rAAV vector genomes.
Molecular forms of rAAV vectors and vector genome copy numbers in the liver were analyzed by Southern blot analysis as previously described (20, 32). Vector genome copy numbers in each cell were expressed as double-stranded vector genome copy numbers per diploid genomic equivalent (ds-vg/dge).
Generation of rAAV vector integration site plasmid libraries and sequencing of plasmid clones.
rAAV vector integration site plasmid libraries were constructed using AAV8-ISceI.AO3 vector-transduced mouse liver genomic DNA as previously described, with modifications (19). In our original method, we mixed mouse genomic DNA and yeast genomic DNA (Saccharomyces cerevisiae S288C) at a 1:1 ratio at the beginning of library construction. Yeast genomic DNA mixed with sample DNA served as a tag sequence of unwanted recombination events that do not represent true rAAV integration events that had occurred in mice. To date, we have analyzed over 2,000 plasmid clones in rAAV vector integration libraries constructed by this method; however, none of the plasmid clones contained such a recombination. Based on this observation, we have concluded that unwanted intermolecular recombination in our assay system is an extremely rare event. Therefore, the libraries were constructed without adding the yeast genomic DNA to samples in the present study. To make the assay quantitative, we modified the method as follows: (i) E. coli ElectroMax DH10B cells (Invitrogen) were transformed with a fixed amount of 3 μg of mouse liver genomic DNA for the construction of all libraries including control libraries, (ii) the same lot of DH10B was used in the experiment, (iii) the bacterial transformation efficiency of DH10B cells was determined using a control mouse liver genomic DNA containing a known number of six representative AAV8-ISceI.AO3 vector integrations, (iv) we determined the sequences of all the plasmid clones that were present in the libraries, and (v) the experiments were repeated twice independently.
The control rAAV vector integration site plasmid libraries were generated in the following manner. First, we generated a control mouse liver genomic DNA containing each of six different rAAV integrations at chromosome number 4 (chr 4; coordinate 11970371), chr 6 (116170657), chr 7 (102785454), chr 10 (38206964), chr 10 (83496903), and chr 19 (53122747) at a frequency of 1.67 for each integration event per 100 diploid genomic equivalents (i.e., 1 integration event at any of the 6 integration sites per 10 diploid genomic equivalents). This control genomic DNA was generated by spiking an equal molar of six plasmids, each of which contained an rAAV vector-cellular DNA junction and was derived from an actual rAAV vector integration site plasmid library that we constructed in our previous study (19). The size range of these plasmids was between 2.4 and 7.8 kb. We used this control DNA containing a known number of rAAV integration events as standard mouse liver genomic DNA to determine the DH10B transformation efficiency with rAAV integration site-containing plasmids. For this, 10 μg of the control DNA was digested with a combination of BamHI and BglII (4 units per μg DNA; New England BioLabs) at 37°C for 4 h. DNA was purified by phenol-chloroform extraction, precipitated with ammonium acetate and ethanol, dissolved in 20 μl of Tris-EDTA (10 mM Tris-HCl [pH 8.0], 1 mM EDTA), and quantified by spectrophotometry. Three micrograms of DNA was then self-ligated with T4 DNA ligase (New England BioLabs), treated with ATP-dependent exonuclease (Plasmid Safe; Epicentre), and used for bacterial transformation in exactly the same manner as that for the rAAV-transduced liver samples. Plasmid DNA recovered from each colony was characterized by BstYI digestion (19), which provided BstYI-restricted DNA fragment patterns unique for each of the six different rAAV integration events contained in the control mouse liver genomic DNA.
Plasmid DNA was prepared from each E. coli colony, and each plasmid DNA sequence was determined as previously described with a 3730x DNA analyzer (Applied Biosystems) and sequencing primers OriP2 and 36-39 BamHI-1 (19). Sequencing with primer OriP2 can determine rAAV vector recombination sites, while primer 36-39 BamHI sequences plasmid DNA toward the recombination site from the opposite side.
Direct sequencing of bisulfite-modified plasmid DNA template.
It is essential to know how many rAAV integration plasmid clones could be isolated in a library to make the assay quantitative. It should be noted that not all the bacterial clones in a library contain an rAAV integration plasmid (19), and not all the plasmid clones can be successfully sequenced by the method described above. With the standard sequencing technique described above, it was often impossible to sequence through AAV ITRs. The occasional failure of sequencing was attributed to a DNA polymerase stall in the AAV ITRs. In our previous studies, we did not further investigate each of the plasmid clones that we failed to sequence due to the high-throughput nature of the analysis (19, 38).
To overcome this problem, we established a method for the direct sequencing of bisulfite-modified plasmid DNA template that does not require a bisulfite-PCR followed by cloning of PCR products into a plasmid. Approximately 10 μg of plasmid DNA was chemically modified with sodium hydrogen sulfite (Sigma-Aldrich) as previously described (20), and a quarter of the chemically modified DNA was subjected to the standard cycle sequence procedure with a sequencing primer, 56-23 OriP2 (5′-ATATATTTTTCCATACCTTACAAAAT-3′). With this method, we successfully sequenced all the plasmid clones that the standard method failed to sequence.
Bioinformatics.
rAAV integration sites were mapped to the mouse genome as described in detail in our previous publication (19). In the present study, we used the mouse genome database mm9, July 2007 Freeze. To map bisulfite-modified sequences to the mouse genome, we created a bisulfite-modified mouse genome database by converting all C's to T's on the upper strand of the genome and on the reverse complement genome of the upper strand (i.e., lower strand). A BLAT search of chemically modified query sequences was done against both the upper and lower strands of the modified mouse genome database. DNA palindrome analysis was performed as previously described (19).
Nucleotide sequence accession numbers.
The GenBank accession numbers for rAAV vector genome-host cellular DNA junction sequences reported in this paper are FI136228 to FI136263.
RESULTS
Neonatal intraperitoneal injection of 1.0 × 1011 vg of rAAV8 vector transduced virtually all the hepatocytes 5 days postinjection, with a vector genome copy number of >100 vg/cell.
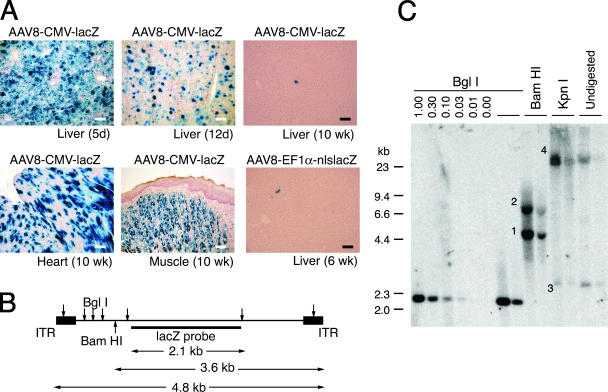
In our preliminary study, we injected neonatal wild-type C57BL/6J mice intraperitoneally with 1.0 × 1011 vg of AAV8-CMV-lacZ. Vector AAV8-CMV-lacZ was initially used to investigate vector transduction not only in the liver but also in other organs such as skeletal muscles and heart. A majority of hepatocytes was transduced 5 days postinjection (Fig. 1A). The vector genome copy number in the liver was 261 ds-vg/dge on average (n = 2). The transduction efficiency substantially declined thereafter in the liver, but this was not the case in the skeletal muscle and heart as reported previously (6, 45) (Fig. 1A). The rapid decline of transgene expression in the liver in AAV8-CMV-lacZ-injected animals could be interpreted by the inactivation of the cytomegalovirus enhancer/promoter in the liver; however, the vector genome copy numbers also substantially decreased by more than 100-fold over a period of 10 weeks (0.76 ± 0.93 ds-vg/dge [mean ± SD] [n = 7]). Neonatal wild-type C57BL/6J mice were also injected intraperitoneally with 2.0 × 1011 vg of vector AAV8-EF1α-nlslacZ. A similar decline in liver transduction was observed as animals grew, and only a small portion of hepatocytes remained X-Gal positive 6 weeks postinjection (Fig. 1A).
FIG. 1.
Liver, heart, and skeletal muscle transduction following intraperitoneal injection of AAV8-CMV-lacZ at a dose of 1.0 × 101 vg/mouse. (A) Representative photomicrographs of sections of the liver (5 days, 12 days, 6 weeks, and 10 weeks postinjection), heart (10 weeks postinjection), and skeletal muscle (tongue at 10 weeks postinjection). Each section was stained with X-Gal and counterstained with hematoxylin. Scale bars represent 100 μm. (B) AAV8-EF1α-nlslacZ vector map. Locations of restriction enzyme sites and the lacZ probe used for the Southern blot analysis are indicated. (C) Southern blot analysis of AAV8-EF1α-nlslacZ vector genomes in livers 6 weeks postinjection. Ten micrograms of total liver genomic DNA was digested with BglI, BamHI, or KpnI; separated on an 0.8% agarose gel together with undigested sample DNA; transferred onto a nylon membrane; and probed with a lacZ sequence-specific probe. BglI cuts the vector genomes at multiple sites, BamHI cuts near the left terminus of the genome, and KpnI does not cut the vector genome. Head-to-tail molecules, tail-to-tail molecules, supercoiled circular monomer genomes, and large concatemers are indicated with the numbers 1 to 4, respectively. 0.00-1.00, rAAV vector genome copy number standards (ds-vg/dge). Each lane represents individual mouse samples.
A considerable number of extrachromosomal rAAV vector genomes could be carried over in hepatocytes in developing mouse livers.
Molecular forms of vector genomes in the AAV8-EF1α-lacZ-transduced mouse livers 6 weeks after neonatal vector injection were determined by Southern blot analysis. Both high-molecular-weight concatemeric genomes and supercoiled circular monomers were observed at a considerable level in the liver (Fig. 1B and C). Although Southern blot analysis cannot discriminate between integrated concatemeric vector genomes and extrachromosomal ones, the presence of circular monomers demonstrates that a considerable number of extrachromosomal genomes could be carried over during liver growth. The presence of a considerable amount of extrachromosomal genomes in the livers was also demonstrated by a two-thirds partial hepatectomy experiment performed 10 weeks postinjection in the mice injected with AAV-CMV-lacZ at birth. Vector genome copy numbers in the liver were further dropped by 75% ± 11% (mean ± SD) (n = 5) following a hepatectomy. All of these observations indicate that it would not be possible to accurately quantify rAAV vector genome integration that has occurred in neonates by simply assessing rAAV transduction efficiency at an adult age based on an assumption that a majority of rAAV transduction should be attributed to rAAV integration in adult mice injected with rAAV8 at birth.
Low-level persistent liver transduction could be achieved by intraperitoneal injection of 2.0 × 1011 vg of AAV8-EF1α-nlslacZ into wild-type and 44Bri mice.
Based on the observations of our preliminary experiment described above, we designed a subsequent experiment aimed at determining the rAAV integration efficiency (Table 1). In the following experiment, we used vector AAV8-EF1α-nlslacZ to focus on long-term liver transduction, 44Bri mice in addition to wild-type mice to eliminate extrachromosomal vector genomes in the liver as much as possible and facilitate the quantification of rAAV integration events, and vector AAV9-LSP-hF.IX, which was injected 17 days prior to the partial hepatectomy serving as an internal control for the vector genome copy number decrease following a hepatectomy.
TABLE 1.
Experimental groups
| Group | Treatmentb
|
No. of animals
|
|||
|---|---|---|---|---|---|
| AAV8-EF1α-nlslacZ (at birth) | AAV9-LSP-hF.IX (17 days prior to PHx) | PHxa (at 4-7 mo) | Wild type | 44Bri | |
| 1 | + | NA | NA | 16 | 10 |
| 2 | + | + | + | 10 | 11 |
| 3 | + | + | − | 7 | 7 |
| 4 | + | − | + | 6 | 6 |
| 5 | − | + | + | 4 | 3 |
| 6 | − | + | − | 11 | 13 |
Two-thirds partial hepatectomy.
+, treated; −, untreated; NA, not applicable. All the mice in group 1 were sacrificed earlier than the other groups.
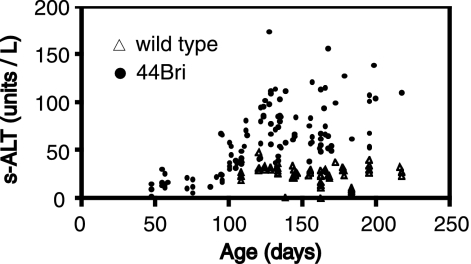
Thirty-nine female wild-type mice and 34 female 44Bri mice injected intraperitoneally with 2.0 × 1011 vg of AAV8-EF1α-nlslacZ at birth were used for the study. We also had 15 wild-type mice and 16 44Bri mice as uninjected controls. As expected, serum transaminase levels began to elevate at the age of approximately 3 months (Fig. 2). To assess the immune response against β-galactosidase, all 73 mice injected with AAV8-EF1α-nlslacZ and 5 wild-type and 44Bri mice each that were not injected with the vector were screened for the presence of anti-β-galactosidase antibody in sera at the age of between 10 and 26 weeks. Approximately 20% of animals elicited a humoral immune response against β-galactosidase irrespective of the strain (Table 2). All the antibody-positive animals (eight wild-type and five 44Bri mice), excluding marginally positive mice (two wild-type and three 44Bri mice), were sacrificed and excluded from the subsequent study. Eight wild-type and five 44Bri litter-matched anti-β-galactosidase antibody-negative mice were also sacrificed at the same time. Liver transduction efficiencies were compared by a histological analysis on X-Gal-stained liver sections between antibody-positive and -negative animals. The liver transduction efficiency was reduced by 2.5-fold in the presence of the antibody in wild-type animals (0.11% ± 0.08% and 0.27% ± 0.14% in antibody-positive and -negative animals, respectively [mean ± SD]; P < 0.05 by a Student's t test), while in 44Bri mice, the difference was not significant (0.07% ± 0.10% and 0.08% ± 0.07% in antibody-positive and -negative animals, respectively [mean ± SD]). AAV8-EFα-nlslacZ-transduced hepatocytes could barely be found in all three animals with a high antibody titer (>4 ng/ml of the standard antibody; 4% of all vector-injected animals). However, low-level persistent liver transduction could be achieved in all the antibody-negative and weakly positive animals.
FIG. 2.
s-ALT levels in wild-type and 44Bri mice at various ages. Sixty-four and 110 serum samples were collected from 64 wild-type mice and 60 44Bri mice at various ages, respectively. s-ALT levels in these samples were measured and plotted as a function of animal age. The results indicate that hepatocellular injury developed at ∼3 months of age and continued thereafter in 44Bri mice, while wild-type mice did not show elevated s-ALT levels.
TABLE 2.
Humoral immune responses against β-galactosidase and AAV8 capsids in wild-type and 44Bri mice injected with AAV8-EF1α-nlslacZ at birth
| Type of antibody | Antibody positivitya | Frequency of antibody positivity/negativity (%)
|
||
|---|---|---|---|---|
| Wild type (39 mice) | 44Bri (34 mice) | Total (73 mice) | ||
| Anti-β- | + | 21 | 15 | 17 |
| galactosidase | +/− | 5 | 9 | 7 |
| − | 74 | 76 | 75 | |
| Anti-AAV8 | + | 48 | 56 | 52 |
| neutralizing | +/− | 15 | 12 | 14 |
| − | 36 | 32 | 34 | |
Antibody positivity (+), marginal positivity (+/−), and negativity (−) are arbitrarily categorized based on the actual values obtained in the experiment. +, >189 pg/ml; +/−, 106 to 123 pg/ml; −, <95 pg/ml of the standard anti-β-galactosidase antibody. +, >90%; +/−, 53 to 85%; −, <50% inhibition of 293 cell transduction for anti-AAV8 neutralizing antibody.
Anti-AAV8 neutralizing antibodies developed in half of the animals injected with rAAV8 at birth.
In our approach, we planned to inject rAAV8-treated mice with vector AAV9-LSP-hF.IX at an adult age prior to the hepatectomy to deliver an internal control molecule for liver regeneration to the liver. We anticipated that some of the mice in our study could develop anti-AAV8 neutralizing antibodies, which might somewhat inhibit rAAV9-mediated gene transfer. In fact, a screening test revealed that approximately half of the animals injected with rAAV8 at birth developed anti-AAV8 neutralizing antibodies irrespective of the strain (Table 2). There was no association between the development of anti-β-galactosidase antibodies and that of anti-AAV8-neutralizing antibodies (chi-square value of 0.44; degree of freedom, 1). Based on this observation, we included anti-AAV8 neutralizing antibody-positive and -negative animals equally in each of the hepatectomy and no-hepatectomy groups, although we found retrospectively that the presence of anti-AAV8 antibodies did not influence the experimental outcomes.
An rAAV9 vector efficiently transduced the liver regardless of the presence of ongoing active liver damage or the presence of anti-AAV8 neutralizing antibodies.
At the age of 4 to 7 months, 17 wild-type and 18 44Bri mice injected with rAAV8 at birth and 15 wild-type and 16 44Bri rAAV8-untreated mice were injected with 2.5 × 1010 vg of AAV9-LSP-hF.IX via the tail vein. We measured plasma hF.IX levels 10 days and 17 days postinjection, which showed no significant difference in plasma hF.IX levels between these two time points. The presence of anti-AAV8 neutralizing antibody did not affect liver transduction efficiency by rAAV9. Plasma hF.IX levels after rAAV9 vector injection at day 17 were 15.8 ± 8.9 μg/ml (plus antibody) (n = 6) versus 21.2 ± 7.4 μg/ml (mice antibody) (n = 10) in wild-type mice and 29.1 ± 13.3 μg/ml (plus antibody plus) (n = 9) versus 37.2 ± 8.9 μg/ml (minus antibody) (n = 6) in 44Bri mice (mean ± SD). Interestingly, the liver transduction efficiency was initially higher in 44Bri mice at day 17 following rAAV9 vector injection (20.6 ± 7.0 μg/ml [n = 32] in wild-type mice versus 36.9 ± 14.5 μg/ml [n = 9] in 44Bri mice [mean ± SD]; P < 0.001 by a Student's t test), and the levels in 44Bri mice then declined below the wild-type mouse levels (18.7 ± 4.9 μg/ml [n = 18] in wild-type mice versus 10.0 ± 5.2 μg/ml [n = 20] in 44Bri mice [mean ± SD]; P < 0.001 by a Student's t test).
Approximately 0.2 double-stranded vg integrated into the host genome in liver transduced with rAAV8 at birth.
Finally, we performed a two-thirds partial hepatectomy on mice injected or not injected with rAAV8 at birth 17 days after rAAV9 injection (groups 2 and 5) (Table 1). The mice were sacrificed for analyses 6 weeks after hepatectomy. Control mice did not undergo a hepatectomy and were sacrificed at the same time when hepatectomized mice were sacrificed (groups 3 and 6) (Table 1). We also included a control group that did not receive an rAAV9 vector but underwent a hepatectomy (group 4) (Table 1). Group 4 was included to investigate whether rAAV9 vector injection could affect the liver transduction with AAV8-EFα1-nlslacZ before and/or after the hepatectomy. A comparison of the experimental results from group 2 and those from group 4 revealed that rAAV9 vector injection had no effect on the rAAV8 transduction efficiency. Liver regeneration following a two-thirds partial hepatectomy was nearly complete in both wild-type and 44Bri mice. The total liver weights of nonhepatectomized and hepatectomized mice at the time of sacrifice were 904 ± 92 mg (n = 18) and 817 ± 73 mg (n = 14) in wild-type mice and 967 ± 157 mg (n = 20) and 863 ± 117 mg (n = 6) in 44Bri mice, respectively (mean ± SD). Macroscopically detectable liver tumors were not observed in any of the mice during the course of the experiment.
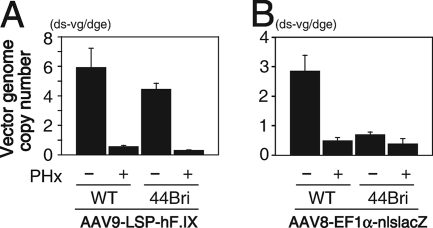
As expected, plasma hF.IX levels and the AAV9-LSP-hF.IX vector copy number in the liver substantially dropped by ∼90% following partial hepatectomy in both rAAV8-injected and noninjected animals (Fig. 3A), which validated our approach. In the same livers that lost ∼90% extrachromosomal rAAV9 genomes following hepatectomy, rAAV8 vector genomes delivered to the liver at birth dropped by only 44% on average in 44Bri mice (Fig. 3B). In 44Bri mice, the rAAV8 vector genome copy number dropped from 0.70 ± 0.31 ds-vg/dge (n = 15) to 0.39 ± 0.65 ds-vg/dge (n = 15) by hepatectomy, while rAAV9 vector genomes dropped from 4.44 ± 1.30 ds-vg/dge (n = 9) to 0.30 ± 0.07 ds-vg/dge (n = 5) (values are means ± SD). The high standard deviation observed in the rAAV8 vector genomes after the hepatectomy in 44Bri mice was in part attributed to the observation that rAAV vector genomes did not decrease or even did increase following partial hepatectomy in some mice. rAAV8 and rAAV9 vector copy numbers that were measured in the same liver samples on the same Southern blot membrane by rehybridization are summarized in Table 3. Nonetheless, the coincidence of the substantial decrease in rAAV9 vector genome copy numbers and significantly mitigated rAAV8 vector genome copy number drops in the same regenerated livers provides strong evidence that a substantial portion of rAAV8 vector genomes that remained following a hepatectomy in our approach represent integrated vector genomes. When we calculate the average rAAV8 vector genome copy numbers after the hepatectomy by excluding mouse 63 in Table 3, in which rAAV8 vector genome copy numbers considerably increased after hepatectomy, the copy number was 0.22 ± 0.13 ds-vg/dge (mean ± SD). Considering that a partial hepatectomy could eliminate ∼90% of extrachromosomal vector genomes, our observations indicate that approximately 0.2 ds-vg/dge [i.e., 0.7 (0.7 to 0.22)/0.9 = 0.17] of rAAV8 should have integrated into the host genome. In wild-type livers, rAAV8 vector genome copy numbers dropped by 83% (Fig. 3B). The copy numbers were 2.83 ± 1.87 ds-vg/dge (n = 13) and 0.49 ± 0.30 ds-vg/dge (n = 9) before and after the hepatectomy, respectively (values are means ± SD). The degree of the rAAV8 vector genome copy number decrease in wild-type mice was greater than that in 44Bri mice but was mitigated compared to the decrease in internal rAAV9 vector genome copy numbers, suggesting that rAAV8 vector genomes had integrated into the host genome in wild-type mouse livers at levels detectable by the partial hepatectomy approach.
FIG. 3.
rAAV vector genome copy numbers in livers before and after a two-thirds partial hepatectomy (PHx) in wild-type (WT) and 44Bri mice. Wild-type and 44Bri mice were injected with 2.0 × 1011 vg of vector AAV8-EF1α-nlslacZ at birth and again injected with 2.5 × 1010 vg of vector AAV9-LSP-hF.IX at the age of 4 to 7 months. Seventeen days after the rAAV9 vector injection, a subset of the mice underwent a two-thirds partial hepatectomy. All the mice were sacrificed 6 weeks after partial hepatectomy. Vector genome copy numbers of each rAAV8 and rAAV9 vector were determined by Southern blot analysis. Partial hepatectomy (−) samples include liver samples collected at the hepatectomy and those from not-hepatectomized mice. Vertical bars represent standard errors.
TABLE 3.
Coincidental changes in genome copy numbers of rAAV8 injected at birth and rAAV9 injected in adulthood in the same regenerated 44Bri mouse livers following partial hepatectomya
| Mouse | AAV8-EF1α-nlslacZ at birth
|
AAV9-LSP-hF.IX at 4-7 mo
|
||||
|---|---|---|---|---|---|---|
| Vector genome copy number in liver (ds-vg/dge)
|
% Remaining | Vector genome copy number in liver (ds-vg/dge)
|
% Remaining | |||
| At PHx | Post-PHx | At PHx | Post-PHx | |||
| 52 | 0.69 | 0.18 | 26 | 4.76 | 0.41 | 9 |
| 53 | 0.29 | 0.17 | 59 | 4.88 | 0.24 | 5 |
| 60 | 0.31 | 0.32 | 104 | 4.91 | 0.33 | 7 |
| 63 | 0.28 | 2.71 | 969 | 3.52 | 0.26 | 7 |
| 64 | 0.75 | 0.23 | 31 | 4.28 | 0.28 | 6 |
PHx, partial hepatectomy. Vector genome copy numbers were determined using the same Southern blot membrane by rehybridization.
Approximately 0.01% of hepatocytes had a transgene-expressible rAAV integration event.
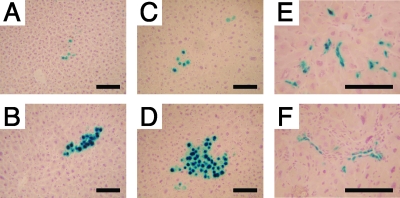
Histology sections were prepared from the livers harvested at the time of partial hepatectomy and at the end of the experiment 6 weeks after partial hepatectomy and analyzed for transduction efficiency by X-Gal staining. Foci of X-Gal-positive hepatocyte clusters comprising up to 200 hepatocyte nuclei were frequently observed in liver sections (Fig. 4). The results obtained from the samples at the time of partial hepatectomy and those from the no-hepatectomy group were combined for the analysis. Approximately 0.2 to 0.3% hepatocytes were found to be expressing β-galactosidase in any of the groups, and this efficiency did not decline during the 6 weeks following partial hepatectomy (Table 4). Foci of X-Gal-positive hepatocytes including the foci containing only a single positive cell emerged at a frequency of approximately 0.1% of hepatocytes on histology sections, and this frequency did not change significantly with hepatectomy (Table 4). Although the formation of a hepatocyte cluster is indicative of an rAAV vector integration event, the frequency of cluster formation in a two-dimensional assay is likely to result in overestimation. Therefore, we estimated the actual frequency of rAAV integration in a random distribution model as detailed in Materials and Methods. Using this statistical method, we estimated that transgene-expressible rAAV integration events occurred at a frequency of approximately 0.01% of total hepatocytes (Table 4).
FIG. 4.
X-Gal-positive hepatocyte clusters and nonparenchymal cell clusters in livers transduced with vector AAV8-EF1α-nlslacZ at birth. Representative photomicrographs of X-Gal-positive hepatocyte clusters (A to D) and nonparenchymal cell clusters (E and F) in liver sections are shown. (A) Wild-type liver with no hepatectomy; (B) wild-type liver after hepatectomy; (C) 44Bri liver with no hepatectomy; (D, E, and F) 44Bri liver after hepatectomy. These nonparenchymal cells appear to be endothelial cells, although the exact identities of these cells have yet to be determined. Each section was stained with X-Gal and counterstained with hematoxylin. Scale bars represent 100 μm.
TABLE 4.
Persistent liver transduction efficiency in wild-type and 44Bri mice injected with 2.0 × 1011 vg of AAV8-EF1α-lacZ at birth determined by X-Gal stainingc
| Mouse | Liver transduction efficiency (% hepatocytes) ± SD determined by:
|
|||||
|---|---|---|---|---|---|---|
| Total no. of transduced cells
|
Total no. of foci of transduced cellsa (2-dimensional assessment)
|
Total no. of transduced cell clustersb (3-dimensional assessment)
|
||||
| No PHx | Post-PHx | No PHx | Post-PHx | No PHx | Post-PHx | |
| Wild type | 0.20 ± 0.11 | 0.19 ± 0.10 | 0.09 ± 0.05 | 0.06 ± 0.03 | 0.011 ± 0.006 | 0.009 ± 0.005 |
| 44Bri | 0.18 ± 0.10 | 0.28 ± 0.10 | 0.08 ± 0.04 | 0.07 ± 0.03 | 0.008 ± 0.005 | 0.009 ± 0.003 |
The foci include solitary X-Gal-positive hepatocytes.
Only hepatocyte clusters comprising three or more positive nuclei on the liver sections were included.
PHx, partial hepatectomy.
A coincidental, interesting finding in the histological analysis of X-Gal-stained liver sections is that clusters of X-Gal-positive nonparenchymal cells were occasionally found by neonatal gene transfer (Fig. 4), although rAAV8 vectors exclusively transduce hepatocytes in adult mouse livers. On average, 1 of every 13 X-Gal-positive clusters containing three or more X-Gal-positive cells or nuclei was composed of nonparenchymal cells. The size of these nonparenchymal cell clusters was of up to 55 cells per cluster in a two-dimensional assessment. Although identities of the cluster-forming nonparenchymal cells have yet to be determined, some of them appeared to be vascular endothelial cells (Fig. 4).
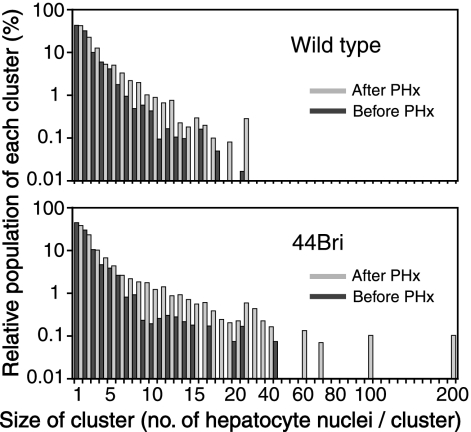
The size of transgene-expressing hepatocyte clusters increased following the hepatectomy: evidence of rAAV integration in many X-Gal-positive hepatocyte clusters in 44Bri mice.
An increase in the total liver mass due to liver regeneration following hepatectomy in theory increases the number of X-Gal-positive hepatocytes in each X-Gal-positive hepatocyte cluster if a cluster comprises hepatocytes with integrated rAAV genomes. This was more clearly evident in 44Bri mice than in wild-type mice (Fig. 5). This was concordant with the more prominent vector genome copy number decrease in wild-type mice than in 44Bri mice. This observation indicates that many X-Gal-positive hepatocyte clusters in 44Bri mice in fact represent rAAV integration events, while X-Gal-positive hepatocyte clusters observed in wild-type mice still contained extrachromosomal genomes. This supports our conclusion that rAAV8 vector injection at a dose of 2.0 × 1011 vg/mouse results in transgene-expressible rAAV integrations in ∼0.01% of hepatocytes.
FIG. 5.
Histograms of the sizes of X-Gal-positive hepatocyte clusters in wild-type and 44Bri mice before and after partial hepatectomy. The relative population of each X-Gal-positive hepatocyte cluster (percent) is plotted against the size of cluster. Black and gray bars represent histograms before and after a two-thirds partial hepatectomy. The size of X-Gal-positive hepatocyte clusters increased after partial hepatectomy, which was obvious in 44Bri mice.
Establishment of a quantitative integrated rAAV vector genome rescue assay.
To further investigate rAAV integration, we established a new assay system that can quantify rAAV integration events in rAAV-transduced tissues. The system used our AAV-ISceI.AO3 vector designed specifically for the isolation of rAAV vector genome-host cellular DNA junctions (19), and it does not rely on liver regeneration. In this approach, the number of rAAV integration events contained in a known amount of rAAV-transduced mouse liver genomic DNA is estimated by a comparison with the results obtained from a control mouse liver genomic DNA containing a known number of rAAV integration events.
For this, we first established a method to sequence through any rAAV vector genome recombination site, which has not been possible using a standard cycle sequencing procedure. Due to DNA polymerase stalling at AAV ITRs in complex structures formed at rAAV vector genome recombination sites, sequences beyond the AAV ITRs could not be determined in 26% of plasmid clones in rAAV integration site libraries in our previous study (19). In the present study, we overcame this problem by performing direct sequencing of bisulfite-modified plasmid DNA templates and searching the sequence results against the mouse genome database that has undergone a virtual sodium bisulfite chemical modification. With this new method, we successfully sequenced all the samples that were difficult to sequence through AAV ITRs.
Second, we determined integrated rAAV vector genome rescue efficiency by the transformation of DH10B, an E. coli strain that we used for the construction of rAAV integration site plasmid libraries. We constructed control mouse liver genomic DNA containing six different AAV-ISceI.AO3 integrations at a frequency of 1 integration in every 10 diploid genomic equivalents (45,000 integrations in 3 μg of genomic DNA), as described in Materials and Methods. The transformation of 3 μg of this control DNA yielded 3,034 and 2,744 bacterial colonies (2,889 on average) in two independent experiments. Plasmid DNA recovered from 100 colonies from one of the two libraries was analyzed for irrelevant plasmid contamination and representativeness of the libraries with BstYI digestion. The result showed that there was no irrelevant plasmid contamination, and the original ratio was fairly retained in the library. The six different rAAV integrations were recovered in the following percentages: 15%, 8%, 17%, 15%, 16%, and 29%. The sizes of rAAV integration site plasmids containing the above-mentioned six integration sites were between 2.4 and 7.8 kb, the sizes that can be efficiently recovered in bacteria at a comparable efficiency (19). Thus, we determined that the integrated rAAV vector genome rescue efficiency in our system is 6.4% (2,889/45,000). This efficiency was used to indirectly quantify rAAV integration events in rAAV8 vector-transduced mouse liver samples. It should be noted that the frequency determined by this method might somewhat underestimate the actual integration frequency and therefore represents the minimum frequency. This is because not all integrated rAAV vector genomes might be rescuable in bacteria. However, the degree of underestimation should not be considerable, as discussed in Discussion.
At least 0.05% of hepatocytes had integrated rAAV genomes as determined by a quantitative rAAV vector genome rescue assay.
To quantify rAAV8 integration frequency in the neonatal liver, 2.0 × 1011 vg of vector AAV8-ISceI.AO3 was injected intraperitoneally into newborn C57BL/6J mice. Five days and 6 weeks postinjection, the mice were sacrificed, and liver samples were collected. Using the same lot of DH10B E. coli and the same experimental procedures, we constructed rAAV8 vector integration site plasmid libraries from 3 μg of genomic DNA isolated from the liver of a mouse sacrificed 6 weeks postinjection. Two libraries were constructed independently. We characterized all the plasmid clones but one in the libraries. As a result, 13 rAAV integration events were identified in each library (total of 26 rAAV integration events). One plasmid clone could not be characterized due to the presence of two sequencing primer-binding sites in a plasmid and was not investigated further. Among the 26 rAAV vector integration site plasmids, four rAAV vector genome-cellular DNA junction sequences were determined by direct sequencing of bisulfite-modified plasmids. Given that 13 integration events in the average could be identified in 3 μg of genomic DNA and integrated rAAV vector genome rescue efficiency in our system was 6.4%, 203 (i.e., 13/0.064 = 203) integrations should have occurred in 450,000 diploid genomic equivalents (i.e., 0.045% of diploid genomic equivalents). Since hepatocytes in neonates are mostly diploid, we have concluded that rAAV integration that was rescuable in bacteria in our system occurred in ∼0.05% of hepatocytes in neonatal livers following 2.0 × 1011 vg of rAAV8. We also constructed rAAV integration site plasmid libraries using liver genomic DNA from mice sacrificed 5 days postinjection, which contained 278 ds-vg/dge vector genomes. Due to the presence of abundant large concatemers in complex structures, we could not remove concatemeric vector genomes efficiently in the experimental procedures, resulting in a very high background of colonies in the libraries, as we previously experienced with the liver samples transduced with 7.2 × 1012 vg of rAAV8 (19). We identified eight rAAV integration sites by sequencing two-thirds of the colonies that we characterized in one of the libraries. The frequency of eight integrations from two-thirds of the plasmid clones in a library was similar to the frequency determined 6 weeks postinjection. This suggested that most rAAV integration events should have completed in an early stage of rAAV-mediated neonatal liver transduction. Because not all rAAV integration events might be rescuable in bacteria, we concluded that at least ∼0.05% of hepatocytes had integrated rAAV genomes.
The rAAV8 vector integrated preferentially in genes but not in or near the mir-341 locus.
With a data set of 34 rAAV integration sites identified in mouse livers transduced with rAAV8 at birth, we investigated the integration site spectrum in the liver. Of 34 integration sites, 2 were not mapped to a unique site but could be determined whether they occurred in genic or intergenic regions; therefore, they were included in our analysis. Twenty-three integration sites (68%) were found in genes (Table 5), and three (9%) integrations each occurred within ±5 kb from the transcription start site or from CpG islands. This trend was the same as what we previously reported for adult mouse livers. However, there was no preference toward integration at or near DNA palindromes (19) in the present data set. Integration in or near the mir-431 locus, the recently identified common rAAV integration site found in mouse hepatocellular carcinoma (8), was not identified in our study.
TABLE 5.
rAAV8 vector integration sites in neonatal mouse liver
| Sample | Time at identificationa | Integration site (mm9)
|
RefSeq gene designationb | Descriptionc | ||
|---|---|---|---|---|---|---|
| Chr | Coordinate | Orientation of rAAV8 vector integration | ||||
| 1 | 6 wk | 1 | 89843492 | − | Usp40 | |
| 2 | 6 wk | 1 | 187600030 | − | ||
| 3 | 6 wk | 4 | 114649807 | − | Cmpk | |
| 4 | 5 days | 5 | 23011422 | − | Srpk2 | |
| 5 | 6 wk | 5 | 65001720 | + | ||
| 6d | 6 wk | 5 | 90902375 | − | Alb1 | |
| 7d | 6 wk | 5 | 90902835 | − | Alb1 | |
| 8 | 6 wk | 5 | 107913490 | − | ||
| 9 | 6 wk | 5 | 117669684 | + | Taok3 | |
| 10 | 6 wk | 5 | 146809177 | + | Cyp3a25 | |
| 11 | 6 wk | 6 | 49214325 | + | Tx < 5 kb; CpG < 5 kb | |
| 12 | 6 wk | 6 | 124889714 | − | ||
| 13 | 5 days | 6 | 142591297 | + | Abcc9 | |
| 14 | 6 wk | 6 | 123447257 or 123564480 | + | Vmn2r22 | |
| 15 | 5 days | 8 | 45886414 | − | ||
| 16 | 6 wk | 8 | 130986565 | + | Nrp1 | |
| 17 | 6 wk | 9 | 19452163 | − | Zfp75 | |
| 18 | 6 wk | 9 | 55404340 | + | Scaper | |
| 19 | 5 days | 9 | 103124021 | + | Trf | |
| 20 | 6 wk | 9 | 108511051 | + | Arih2 | |
| 21 | 5 days | 10 | 76682203 | − | ||
| 22 | 5 days | 11 | 7101461 | + | Igfbp1 | |
| 23d | 6 wk | 11 | 11844864 | − | Grb10 | |
| 24d | 6 wk | 11 | 11884763 | + | Grb10 | |
| 25 | 6 wk | 11 | 84663048 | + | Ggnbp2 | |
| 26 | 6 wk | 11 | 121478785 | + | B3gnt | |
| 27 | 6 wk | 12 | 112590478 | − | Cdc42bpb | |
| 28 | 6 wk | 13 | 11616234 | + | ||
| 29 | 6 wk | 14 | 123285098 | − | Pcca | |
| 30 | 6 wk | 15 | 26511925 | − | Fbxl7 | |
| 31 | 6 wk | 15 | 31465196 | − | Tx < 5 kb; CpG < 5 kb | |
| 32 | 5 days | 15 | 79921903 | + | Syngr1 | Tx < 5 kb; CpG < 5 kb |
| 33 | 5 days | 16 | 45193718 | − | ||
| 34 | 6 wk | Y | Multiple hits | Integration into LINE-L1 | ||
rAAV8 integration sites in mouse liver were identified either 5 days or 6 weeks postinjection.
Blank cells indicate integrations into intergenic regions.
Tx < 5 kb, integrations within ±5 kb from transcription start sites; CpG < 5 kb, integrations within ±5 kb from CpG islands.
Samples 6 and 7, and 23 and 14, were identified in two separate libraries; therefore, they are considered to be independent integration events.
DISCUSSION
In the present study, we aimed to determine rAAV8 vector integration frequency and integration site spectrum in the liver when the vector is systemically administered into neonatal mice. Different vectors show distinct integration frequencies and target site preferences in vitro and in vivo (19, 29, 30). The integration of gene therapy vectors into the host chromosomal DNA is an attractive feature for many clinical applications; however, vector genome integration has shown to have a possibility to pose a risk of insertional mutagenesis as clearly demonstrated in the SCID-X1 gene therapy (16). Although it has yet to be established what risks rAAV integration might pose when delivered into newborns, our study has revealed that the neonatal injection of 2.0 × 1011 vg of rAAV8 integrated preferentially into genes at a frequency of at least 1 in 2,000 hepatocytes at a level of ∼0.2 ds-vg/dge on average.
The quantification of rAAV integration events that occur in rAAV-transduced animal tissues has been a significant challenge. Quantitative PCR-based methods have been used to quantify the vector genome integration frequency in plasmid DNA or rAAV vector-injected animal tissues (21, 25, 42). These PCR-based methods could be used for the demonstration that the integration frequencies were below the sensitivity of the assays (21, 25, 42); however, they would not be the most appropriate for the demonstration and quantification of actual rAAV vector genome integrations that occur in rAAV-transduced animal tissues. In the liver, the most common approach to estimate rAAV genome integration frequency is to perform a two-thirds partial hepatectomy (39, 43). In this approach, the relative amount of integrated vector genomes compared to the total vector genomes is estimated by the degree of the decrease in vector genome copy numbers per cell following a hepatectomy. This estimation is possible because integrated vector genomes should not be diluted by undergoing cell divisions. A limitation of this approach, however, is that it provides relatively accurate measurements only when rAAV vector genomes integrate into the host genome at a considerably high efficiency (e.g., 40% or more of rAAV genomes in the liver) (43). This is because one or two cell cycles induced by a partial hepatectomy are not sufficient to eliminate all extrachromosomal vector genomes present in hepatocytes. Five percent to 20% of extrachromosomal genomes could remain in cells following a hepatectomy and may contribute to transgene expression. Therefore, although a vector copy number decline following a hepatectomy could give us an idea of at least how many rAAV genomes are extrachromosomal and at most how many rAAV genomes represent integrated genomes, it would not tell us the actual or minimum integration frequency.
When rAAV vectors are delivered to developing neonatal livers, a substantial amount of extrachromosomal vector genomes can be eliminated due to hepatocyte cell divisions associated with liver growth (6, 45). Mouse liver weight increases by a factor of 25 during the first 8 weeks of age (6), which would in theory require five cycles of cell divisions if the size of hepatocytes did not change during liver growth. Therefore, one can imagine that it may be possible to quantify rAAV vector genome integrations that occur in neonatal livers by a simple means, i.e., by analyzing the vector genome copy numbers and transgene expression at an adult age. However, our present study has indicated that the quantification of rAAV integration in neonatal gene transfer should not be that simple, because extrachromosomal vector genomes still remained at a level that makes an interpretation of the experimental results difficult. Our observations have indicated that hepatocytes could divide or enter the S phase of the cell cycle only several times or less in developing livers after birth because the sizes of X-Gal-positive hepatocyte clusters were mostly small unless the livers underwent compensatory regeneration (Fig. 4).
In the present study, to overcome these potential problems in the liver regeneration-based approach to quantify rAAV integrations in mouse livers, we developed two completely different methods. In the first approach, we further eliminated extrachromosomal vector genomes by exploiting both continuous and transient liver regeneration in addition to the normal growth in developing livers. In the second approach, we used a newly established integrated rAAV vector genome rescue assay.
In the first method, we used 44Bri mice (5), a well-established chronic hepatitis animal model. In this transgenic strain, chronic liver injury develops at ∼3 months of age and continues thereafter in the entire life of animals (5). Therefore, the influence of the liver damage on the rAAV vector integration process in neonatal livers of 44Bri mice should be considered to be minimum. The results in the present study supported a notion that the rAAV8 vector integration process completes at an early stage of rAAV8 vector transduction in neonatal livers. Although it is possible that rAAV vector genomes could integrate after the continuous liver damage developed at 3 months of age in 44Bri mice, we presume that the chance of rAAV integration in a later stage was not as high as that in the early stage because in a later stage, rAAV vector genomes were mostly in the form of stable circular monomers and concatemers, which are presumably not the intermediates toward integration. A study has indicated that circular monomer genomes are recombinogenic intermediates (9); however, our observations in mouse liver did not support this notion (31). Nonetheless, our first approach, in which we included various control groups and delivered internal control rAAV genomes to the liver, allowed us to comprehensively quantify rAAV vector integrations.
In the second method, we injected vector AAV8-ISceI.AO3 and determined how many rAAV integrations occurred in 3 μg of liver genomic DNA corresponding to 450,000 diploid hepatocytes. We found that in 450,000 diploid hepatocytes, 203 rAAV vector genome integration events that resulted in the formation of rAAV proviral genomes that are rescuable in bacteria should have occurred. The actual number of rAAV integration events might be higher than 203 out of 450,000 due to the nature of our integrated rAAV vector genome rescue assay. However, the degree of underestimation should not be considerable based on the following observations. The size of AAV8-ISceI.AO3 was 3.1 kb in length, and deletions of vector genome sequences of up to 0.6 kb from the right vector terminus could result in the formation of vector genomes that are rescuable in bacteria in our system. If rAAV vector genome-cellular DNA recombination occurred outside of this 0.6-kb region, such integration events could not be identified in our system, which underestimates integration frequency to some extent. In our previous study using AAV2-EF1α-hFAH.AOS, we identified 576 rAAV vector genome-cellular DNA recombinations that occurred within the 0.4-kb region from the vector genome termini (38). Vector genome terminal deletions of up to 0.4 kb did not impair the ability of the AAV2-EF1α-hFAH.AOS vector genome to be rescued in bacteria. Of the 576 recombinations that occurred within the 0.4-kb vector-terminal region, 5 recombinations occurred in the 3/8 (0.15-kb) region to the center, while 571 (99%) recombinations were found in the 5/8 (0.25-kb) region to the vector genome terminus. Assuming that only five integrations occur in any 0.15-kb region of the AAV8-ISceI.AO3 (3.1 kb) genome except for the 0.25-kb right terminal region when 571 integrations occur in the 0.25-kb right terminal region, 83 integrations [5 × (3.1 to 0.25)/0.15 to 5 × (0.6 to 0.25)/0.15 = 83] would be expected to form unrescuable rAAV proviral genomes. Therefore, 13% [83/(571 + 83) = 0.13] of rAAV integrations would represent those that could not be identified by our method if this assumption is appropriate. Nonetheless, our observations in the present study indicate that rAAV integration occurs at a frequency of 1 in approximately 103 hepatocytes when newborns are injected with 2.0 × 1011 vg of rAAV8.
There was a significant discrepancy between the total number of integrated vector genomes (i.e., ∼0.2 ds-vg/dge) and the total number of integration events (∼10−3 integration events/dge). Although this discrepancy might be attributed to the fact that these two numbers were determined by two different experiments, the most likely interpretation is that rAAV vector genomes integrate as large concatemers when the vector genome load per cell is substantially high (e.g., >100 ds-vg/dge). Although we could not characterize in detail integrated rAAV vector genome concatemers by Southern blot analysis in the present study due to the low vector genome copy numbers in the regenerative liver DNA samples, we found in a separate study that integrated vector genomes in clonally expanded neoplastic hepatocytes in liver tumors that developed in two mice injected with 7.2 × 1012 vg/mouse of rAAV8 at an adult age were both in the form of large concatemers (H. Nakai et al., unpublished results). Repetitive genomic sequences are targets for gene silencing via various mechanisms including pathways involving small interfering RNA (13, 24, 26, 44), and exogenously introduced transgenes that form concatemers have shown to be less transcriptionally active in mouse livers (4, 35). Thus, our observation that only a small portion of rAAV integration events could lead to persistent transgene expression is consistent with our inference that many rAAV provirus genomes are in the form of large concatemers, and transgene expression from concatemeric rAAV genomes was suppressed.
Recently, a study by Donsante et al. showed that rAAV integration at or near the mir-341 locus could lead to hepatocarcinogenesis in mice injected with rAAV2 vectors at birth. This has been evidenced by the observation that rAAV integration occurred within a narrow 6-kb region in the vicinity of the mir-341 locus in at least four of six liver tumors in mice injected with rAAV (8). A mechanism by which rAAV integration results in the overexpression of neighboring genes with multiple small nucleolar RNAs and microRNAs, which could dysregulate host gene expression significantly, leading to hepatocarcinogenesis, has been proposed. We have identified several hot spots for rAAV integration in mouse livers, among which is the Albumin 1 gene, a gene known to be strongly expressed in the liver. Five of 603 and 2 of 34 integrations occurred within this gene in our previous (19) and present (Table 5) studies. The mir-341 locus, however, does not appear to be a hot spot for rAAV integration as far as we analyzed 34 rAAV integration sites in rAAV8-tranduced mouse livers. Therefore, it is likely that the rAAV-mediated insertional mutagenesis is a consequence of relatively random rAAV vector integration followed by selection due to an acquired growth advantage. Thus, the observation by Donsante et al. can be explained only if the rAAV2 vector could have integrated into the host genome at a considerably high frequency at which at least four of six mice could have rAAV vector integration within a very narrow specific genomic region in the livers. Assuming that the probability of rAAV integration into the 6-kb mir-341 region is 2 × 10−6 (6,000 bp/3 × 109 bp) based on a random model, the probability that at least four of six liver tumors carrying integrated rAAV genomes have rAAV integration into the same 6-kb region when the total number of rAAV integration events in the whole mouse liver is “N” follows the following binomial equation:
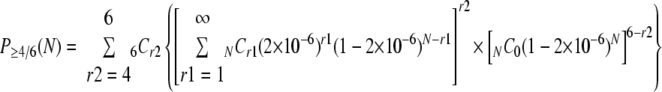 |
Here, P≥4/6(1.6 × 105) equals 0.05, P≥4/6(1 × 106) equals 0.96, and P≥4/6(2 × 106) equals 1.00. Therefore, at least 0.16 million integrations should have occurred in mouse liver to accommodate this frequency (P < 0.05). In the study by Donsante et al., they failed to identify whether the cancer cells had in fact rAAV integration in two of the six liver tumors by their method (8). If there was no rAAV integration in the two liver tumors, P4/4(3.2 × 105) equals 0.05, P4/4(1 × 106) equals 0.56, P4/4(2 × 106) equals 0.92, and P4/4(3 × 106) equals 0.99. Therefore, there should have been over 0.32 million integrations per mouse liver in this case (P < 0.05). Thus, the rAAV2 vector integration frequency in the study by Donsante et al. might be comparable to or even more than that of rAAV8 vector integration observed in the present study.
In any approaches using rAAV8 in neonates, whether or not the liver is the target for gene transfer, vector genomes are destined to be delivered to hepatocytes with considerable efficiency, and a portion of the genomes integrate into the host genome at a frequency of ∼10−3 hepatocytes when injected with a dose range of ∼1011 vg/mouse. It remains elusive whether rAAV vector integration frequency proportionately increases or decreases with increasing or decreasing vector doses, respectively. Accumulated observations by us and others have indirectly implied that this might not be the case. In our previous study, we could identify many rAAV2 vector integration sites in mouse livers at an efficiency comparable to that with the rAAV8 vector even with a much lower vector genome load in hepatocytes (19). In addition, we have an impression in our previous study (19) that a substantially high vector genome load in hepatocytes conversely decreases integration frequency, presumably by activating DNA repair pathways robustly. The robust activation of various DNA repair pathways triggered by a very high vector genome load might repair chromosome breakage sites, depleting the platforms for rAAV integration (28). This might explain why we did not see preferential integration at DNA palindromes when rAAV8 vectors were injected into newborn mice or when adult mouse livers were exposed to a very high dose of rAAV8 vectors. If this is true, a decrease in rAAV8 vector doses or the use of the rAAV2 vector might further increase rAAV vector integration frequency, which may increase a chance of insertional mutagenesis in rAAV-mediated neonatal gene transfer. Thus, more detailed dose-response studies are required to translate this promising approach into clinical applications.
Acknowledgments
We thank Guangping Gao and James M. Wilson for providing the AAV8 and AAV9 packaging plasmids and Mark A. Kay for helpful discussions of the experimental results.
This work was supported by Public Health Service grants DK68636 and DK78388 from the National Institutes of Health (to H.N.), a career development award from the National Hemophilia Foundation (to H.N.), and, at least in part, the National Cancer Institute, DHHS, under contract number N01-CO-12400 with SAIC-Frederick, Inc., and Public Health Service grant HL64274 from the National Institutes of Health.
The contents of this publication do not necessarily reflect the views or policies of the DHHS, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Published ahead of print on 9 July 2008.
REFERENCES
- 1.Bilbao, R., D. P. Reay, J. Li, X. Xiao, and P. R. Clemens. 2005. Patterns of gene expression from in utero delivery of adenoviral-associated vector serotype 1. Hum. Gene Ther. 16678-684. [DOI] [PubMed] [Google Scholar]
- 2.Bostick, B., A. Ghosh, Y. Yue, C. Long, and D. Duan. 2007. Systemic AAV-9 transduction in mice is influenced by animal age but not by the route of administration. Gene Ther. 141605-1609. [DOI] [PubMed] [Google Scholar]
- 3.Bouchard, S., T. C. MacKenzie, A. P. Radu, S. Hayashi, W. H. Peranteau, N. Chirmule, and A. W. Flake. 2003. Long-term transgene expression in cardiac and skeletal muscle following fetal administration of adenoviral or adeno-associated viral vectors in mice. J. Gene Med. 5941-950. [DOI] [PubMed] [Google Scholar]
- 4.Chen, Z. Y., C. Y. He, L. Meuse, and M. A. Kay. 2004. Silencing of episomal transgene expression by plasmid bacterial DNA elements in vivo. Gene Ther. 11856-864. [DOI] [PubMed] [Google Scholar]
- 5.Chisari, F. V., K. Klopchin, T. Moriyama, C. Pasquinelli, H. A. Dunsford, S. Sell, C. A. Pinkert, R. L. Brinster, and R. D. Palmiter. 1989. Molecular pathogenesis of hepatocellular carcinoma in hepatitis B virus transgenic mice. Cell 591145-1156. [DOI] [PubMed] [Google Scholar]
- 6.Cunningham, S. C., A. P. Dane, A. Spinoulas, and I. E. Alexander. 2008. Gene delivery to the juvenile mouse liver using AAV2/8 vectors. Mol. Ther. 161081-1088. [DOI] [PubMed] [Google Scholar]
- 7.Daly, T. M., K. K. Ohlemiller, M. S. Roberts, C. A. Vogler, and M. S. Sands. 2001. Prevention of systemic clinical disease in MPS VII mice following AAV-mediated neonatal gene transfer. Gene Ther. 81291-1298. [DOI] [PubMed] [Google Scholar]
- 8.Donsante, A., D. G. Miller, Y. Li, C. Vogler, E. M. Brunt, D. W. Russell, and M. S. Sands. 2007. AAV vector integration sites in mouse hepatocellular carcinoma. Science 317477. [DOI] [PubMed] [Google Scholar]
- 9.Duan, D., P. Sharma, J. Yang, Y. Yue, L. Dudus, Y. Zhang, K. J. Fisher, and J. F. Engelhardt. 1998. Circular intermediates of recombinant adeno-associated virus have defined structural characteristics responsible for long-term episomal persistence in muscle tissue. J. Virol. 728568-8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elliger, S. S., C. A. Elliger, C. Lang, and G. L. Watson. 2002. Enhanced secretion and uptake of beta-glucuronidase improves adeno-associated viral-mediated gene therapy of mucopolysaccharidosis type VII mice. Mol. Ther. 5617-626. [DOI] [PubMed] [Google Scholar]
- 11.Gao, G., L. H. Vandenberghe, M. R. Alvira, Y. Lu, R. Calcedo, X. Zhou, and J. M. Wilson. 2004. Clades of adeno-associated viruses are widely disseminated in human tissues. J. Virol. 786381-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao, G. P., M. R. Alvira, L. Wang, R. Calcedo, J. Johnston, and J. M. Wilson. 2002. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc. Natl. Acad. Sci. USA 9911854-11859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garrick, D., S. Fiering, D. I. Martin, and E. Whitelaw. 1998. Repeat-induced gene silencing in mammals. Nat. Genet. 1856-59. [DOI] [PubMed] [Google Scholar]
- 14.Ghosh, A., Y. Yue, C. Long, B. Bostick, and D. Duan. 2007. Efficient whole-body transduction with trans-splicing adeno-associated viral vectors. Mol. Ther. 15750-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grimm, D., S. Zhou, H. Nakai, C. E. Thomas, T. A. Storm, S. Fuess, T. Matsushita, R. Suroski, M. Lochrie, L. Meuse, A. McClelland, P. Colosi, and M. A. Kay. 2003. Pre-clinical evaluation of pseudotyped adeno-associated virus (AAV) vectors for liver gene therapy. Blood 1022412-2419. [DOI] [PubMed] [Google Scholar]
- 16.Hacein-Bey-Abina, S., C. Von Kalle, M. Schmidt, M. P. McCormack, N. Wulffraat, P. Leboulch, A. Lim, C. S. Osborne, R. Pawliuk, E. Morillon, R. Sorensen, A. Forster, P. Fraser, J. I. Cohen, G. de Saint Basile, I. Alexander, U. Wintergerst, T. Frebourg, A. Aurias, D. Stoppa-Lyonnet, S. Romana, I. Radford-Weiss, F. Gross, F. Valensi, E. Delabesse, E. Macintyre, F. Sigaux, J. Soulier, L. E. Leiva, M. Wissler, C. Prinz, T. H. Rabbitts, F. Le Deist, A. Fischer, and M. Cavazzana-Calvo. 2003. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 302415-419. [DOI] [PubMed] [Google Scholar]
- 17.Hartung, S. D., J. L. Frandsen, D. Pan, B. L. Koniar, P. Graupman, R. Gunther, W. C. Low, C. B. Whitley, and R. S. McIvor. 2004. Correction of metabolic, craniofacial, and neurologic abnormalities in MPS I mice treated at birth with adeno-associated virus vector transducing the human alpha-L-iduronidase gene. Mol. Ther. 9866-875. [DOI] [PubMed] [Google Scholar]
- 18.Inagaki, K., S. Fuess, T. A. Storm, G. A. Gibson, C. F. McTiernan, M. A. Kay, and H. Nakai. 2006. Robust systemic transduction with AAV9 vectors in mice: efficient global cardiac gene transfer superior to that of AAV8. Mol. Ther. 1445-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inagaki, K., S. M. Lewis, X. Wu, C. Ma, D. J. Munroe, S. Fuess, T. A. Storm, M. A. Kay, and H. Nakai. 2007. DNA palindromes with a modest arm length of ≲20 base pairs are a significant target for recombinant adeno-associated virus vector integration in the liver, muscles, and heart in mice. J. Virol. 8111290-11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inagaki, K., C. Ma, T. A. Storm, M. A. Kay, and H. Nakai. 2007. The role of DNA-PKcs and Artemis in opening viral DNA hairpin termini in various tissues in mice. J. Virol. 8111304-11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ledwith, B. J., S. Manam, P. J. Troilo, A. B. Barnum, C. J. Pauley, T. G. Griffiths II, L. B. Harper, C. M. Beare, W. J. Bagdon, and W. W. Nichols. 2000. Plasmid DNA vaccines: investigation of integration into host cellular DNA following intramuscular injection in mice. Intervirology 43258-272. [DOI] [PubMed] [Google Scholar]
- 22.Lipshutz, G. S., D. Titre, M. Brindle, A. R. Bisconte, C. H. Contag, and K. M. Gaensler. 2003. Comparison of gene expression after intraperitoneal delivery of AAV2 or AAV5 in utero. Mol. Ther. 890-98. [DOI] [PubMed] [Google Scholar]
- 23.Mah, C., K. O. Cresawn, T. J. Fraites, Jr., C. A. Pacak, M. A. Lewis, I. Zolotukhin, and B. J. Byrne. 2005. Sustained correction of glycogen storage disease type II using adeno-associated virus serotype 1 vectors. Gene Ther. 121405-1409. [DOI] [PubMed] [Google Scholar]
- 24.Martienssen, R. A. 2003. Maintenance of heterochromatin by RNA interference of tandem repeats. Nat. Genet. 35213-214. [DOI] [PubMed] [Google Scholar]
- 25.Martin, T., S. E. Parker, R. Hedstrom, T. Le, S. L. Hoffman, J. Norman, P. Hobart, and D. Lew. 1999. Plasmid DNA malaria vaccine: the potential for genomic integration after intramuscular injection. Hum. Gene Ther. 10759-768. [DOI] [PubMed] [Google Scholar]
- 26.McBurney, M. W., T. Mai, X. Yang, and K. Jardine. 2002. Evidence for repeat-induced gene silencing in cultured mammalian cells: inactivation of tandem repeats of transfected genes. Exp. Cell Res. 2741-8. [DOI] [PubMed] [Google Scholar]
- 27.McCarty, D. M., S. M. Young, Jr., and R. J. Samulski. 2004. Integration of adeno-associated virus (AAV) and recombinant AAV vectors. Annu. Rev. Genet. 38819-845. [DOI] [PubMed] [Google Scholar]
- 28.Miller, D. G., L. M. Petek, and D. W. Russell. 2004. Adeno-associated virus vectors integrate at chromosome breakage sites. Nat. Genet. 36767-773. [DOI] [PubMed] [Google Scholar]
- 29.Miller, D. G., G. D. Trobridge, L. M. Petek, M. A. Jacobs, R. Kaul, and D. W. Russell. 2005. Large-scale analysis of adeno-associated virus vector integration sites in normal human cells. J. Virol. 7911434-11442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitchell, R. S., B. F. Beitzel, A. R. Schroder, P. Shinn, H. Chen, C. C. Berry, J. R. Ecker, and F. D. Bushman. 2004. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2E234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakai, H., S. Fuess, T. A. Storm, L. A. Meuse, and M. A. Kay. 2003. Free DNA ends are essential for concatemerization of synthetic double-stranded adeno-associated virus vector genomes transfected into mouse hepatocytes in vivo. Mol. Ther. 7112-121. [DOI] [PubMed] [Google Scholar]
- 32.Nakai, H., S. Fuess, T. A. Storm, S. Muramatsu, Y. Nara, and M. A. Kay. 2005. Unrestricted hepatocyte transduction with adeno-associated virus serotype 8 vectors in mice. J. Virol. 79214-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakai, H., R. W. Herzog, J. N. Hagstrom, J. Walter, S. H. Kung, E. Y. Yang, S. J. Tai, Y. Iwaki, G. J. Kurtzman, K. J. Fisher, P. Colosi, L. B. Couto, and K. A. High. 1998. Adeno-associated viral vector-mediated gene transfer of human blood coagulation factor IX into mouse liver. Blood 914600-4607. [PubMed] [Google Scholar]
- 34.Nakai, H., Y. Iwaki, M. A. Kay, and L. B. Couto. 1999. Isolation of recombinant adeno-associated virus vector-cellular DNA junctions from mouse liver. J. Virol. 735438-5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakai, H., E. Montini, S. Fuess, T. A. Storm, L. Meuse, M. Finegold, M. Grompe, and M. A. Kay. 2003. Helper-independent and AAV-ITR-independent chromosomal integration of double-stranded linear DNA vectors in mice. Mol. Ther. 7101-111. [DOI] [PubMed] [Google Scholar]
- 36.Nakai, H., T. A. Storm, and M. A. Kay. 2000. Increasing the size of rAAV-mediated expression cassettes in vivo by intermolecular joining of two complementary vectors. Nat. Biotechnol. 18527-532. [DOI] [PubMed] [Google Scholar]
- 37.Nakai, H., C. E. Thomas, T. A. Storm, S. Fuess, S. Powell, J. F. Wright, and M. A. Kay. 2002. A limited number of transducible hepatocytes restricts a wide-range linear vector dose response in recombinant adeno-associated virus-mediated liver transduction. J. Virol. 7611343-11349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakai, H., X. Wu, S. Fuess, T. A. Storm, D. Munroe, E. Montini, S. M. Burgess, M. Grompe, and M. A. Kay. 2005. Large-scale molecular characterization of adeno-associated virus vector integration in mouse liver. J. Virol. 793606-3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakai, H., S. R. Yant, T. A. Storm, S. Fuess, L. Meuse, and M. A. Kay. 2001. Extrachromosomal recombinant adeno-associated virus vector genomes are primarily responsible for stable liver transduction in vivo. J. Virol. 756969-6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pacak, C. A., C. S. Mah, B. D. Thattaliyath, T. J. Conlon, M. A. Lewis, D. E. Cloutier, I. Zolotukhin, A. F. Tarantal, and B. J. Byrne. 2006. Recombinant adeno-associated virus serotype 9 leads to preferential cardiac transduction in vivo. Circ. Res. 99e3-e9. [DOI] [PubMed] [Google Scholar]
- 41.Rucker, M., T. J. Fraites, Jr., S. L. Porvasnik, M. A. Lewis, I. Zolotukhin, D. A. Cloutier, and B. J. Byrne. 2004. Rescue of enzyme deficiency in embryonic diaphragm in a mouse model of metabolic myopathy: Pompe disease. Development 1313007-3019. [DOI] [PubMed] [Google Scholar]
- 42.Schnepp, B. C., K. R. Clark, D. L. Klemanski, C. A. Pacak, and P. R. Johnson. 2003. Genetic fate of recombinant adeno-associated virus vector genomes in muscle. J. Virol. 773495-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song, S., Y. Lu, Y. K. Choi, Y. Han, Q. Tang, G. Zhao, K. I. Berns, and T. R. Flotte. 2004. DNA-dependent PK inhibits adeno-associated virus DNA integration. Proc. Natl. Acad. Sci. USA 1012112-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, F., N. Koyama, H. Nishida, T. Haraguchi, W. Reith, and T. Tsukamoto. 2006. The assembly and maintenance of heterochromatin initiated by transgene repeats are independent of the RNA interference pathway in mammalian cells. Mol. Cell. Biol. 264028-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, Z., T. Zhu, C. Qiao, L. Zhou, B. Wang, J. Zhang, C. Chen, J. Li, and X. Xiao. 2005. Adeno-associated virus serotype 8 efficiently delivers genes to muscle and heart. Nat. Biotechnol. 23321-328. [DOI] [PubMed] [Google Scholar]