Abstract
The severe acute respiratory syndrome coronavirus (SARS-CoV) ORF7b (also called 7b) protein is an integral membrane protein that is translated from a bicistronic open reading frame encoded within subgenomic RNA 7. When expressed independently or during virus infection, ORF7b accumulates in the Golgi compartment, colocalizing with both cis- and trans-Golgi markers. To identify the domains of this protein that are responsible for Golgi localization, we have generated a set of mutant proteins and analyzed their subcellular localizations by indirect immunofluorescence confocal microscopy. The N- and C-terminal sequences are dispensable, but the ORF7b transmembrane domain (TMD) is essential for Golgi compartment localization. When the TMD of human CD4 was replaced with the ORF7b TMD, the resulting chimeric protein localized to the Golgi complex. Scanning alanine mutagenesis identified two regions in the carboxy-terminal portion of the TMD that eliminated the Golgi complex localization of the chimeric CD4 proteins or ORF7b protein. Collectively, these data demonstrate that the Golgi complex retention signal of the ORF7b protein resides solely within the TMD.
Although the majority of enveloped viruses bud from the plasma membrane, several virus families utilize intracellular compartments for budding; examples include bunyaviruses, which bud into the Golgi region (20, 35, 63); hepadnaviruses, which bud into membranes derived from endoplasmic reticulum (ER)-Golgi intermediate compartments (ERGICs) (38, 56, 80); and flaviviruses, which bud into the ER (32, 40). The structural components of these viruses must have targeting motifs for protein retention at the respective site of budding (9, 12). Viruses have evolved numerous mechanisms for targeting of structural proteins to necessary intracellular compartments. Varicella-Zoster virus glycoprotein I is targeted to the trans-Golgi network (TGN) via two independent motifs within the protein's cytoplasmic tail (86). GN proteins from bunyaviruses localize to the Golgi apparatus in different ways. The GN protein of Uukuniemi virus utilizes a targeting motif contained entirely within the cytoplasmic tail (2), whereas GN proteins from Rift Valley fever and Punta Toro viruses contain targeting sequences mapping to residues spanning the transmembrane domain (TMD) and the adjacent region of the cytoplasmic tail (11, 36). In contrast, the Bunyamwera virus GN protein 18-residue TMD is necessary and sufficient for Golgi complex retention (66).
Members of the Coronaviridae are enveloped viruses with large, positive-stranded RNA genomes that range from 27 to 31 kb in size (34). There is some discrepancy over the precise location of coronavirus budding, but it is generally accepted that budding occurs at membranes within the early secretory pathway, and significant evidence implicates the ERGIC (1, 7, 17, 19, 23, 52, 69, 76, 77). Coronavirus structural proteins are often found in high concentrations at the Golgi apparatus. M proteins from mouse hepatitis virus (MHV), avian infectious bronchitis virus (IBV), porcine transmissible gastroenteritis virus, SARS-CoV, and feline coronavirus all localize to the Golgi complex in cDNA-transfected cells (17, 24, 30, 31, 44, 58, 70), with Golgi complex targeting sequences identified in various locations. The MHV M first and second TMDs and cytoplasmic tail are necessary for Golgi complex retention (25), whereas the first TMD within the IBV M protein is sufficient for cis-Golgi localization (70). The IBV E protein localizes to the Golgi region utilizing a targeting motif contained entirely within the cytoplasmic tail (6); however, the MHV E protein localizes to the ER and ERGIC when expressed independently of M, suggesting that it does not contain an independent Golgi retention sequence (53). Interestingly, the SARS-CoV E protein has been shown to accumulate in either the ER or Golgi region, depending on the cell type analyzed (22, 44). All coronavirus spike (S) proteins identified to date localize primarily to the cell surface, with intracellular accumulations being detectable throughout the secretory pathway. It has been proposed for several coronaviruses that S contains a dibasic motif within the cytoplasmic tail, allowing the recycling of intracellular S between the ER and Golgi complex, which would allow packaging into virions (26, 37).
Coronavirus virion envelopes typically contain three integral membrane proteins, envelope (E), membrane (M), and spike (S); however, the incorporation of accessory genes into the viral envelope has been reported for various coronaviruses. The SARS-CoV genome encodes eight distinct accessory genes (ORF3a, -3b, -6, -7a, -7b, -8a, -8b, and -9b), the most of any coronavirus identified to date (33, 57, 75), with the exception of the recently identified whale coronavirus, which also has eight predicted accessory genes (39). Several of the SARS-CoV accessory proteins, including ORF3a, ORF6, ORF7a, and ORF7b, have been identified as being virion-associated proteins (13-15, 61, 65). The function of these proteins within virus particles remains unclear.
The intracellular localization of several SARS-CoV accessory proteins that are packaged into virus particles has been studied. The ORF3a protein localizes to the Golgi complex and the cell surface (49, 72); the ORF6 protein localizes to numerous intracellular membrane structures, predominantly the ER and Golgi complex (10, 51); and both ORF7a and ORF7b strongly localize to the Golgi region (18, 45, 61, 62). It has been demonstrated that the ORF7a TMD and short cytoplasmic tail are sufficient to confer some degree of Golgi complex localization to a chimeric protein otherwise localized to the cell surface (45). Identifying intracellular targeting motifs of these proteins may help elucidate the mechanisms involved in their packaging into virus particles. In this study, we have identified a Golgi complex localization signal within the putative 22-amino-acid TMD of ORF7b. The ORF7b TMD is necessary for its Golgi complex localization, as replacing it with the TMD from the human endoprotease furin results in aberrant localization. It is also sufficient to redirect the cell surface-targeted glycoprotein CD4 to the Golgi apparatus. The ORF7b amino- and carboxy-terminal sequences do not contribute to subcellular localization, and scanning alanine mutagenesis has implicated TMD residues 13 to 15 and 19 to 22 as being critical for Golgi complex retention.
MATERIALS AND METHODS
Cells.
Vero (ATCC CRL-1586) and 293T (71) cells were cultured in Dulbecco's modified Eagle medium containing 10% fetal bovine serum (Atlanta Biologicals), 1 mM glutamine, 1 mM sodium pyruvate (Invitrogen), 100 U/ml penicillin (Invitrogen), and 100 μg/ml streptomycin (Invitrogen). Cells were incubated in a 5% CO2 humidified incubator at 37°C.
Plasmids.
Plasmid pCAGGS (48) encoding either cDNA for the native full-length SARS-CoV ORF7b protein or ORF7b with a carboxy-terminal c-Myc epitope tag was described previously (61). Amino- or carboxy-terminal deletions or alanine substitutions detailed in Fig. 1 were introduced into ORF7b by overlap PCR amplification using primers containing the desired mutations. An ER-restricted ORF7b mutant was generated by the addition of nucleotide sequence-encoding residues KKAA, a canonical ER retrieval motif (16, 67), to the C terminus of ORF7b-Myc. ORF7b TMD alanine mutants at residues 21 to 23 and 27 to 30 were similarly generated by overlap PCR. The resulting cDNAs were digested with EcoRI and XhoI, ligated into similarly digested vector pCAGGS, and sequenced.
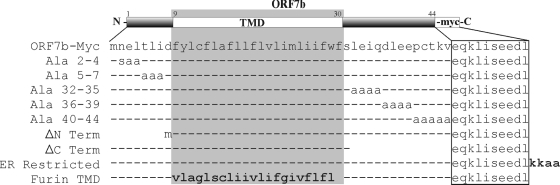
FIG. 1.
Schematic diagram of ORF7b-Myc proteins. ORF7b-Myc constructs included N-terminal mutants (mutants at residues 2 to 4 and 5 to 7 and ΔN), C-terminal mutants (mutants at residues 32 to 35, 36 to 39, and 40 to 44 and ΔC), and an ER-restricted mutant containing a canonical ER retrieval motif, KKAA, at the C terminus. The native ORF7b TMD was also replaced with the TMD from human furin. Numbers represent amino acid positions within the ORF7b protein, and dashes represent amino acids that have not been altered.
ORF7b-Myc containing the human furin TMD was generated by overlap PCR. The ORF7b nucleotide sequence encoding the transmembrane domain was replaced with sequence encoding the entire 21-residue TMD of furin (GenBank accession number BC012181.1). The cDNA was digested and ligated into pCAGGS as described above and sequenced.
The human CD4 cDNA (courtesy of Ken Murphy, Washington University) was cloned into expression vector pCAGGS by utilizing EcoRI and XhoI restriction sites. The CD4 TMD was replaced with the ORF7b TMD by overlap PCR. Scanning alanine mutagenesis was performed on the CD4 ORF7b TMD or ORF7b as detailed in Fig. 4. All TMD mutations were introduced by overlap PCR. CD4 mutants were digested with EcoRI and XhoI, ligated into pCAGGS, and sequenced. All primer sequences are available upon request.
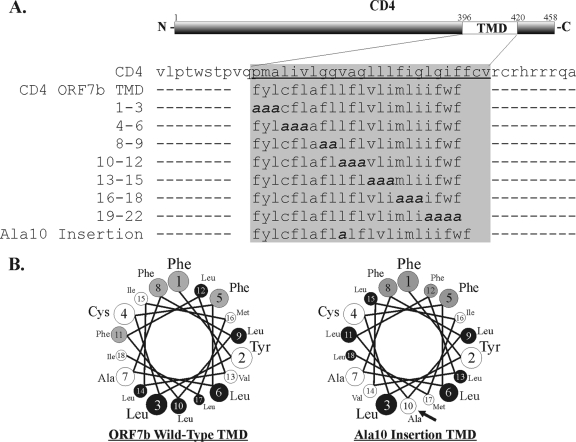
FIG. 4.
Diagram and amino acid sequence of CD4 ORF7b TMD mutants. (A) The TMD of human CD4 was replaced with the TMD of SARS-CoV ORF7b. Additional scanning alanine mutagenesis was performed within the ORF7b TMD, generating mutants at residues 1 to 3, 4 to 6, 8 to 9, 10 to 12, 13 to 15, 16 to 18, and 19 to 22, with numbers representing amino acid locations within the ORF7b TMD. To disrupt the predicted α-helical structure of the TMD, a single alanine was inserted at position 10 within the TMD (Ala10 insertion). (B) Helical wheel depiction of the predicted α-helices of residues 1 to 18 from the wild-type ORF7b TMD (left) and the Ala10 insertion TMD (right). Phenylalanine resides are highlighted in gray, and leucine residues are highlighted in black.
The S15-green fluorescent protein (GFP) cDNA contains the GFP open reading frame fused to the first 15 amino acids of Src protein tyrosine kinase and was described previously (55).
Transient transfection.
Transfection experiments were performed using the LT-1 transfection reagent (Mirus) according to the manufacturer's protocol. For Western blotting or fluorescence-activated cell sorter (FACS) analysis, 1.5 × 105 293T cells were plated into each well of a six-well plate (Corning) and allowed to attach for 24 h at 37°C. The cells were transfected with 1 μg of DNA mixed with 2 μl of LT-1 transfection reagent and incubated for 18 h at 37°C prior to lysis or fixation. For confocal microscopy, 7.5 × 104 Vero cells were plated into each well of a 12-well plate (Corning) containing glass coverslips and allowed to attach for 24 h at 37°C. The cells were transfected with 0.5 μg of DNA mixed with 2 μl LT-1 and incubated for 18 h at 37°C.
Flow cytometry.
For flow cytometry, transfected 293T cells were detached from tissue culture dishes using 100 mM EDTA and washed three times with phosphate-buffered saline (PBS). For permeabilization, the cells were immediately fixed with 2% paraformaldehyde in PBS for 10 min at room temperature, washed three times with PBS, resuspended in PBS plus 0.1% Triton X-100 for 5 min, and washed two times with PBS. For the detection of cell surface protein expression, the primary antibody incubation was performed on ice. Cells were blocked with block buffer (PBS plus 3% NGS [Sigma] plus 0.5% bovine serum albumin) for 10 min. The cells were then incubated with anti-CD4 mouse monoclonal antibody (MAb) (1-μg/ml final concentration) (mAb379; R&D Systems) diluted in blocking buffer. Cells were washed three times with PBS and fixed with 2% methanol-free formaldehyde in PBS for 10 min. Cells were then incubated with goat anti-mouse immunoglobulin G (IgG) antibody conjugated to Alexa Fluor 488 (4-μg/ml final concentration in blocking buffer) (Molecular Probes) for 30 min and washed three times with PBS. The cells were analyzed by flow cytometry (FACSCalibur dual-laser flow cytometer; Becton Dickinson), and data were collected using CellQuest software.
Indirect immunofluorescence and confocal microscopy.
For indirect immunofluorescence analysis, cells were washed three times with PBS and either (i) fixed with 2% paraformaldehyde in PBS for 10 min for intracellular protein localization or (ii) placed on ice for 10 min to assess plasma membrane localization. For intracellular immunostaining, 0.1% saponin was added to all subsequent antibody dilutions and wash buffers.
Cells were blocked with block buffer (PBS plus 3% NGS [Sigma] plus 0.5% bovine serum albumin) for 10 min. The cells were then incubated with antibody diluted in blocking buffer for 1 h, followed by three washes in PBS and fixation with 2% paraformaldehyde in PBS for 10 min. Primary antibodies used were rabbit anti-ORF7b polyclonal sera (1:1,000 dilution) (61), anti-GM130 mouse MAb (2.5-μg/ml final concentration) (BD Biosciences), rabbit anti-calnexin IgG (1:500 dilution) (Sigma), rabbit anti-ERGIC53 IgG (5-μg/ml final concentration) (Sigma), anti-LAMP1 mouse MAb H4A3 (5-μg/ml final concentration) (Developmental Studies Hybridoma Bank, NICHD), anti-c-Myc mouse MAb (1:100 dilution) (supernatant from hybridoma 9E10; ATCC CRL-1729), anti-c-Myc rabbit antibody (0.5-μg/ml final concentration) (Sigma), and anti-CD4 mouse MAb (2-μg/ml final concentration) (mAb379; R&D Systems). Wheat germ agglutinin conjugated to Alexa Fluor 555 (2-μg/ml final concentration) (Molecular Probes) was added to primary antibody dilutions in nonpermeabilized conditions to highlight the plasma membrane. Anti-CD4 antibody was directly conjugated to Alexa Fluor 488 (Molecular Probes). Cells were washed three times with PBS, incubated with secondary antibody (Alexa Fluor 488-conjugated goat anti-mouse IgG, Alexa Fluor 555-conjugated goat anti-mouse IgG, Alexa Fluor 555-conjugated goat anti-rabbit IgG, or Alexa Fluor 647-conjugated goat anti-rabbit IgG [4-μg/ml final concentration; Molecular Probes]) as appropriate. Nuclei were counterstained concurrently with TO-PRO-3 iodide (Molecular Probes). Coverslips were mounted onto microscope slides using Prolong Antifade Gold (Molecular Probes) and visualized on a Zeiss LSM 510 confocal microscope. Images shown are representative of the majority of cells expressing a particular mutated protein. All colocalization experiments were done at least twice, and dozens of cells were analyzed before representative images were chosen.
SDS-PAGE and Western blotting.
Cells were lysed in 1% sodium dodecyl sulfate (SDS) in water, and nucleic acid was sheared with 10 strokes through a 26-gauge needle and syringe, sonicated for 5 min, mixed at a 1:1 ratio with Laemmli 2× SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer with 200 mM dithiothreitol, and boiled for 5 min (50). Samples were loaded onto either 10% or 15% polyacrylamide gels (Mini Trans-Blot; Bio-Rad).
For Western blotting, separated polypeptides were transferred onto polyvinylidene difluoride membranes (Millipore) and blocked in PBS containing 0.3% Tween 20 and 5% nonfat dry milk (block buffer). Membranes were incubated with the following antibodies diluted in block buffer: anti-β-actin mouse MAb (0.25-μg/ml final concentration) (Abcam), anti-c-Myc mouse MAb (1:100 dilution, supernatant from hybridoma 9E10; ATCC CRL-1729), anti-CD4 mouse MAb (2-μg/ml final concentration) (mAb379; R&D Systems), and rabbit anti-ORF7b antiserum (1:1,000 dilution) (61). Primary antibodies were detected using species-specific IgG secondary antibodies coupled to horseradish peroxidase (Jackson Laboratories). The blots were soaked in chemiluminescent reagent (ECL Plus Pico; Amersham Biosciences) and imaged using either chemiluminescence and exposure to X-ray film (Molecular Technologies) or chemifluorescence followed by phosphorimager analysis (FujiFilm FLA-5000) to quantify the signal intensity.
Peptide N-glycosidase F and Endo H digestions.
At 20 h posttransfection, 293T cells were lysed in extraction buffer (50 mM Tris [pH 7.4], 0.5% SDS), sheared with 10 strokes through a 26-gauge needle and syringe, sonicated, and boiled for 10 min. For peptide-N-glycosidase F (PNGase F) digestion, 80 μl of lysate was mixed with a solution containing 10 μl of 10× G7 reaction buffer, 10 μl of 10% NP-40, and 750 units of PNGase F (New England BioLabs) and incubated for 16 h at 37°C. For endoglycosidase H (Endo H) digestion, 80 μl of lysate was mixed with a solution containing 10 μl of 10× G5 reaction buffer, 10 μl of PBS, and 750 units of Endo H (New England BioLabs) and incubated 16 h at 37°C. The reaction mixtures were analyzed on 10% SDS-PAGE gels followed by Western blotting.
Viruses.
Recombinant SARS-CoV containing GFP in place of the ORF7 coding region was kindly provided by Ralph Baric, University of North Carolina. Briefly, nucleotides 27276 to 27643 (GenBank accession number A278741) were replaced with the open reading frame for GFP, and recombinant viruses were generated as previously described (84). Virus stocks were generated, and cells were infected as described previously (61, 62).
RESULTS
Intracellular localization of ORF7b N- and C-terminal scanning alanine mutants.
The ORF7b protein colocalizes with markers of the Golgi apparatus when expressed either during viral infection or from cDNA (61, 62). Because subcellular localization is not altered in the presence of other viral proteins, it is likely that the ORF7b protein contains a Golgi complex targeting sequence. As illustrated in Fig. 1, ORF7b is predicted to have an eight-residue amino (N) terminus, a 22-residue TMD, and a 14-residue carboxy (C)-terminal tail. The protein does not contain a signal sequence, and the C terminus is exposed to the cytosol, suggesting that the ORF7b protein is a type III integral membrane protein (61). The ORF7b protein has no significant homology with any other identified viral or cellular protein.
To determine if residues within the N- or C-terminal regions of ORF7b are required for Golgi complex targeting, a series of scanning alanine mutants was constructed (Fig. 1). Three to five consecutive residues were mutated to Ala in ORF7b containing a C-terminal Myc epitope tag. To further identify if the termini contribute to protein localization, tail truncation mutants were generated as depicted in Fig. 1. Seven of the eight N-terminal residues (ΔN) or 13 of the 14 C-terminal residues (ΔC) were removed. Additionally, a canonical dilysine ER retrieval motif (KKAA) was added to the C terminus of ORF7b-Myc (16, 67) to determine if the native Golgi complex targeting of ORF7b can be overcome by a known localization sequence. The dilysine motif, found near the terminus of some integral membrane protein cytoplasmic tails, interacts with the COPI cytosolic coat protein complex and facilitates the retrieval of the protein from the Golgi complex to the ER (reviewed in reference 73).
The intracellular localization of the scanning alanine, tail deletion, and ER retrieval ORF7b mutants was analyzed by confocal immunofluorescence microscopy (Fig. 2). The results revealed that the N terminus is not required for Golgi complex localization. Both scanning alanine mutants at residues 2 to 4 and 5 to 7 resembled wild-type ORF7b-Myc and retained a high level of colocalization with the cis-Golgi marker GM130 (Fig. 2Ac, f, and i). In fact, the deletion of the entire N terminus had no significant impact on subcellular localization, as ΔN maintained Golgi complex localization (Fig. 2Aj to l). Together, these data suggest that the predicted luminal domain of ORF7b does not contribute to Golgi complex retention.
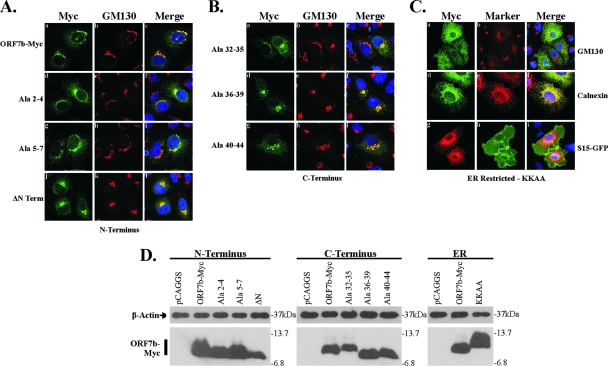
FIG. 2.
Expression and subcellular localization of ORF7b N- and C-terminal tail mutants. Vero cells grown on coverslips were transfected with plasmids containing the designated ORF7b mutant cDNAs. At 18 h posttransfection, cells were analyzed by immunofluorescence confocal microscopy. Nuclei were counterstained with TO-PRO-3 (blue). (A) ORF7b-Myc wild-type and N-terminal tail mutant subcellular localization. Cells were immunostained with antibody against the Myc epitope tag and antibody against the cis-Golgi marker GM130. All three N-terminal mutated proteins resembled wild-type ORF7b-Myc and colocalized with GM130. Deletion of the entire lumenal domain of ORF7b (ΔN) resulted in no changes in intracellular localization. (B) ORF7b-Myc C-terminal tail mutant subcellular localization. Cells were immunostained as described above for ORF7b-Myc and GM130. All three mutated proteins colocalized with GM130. The C-terminal tail deletion construct (ΔC) was not detectable (data not shown). (C) The addition of KKAA, an ER retrieval motif, results in the mislocalization of ORF7b-Myc to the ER. Cells expressing ORF7b-Myc KKAA were immunostained for ORF7b-Myc and the Golgi protein GM130 or the ER-associated protein calnexin. In addition, cells were cotransfected with plasmids expressing a plasma membrane-targeted GFP construct (S15-GFP) and ORF7b-Myc KKAA. All images were obtained with a 63× oil immersion objective lens and represent a z-stack projection of 0.5-μm slices obtained by confocal microscopy. (D) 293T cells were transfected with plasmids encoding the indicated cDNAs, lysed 18 h posttransfection, and analyzed for ORF7b-Myc mutant and β-actin expression by Western blotting. All data are representative of three independent experiments.
Similar results were obtained with the C-terminal scanning alanine mutants. All C-terminal alanine mutants retained strong colocalization with GM130 (Fig. 2Bc, f, and i), suggesting that the cytoplasmic tail contained no Golgi complex targeting sequences. Deletion of the entire C terminus resulted in a loss of protein expression, suggesting that the stability of the tail deletion mutant was likely altered, allowing either rapid degradation or processing such that the Myc epitope is no longer recognized by the anti-Myc antibody (data not shown).
The addition of the dilysine ER retrieval motif was sufficient to redirect ORF7b-Myc to the ER, as shown by colocalization with calnexin (Fig. 2Cf). Little Golgi complex colocalization remained (Fig. 2Ca to c), and no protein was trafficked to the cell surface (Fig. 2Cg to i). These data indicates that the signals mediating ORF7b Golgi complex localization can be overcome by an ER retrieval signal, resulting in cycling between the Golgi complex and the ER.
All scanning alanine constructs, the ORF7b-Myc ΔN, and the KKAA mutant were expressed at similar levels in 293T cells transfected with each cDNA, as determined by Western blotting (Fig. 2D). Slight migration differences are likely due to changes in amino acid composition. These results suggest that the N and C termini of ORF7b are not required for Golgi complex targeting and that the protein can be redirected to a different compartment with the addition of an appropriate motif.
The intracellular localization of ORF7b containing a chimeric TMD.
To delineate the contribution of the TMD to Golgi complex localization, an ORF7b chimera was generated by replacing the ORF7b TMD with the 21-residue TMD from the human endoprotease furin (Fig. 1). An acidic amino acid sequence and a tyrosine-containing sequence within the furin cytoplasmic tail are responsible for targeting furin to the trans-Golgi network; the deletion of the furin cytoplasmic tail results in protein localizing to lysosome-like vesicles and to the plasma membrane (3, 79). Thus, the furin TMD should not contribute to ORF7b Golgi complex localization.
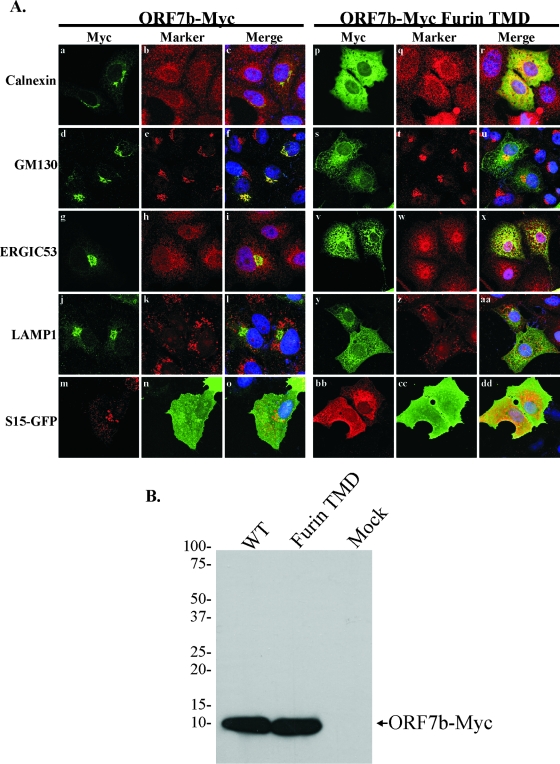
Vero cells were transfected with plasmids expressing either ORF7b-Myc or the furin TMD mutant and analyzed by confocal microscopy (Fig. 3A). ORF7b-Myc colocalized strongly only with GM130 (Fig. 3Af) while demonstrating little or no colocalization with calnexin (Fig. 3Ac), the ERGIC marker ERGIC53 (Fig. 3Ai), the lysosomal marker LAMP1 (Fig. 3Al), or the plasma membrane highlighted by S15-GFP (Fig. 3Ao). In contrast, the ORF7b-Myc furin TMD protein showed partial colocalization with both calnexin and ERGIC53 (Fig. 3Ar and x, respectively) and a significant decrease in Golgi complex localization. The mutant protein was not present at significant levels in lysosomes (Fig. 3Aaa) or at the plasma membrane (Fig. 3Add). The ORF7b-Myc and ORF7b-Myc furin TMD mutants were expressed at equivalent levels as judged by Western blotting of 293T cell lysates (Fig. 3B). These data indicate that the ORF7b TMD is essential for the localization of the protein to the Golgi region.
FIG. 3.
The ORF7b TMD is required for Golgi complex localization. (A) Vero cells grown on coverslips were transfected with plasmids containing ORF7b-Myc wild-type (WT) or furin TMD cDNAs; designated wells were cotransfected with a plasmid expressing a plasma membrane-targeted GFP construct (S15-GFP). At 18 h posttransfection, cells were immunostained with antibody against the Myc epitope tag and antibody against calnexin, GM130, the ER-to-Golgi complex intermediate compartment (ERGIC53), or the lysosomal marker LAMP1. (B) 293T cells were transfected with plasmids encoding the indicated cDNAs, lysed 18 h posttransfection, and analyzed for ORF7b-Myc wild-type or furin TMD expression by Western blotting. Images are representative of three independent experiments.
The ORF7b TMD is sufficient to retain CD4 at the Golgi complex.
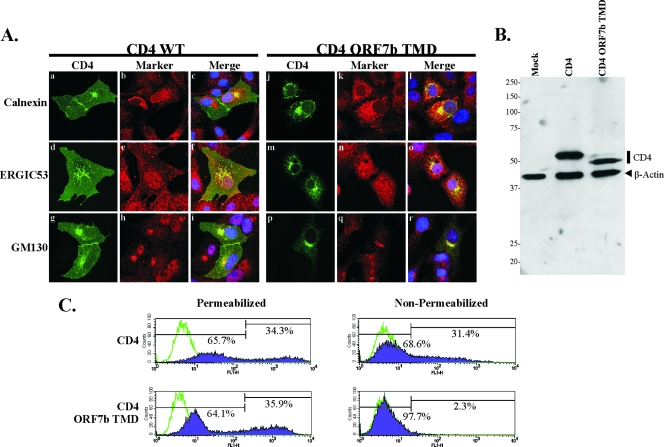
To determine if the ORF7b TMD is sufficient to alter the localization of an integral membrane protein normally expressed on the plasma membrane, a chimeric human CD4 construct was generated (Fig. 4A). CD4 is a single-TMD glycoprotein that is efficiently transported to the plasma membrane. The membrane-spanning domain of human CD4 was replaced with that from ORF7b (CD4 ORF7b TMD), and cDNA expressing the chimeric protein was transiently transfected into Vero cells. At 18 h posttransfection, cells were fixed, costained with a mouse MAb recognizing the CD4 extracellular domain and for various intracellular markers, and analyzed by indirect immunofluorescence confocal microscopy (Fig. 5A). As expected, wild-type CD4 was clearly present at the cell surface, highlighting the plasma membrane of expressing cells (Fig. 5Aa, d, and g). Some CD4 colocalized with calnexin, ERGIC53, and GM130, presumably due to its transient presence in those compartments during transport to the plasma membrane. The replacement of the CD4 TMD with that from ORF7b altered the subcellular localization of the protein such that little or no surface staining was visible by microscopy. The mutant protein was retained in a juxtanuclear location, colocalizing predominantly with GM130 (Fig. 5Al, o, and r), suggesting that the ORF7b TMD is sufficient to not only prevent the trafficking of CD4 to the plasma membrane but also retain it at the Golgi apparatus.
FIG. 5.
The ORF7b TMD is sufficient for retaining CD4 at the Golgi complex. (A) Vero cells grown on coverslips were transfected with a plasmid encoding either CD4 or CD4 ORF7b TMD. At 18 h posttransfection, cells were fixed and immunostained with antibodies against CD4 (green) or markers for intracellular compartments (red), including the ER (calnexin), ER-to-Golgi intermediate compartment (ERGIC53), or cis-Golgi compartment (GM130). Wild-type CD4 was clearly visible at the plasma membrane, whereas CD4 ORF7b TMD colocalized predominantly with GM130. All images were obtained with a 63× oil immersion objective lens and represent a z-stack projection of 0.5-μm slices obtained by laser scanning confocal microscopy. (B) 293T cells were transfected with empty vector pCAGGS (Mock) or plasmids encoding either CD4 or CD4 ORF7b TMD. At 18 h posttransfection, levels of CD4 and β-actin protein expression were determined by SDS-PAGE and Western blotting. (C) Surface expression of CD4 or CD4 ORF7b TMD was analyzed by flow cytometry. 293T cells were transfected with plasmids expressing either CD4 or CD4 ORF7b TMD and stained with anti-CD4 antibody at 18 h posttransfection under either permeabilized or nonpermeabilized conditions. The ORF7b TMD sufficiently retained most of the CD4 protein in an intracellular compartment, as little was observed on the cell surface. Transfected cells stained with secondary antibody alone are represented by green lines; CD4-positive cells are shown in purple. The percentage of total cells within each gate is shown. Data are representative of five independent experiments. WT, wild type.
The surface expression patterns of wild-type and chimeric CD4 were confirmed by flow cytometry (Fig. 5C). 293T cells were transfected with plasmids expressing CD4 or CD4 ORF7b TMD. Eighteen hours posttransfection, cells were stained under permeabilizing or nonpermeabilizing conditions using an anti-CD4 antibody, and protein expression was quantified by FACS analysis. As expected, CD4 was detected in similar percentages of permeabilized and nonpermeabilized cells (34.3% and 31.4%, respectively; mean fluorescences, 747.4 and 69.1), suggesting that nearly all CD4-expressing cells have surface-expressed protein. In contrast, CD4 ORF7b TMD-transfected cells expressed intracellular protein, but little or no CD4 was detectable on the surface of nonpermeabilized cells (35.9% and 2.3%, respectively; mean fluorescences, 399.0 and 6.37), confirming the results obtained by microscopy. The expression levels of both proteins were analyzed by Western blotting; no significant differences were observed (Fig. 5B), consistent with the flow cytometry data. The CD4 ORF7b TMD construct appeared to have a faster-migrating band than wild-type CD4, and a small doublet band was observed with slightly slower migration. Given the subcellular localization data, this is presumably due to differences in N-linked glycosylation acquired during the secretory process. These observations demonstrate that the TMD of ORF7b is sufficient to retain a plasma membrane protein in the Golgi region.
Identification of Golgi complex targeting sequences in the CD4 ORF7b TMD protein.
Analysis of the Golgi retention properties of various cellular Golgi-resident enzyme TMDs has not revealed any consensus sequence homology or conserved motifs (41-43, 70) that are responsible for the intracellular retention. It was further suggested that the physical properties, including the structure and length of the membrane-spanning domain, may contribute to Golgi complex retention activity (reviewed in reference 4). Statistical analysis suggested that Golgi complex-resident enzymes contain short TMDs, averaging 15 residues in length, compared to the approximately 20-residue average for TMDs from plasma membrane proteins (4). This is clearly not a steadfast rule, as both the ORF7b and IBV M TMDs are predicted to be 22 residues in length, and both are sufficient to confer Golgi complex localization to a plasma membrane protein (Fig. 5A) (70). It was also previously reported that TMDs of mammalian resident Golgi complex enzymes may contain more of the bulky residue phenylalanine than other membrane-spanning domains of non-Golgi complex-localized proteins (4, 21). The ORF7b TMD has a total of six phenylalanine residues, significantly more than the 5.3% average reported for plasma membrane protein TMDs.
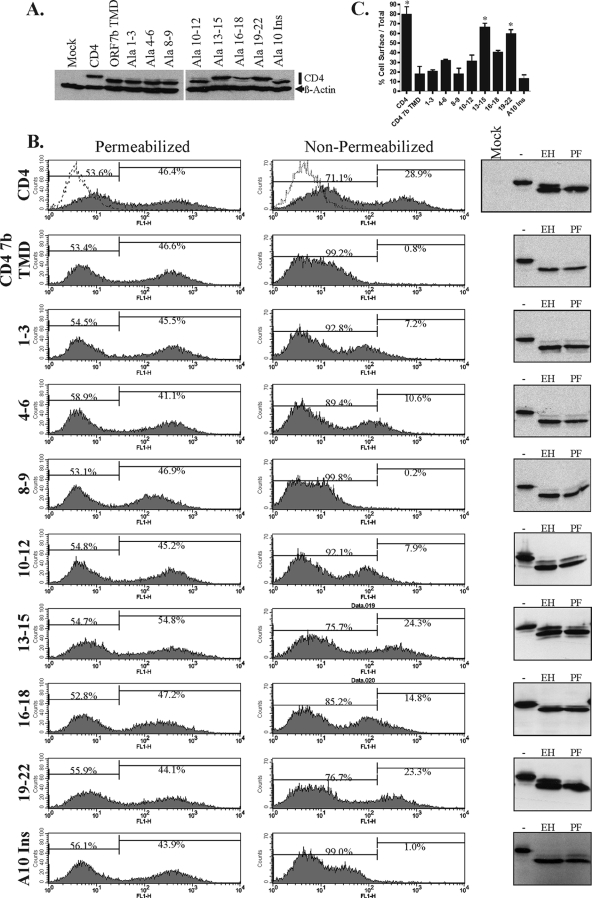
To define specific residues in the ORF7b TMD that are critical for Golgi complex retention, scanning alanine mutagenesis was performed on the membrane-spanning domain of CD4 ORF7b TMD. As detailed in Fig. 4A, alanine substitutions were generated throughout the entire TMD in sets of 2 to 4. Nomenclature denotes the residue number within the putative TMD. The seventh residue of the ORF7b TMD is an alanine and was left unchanged in the scanning alanine mutagenesis. When residues 1 to 18 of the ORF7b TMD are projected in a helical wheel pattern, it is apparent that one face of the TMD is rich in leucine residues (Fig. 4B, left). It was recently demonstrated that a leucine zipper-like structure within the transmembrane segment of the erythropoietin receptor mediates its homotypic interactions (59). To determine if this face or the overall helical structure of the TMD is required for Golgi complex retention, a single alanine residue was inserted at position 10 within the TMD (Ala10 insertion) This insertion disrupts the wild-type helix, redistributing the leucines across multiple faces of the TMD (Fig. 4B, right).
Substitution or insertion mutants were cloned into expression vector pCAGGS, transfected into 293T cells, and analyzed for cell surface expression. Mutations that disrupt the Golgi complex retention properties of the TMD should allow the CD4 chimera to traffic to the plasma membrane. Transfected cells were lysed, and expression was analyzed by Western blotting (Fig. 6A); all the proteins appeared to be expressed to similar levels. Two CD4 bands were visible for each of the constructs, presumably due to differences in glycosylation. Transfected 293T cells were then immunostained using permeabilized or nonpermeabilized conditions with an antibody recognizing an epitope within the CD4 extracellular domain and analyzed by flow cytometry (Fig. 6B). The percentage of cell surface-expressed protein was calculated as the mean fluorescent intensity of nonpermeabilized cells divided by the mean fluorescent intensity of permeabilized cells (Fig. 6C). As expected, wild-type CD4 was efficiently trafficked to the plasma membrane, as nonpermeabilized cells had more than 80% of the fluorescent intensity of the permeabilized cells. The CD4 ORF7b TMD was retained almost entirely intracellularly. Mutants at residues 8 to 9 and the Ala10 insertion resembled the CD4 ORF7b TMD, demonstrating no significant change in cell surface expression (P = 0.994 and 0.629, respectively). Mutants at residues 1 to 3, 4 to 6, 10 to 12, and 16 to 18 had only a moderate but statistically insignificant increase in cell surface expression (P = 0.756, 0.168, 0.279, and 0.058, respectively). In contrast, mutants at residues 13 to 15 and 19 to 22 had high levels of surface expression, suggesting that these two regions were critically important for Golgi complex retention (P = 0.007 and 0.012, respectively).
FIG. 6.
ORF7b TMD residues 13 to 15 and 19 to 22 are critical for intracellular retention. (A) 293T cells were transfected with plasmids encoding the indicated cDNAs, lysed 18 h posttransfection, and analyzed for CD4 wild-type or TMD mutant expression by Western blotting. (B) Surface expression of CD4 was analyzed by flow cytometry utilizing an anti-CD4 antibody targeting an epitope within the CD4 extracellular domain. 293T cells were transfected with plasmids encoding the indicated cDNAs and analyzed at 18 h posttransfection by flow cytometry under permeabilized or nonpermeabilized conditions. Total CD4 protein expression (permeabilized conditions) and cell surface-expressed CD4 (nonpermeabilized conditions) were quantified. Mock-transfected cells are represented by the dotted line. The assay was repeated three times; data are from one representative experiment, with the percentage of gated cells shown. (Right) 293T cells were transfected with the designated plasmid. Whole-cell lysates were mock treated or digested with either Endo H or PNGase F to analyze N-linked glycosylation of the CD4 mutants. Digestions were analyzed by SDS-PAGE and Western blotting. (C) Percentage of cell surface-localized CD4 was calculated as the ratio of the mean fluorescence intensity of stained, nonpermeabilized cells to the intensity of stained, permeabilized cells. Data are the averages of three independent experiments. Means and standard errors are shown. P values of for three experiments were calculated for each construct in comparison with the CD4 ORF7b TMD surface expression ratio. *, P < 0.05.
To confirm the transport of the CD4 ORF7b TMD mutants beyond the cis-Golgi complex, whole-cell lysates from transfected 293T cells were mock treated or treated with either Endo H or PNGase F. Endo H cleaves mannose-rich N-linked glycans that have not been processed by mannosidase enzymes in the medial Golgi complex, whereas PNGase F cleaves all N-linked sugar moieties. After overnight glycosidase digestion, lysates were analyzed by Western blotting with anti-CD4 antibody (Fig. 6B, right column). Wild-type CD4 had a significant Endo H-resistant population, representing cell surface and Golgi complex protein. The CD4 ORF7b TMD, however, was nearly 100% sensitive to Endo H digestion. While Ala mutants at residues 1 to 3, 4 to 6, 8 to 9, 10 to 12, and 16 to 18 and the Ala10 insertion were predominantly Endo H sensitive, mutants at residues 13 to 15 and 19 to 22 displayed a significant Endo H-resistant population, further confirming the trafficking of these proteins beyond the Golgi region.
Subcellular localization of TMD mutants.
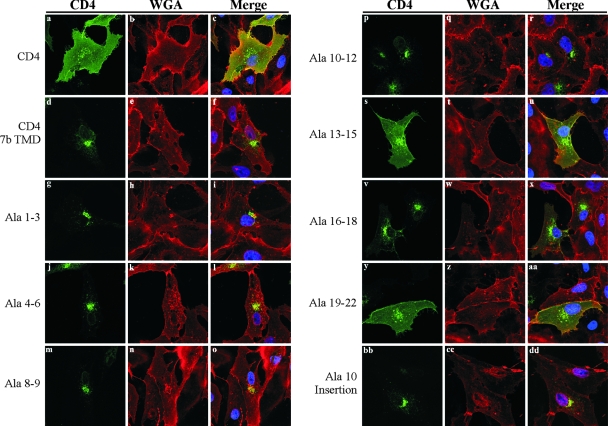
Vero cells were transfected with their respective plasmids, and cells were analyzed by indirect immunofluorescence confocal microscopy (Fig. 7). To highlight the cell surface, cells were first incubated under nonpermeabilized conditions on ice with wheat germ agglutinin conjugated to Alexa Fluor 555, followed by fixation, permeabilization, and immunostaining with anti-CD4 antibody. Mutants at residues 1 to 3, 4 to 6, 8 to 9, and 10 to 12 and the Ala10 insertion all resembled the CD4 ORF7b TMD, with predominant juxtanuclear staining and little plasma membrane staining evident (Fig. 7f, i, l, o, r, and dd). Mutants at residues 13 to 15 and 19 to 22 resembled wild-type CD4, with significant plasma membrane localization and intracellular staining reminiscent of secretory pathway organelles (Fig. 7c, u, and aa). The mutant at residues 16 to 18 had predominantly juxtanuclear staining, with light plasma membrane presence detectable (Fig. 7x). These results confirmed the FACS analysis data.
FIG. 7.
ORF7b TMD residues 13 to 15 and 19 to 22 are required for Golgi complex retention. Vero cells grown on coverslips were transfected with plasmids encoding the indicated proteins. At 18 h posttransfection, cells were incubated with wheat germ agglutinin (WGA) conjugated to Alexa Fluor 555 to highlight the cellular plasma membranes (red). Cells were subsequently fixed and immunostained with anti-CD4 antibody (green). Staining patterns of TMD mutants at residues 13 to 15 and 19 to 22 resembled that of wild-type CD4, with significant plasma membrane localization. The mutant at residues 16 to 18 had predominantly Golgi complex localization, with low levels of CD4 visible at the cell surface; all other mutants had no visible plasma membrane staining. All images were obtained with a 63× oil immersion objective lens and represent a z-stack projection of 0.5-μm slices obtained by laser scanning confocal microscopy.
These data demonstrate that the C-terminal region of the ORF7b TMD is critical for the Golgi complex retention phenotype. In particular, mutation of TMD residues 13 to 15 and 19 to 22 had a significant impact on intracellular retention; the mutation of the intervening residues 16 to 18 had a more modest but detectable effect on Golgi complex localization. The alpha-helical structure of the transmembrane region gave no significant contribution to the retention motif, as disrupting the helical conformation resulted in no increased trafficking to the plasma membrane.
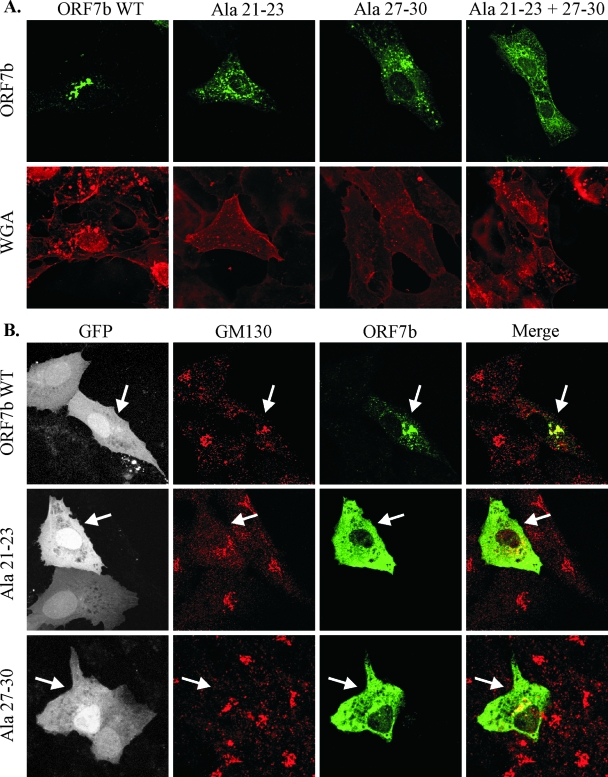
To confirm that residues 13 to 15 and 19 to 22 within the ORF7b TMD are important for Golgi complex retention, the mutations were introduced into wild-type ORF7b cDNA. A double mutant containing both mutations was also generated. TMD residues 13 to 15 and 19 to 22 are located at positions 21 to 23 and 27 to 30, respectively, within the native ORF7b protein. To avoid nomenclature confusion, the mutations within native ORF7b are referred to by location within the full-length protein. Transfected 293T cells were lysed, and expression was analyzed by Western blotting; all constructs appeared to express to similar levels (data not shown). Vero cells were transfected with constructs of wild-type ORF7b and mutants at residues 21 to 23, 27 to 30, or 21 to 23 and 27 to 30 and analyzed by indirect immunofluorescence confocal microscopy (Fig. 8A). Results demonstrate that these residues within the native TMD are in fact critical for the Golgi complex retention phenotype. The mutation of residues 21 to 23 or 27 to 30 or all simultaneously resulted in aberrant subcellular localization. The mutant proteins did not colocalize with LAMP1 or GM130 or localized to the plasma membrane; although the precise location of the mutant proteins is not clear, it is apparent that these residues are critical for Golgi complex retention.
FIG. 8.
Subcellular localization of ORF7b containing TMD alanine mutations at residues 21 to 23 and 27 to 30. Vero cells grown on coverslips were transfected with plasmids expressing the indicated cDNAs. (A) Cells were placed on ice and stained with wheat germ agglutinin (WGA) to highlight the plasma membrane (right column). Cells were then fixed, immunostained with polyclonal anti-ORF7b serum (left column), and analyzed by confocal microscopy. (B) Vero cells were infected with rSARS-CoV GFPΔORF7ab virus at a multiplicity of infection of approximately 5.0. Thirty minutes postinfection, cells were transfected with the indicated cDNAs. Eighteen hours posttransfection, cells were fixed, immunostained with anti-ORF7b serum and anti-GM130 MAb, and analyzed by confocal microscopy. Virus-infected cells are indicated by GFP expression. Arrows indicate cells that are expressing both GFP and the indicated transfected cDNA. All images were obtained with a 63× oil immersion objective lens and represent a z-stack projection of 0.5-μm slices obtained by laser scanning confocal microscopy. WT, wild type.
The subcellular localization of the ORF7b TMD alanine mutants was further analyzed in virus-infected cells. Vero cells were infected with rSARS-CoV GFPΔORF7ab, a recombinant virus strain encoding GFP in place of ORF7 (62, 68, 84, 85). Cells were subsequently transfected with cDNA expressing either ORF7b, ORF7b Ala residues 21 to 23, or ORF7b Ala residues 27 to 30. Cells were fixed 18 h posttransfection, and the percentage of infected and transfected cells was quantified by flow cytometry (data not shown). The percentage of cells expressing both viral antigen and transfected protein was very low (3.6% to 8.6% of total cell populations), preventing the quantitation of trans-expressed ORF7b protein packaged into viral particles. The intracellular localization of ORF7b and the two mislocalized ORF7b mutant proteins in virus-infected cells was not significantly different from that seen in cDNA-transfected cells, indicating that ORF7b localization is not altered by other viral proteins (Fig. 8B). This suggests that the Golgi complex localization of ORF7b is due to intrinsic factors contained entirely within the ORF7b TMD, and the loss of Golgi apparatus localization due to TMD mutagenesis is not reversible in the presence of other viral structural proteins.
DISCUSSION
Viruses of the Coronaviridae assemble and bud at membranes early in the secretory pathway, likely the ERGIC (1, 7, 17, 19, 23, 52, 69, 76, 77). Immediately after budding, coronavirus particles appear large and annular by electron microscopy. Virions undergo an intracellular postbudding maturation process during their transport through the Golgi complex (54, 60, 78). The mechanisms involved in this maturation process and reasons why the process occurs remain unclear; however, it is clear that the Golgi complex is required for structural maturation to occur (8). Additionally, many of the coronavirus structural proteins localize to the Golgi compartment in transfected and infected cells (reviewed in references 8 and 34).
We previously showed that the SARS-CoV ORF7b accessory protein is expressed in virus-infected cells utilizing a ribosomal leaky scanning mechanism, localizes to the Golgi region in the context of cDNA transfection or virus infection, and is packaged into virus particles (61). The expression of the ORF7b protein has been shown to induce apoptosis in cells, but the significance of this in the virus replication cycle remains unclear (18, 62). ORF7a and ORF7b are not required for virus replication or pathogenicity in vitro in all cell lines examined to date or in vivo in BALB/c mice or Syrian golden hamsters (62, 68, 85). Interestingly, recombinant SARS-CoV strains lacking ORF7a and ORF7b induce early stages of apoptosis in infected Vero cells equivalently to wild-type virus, but cells infected with ΔORF7ab viruses are significantly diminished in ability to undergo oligonucleosomal DNA fragmentation (62). The precise role of ORF7b in the virus life cycle has yet to be elucidated.
We have identified a Golgi complex retention signal within the single membrane-spanning domain of the SARS-CoV ORF7b protein. The amino- and carboxy-terminal sequences of the protein do not appear to contribute to Golgi complex localization. In contrast, replacement of the native TMD with that from human furin resulted in a complete loss of Golgi complex localization. Not only was the ORF7b TMD necessary for Golgi complex localization, but further analysis using the plasma membrane glycoprotein CD4 demonstrated that it was sufficient to retain a single membrane-spanning domain protein at the Golgi region. We have mapped the retention sequence to residues in the C-terminal portion of the 22-amino-acid domain. The mutation of residues 13 to 22 within the TMD resulted in diminished Golgi complex retention, with residues 13 to 15 and 19 to 22 being the most critical. Similar to the MHV E protein, the helical pitch of the TMD alpha-helix is not critical for mediating the Golgi complex localization of the protein despite the disruption of the residues lining one particular face of the helix (83). Interestingly, the IBV M protein also contains Golgi complex targeting information within the TMD; four critical residues that lined one face of the helix were identified. The disruption of that helix by residue insertion resulted in a severe reduction of the Golgi complex retention capacity (29, 70). The four IBV M residues identified were all uncharged, polar amino acids that lined one face of the helix. In comparison, the regions identified for the ORF7b TMD contain all nonpolar (residues 13 to 15 and 19) and aromatic (residues 20 to 22) amino acids.
Golgi complex localization of integral membrane proteins is presumed to occur via either retention or retrieval processes (reviewed in reference 42). Retention is mediated by a static anchoring of a protein in the appropriate compartment, whereas retrieval processes are dynamic and utilize signals in the cytoplasmic tail of the protein. Retrieval motifs have been identified as short, tyrosine-containing or acidic stretches of amino acids (42) and have been identified in a number of TGN proteins, including furin, TGN38, and mannose-6-phosphate receptor (reviewed in reference 41). In contrast, no canonical Golgi complex retention motif has been identified.
Retention at the Golgi complex is hypothesized to occur via one of two mechanisms: the lipid-sorting or “bilayer thickness” model or the oligomerization or “kin recognition” model. The membrane bilayer thickness at intracellular membranes may be altered due to cholesterol and sphingolipid composition, and this may play a significant role in the subcellular trafficking of integral membrane proteins (4, 5, 27, 43). This model suggests that a shorter TMD would be retained intracellularly, where the bilayers is not as thick as it is at the plasma membrane. It has also been proposed that Golgi complex-targeted proteins contain TMDs with higher-than-average concentrations of the residue phenylalanine. Large projecting side chains, such as those contributed by phenylalanine, could be energetically unfavorable in a cholesterol-rich domain because fitting them into the highly ordered bilayer would have increased energy cost (4, 27). The physical properties of the protein TMDs would subsequently serve to exclude it from a cholesterol-sphingolipid-rich vesicle as it is forming, retaining it in the Golgi compartment.
The “kin recognition” model theorizes that resident proteins form large homo- or hetero-oligomers within the Golgi membranes preventing their packaging into transport vesicles for trafficking through the secretory pathway (46, 47, 81). This has been proposed for numerous cellular and viral proteins, including the IBV M protein (81), the cellular p63 protein (64), and the cellular glycosyltransferases β-1,4-galactosyltransferase, N-acetylglucosaminyltransferase I, mannosidase II, and α-2,6-sialyltransferase (28, 46, 74, 82). It remains unclear if the oligomerization contributes to Golgi complex retention or if the formation of oligomers is a consequence of concentration of the proteins in specific membranes due to other retention factors.
The ORF7b protein has a fairly long transmembrane domain at 22 amino acids, significantly longer than the proposed average size of 15 residues for Golgi complex-resident proteins (4). This would suggest that TMD length is not the predominant mechanism holding ORF7b in the Golgi complex. The ORF7b TMD does, however, contain a high concentration of phenylalanine residues, consistent with the theory that protein TMDs containing amino acids with large side chains prefer the more energetically favorable bilayer of the Golgi complex. The mutation of some of the phenylalanine residues within the ORF7b TMD had no effect on Golgi complex retention (TMD residues 1, 5, 8, and 11), although all phenylalanine residues within the TMD were not mutated simultaneously. Two phenylalanine residues are located at the carboxy-most end of the TMD (residues 20 and 22), and the mutation of this region did have a significant impact on the Golgi complex retention capability of the sequence. However, phenylalanine concentration can be ruled out as the sole mechanism for Golgi complex retention, as replacing TMD residues 13 to 15 (VLI) of the CD4 ORF7b TMD chimera with alanines eliminated the Golgi complex retention phenotype.
It is possible that the ORF7b protein utilizes a “kin recognition” mechanism for retention within the Golgi membranes. ORF7b does not form disulfide-linked oligomers (61); however, the oligomerization state of the protein has not been investigated further. It is plausible that the identified residues within the C-terminal region of the TMD are important for homotypic interactions. Further analysis of this may be warranted to fully understand the mechanism of Golgi complex retention. It may also be possible that the identified residues are involved in direct interactions with other Golgi complex-resident protein TMDs, allowing ORF7b monomers to anchor into the Golgi complex region.
While ORF7b is packaged into virions, it is clear that the ORF7b protein is not required for the virus budding process, as gene 7 deletion viruses replicate efficiently in vitro and in vivo. It is not clear whether Golgi complex localization is a requirement for ORF7b packaging into virions. The coronavirus spike and hemagglutinin proteins are not required for virion budding but are copackaged into virus particles via interactions with M (reviewed in reference 8). To identify the mechanism of incorporation of ORF7b, the mutants generated in this study may further be utilized to analyze the requirements not only for Golgi complex localization in virus packaging but also for direct interactions with other structural proteins.
Fundamental mechanisms involved in protein sorting within the exocytic pathway remain an area of intense research. We have identified that the transmembrane region is necessary and sufficient for conferring ORF7b Golgi complex localization and have identified specific regions within the TMD that are critical for this phenotype. Thus, the ORF7b protein may be used as a model Golgi complex-resident protein in future experiments to provide additional insight into mechanisms of protein sorting.
Acknowledgments
We thank all members of the Diamond and Pekosz laboratories for insightful discussions and comments. We thank the Molecular Microbiology Imaging Facility for help with microscopy.
This study was supported by the Markey Pathway (S.R.S.) and National Institute of Health grants T32 HL07317 (S.R.S.) and AI059328 (A.P.). A.P. also acknowledges the support of The Eliasberg Foundation and the Marjorie Gilbert Foundation.
Footnotes
Published ahead of print on 16 July 2008.
REFERENCES
- 1.Afzelius, B. A. 1994. Ultrastructure of human nasal epithelium during an episode of coronavirus infection. Virchows Arch. 424295-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson, A. M., L. Melin, A. Bean, and R. F. Pettersson. 1997. A retention signal necessary and sufficient for Golgi localization maps to the cytoplasmic tail of a Bunyaviridae (Uukuniemi virus) membrane glycoprotein. J. Virol. 714717-4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bosshart, H., J. Humphrey, E. Deignan, J. Davidson, J. Drazba, L. Yuan, V. Oorschot, P. J. Peters, and J. S. Bonifacino. 1994. The cytoplasmic domain mediates localization of furin to the trans-Golgi network en route to the endosomal/lysosomal system. J. Cell Biol. 1261157-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bretscher, M. S., and S. Munro. 1993. Cholesterol and the Golgi apparatus. Science 2611280-1281. [DOI] [PubMed] [Google Scholar]
- 5.Colley, K. J. 1997. Golgi localization of glycosyltransferases: more questions than answers. Glycobiology 71-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corse, E., and C. E. Machamer. 2002. The cytoplasmic tail of infectious bronchitis virus E protein directs Golgi targeting. J. Virol. 761273-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dea, S., S. Garzon, H. Strykowski, and P. Tijssen. 1989. Ultrastructure and protein A-gold immunolabelling of HRT-18 cells infected with turkey enteric coronavirus. Vet. Microbiol. 2021-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Haan, C. A., and P. J. Rottier. 2005. Molecular interactions in the assembly of coronaviruses. Adv. Virus Res. 64165-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garoff, H., R. Hewson, and D. J. Opstelten. 1998. Virus maturation by budding. Microbiol. Mol. Biol. Rev. 621171-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geng, H., Y.-M. Liu, W.-S. Chan, A. W.-I. Lo, D. M.-Y. Au, M. M.-Y. Waye, and Y.-Y. Ho. 2005. The putative protein 6 of the severe acute respiratory syndrome-associated coronavirus: expression and functional characterization. FEBS Lett. 5796763-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerrard, S. R., and S. T. Nichol. 2002. Characterization of the Golgi retention motif of Rift Valley fever virus G(N) glycoprotein. J. Virol. 7612200-12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffiths, G., and P. Rottier. 1992. Cell biology of viruses that assemble along the biosynthetic pathway. Semin. Cell Biol. 3367-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang, C., N. Ito, C. T. Tseng, and S. Makino. 2006. Severe acute respiratory syndrome coronavirus 7a accessory protein is a viral structural protein. J. Virol. 807287-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang, C., C. J. Peters, and S. Makino. 2007. Severe acute respiratory syndrome coronavirus accessory protein 6 is a virion-associated protein and is released from 6 protein-expressing cells. J. Virol. 815423-5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito, N., E. C. Mossel, K. Narayanan, V. L. Popov, C. Huang, T. Inoue, C. J. Peters, and S. Makino. 2005. Severe acute respiratory syndrome coronavirus 3a protein is a viral structural protein. J. Virol. 793182-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson, M. R., T. Nilsson, and P. A. Peterson. 1990. Identification of a consensus motif for retention of transmembrane proteins in the endoplasmic reticulum. EMBO J. 93153-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klumperman, J., J. K. Locker, A. Meijer, M. C. Horzinek, H. J. Geuze, and P. J. Rottier. 1994. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J. Virol. 686523-6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kopecky-Bromberg, S. A., L. Martinez-Sobrido, and P. Palese. 2006. 7a protein of severe acute respiratory syndrome coronavirus inhibits cellular protein synthesis and activates p38 mitogen-activated protein kinase. J. Virol. 80785-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krijnse-Locker, J., M. Ericsson, P. J. Rottier, and G. Griffiths. 1994. Characterization of the budding compartment of mouse hepatitis virus: evidence that transport from the RER to the Golgi complex requires only one vesicular transport step. J. Cell Biol. 12455-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuismanen, E., K. Hedman, J. Saraste, and R. F. Pettersson. 1982. Uukuniemi virus maturation: accumulation of virus particles and viral antigens in the Golgi complex. Mol. Cell. Biol. 21444-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Landolt-Marticorena, C., K. A. Williams, C. M. Deber, and R. A. Reithmeier. 1993. Non-random distribution of amino acids in the transmembrane segments of human type I single span membrane proteins. J. Mol. Biol. 229602-608. [DOI] [PubMed] [Google Scholar]
- 22.Liao, Y., Q. Yuan, J. Torres, J. P. Tam, and D. X. Liu. 2006. Biochemical and functional characterization of the membrane association and membrane permeabilizing activity of the severe acute respiratory syndrome coronavirus envelope protein. Virology 349264-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim, K. P., and D. X. Liu. 2001. The missing link in coronavirus assembly. Retention of the avian coronavirus infectious bronchitis virus envelope protein in the pre-Golgi compartments and physical interaction between the envelope and membrane proteins. J. Biol. Chem. 27617515-17523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Locker, J. K., G. Griffiths, M. C. Horzinek, and P. J. Rottier. 1992. O-glycosylation of the coronavirus M protein. Differential localization of sialyltransferases in N- and O-linked glycosylation. J. Biol. Chem. 26714094-14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Locker, J. K., J. Klumperman, V. Oorschot, M. C. Horzinek, H. J. Geuze, and P. J. Rottier. 1994. The cytoplasmic tail of mouse hepatitis virus M protein is essential but not sufficient for its retention in the Golgi complex. J. Biol. Chem. 26928263-28269. [PubMed] [Google Scholar]
- 26.Lontok, E., E. Corse, and C. E. Machamer. 2004. Intracellular targeting signals contribute to localization of coronavirus spike proteins near the virus assembly site. J. Virol. 785913-5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lundbaek, J. A., O. S. Andersen, T. Werge, and C. Nielsen. 2003. Cholesterol-induced protein sorting: an analysis of energetic feasibility. Biophys. J. 842080-2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma, J., and K. J. Colley. 1996. A disulfide-bonded dimer of the Golgi beta-galactoside alpha2,6-sialyltransferase is catalytically inactive yet still retains the ability to bind galactose. J. Biol. Chem. 2717758-7766. [DOI] [PubMed] [Google Scholar]
- 29.Machamer, C. E., M. G. Grim, A. Esquela, S. W. Chung, M. Rolls, K. Ryan, and A. M. Swift. 1993. Retention of a cis Golgi protein requires polar residues on one face of a predicted alpha-helix in the transmembrane domain. Mol. Biol. Cell 4695-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Machamer, C. E., S. A. Mentone, J. K. Rose, and M. G. Farquhar. 1990. The E1 glycoprotein of an avian coronavirus is targeted to the cis Golgi complex. Proc. Natl. Acad. Sci. USA 876944-6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Machamer, C. E., and J. K. Rose. 1987. A specific transmembrane domain of a coronavirus E1 glycoprotein is required for its retention in the Golgi region. J. Cell Biol. 1051205-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mackenzie, J. M., and E. G. Westaway. 2001. Assembly and maturation of the flavivirus Kunjin virus appear to occur in the rough endoplasmic reticulum and along the secretory pathway, respectively. J. Virol. 7510787-10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marra, M. A., S. J. M. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. N. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 3001399-1404. [DOI] [PubMed] [Google Scholar]
- 34.Masters, P. S. 2006. The molecular biology of coronaviruses. Adv. Virus Res. 66193-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsuoka, Y., S. Y. Chen, and R. W. Compans. 1991. Bunyavirus protein transport and assembly. Curr. Top. Microbiol. Immunol. 169161-179. [DOI] [PubMed] [Google Scholar]
- 36.Matsuoka, Y., S. Y. Chen, and R. W. Compans. 1994. A signal for Golgi retention in the bunyavirus G1 glycoprotein. J. Biol. Chem. 26922565-22573. [PubMed] [Google Scholar]
- 37.McBride, C. E., J. Li, and C. E. Machamer. 2007. The cytoplasmic tail of the severe acute respiratory syndrome coronavirus spike protein contains a novel endoplasmic reticulum retrieval signal that binds COPI and promotes interaction with membrane protein. J. Virol. 812418-2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mhamdi, M., A. Funk, H. Hohenberg, H. Will, and H. Sirma. 2007. Assembly and budding of a hepatitis B virus is mediated by a novel type of intracellular vesicles. Hepatology 4695-106. [DOI] [PubMed] [Google Scholar]
- 39.Mihindukulasuriya, K. A., G. Wu, J. St. Leger, R. W. Nordhausen, and D. Wang. 2008. Identification of a novel coronavirus from a beluga whale using a panviral microarray. J. Virol. 825084-5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mukhopadhyay, S., R. J. Kuhn, and M. G. Rossmann. 2005. A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 313-22. [DOI] [PubMed] [Google Scholar]
- 41.Munro, S. 1995. An investigation of the role of transmembrane domains in Golgi protein retention. EMBO J. 144695-4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Munro, S. 1998. Localization of proteins to the Golgi apparatus. Trends Cell Biol. 811-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Munro, S. 1991. Sequences within and adjacent to the transmembrane segment of alpha-2,6-sialyltransferase specify Golgi retention. EMBO J. 103577-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nal, B., C. Chan, F. Kien, L. Siu, J. Tse, K. Chu, J. Kam, I. Staropoli, B. Crescenzo-Chaigne, N. Escriou, S. van der Werf, K.-Y. Yuen, and R. Altmeyer. 2005. Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S, M and E. J. Gen. Virol. 861423-1434. [DOI] [PubMed] [Google Scholar]
- 45.Nelson, C. A., A. Pekosz, C. A. Lee, M. S. Diamond, and D. H. Fremont. 2005. Structure and intracellular targeting of the SARS-coronavirus Orf7a accessory protein. Structure (Cambridge) 1375-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nilsson, T., M. H. Hoe, P. Slusarewicz, C. Rabouille, R. Watson, F. Hunte, G. Watzele, E. G. Berger, and G. Warren. 1994. Kin recognition between medial Golgi enzymes in HeLa cells. EMBO J. 13562-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nilsson, T., P. Slusarewicz, M. H. Hoe, and G. Warren. 1993. Kin recognition. A model for the retention of Golgi enzymes. FEBS Lett. 3301-4. [DOI] [PubMed] [Google Scholar]
- 48.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108193-199. [DOI] [PubMed] [Google Scholar]
- 49.Padhan, K., C. Tanwar, A. Hussain, P. Y. Hui, M. Y. Lee, C. Y. Cheung, J. S. Peiris, and S. Jameel. 2007. Severe acute respiratory syndrome coronavirus Orf3a protein interacts with caveolin. J. Gen. Virol. 883067-3077. [DOI] [PubMed] [Google Scholar]
- 50.Paterson, R. G., and R. A. Lamb. 1993. The molecular biology of influenza viruses and paramyxoviruses, p. 35-73. In A. J. Davison and R. M. Elliott (ed.), Molecular virology: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 51.Pewe, L., H. Zhou, J. Netland, C. Tangudu, H. Olivares, L. Shi, D. Look, T. Gallagher, and S. Perlman. 2005. A severe acute respiratory syndrome-associated coronavirus-specific protein enhances virulence of an attenuated murine coronavirus. J. Virol. 7911335-11342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qinfen, Z., C. Jinming, H. Xiaojun, Z. Huanying, H. Jicheng, F. Ling, L. Kunpeng, and Z. Jingqiang. 2004. The life cycle of SARS coronavirus in Vero E6 cells. J. Med. Virol. 73332-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raamsman, M. J., J. K. Locker, A. de Hooge, A. A. de Vries, G. Griffiths, H. Vennema, and P. J. Rottier. 2000. Characterization of the coronavirus mouse hepatitis virus strain A59 small membrane protein E. J. Virol. 742333-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Risco, C., M. Muntion, L. Enjuanes, and J. L. Carrascosa. 1998. Two types of virus-related particles are found during transmissible gastroenteritis virus morphogenesis. J. Virol. 724022-4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rodgers, W. 2002. Making membranes green: construction and characterization of GFP-fusion proteins targeted to discrete plasma membrane domains. BioTechniques 321044-1046, 1048, 1050-1051. [DOI] [PubMed] [Google Scholar]
- 56.Roingeard, P., and C. Hourioux. 2008. Hepatitis C virus core protein, lipid droplets and steatosis. J. Viral Hepat. 15157-164. [DOI] [PubMed] [Google Scholar]
- 57.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M.-H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. T. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. M. E. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 3001394-1399. [DOI] [PubMed] [Google Scholar]
- 58.Rottier, P. J., and J. K. Rose. 1987. Coronavirus E1 glycoprotein expressed from cloned cDNA localizes in the Golgi region. J. Virol. 612042-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ruan, W., V. Becker, U. Klingmuller, and D. Langosch. 2004. The interface between self-assembling erythropoietin receptor transmembrane segments corresponds to a membrane-spanning leucine zipper. J. Biol. Chem. 2793273-3279. [DOI] [PubMed] [Google Scholar]
- 60.Salanueva, I. J., J. L. Carrascosa, and C. Risco. 1999. Structural maturation of the transmissible gastroenteritis coronavirus. J. Virol. 737952-7964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schaecher, S. R., J. M. Mackenzie, and A. Pekosz. 2007. The ORF7b protein of severe acute respiratory syndrome coronavirus (SARS-CoV) is expressed in virus-infected cells and incorporated into SARS-CoV particles. J. Virol. 81718-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schaecher, S. R., E. Touchette, J. Schriewer, R. M. Buller, and A. Pekosz. 2007. Severe acute respiratory syndrome coronavirus gene 7 products contribute to virus-induced apoptosis. J. Virol. 8111054-11068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schmaljohn, C., and J. Hooper. 2001. Bunyaviridae: the viruses and their replication, p. 1581-1602. In P. M. H. D. M. Knipe, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 64.Schweizer, A., J. Rohrer, H. P. Hauri, and S. Kornfeld. 1994. Retention of p63 in an ER-Golgi intermediate compartment depends on the presence of all three of its domains and on its ability to form oligomers. J. Cell Biol. 12625-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shen, S., P. S. Lin, Y. C. Chao, A. Zhang, X. Yang, S. G. Lim, W. Hong, and Y. J. Tan. 2005. The severe acute respiratory syndrome coronavirus 3a is a novel structural protein. Biochem. Biophys. Res. Commun. 330286-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shi, X., D. F. Lappin, and R. M. Elliott. 2004. Mapping the Golgi targeting and retention signal of Bunyamwera virus glycoproteins. J. Virol. 7810793-10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shin, J., R. L. Dunbrack, Jr., S. Lee, and J. L. Strominger. 1991. Signals for retention of transmembrane proteins in the endoplasmic reticulum studied with CD4 truncation mutants. Proc. Natl. Acad. Sci. USA 881918-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sims, A. C., R. S. Baric, B. Yount, S. E. Burkett, P. L. Collins, and R. J. Pickles. 2005. Severe acute respiratory syndrome coronavirus infection of human ciliated airway epithelia: role of ciliated cells in viral spread in the conducting airways of the lungs. J. Virol. 7915511-15524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stertz, S., M. Reichelt, M. Spiegel, T. Kuri, L. Martinez-Sobrido, A. Garcia-Sastre, F. Weber, and G. Kochs. 2007. The intracellular sites of early replication and budding of SARS-coronavirus. Virology 361304-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Swift, A. M., and C. E. Machamer. 1991. A Golgi retention signal in a membrane-spanning domain of coronavirus E1 protein. J. Cell Biol. 11519-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Takeda, M., A. Pekosz, K. Shuck, L. H. Pinto, and R. A. Lamb. 2002. Influenza A virus M2 ion channel activity is essential for efficient replication in tissue culture. J. Virol. 761391-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tan, Y.-J., E. Teng, S. Shen, T. H. P. Tan, P.-Y. Goh, B. C. Fielding, E.-E. Ooi, H.-C. Tan, S. G. Lim, and W. Hong. 2004. A novel severe acute respiratory syndrome coronavirus protein, U274, is transported to the cell surface and undergoes endocytosis. J. Virol. 786723-6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Teasdale, R. D., and M. R. Jackson. 1996. Signal-mediated sorting of membrane proteins between the endoplasmic reticulum and the Golgi apparatus. Annu. Rev. Cell Dev. Biol. 1227-54. [DOI] [PubMed] [Google Scholar]
- 74.Teasdale, R. D., F. Matheson, and P. A. Gleeson. 1994. Post-translational modifications distinguish cell surface from Golgi-retained beta 1,4 galactosyltransferase molecules. Golgi localization involves active retention. Glycobiology 4917-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thiel, V., K. A. Ivanov, A. Putics, T. Hertzig, B. Schelle, S. Bayer, B. Weissbrich, E. J. Snijder, H. Rabenau, H. W. Doerr, A. E. Gorbalenya, and J. Ziebuhr. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 842305-2315. [DOI] [PubMed] [Google Scholar]
- 76.Tooze, J., S. Tooze, and G. Warren. 1984. Replication of coronavirus MHV-A59 in sac- cells: determination of the first site of budding of progeny virions. Eur. J. Cell Biol. 33281-293. [PubMed] [Google Scholar]
- 77.Tooze, J., and S. A. Tooze. 1985. Infection of AtT20 murine pituitary tumour cells by mouse hepatitis virus strain A59: virus budding is restricted to the Golgi region. Eur. J. Cell Biol. 37203-212. [PubMed] [Google Scholar]
- 78.Tooze, J., S. A. Tooze, and S. D. Fuller. 1987. Sorting of progeny coronavirus from condensed secretory proteins at the exit from the trans-Golgi network of AtT20 cells. J. Cell Biol. 1051215-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Voorhees, P., E. Deignan, E. van Donselaar, J. Humphrey, M. S. Marks, P. J. Peters, and J. S. Bonifacino. 1995. An acidic sequence within the cytoplasmic domain of furin functions as a determinant of trans-Golgi network localization and internalization from the cell surface. EMBO J. 144961-4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Watanabe, T., E. M. Sorensen, A. Naito, M. Schott, S. Kim, and P. Ahlquist. 2007. Involvement of host cellular multivesicular body functions in hepatitis B virus budding. Proc. Natl. Acad. Sci. USA 10410205-10210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Weisz, O. A., A. M. Swift, and C. E. Machamer. 1993. Oligomerization of a membrane protein correlates with its retention in the Golgi complex. J. Cell Biol. 1221185-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yamaguchi, N., and M. N. Fukuda. 1995. Golgi retention mechanism of beta-1,4-galactosyltransferase. Membrane-spanning domain-dependent homodimerization and association with alpha- and beta-tubulins. J. Biol. Chem. 27012170-12176. [DOI] [PubMed] [Google Scholar]
- 83.Ye, Y., and B. G. Hogue. 2007. Role of the coronavirus E viroporin protein transmembrane domain in virus assembly. J. Virol. 813597-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yount, B., K. M. Curtis, E. A. Fritz, L. E. Hensley, P. B. Jahrling, E. Prentice, M. R. Denison, T. W. Geisbert, and R. S. Baric. 2003. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA 10012995-13000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yount, B., R. S. Roberts, A. C. Sims, D. Deming, M. B. Frieman, J. Sparks, M. R. Denison, N. Davis, and R. S. Baric. 2005. Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J. Virol. 7914909-14922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhu, Z., Y. Hao, M. D. Gershon, R. T. Ambron, and A. A. Gershon. 1996. Targeting of glycoprotein I (gE) of varicella-zoster virus to the trans-Golgi network by an AYRV sequence and an acidic amino acid-rich patch in the cytosolic domain of the molecule. J. Virol. 706563-6575. [DOI] [PMC free article] [PubMed] [Google Scholar]