Abstract
The relationship between immunosenescence and the host response to virus infection is poorly understood at the molecular level. Two different patterns of pulmonary host responses to virus were observed when gene expression profiles from severe acute respiratory syndrome coronavirus (SARS-CoV)-infected young mice that show minimal disease were compared to those from SARS-CoV-infected aged mice that develop pneumonitis. In young mice, genes related to cellular development, cell growth, and cell cycle were downregulated during peak viral replication, and these transcripts returned to basal levels as virus was cleared. In contrast, aged mice had a greater number of upregulated immune response and cell-to-cell signaling genes, and the expression of many genes was sustained even after viral clearance, suggesting an exacerbated host response to virus. Interestingly, in SARS-CoV-infected aged mice, a subset of genes, including Tnfa, Il6, Ccl2, Ccl3, Cxcl10, and Ifng, was induced in a biphasic pattern that correlated with peak viral replication and a subsequent influx of lymphocytes and severe histopathologic changes in the lungs. We provide insight into gene expression profiles and molecular signatures underlying immunosenescence in the context of the host response to viral infection.
The fastest-growing group in the U.S. population comprises individuals over the age of 65 years (42), with the maximum life span of an individual currently estimated at 120 years (7). While the elderly population steadily increases, this group also experiences enhanced severity of bacterial and viral infections, including pneumococcal, respiratory syncytial virus, and influenza virus infections. Influenza virus alone causes significant morbidity in persons ≥65 years of age, accounting for ca. 90% of the estimated 36,000 deaths yearly (38). More recently, severe acute respiratory syndrome coronavirus (SARS-CoV) emerged in the human population, disproportionately affecting the elderly, with up to 50% of all deaths occurring in persons >65 years of age (10). The immune response of the aging host could play a role in the enhanced susceptibility of the elderly to development of severe respiratory tract disease after infection with these pathogens. This aging or “remodeling” of the immune response is designated “immunosenescence” and is found in both long- and short-lived species as a function of their age relative to life expectancy rather than as chronologic time. For example, although the immune system of a 12-month-old child is literally in its infancy and becomes more competent with each passing day, the immune system of a 12-month-old mouse shows signs of immunosenescence and could be used to investigate an aged, yet naive, host response to a new pathogen.
Mice have been used to investigate SARS-CoV infection (12, 35, 41). The virus replicates in the upper and lower respiratory tract of young mice and is cleared rapidly with transient mild pneumonitis (29). Although an adaptive immune response that can protect from reinfection occurs in mice, the innate immune response appears to be sufficient for viral clearance. Selected strains of mice with targeted genetic defects have been used to probe pathways in the innate and adaptive immune response during SARS-CoV infection. CD1−/− mice and Rag1−/− mice (12) cleared the virus like wild-type mice, indicating that young mice do not require adaptive immunity or natural killer (NK) cell function for viral clearance. Stat1−/− mice (14) showed high levels of viral replication in the lungs, severe pulmonary disease with inflammation of small airways and alveoli, and systemic spread to the liver and spleen, indicating the importance of the effects of interferon, and the innate immune response. Based on the heightened susceptibility of elderly humans to severe SARS, it was hypothesized that aged mice might be more susceptible to disease than young mice. The study demonstrated that SARS-CoV replication in 12-month-old BALB/c mice was associated with clinical illness and pneumonia, demonstrating an age-related susceptibility to SARS disease in mice that parallels the human experience (30).
In the present study, we investigated the response to SARS-CoV infection by comparing gene expression profiles in pulmonary samples from young and old SARS-CoV-infected mice. Young mice cleared the virus, showed mild lung pathology, and demonstrated a limited number of differentially expressed genes. Older mice demonstrated delayed clearance of virus and developed histopathological lesions similar to those observed in human cases of SARS (11, 21, 26, 27, 39). The aged mice also demonstrated many strongly differentially expressed genes that remained throughout the infection and may be indicative of an exacerbated host response to the virus. The present study represents an endeavor to define molecular signatures underlying senescence and the effects of the remodeling of the immune system, driven by aging, by the course of viral infection, viral clearance, and pathology associated with viral infection.
MATERIALS AND METHODS
Animal studies.
Animal experiments were previously conducted for initial characterization studies in aged and young BALB/c mice. Briefly, 105 50% tissue culture infective doses of SARS-CoV (Urbani isolate) were administered intranasally to either 8-week-old (young) or 12- to 14-month-old (aged) BALB/c mice as previously described (12, 30, 41). Four mice in each age group were euthanized on days 1, 2, 5, and 7 postinfection. Five mock-infected mice in each age group were inoculated with phosphate-buffered saline and euthanized on day 7, serving as a negative control group. At necropsy, lungs were removed and stored at −70°C until the end of the study.
RNA isolation and genomic precharacterization.
One lung from each mouse was transferred to Solution D (6) to be homogenized, and RNA was extracted and purified according to our standard protocol (17). One aged mock-infected reference and one young mock-infected reference were created by pooling equal mass quantities of total RNA extracted from five aged and five young mock-infected mice, respectively. Equal mass quantities of total RNA extracted from four virus-infected mice at each time point were pooled to generate four virus-infected samples (days 1, 2, 5, and 7) for each age group (i.e., each virus-infected sample was composed of lungs from four individual mice). To assure that pooling was an appropriate strategy for generating experimental samples for microarray analysis, individual infected aged BALB/c mouse samples obtained at day 2 were checked for similarity of viral and cellular transcripts by using a panel of TaqMan primer/probes (e.g., Irf1, Irf7, Ifnb1, Ccl4, Stat1, Gapdh, SARS-CoV M, and SARS-CoV NP). This “proof of concept” characterization showed that samples from these inbred infected BALB/c mice were similar in viral and cellular transcripts (see Fig. S1 in the supplemental material), so each four-mouse set was pooled to create an experimental microarray sample. To confirm that the extracted total RNA was pure and intact, the pooled samples were checked by using an Agilent 2100 bioanalyzer (Agilent Technologies, Inc., Palo Alto, CA); all samples were judged pristine.
Oligonucleotide microarray analysis.
Each pooled sample was used to produce a cRNA probe using an Agilent low RNA input fluorescent linear amplification kit. Each experimental sample from the pooled infected aged or the pooled infected young mice was then cohybridized with a reference sample from the pooled mock-infected young mice, designated “young” so that these aged-young SARS-CoV-infected mouse samples could be directly compared to data from earlier experiments in our historical microarray database that includes data from experiments that were performed in young BALB/c mice. In addition, one microarray experiment was performed comparing the pooled young mock-infected reference to the pooled mock-infected aged reference so that differences in basal levels that might indicate immunosenescence could be determined. This young-aged mock reference comparison would also allow us to re-ratio (31) the microarray experiments to obtain an in silico version of each experimental aged mice versus a mock-infected aged reference. The experimental design is shown in Fig. S2 in the supplemental material. All microarray slide hybridization was performed with mouse oligonucleotide arrays (G4121B; Agilent Technologies). Slides were scanned with an Agilent DNA microarray scanner, and image analysis was performed by using Agilent feature extractor software. Each microarray experiment was done with four technical replicates by reversing dye hybridization for experimental and reference samples (19).
All data were entered into a custom-designed database, Expression Array Manager, and then uploaded into Resolver 4.0 (Rosetta Biosoftware, Seattle, WA) for gene expression analysis. For functional gene expression mining, DecisionSite for Functional Genomics (Spotfire, Inc., Somerville, MA), and Ingenuity Pathway Analysis (Ingenuity Systems, Redwood City, CA) were used to generate heat maps and group genes according to biological processes and canonical pathways. Finally, biological gene sets (referred to as biosets) were compiled for key cellular processes by selecting genes of interest that were represented on the microarray and also had Gene Ontology (GO) annotation (13). In accordance with proposed standards (1), all of the data described here, including sample information, intensity measurements, gene lists, error analysis, microarray content, and slide hybridization conditions, are available in the public domain through Expression Array Manager at http://expression.viromics.washington.edu.
Quantitative and absolute quantification real-time RT-PCR.
Relative quantitative real-time RT-PCR (qRT-PCR) was performed to detect SARS-CoV in lung tissue samples from infected mice and to validate a number of gene expression changes of cellular genes (e.g., Cdc2a, Pcna, Top2a, Cxcl10, Ddx58, Irf7, Isgf3g, Stat1, Stat2, and TNF) as detected on the microarrays. Total RNA samples were treated with DNase using a DNA-free DNase kit (Ambion, Inc., Austin, TX). cDNA was generated by using reverse transcription reagents and random hexamers (Applied Biosystems, Foster City, CA). qRT-PCR was run on the ABI 7500 PCR system, using TaqMan chemistry (Applied Biosystems, Foster City, CA). Primers and probe specific for the nucleoprotein gene of SARS-CoV have been described earlier (22), and primers and probes specific for murine cellular genes were purchased from Applied Biosystems as “gene expression assays.” Targets were run in quadruplicate following protocols supplied by Applied Biosystems. Absolute qRT-PCR was used to obtain viral copy numbers per nanogram of total RNA. These data were generated by using absolute qRT-PCR techniques, combined with the creation of a standard curve using known quantities of SARS armored RNA (developed jointly by Ambion, Inc., and Cenetron Diagnostics, LLC [catalog no. 42091]). SARS armored RNA can be used as a viral mRNA positive control and is stable upon storage because the control has a defined viral SARS-CoV sequence that is packaged in bacteriophage coat proteins to protect and stabilize the RNA. Armored RNA controls are highly uniform, precisely calibrated, and entirely noninfectious.
BD cytometric bead array and enzyme-linked immunosorbent assay.
Supernatants of 10% (wt/vol) lung homogenates (J. Chen et al., unpublished data) were used for detection of cytokine and chemokine protein expression using BD CBA mouse inflammation and Th1/Th2 kits (BD Biosciences, San Diego, CA) and murine Quantikine immunoassay kits (R&D, Minneapolis, MN) according to the manufacturer's protocols. The data are expressed as nanograms of protein per gram of tissue.
RESULTS
Comparison of aged and young BALB/c mouse models for SARS.
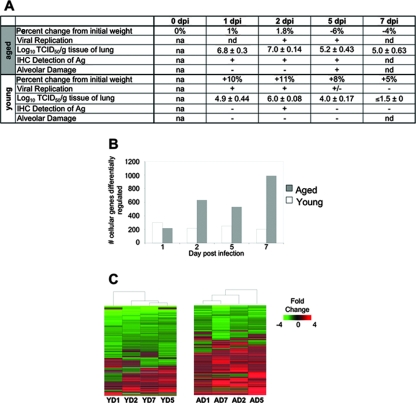
Previous studies have demonstrated that young BALB/c mice infected with SARS-CoV showed pulmonary viral replication with no clinical illness (35, 41), while aged BALB/c mice infected with SARS-CoV showed pulmonary viral replication associated with clinical illness and significant pathological changes in the lungs (30). Clinical illness was characterized by significant weight loss, hunching, ruffled fur, and slight dehydration. Pulmonary pathology was characterized by pneumonitis, alveolar damage, hyaline membrane formation, edema, and persistence of fibrotic foci. We compared aged and young mice infected with SARS-CoV (Urbani) and detected high titers of virus at days 1 and 2 in young mice and at days 2 and 5 in old mice (Fig. 1A), with titers in aged mice being 10-fold higher than in young mice on day 2.
FIG. 1.
Aged mice show a delay in viral clearance and a greater number of differentially regulated host cellular genes than young mice after SARS-CoV infection. (A) Characterization of SARS-CoV infection in young and old mice as reported previously by Subbarao et al. (35) and Roberts et al. (30). Plus signs indicate the presence of a finding, and minus signs indicate the absence of a finding. na, not applicable; nd, not done. The mean virus titer is expressed as the log10 50% tissue culture infective dose per gram of tissue. In aged mice, day 1 titers are typically 1-log lower than day 2 titers and day 7 titers are near or below the limit of detection. (B) The number of cellular genes differentially expressed in lung tissues is the result of comparing gene expression in lungs of experimentally infected mice (pooled, n = 4) with gene expression in lungs of mock-infected mice (pooled, n = 5); genes were included (y axis) if they met the criteria of an absolute fold change of ≥2 and P ≤ 0.01 for young (open) and old (gray) SARS-CoV-infected mice on the indicated days (x axis). (C) Global gene expression profiles are the results of comparing gene expression in lungs of experimentally infected mice (pooled, n = 4) with gene expression in lungs of mock-infected mice (pooled, n = 5); genes were included if they met the criteria of an absolute fold change of ≥2 and P ≤ 0.01 in at least one experiment. The data are presented for young (Y) and aged (A) mice on indicated days (D) 1, 2, 5, and 7 postinfection. These data were plotted as a heat map, where each matrix entry represents a gene expression value. Red corresponds to higher gene expression than that of the reference; green corresponds to lower gene expression. Dendrograms (trees) of the heat map represent the degree of relatedness between the samples, with short branches denoting a high degree of similarity and long branches denoting a low degree of similarity. Heat maps from young (541 sequences) and aged (1,735 sequences) mice were created individually because each set was compared to its age-matched mock-infected reference.
We next investigated differential host cellular gene transcripts in response to virus infection. First, we examined each gene expression experiment separately to see whether a general host response to virus infection could be identified; genes were included if they showed an absolute fold change of ≥2 and a P value of ≤0.01. With the exception of day 1 postinfection, young SARS-CoV-infected mice had fewer differentially expressed genes than older mice, but the number of genes stayed the same over the course of the infection in young mice (Fig. 1B). In contrast, from day 2 postinfection onward, along with viral replication, the number of differentially expressed genes in the lungs of SARS-CoV-infected aged mice approximately tripled, demonstrating a more robust host response in aged mice. In order to examine how timing after virus administration influenced gene expression, global expression profiling was performed. Hierarchical clustering methods were used to order rows (genes) and columns (samples) and identify groups of genes or samples with similar expression patterns (Fig. 1C) (5, 32). These analyses yielded 541 gene sequences for the young SARS-CoV-infected mice and 1,735 gene sequences for the aged SARS-CoV-infected mice. These cluster diagrams suggest that aged mice show a more zealous response to virus infection, as indicated by the greater number of matrix entries (genes), as well as the brighter intensities of the matrix entries (fold changes in transcripts) included in the heat map for aged mice. The results did not cluster by time from virus administration or by number of differentially expressed cellular genes, as indicated by the dendrogram at the top of each heat map. Dendrograms (trees) of the heat map represent the degree of relatedness between the samples, with short branches denoting a high degree of similarity and long branches denoting a low degree of similarity. In addition, the predominant pattern of gene expression in aged mice was of upregulated genes, which is suggestive of an active host response to viral infection, while the predominant pattern in young mice was of downregulated genes, a finding suggestive of a reduction of transcription.
Prominent host gene expression profiles.
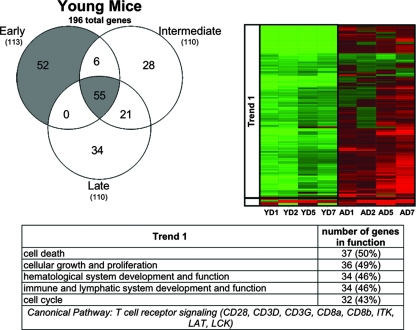
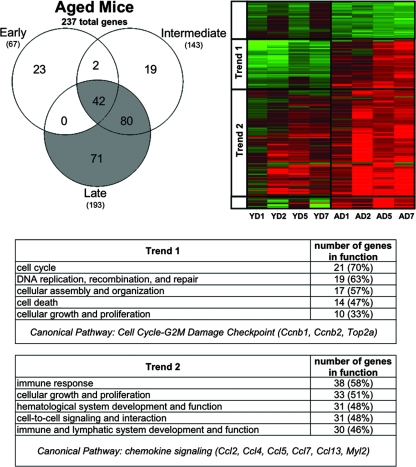
Because more than 1,000 genes were differentially expressed in SARS-CoV-infected aged mice, we stratified the results into early, intermediate, and late phases over the course of infection to dissect and more fully understand expression profiles of host cellular genes. Early genes were defined as showing an absolute fold change of ≥2 and a P value of ≤0.01 on both days 1 and 2 postinfection. Intermediate genes were defined by the same criteria on both days 2 and 5 postinfection. Late genes were defined by the same criteria on both days 5 and 7 postinfection. The terms “early,” “intermediate,” and “late” are not based on physiologic criteria; they only refer to gene expression within the context of the time points included in our experiments. The requirement that each gene be regulated at more than one time point increases the likelihood that the genes are consistently affected in the course of SARS-CoV infection, and therefore these sets of genes will constitute a smaller subset of the global gene expression present in Fig. 1C. Using this strategy, 196 total genes were differentially expressed in young SARS-CoV-infected mice (Fig. 2), while 237 total genes were differentially expressed in aged SARS-CoV-infected mice (Fig. 3). We generated Venn diagrams to determine the extent of overlap (i.e., similar response) at early, intermediate, and late time points (heat maps of all Venn diagram permutations are supplied (see Fig. S3 and S4 in the supplemental material).
FIG. 2.
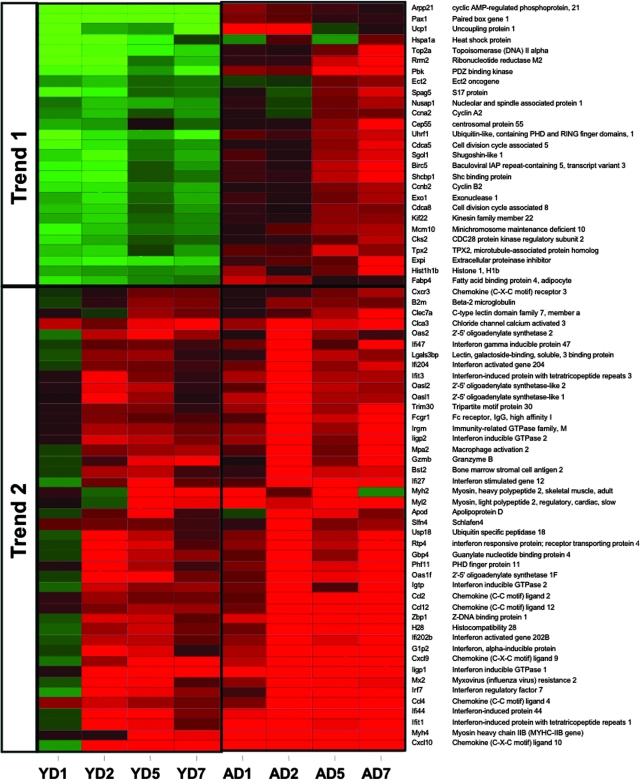
Young SARS-CoV-infected mice showed a prominent response at early time points and throughout the course of infection. Early, intermediate, and late genes were defined in the text. Briefly, genes shown in the Venn diagram and subsequent heat map met the criteria of an absolute fold change of ≥2 and P ≤ 0.01 in at least two time points. Early genes met the criteria on both days 1 and 2, intermediate genes met the criteria on both days 2 and 5, and late genes met the criteria on both days 5 and 7 postinfection. Therefore, this representation is a subset of the global gene expression seen in Fig. 1C. Gene expression profiles are the result of comparing gene expression in the lungs of experimentally infected mice (pooled, n = 4) with gene expression in the lungs of mock-infected mice (pooled, n = 5) The Venn diagram uses three circles, representing each time definition, to show the degree of overlap of gene expression. A total of 196 genes is shown in the Venn diagram of the young SARS-CoV-infected mice, with the most prominent gene responses seen at early time points (52 sequences, no overlap) and throughout the course of infection (55 sequences, overlap of early, intermediate, and late). The data for these most-prominent responses (colored gray in the Venn diagram) are next presented as a heat map for young (Y) mice on indicated days (D) 1, 2, 5, and 7 postinfection. The same genes for aged (A) mice are also included in the heat map for comparison. These genes in the heat map are primarily downregulated (trend 1) in young mice and are inversely correlated in aged mice. Predominant functional annotations, as well as the top canonical pathways, are shown in the trend 1 table. Functional annotation was generated by using Ingenuity Pathway Analysis as described in the text. The percentage was calculated by using the total number of genes in each functionality group and the total number of genes that could be functionally annotated. Heat maps of all of the areas in the Venn diagram are given in Fig. S3 in the supplemental material.
FIG. 3.
Aged SARS-CoV-infected mice showed a prominent response at late time points and throughout the course of infection. Early, intermediate, and late genes were defined in the text. Briefly, genes shown in the Venn diagram and subsequent heat map each met the criteria of an absolute fold change of ≥2 and P ≤ 0.01 in at least two time points. Early genes met the criteria on both days 1 and 2, intermediate genes met the criteria on both days 2 and 5, and late genes met the criteria on both days 5 and 7 postinfection. Therefore, this representation is a subset of the global gene expression present in Fig. 1C. Gene expression profiles are the result of comparing gene expression in RNA pooled from the lungs of four experimentally infected mice at each time point with gene expression in the lungs of mock-infected mice (pooled, n = 5). The Venn diagram uses three circles, representing each time definition, to show the degree of overlap of gene expression. A total of 237 genes are shown in the Venn diagram of the SARS-CoV-infected aged mice, with the most prominent gene responses seen at late time points (80 gene sequences, overlap of intermediate and late; and 71 sequences, no overlap) and throughout the course of infection (42 sequences, overlap of early, intermediate, and late). The data for these most prominent responses (colored gray in the Venn diagram) are next presented as a heat map for aged (A) mice on indicated days (D) 1, 2, 5, and 7 postinfection. The same genes for young (Y) mice are also included in the heat map for comparison. Trend 1 shows an inverse correlation between the responses of aged and young mice to SARS-CoV infection, and the predominant functional annotations and top canonical pathways are shown in the trend 1 table. Trend 2 shows a correlation between the responses of aged and young mice to SARS-CoV infection, and the predominant functional annotations and top canonical pathways are shown in the trend 2 table. Functional annotation was generated by using Ingenuity Pathway Analysis as described in the text. The percentage was calculated by using the total number of genes in each functionality group and the total number of genes that could be functionally annotated. Heat maps of all of the areas in the Venn diagram are found in Fig. S4 in the supplemental material.
Notably, in young SARS-CoV-infected mice, the greatest number of genes was differentially expressed (gray area in the Venn diagram) at early time points (52 genes) or throughout the course of the infection (early/intermediate/late, 55 genes) (Fig. 2). Surprisingly, a majority of these genes were downregulated, and their function corresponded to cellular development, cell growth, and cell cycle. In contrast, in aged SARS-CoV-infected mice the greatest number of genes was differentially expressed (gray area in the Venn diagram) at late (71 genes), late/intermediate (80 genes), and throughout the course of infection (early/intermediate/late, 42 genes) (Fig. 3). Two trends were observed. One set of genes that comprised cell cycle, DNA repair, and cell death functions was inversely correlated in the two groups of mice, with downregulation prevalent in young mice and upregulation in aged mice. The second trend was seen with genes that were upregulated in both sets of mice. In the aged mice, these genes were upregulated at day 1, became more strongly upregulated on day 2 corresponding to peak viral replication, and were expressed at high levels to day 7. In contrast, the young mice showed very little upregulation on day 1 but had a prominent response on day 2, also corresponding to peak viral replication, followed by a regression toward basal levels. This set comprised strong immune response and cell-to-cell signaling functions. In addition, the 196 genes differentially expressed in the young SARS-CoV-infected mice (Fig. 2) and the 237 genes differentially expressed in the aged SARS-CoV-infected mice (Fig. 3) were manipulated to find the overlap between the two groups. These groups had 71 genes in common (Fig. 4), again showing a strong presence of genes involved in cell cycle and immune response.
FIG. 4.
A small set of genes are affected in both aged and young mice during SARS-CoV infection The heat map shows genes that met the criteria of an absolute fold change of ≥2 and P ≤ 0.01 criteria in both the aged 237 gene set and the young 196 gene set. Gene expression profiles are the result of comparing gene expression in RNA pooled from lungs of four experimentally infected mice at each time point with gene expression in the lungs of mock-infected mice (pooled, n = 5). The data are presented for young (Y) and aged (A) mice on indicated days (D) 1, 2, 5, and 7 postinfection. Genes that are predominantly related to cell cycle functions were inversely correlated between aged and young mice in trend 1. Trend 2 shows the correlation between aged and young, with the predominant function being immune response.
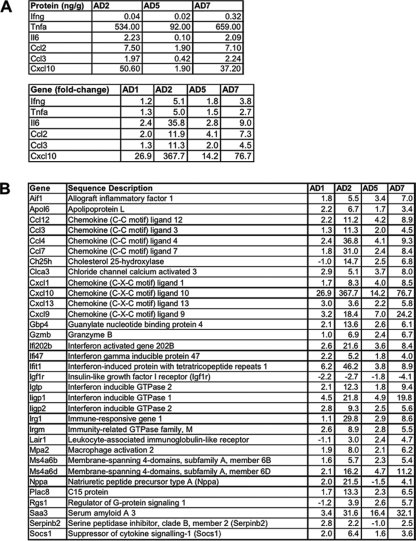
We also wanted to highlight the differentially expressed genes within the context of the immune response of aged mice. Protein levels of Tnfa (534.00, 92.00, and 659.00 ng/g), Il6, (2.23, 0.10, and 2.09 ng/g), Ccl2 (Mcp1) (7.50, 1.90, and 7.10 ng/g), Ccl3 (Mip1a) (1.97, 0.42, and 2.24 ng/g), and Cxcl10 (Ip10) (50.60, 1.90, and 37.20 ng/g), but not Ifng (0.04, 0.02, and 0.32 ng/g), were elevated in a biphasic pattern in aged SARS-CoV-infected mice (Chen et al., unpublished). mRNA levels of these genes, as well as Ifng, showed a biphasic pattern (Fig. 5A). To identify other genes regulated in this manner, we evaluated the 196 genes of young SARS-CoV-infected mice (Fig. 2) and 237 genes of aged SARS-CoV-infected mice (Fig. 3) for genes that showed a fold change of ≥2 at day 2, a decrease in fold change at day 5, and a fold changeday 7/fold changeday 5 ratio of ≥2. Interestingly, 33 genes were identified in aged SARS-CoV-infected mice (Fig. 5B) that showed this biphasic pattern. These criteria were not met by gene expression in young mice. The first phase of the biphasic response correlates well with peak virus titer and migration and activation of NK cells, macrophages, and plasmacytoid dendritic cells (pDCs) into the lungs (Chen et al., unpublished). The second phase of the biphasic response occurs with viral clearance; an influx and activation of T lymphocytes, particularly CD8+ T cells; and severe histopathologic changes in the lungs (Chen et al., unpublished).
FIG. 5.
Biphasic induction of mRNA in aged SARS-CoV-infected mice. (A) Protein and mRNA levels of indicated cytokines and chemokines in aged SARS-CoV-infected mice on the indicated days postinfection. The data are presented for aged (A) mice on indicated days (D) 1, 2, 5, and 7 postinfection. Gene transcripts (fold change) showed biphasic responses similar to those observed with protein levels (ng of cytokine per g of lung tissue). Although Ifng did not show a biphasic protein response, the gene showed biphasic transcript. Fold changes of transcripts are the result of comparing the gene expression in the lungs of experimentally infected mice (pooled, n = 4) with the gene expression in the lungs of mock-infected mice (pooled, n = 5). Concentration of cytokines are the results of cytokine bead assays. (B) Graph showing the fold change in genes exhibiting a biphasic response on the indicated days postinfection. Each line represents an individual gene. Using the criteria of a fold change of ≥2 at day 2, a decrease in fold change at day 5, and a fold changeday 7/fold changeday 5 ratio of ≥2, 33 genes were defined as showing a biphasic response.
Because the same innate immune response genes are operative in young and aged SARS-CoV-infected mice, we sought to understand these responses in the context of senescence. We directly compared lung samples from mock-infected aged mice to lung samples from mock-infected young mice in order to identify basal gene expression and to put the host response to viral infection within the context of aging (see Fig. S5 in the supplemental material). Immunoglobulin-related genes showed a general trend of upregulation in aged mice, and genes involved in immunoglobulin recombination were suppressed. These trends might reflect a more mature host immune system that has encountered several antigens. Genes involved in T-cell receptor signaling were also suppressed in the aged mice. This observation parallels what was reported previously using peripheral blood mononuclear cells from aged mice and aged humans (16). The suppression of these genes may represent a more limited population of naive T cells found in aged mice. In addition, a number of cell cycle and cyclin transcripts that may be markers of a general cellular senescence were downregulated. Future studies in aging mice will be important to further define this cellular senescence that is postulated to be a unique state of arrest that is distinct from apoptosis or necrosis (20).
Detected networks and gene interactions.
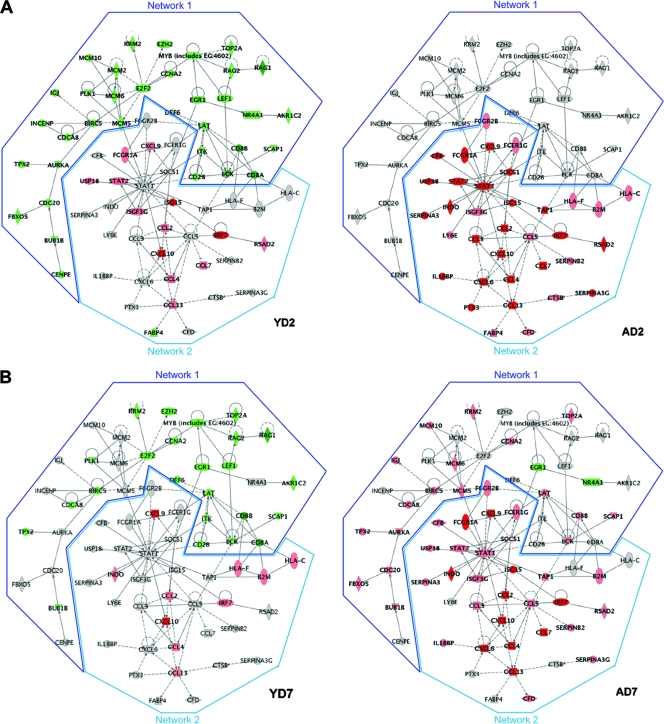
In order to assess the interplay between genes during the host response to virus infection, we used a functional analysis approach to construct a graphical network of biologically related genes derived from known relationships present in the Ingenuity Pathways Knowledge Base. Many networks can be constructed, but we limited our investigation to the networks that were most prevalent at the time of peak viral replication (i.e., day 2 for both sets of mice) and to networks that showed a significant over-representation of focus genes. Because responses of young and old SARS-CoV-infected mice differed more than they resembled one another, the top functional network identified in young mice was not the same as the top functional network identified in aged mice. In young SARS-CoV-infected mice, genes participating in cell cycle, cellular development, and hematological system development and function were computationally postulated to show regulatory interactions. In aged mice, genes participating in immune response, inflammatory disease, and cell-to-cell signaling and interaction were computationally postulated to show regulatory interactions. Although each of these networks was identified and represents the most prevalent interaction occurring in young or aged mice, the networks are not exclusive to either group.
To get a more complete comparison using both groups of mice, we combined the top young network (dark blue outline) and the top aged network (light blue outline) to construct a merged network, shown in Fig. 6. The network analyses reveal that differential gene regulation does not happen in isolation, but genes within pathways show interactions that constitute a network. The network diagram shows these interactions: both direct (solid lines) and indirect (dashed lines) interactions among these genes. The left panel in Fig. 6A illustrates that during peak viral replication in young SARS-CoV-infected mice, there is a general downregulation of “cell cycle”-type interactions but some upregulation of “immune response”-type interactions that we originally identified using data from aged mice. The aged SARS-CoV-infected mice show a much more robust upregulation of the “immune response” network (many showing ≥5-fold upregulation) without any downregulation in the “cell cycle” network during peak viral replication (Fig. 6A). The presence of so many highly upregulated immune genes in the aged SARS-CoV-infected mice suggests an uncontrolled response to virus infection. Interestingly, although the young SARS-CoV-infected mice seem to show a greater number of differentially expressed “cell cycle”-type genes than aged SARS-CoV-infected mice, when basal levels of these genes are evaluated in mock-infected aged mice, they are already downregulated >2-fold compared to levels in young mice, with the exclusion of Egr1 and Nr4a1 (unchanged) and Igj (upregulated) (see Fig. S6 in the supplemental material). Possibly, these genes cannot be further downregulated in aged mice, even though in other systems, coronaviruses are capable of inhibiting cell cycle, leading to an accumulation of infected cells in the G0/G1 phase (4, 36, 37).
FIG. 6.
(A) At the time of peak viral replication, aged and young mice present different “top” networks. A network is a group of biologically related genes that is derived from known relationships present in the Ingenuity Pathways Knowledge Base. These diagrams, which comprise the two top networks, represent the interactions, both direct (solid lines) and indirect (dashed lines), between genes and gene products identified during peak viral replication. Network 1 was the most prominent network detected at day 2 in young SARS-CoV-infected mice and was composed of cell cycle, cellular development, and hematological system development functions. Network 2 was the most prominent network detected at day 2 in aged SARS-CoV-infected mice and was composed of immune response, inflammatory response, and cell-to-cell signaling functions. Young mice show network 1 to be downregulated, and although aged mice show a very strong induction of network 2, young mice also induce this network. Dark red indicates a fold change of ≥5, pink indicates a fold change of ≥2, and gray indicates a fold change of <2. Dark green indicates a fold change of ≥5, light green indicates a fold change of ≥2, and gray indicates an absolute fold change of <2. Gene expression profiles are the result of comparing gene expression in the lungs of experimentally infected mice (pooled, n = 4) with gene expression in the lungs of mock-infected mice (pooled, n = 5). (B) Network interactions at day 7 postinfection when SARS-CoV is cleared from the lungs. This is the same merged network shown in Fig. 6A. Again, young mice showed a prominent downregulation at day 7 but also seem to show a trend of returning to levels seen in mock-infected animals. Aged SARS-CoV-infected mice show an increase of transcripts in network 1 and a continued strong induction of network 2 at day 7. Dark red indicates a fold change of ≥5, pink indicates a fold change of ≥2, and gray indicates a fold change of <2. Dark green indicates a fold change of ≥5, light green indicates a fold change of ≥ 2, and gray indicates an absolute fold change of <2. Gene expression profiles are the result of comparing gene expression in the lungs of experimentally infected mice (pooled, n = 4) with gene expression in the lungs of mock-infected mice (pooled, n = 5).
The same merged network is presented in Fig. 6B to show how interactions are affected as the virus is cleared. In the left panel, young mice show a resolution of downregulated “cell cycle”-type genes (∼30% less) and resolution of upregulated “immune response”-type genes as virus infection resolves. Notably in this phase, Hla-f, Hla-c, and B2m are now upregulated, while Cd8a and Cd8b still show a strong absence of transcripts, a finding perhaps indicative of a controlled T-cell receptor signaling progression (this trend is also seen at day 5). The right panel shows how the immune response affects cell cycle genes. The immune response is still induced robustly in aged mice and now at 7 days postinfection, genes involved in cell cycle, cellular development, and hematological system development and function are also beginning to be induced; this profile mimics the histologic evidence of lung repair on days 5 and 7 (30). Cxcl10 (Ip-10), Ccl2 (Mip-1), Irf7, and Isg15 (G1p2) are still highly upregulated, even as virus is cleared, and are indicators that this heightened immune response is not required for viral clearance and may be responsible for immunopathology.
DISCUSSION
The present study was motivated by our poor understanding of the relationship between ageing and the host response to virus infection. It has been long known that young and old mice respond quite differently to infection with different viruses (15, 25, 43). We therefore compared gene expression profiles from the lungs of young and old mice infected with SARS-CoV to define the extent to which the host response determined the outcome of infection and identified differences in the pattern, magnitude, and duration of the host response to SARS-CoV infection in the two groups of mice. A comparison of gene expression profiles in mock-infected young and old mice provided additional insights into the effect of age on gene expression, and the comparison of SARS-CoV-infected mice from the two age groups allowed us a glimpse of the immune response to a respiratory pathogen within the framework of senescence.
The aged BALB/c mouse is a valuable model for SARS because it allows the analysis of four outcome measures: quantitative virology, clinical symptoms, histopathological changes, and immunohistochemistry in the lungs (30, 40). Of these outcomes, only quantitative virology can be studied in young SARS-CoV-infected BALB/c mice. Our study provides host gene expression as an additional outcome measure in the context of a head-to-head comparison of aged and young BALB/c mouse models of SARS. Gene expression analysis revealed a transcriptional profile in aged mice that indicates a more robust response to virus infection than in young mice: a greater number of genes are transcribed, and transcripts have a greater magnitude of change. The peak immune response in young mice corresponds to the peak in viral replication, and the transcripts regress to basal levels as the virus is cleared. Aged mice also exhibit a peak host immune response corresponding to the peak in viral replication, and the delay in viral clearance in aged mice is accompanied by a delay in the return of gene expression to basal levels. In addition, a subset of genes is induced in a second wave of a biphasic response, corresponding to the influx and activation of T lymphocytes and severe histopathologic changes in the lungs (Chen et al., unpublished).
The changes associated with senescence that have been described in the literature mainly pertain to adaptive immune responses; the components of the innate response have not been studied as well. Innate immunity may hold clues to explain the increased morbidity in the elderly since this arm of the immune system may become progressively more important in aging individuals as a means to fill the “immunologic gap” that appears as adaptive immunity wanes. In addition to waning immune responses with advancing age, changes in lung structure and function, such as loss of elastin fibers, decreases in the number of capillaries per alveolus, decreases in the efficiency of mucociliary clearance, or decreased respiratory capacity, likely play a role in host response to respiratory infection (24). Our findings in aged and young SARS-CoV-infected mice are consistent with what has been seen in previous studies of primary viral infection in aged mice, with influenza virus, lymphocytic choriomeningitis virus, Friend virus, E55+ murine leukemia virus, and respiratory syncytial virus, where clearance of virus in aged mice was delayed compared to young mice. These studies have led investigators to conclude that the aging immune system is not abated but is simply functioning at a level different than that of young mice (15, 25, 43). In young mice, type I interferon was detected within hours of primary viral infection with influenza virus, lymphocytic choriomeningitis virus, and E55+ murine leukemia virus (15, 25). In aged mice, the detection of type I interferon was not as straightforward, and the interferon response that was detected may or may not have been functional (15, 25). Type I interferon has previously been identified as an enhancer of NK cell cytotoxicity in other studies of primary infection in aged animals and may play a role in depletion of nonspecific T cells to initiate expansion of antigen-specific T cells (16). NK cells are critical to the removal of intracellular pathogens, and nonspecific T-cell depletion may be necessary to provide “space” for the proliferation and expansion of T cells specific to newly introduced pathogen (16, 25, 28). Both in mice and in humans, the ability of type I interferon to enhance cytotoxicity of NK cells, deplete nonspecific T cells, and expand antigen-specific T cells decreases with age (16, 25, 28, 34). The remodeled immune system of the aged could play a role in delaying clearance of virus.
In our gene expression study of SARS-CoV-infected mice, beta-interferon transcription was more prevalent in aged mice than young. Although we did not measure beta-interferon activity levels in the present study, the upregulated transcription of interferon-stimulated genes suggests that the beta interferon produced by aged mice is functionally active (see Fig. S7 in the supplemental material). High levels of beta interferon may be necessary to clear virus in the aged mice infected with SARS-CoV. In aged mice, Ifnb mRNA showed the greatest upregulation at day 2, which corresponded with an approximate doubling of the number of pDCs detected in the lungs (22.2 × 104/lung, ∼11% of total CD3− CD11b− CD19− lung leukocytes) (Chen et al., unpublished). pDCs are considered first-line sentinels in immune surveillance and are capable of producing large amounts of type I interferon (2, 8, 9, 23, 33) and have been found to produce type I interferon during coronavirus infection (3).
We speculate that the immune dysregulation observed in aged mice may contribute to the damage detected in the lungs by an immunopathological mechanism. The aged mice, experiencing infection with SARS-CoV, show an upregulation of an abundance of host cellular genes in response to the virus but fail to bring this response under control even as the virus is cleared, whereas young mice are able to clear SARS-CoV rapidly and show a controlled host response. Because RNA from the infected lungs was used for gene expression analyses, it is certainly possible that some of the changes occurred in cells that were productively infected and other changes were in activated bystander cells or in cells that phagocytosed viral antigen but were not productively infected. Even if these different types of cells could be sorted and these experiments were not constrained by SARS biosafety containment protocols, it is unlikely differences could be distinguished among these cellular subgroupings. Even with this caveat, the virus leaves behind a unique functional genomics “fingerprint” in the aged host compared to the young host that may be a signature of imminent or ongoing morbidity. A similar association of immune dysregulation with lung damage was seen in BALB/c mice infected with the highly virulent reconstructed 1918 influenza virus. Using our laboratory's historical microarray database, we compared the data from SARS-CoV-infected mice in this experiment to a set of data from young BALB/c mice that were infected with a human influenza A virus (A/Texas/36/91) isolated during a seasonal epidemic of influenza or reconstructed highly virulent human influenza A virus (r1918) (18). Although the mechanism by which these viruses elicit host responses may differ because SARS-CoV-infected mice have a prominent infiltration of NK cells, macrophages, and pDCs, while influenza-infected mice have a prominent infiltration of neutrophils, trends in host response to different respiratory viruses may be informative. Aged SARS-CoV mice show a more similar host response to young mice infected with the highly virulent reconstructed 1918 influenza virus that replicated to high titer and was associated with prominent pulmonary pathology and death by day 5 postinfection (see Fig. S8 in the supplemental material) than to mice infected with a nonlethal epidemic influenza virus. The similarity of the genomic profiles of mice infected with a nonlethal SARS coronavirus to those of mice infected with a highly virulent and lethal influenza virus suggests that the sustained response to infection seen in aged SARS-CoV-infected mice may play a role in the pathogenesis of pulmonary disease.
Translating this molecular signature into a common denominator within the context of other respiratory virus infection and morbidity in an aged host is an important question to answer with future studies. The continued use of older animals to examine the host response to different respiratory pathogens will provide an important framework within which the impact of senescence (e.g., immune, cellular, structural (24) on disease progression and the mechanisms of increased severity of respiratory tract infections in the elderly can be better understood.
Supplementary Material
Acknowledgments
This study was supported in part by the Intramural Research Program of NIAID, NIH (K.S.); P51 RR00166-45, NCRR, NIH, Regional Primate Research Center (Core Grant), Division of Functional Genomics and Infectious Disease (M.G.K., Division Head); and R01 HL080621-01, NHLBI, NIH, Macaque Model and Gene Expression Profiling of SARS (M.G.K.).
Footnotes
Published ahead of print on 16 July 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Brazma, A., P. Hingamp, J. Quackenbush, G. Sherlock, P. Spellman, C. Stoeckert, J. Aach, W. Ansorge, C. A. Ball, H. C. Causton, T. Gaasterland, P. Glenisson, F. C. Holstege, I. F. Kim, V. Markowitz, J. C. Matese, H. Parkinson, A. Robinson, U. Sarkans, S. Schulze-Kremer, J. Stewart, R. Taylor, J. Vilo, and M. Vingron. 2001. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat. Genet. 29365-371. [DOI] [PubMed] [Google Scholar]
- 2.Cella, M., D. Jarrossay, F. Facchetti, O. Alebardi, H. Nakajima, A. Lanzavecchia, and M. Colonna. 1999. Plasmacytoid monocytes migrate to inflamed lymph nodes and produce large amounts of type I interferon. Nat. Med. 5919-923. [DOI] [PubMed] [Google Scholar]
- 3.Cervantes-Barragan, L., R. Zust, F. Weber, M. Spiegel, K. S. Lang, S. Akira, V. Thiel, and B. Ludewig. 2007. Control of coronavirus infection through plasmacytoid dendritic-cell-derived type I interferon. Blood 1091131-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, C. J., and S. Makino. 2004. Murine coronavirus replication induces cell cycle arrest in G0/G1 phase. J. Virol. 785658-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chipman, H., T. J. Hastie, and R. Tibshirani. 2003. Clustering microarray data. CRC Press, New York, NY.
- 6.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162156-159. [DOI] [PubMed] [Google Scholar]
- 7.Coles, L. S. 2004. Demography of human supercentenarians. J. Gerontol. A Biol. Sci. Med. Sci. 59B579-B586. [DOI] [PubMed] [Google Scholar]
- 8.Colonna, M., G. Trinchieri, and Y. J. Liu. 2004. Plasmacytoid dendritic cells in immunity. Nat. Immunol. 51219-1226. [DOI] [PubMed] [Google Scholar]
- 9.Demedts, I. K., K. R. Bracke, T. Maes, G. F. Joos, and G. G. Brusselle. 2006. Different roles for human lung dendritic cell subsets in pulmonary immune defense mechanisms. Am. J. Respir. Cell Mol. Biol. 35387-393. [DOI] [PubMed] [Google Scholar]
- 10.Donnelly, C. A., A. C. Ghani, G. M. Leung, A. J. Hedley, C. Fraser, S. Riley, L. J. Abu-Raddad, L. M. Ho, T. Q. Thach, P. Chau, K. P. Chan, T. H. Lam, L. Y. Tse, T. Tsang, S. H. Liu, J. H. Kong, E. M. Lau, N. M. Ferguson, and R. M. Anderson. 2003. Epidemiological determinants of spread of causal agent of severe acute respiratory syndrome in Hong Kong. Lancet 3611761-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franks, T. J., P. Y. Chong, P. Chui, J. R. Galvin, R. M. Lourens, A. H. Reid, E. Selbs, C. P. McEvoy, C. D. Hayden, J. Fukuoka, J. K. Taubenberger, and W. D. Travis. 2003. Lung pathology of severe acute respiratory syndrome (SARS): a study of 8 autopsy cases from Singapore. Hum. Pathol. 34743-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glass, W. G., K. Subbarao, B. Murphy, and P. M. Murphy. 2004. Mechanisms of host defense following severe acute respiratory syndrome-coronavirus (SARS-CoV) pulmonary infection of mice. J. Immunol. 1734030-4039. [DOI] [PubMed] [Google Scholar]
- 13.Harris, M. A., J. Clark, A. Ireland, J. Lomax, M. Ashburner, R. Foulger, K. Eilbeck, S. Lewis, B. Marshall, C. Mungall, J. Richter, G. M. Rubin, J. A. Blake, C. Bult, M. Dolan, H. Drabkin, J. T. Eppig, D. P. Hill, L. Ni, M. Ringwald, R. Balakrishnan, J. M. Cherry, K. R. Christie, M. C. Costanzo, S. S. Dwight, S. Engel, D. G. Fisk, J. E. Hirschman, E. L. Hong, R. S. Nash, A. Sethuraman, C. L. Theesfeld, D. Botstein, K. Dolinski, B. Feierbach, T. Berardini, S. Mundodi, S. Y. Rhee, R. Apweiler, D. Barrell, E. Camon, E. Dimmer, V. Lee, R. Chisholm, P. Gaudet, W. Kibbe, R. Kishore, E. M. Schwarz, P. Sternberg, M. Gwinn, L. Hannick, J. Wortman, M. Berriman, V. Wood, N. de la Cruz, P. Tonellato, P. Jaiswal, T. Seigfried, and R. White. 2004. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 32D258-D261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hogan, R. J., G. Gao, T. Rowe, P. Bell, D. Flieder, J. Paragas, G. P. Kobinger, N. A. Wivel, R. G. Crystal, J. Boyer, H. Feldmann, T. G. Voss, and J. M. Wilson. 2004. Resolution of primary severe acute respiratory syndrome-associated coronavirus infection requires Stat1. J. Virol. 7811416-11421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang, J., F. Anaraki, K. J. Blank, and D. M. Murasko. 2003. Cutting edge: T cells from aged mice are resistant to depletion early during virus infection. J. Immunol. 1713353-3357. [DOI] [PubMed] [Google Scholar]
- 16.Jiang, J., D. Gross, S. Nogusa, P. Elbaum, and D. M. Murasko. 2005. Depletion of T cells by type I interferon: differences between young and aged mice. J. Immunol. 1751820-1826. [DOI] [PubMed] [Google Scholar]
- 17.Kash, J. C., C. F. Basler, A. Garcia-Sastre, V. Carter, R. Billharz, D. E. Swayne, R. M. Przygodzki, J. K. Taubenberger, M. G. Katze, and T. M. Tumpey. 2004. Global host immune response: pathogenesis and transcriptional profiling of type A influenza viruses expressing the hemagglutinin and neuraminidase genes from the 1918 pandemic virus. J. Virol. 789499-9511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kash, J. C., T. M. Tumpey, S. C. Proll, V. Carter, O. Perwitasari, M. J. Thomas, C. F. Basler, P. Palese, J. K. Taubenberger, A. Garcia-Sastre, D. E. Swayne, and M. G. Katze. 2006. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature 443578-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kerr, M. K., and G. A. Churchill. 2001. Statistical design and the analysis of gene expression microarray data. Genet. Res. 77123-128. [DOI] [PubMed] [Google Scholar]
- 20.Kipling, D. 2001. Telomeres, replicative senescence and human ageing. Maturitas 3825-38. [DOI] [PubMed] [Google Scholar]
- 21.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, and L. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 3481953-1966. [DOI] [PubMed] [Google Scholar]
- 22.Kuiken, T., R. A. Fouchier, M. Schutten, G. F. Rimmelzwaan, G. van Amerongen, D. van Riel, J. D. Laman, T. de Jong, G. van Doornum, W. Lim, A. E. Ling, P. K. Chan, J. S. Tam, M. C. Zambon, R. Gopal, C. Drosten, S. van der Werf, N. Escriou, J. C. Manuguerra, K. Stohr, J. S. Peiris, and A. D. Osterhaus. 2003. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet 362263-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masten, B. J., G. K. Olson, C. A. Tarleton, C. Rund, M. Schuyler, R. Mehran, T. Archibeque, and M. F. Lipscomb. 2006. Characterization of myeloid and plasmacytoid dendritic cells in human lung. J. Immunol. 1777784-7793. [DOI] [PubMed] [Google Scholar]
- 24.Meyer, K. C. 2005. Aging. Proc. Am. Thorac. Soc. 2433-439. [DOI] [PubMed] [Google Scholar]
- 25.Murasko, D. M., and J. Jiang. 2005. Response of aged mice to primary virus infections. Immunol. Rev. 205285-296. [DOI] [PubMed] [Google Scholar]
- 26.Nicholls, J. M., L. L. Poon, K. C. Lee, W. F. Ng, S. T. Lai, C. Y. Leung, C. M. Chu, P. K. Hui, K. L. Mak, W. Lim, K. W. Yan, K. H. Chan, N. C. Tsang, Y. Guan, K. Y. Yuen, and J. S. Peiris. 2003. Lung pathology of fatal severe acute respiratory syndrome. Lancet 3611773-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peiris, J. S., S. T. Lai, L. L. Poon, Y. Guan, L. Y. Yam, W. Lim, J. Nicholls, W. K. Yee, W. W. Yan, M. T. Cheung, V. C. Cheng, K. H. Chan, D. N. Tsang, R. W. Yung, T. K. Ng, and K. Y. Yuen. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 3611319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plackett, T. P., E. D. Boehmer, D. E. Faunce, and E. J. Kovacs. 2004. Aging and innate immune cells. J. Leukoc. Biol. 76291-299. [DOI] [PubMed] [Google Scholar]
- 29.Roberts, A., D. Deming, C. D. Paddock, A. Cheng, B. Yount, L. Vogel, B. D. Herman, T. Sheahan, M. Heise, G. L. Genrich, S. R. Zaki, R. Baric, and K. Subbarao. 2007. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog. 3e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roberts, A., C. Paddock, L. Vogel, E. Butler, S. Zaki, and K. Subbarao. 2005. Aged BALB/c mice as a model for increased severity of severe acute respiratory syndrome in elderly humans. J. Virol. 795833-5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosetta-Biosoftware. 2003. Re-ratio and ratio split processing: supplement user information: technical note. Rosetta-Biosoftware, Seattle, WA.
- 32.Shannon, W., R. Culverhouse, and J. Duncan. 2003. Analyzing microarray data using cluster analysis. Pharmacogenomics 441-52. [DOI] [PubMed] [Google Scholar]
- 33.Siegal, F. P., N. Kadowaki, M. Shodell, P. A. Fitzgerald-Bocarsly, K. Shah, S. Ho, S. Antonenko, and Y. J. Liu. 1999. The nature of the principal type 1 interferon-producing cells in human blood. Science 2841835-1837. [DOI] [PubMed] [Google Scholar]
- 34.Solana, R., G. Pawelec, and R. Tarazona. 2006. Aging and innate immunity. Immunity 24491-494. [DOI] [PubMed] [Google Scholar]
- 35.Subbarao, K., J. McAuliffe, L. Vogel, G. Fahle, S. Fischer, K. Tatti, M. Packard, W. J. Shieh, S. Zaki, and B. Murphy. 2004. Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. J. Virol. 783572-3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Surjit, M., B. Liu, V. T. Chow, and S. K. Lal. 2006. The nucleocapsid protein of severe acute respiratory syndrome-coronavirus inhibits the activity of cyclin-cyclin-dependent kinase complex and blocks S phase progression in mammalian cells. J. Biol. Chem. 28110669-10681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang, B. S., K. H. Chan, V. C. Cheng, P. C. Woo, S. K. Lau, C. C. Lam, T. L. Chan, A. K. Wu, I. F. Hung, S. Y. Leung, and K. Y. Yuen. 2005. Comparative host gene transcription by microarray analysis early after infection of the Huh7 cell line by severe acute respiratory syndrome coronavirus and human coronavirus 229E. J. Virol. 796180-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson, W. W., D. K. Shay, E. Weintraub, L. Brammer, N. Cox, L. J. Anderson, and K. Fukuda. 2003. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA 289179-186. [DOI] [PubMed] [Google Scholar]
- 39.Tsang, K. W., P. L. Ho, G. C. Ooi, W. K. Yee, T. Wang, M. Chan-Yeung, W. K. Lam, W. H. Seto, L. Y. Yam, T. M. Cheung, P. C. Wong, B. Lam, M. S. Ip, J. Chan, K. Y. Yuen, and K. N. Lai. 2003. A cluster of cases of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 3481977-1985. [DOI] [PubMed] [Google Scholar]
- 40.Vogel, L. N., A. Roberts, C. D. Paddock, G. L. Genrich, E. W. Lamirande, S. U. Kapadia, J. K. Rose, S. R. Zaki, and K. Subbarao. 2007. Utility of the aged BALB/c mouse model to demonstrate prevention and control strategies for severe acute respiratory syndrome coronavirus (SARS-CoV). Vaccine 252173-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wentworth, D. E., L. Gillim-Ross, N. Espina, and K. A. Bernard. 2004. Mice susceptible to SARS coronavirus. Emerg. Infect. Dis. 101293-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yancik, R. 1997. Cancer burden in the aged: an epidemiologic and demographic overview. Cancer 801273-1283. [PubMed] [Google Scholar]
- 43.Zhang, Y., Y. Wang, X. Gilmore, K. Xu, P. R. Wyde, and I. N. Mbawuike. 2002. An aged mouse model for RSV infection and diminished CD8+ CTL responses. Exp. Biol. Med. 227133-140. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.