Abstract
Wild-type strains of equine infectious anemia virus (EIAV) prevent superinfection of previously infected cells. A variant strain of virus that spontaneously arose during passage, EIAVvMA-1c, can circumvent this mechanism in some cells, such as equine dermis (ED) cells, but not in others, such as equine endothelial cells. EIAVvMA-1c superinfection of ED cells results in a buildup of unintegrated viral DNA and rapid killing of the cell monolayer. Here, we examined the mechanism of resistance that is used by EIAV to prevent superinfection and explored the means by which EIAVvMA-1c overcomes this restriction. We found that the cellular receptor used by EIAV, equine lentivirus receptor 1 (ELR1), remains on the surface of cells chronically infected with EIAV, suggesting that wild-type EIAV interferes with superinfection by masking ELR1. The addition of soluble wild-type SU protein to the medium during infection blocked infection by wild-type strains of virus, implicating SU as the viral protein responsible for interfering with virion entry into previously infected cells. Additionally, interference of wild-type EIAV binding to ELR1 by the addition of either anti-ELR1 antibodies or the ELR1 ectodomain prevented entry of the wild-type strains of EIAV into two permissive cell populations. Many of these same interference treatments prevented EIAVvMA-1c infection of endothelial cells but only modestly affected the ability of EIAVvMA-1c to enter and kill previously infected ED cells. These findings indicate that EIAVvMA-1c retains the ability to use ELR1 for entry and suggest that this virus can interact with an additional, unidentified receptor to superinfect ED cells.
Superinfection occurs when a virally infected cell becomes reinfected by the same or a similar virus. Many viruses have evolved mechanisms to prevent superinfection, suggesting that viral superinfection reduces viral fitness. Inhibition of virus superinfection is called superinfection resistance or receptor interference.
A variety of superinfection resistance mechanisms have been described. Some retroviruses mask their receptor on the surface of the cell, preventing further use of that receptor. Receptor masking is the simplest form of superinfection resistance and in many cases is accomplished by the viral surface glycoprotein (SU) binding to its receptor on the cell's surface (20). Both avian reticuloendotheliosis virus and murine leukemia virus (MuLV) undergo receptor masking through the expression of soluble SU protein, which is sufficient for receptor interference (7, 10, 26). Alternatively, some retroviruses, such as human immunodeficiency virus (HIV), reduce or eliminate their receptors from the plasma membrane, preventing newly synthesized virions from binding to the infected cell (11, 29). HIV utilizes multiple mechanisms to down-modulate CD4. These mechanisms include viral glycoprotein-CD4 interactions that trap CD4 within the endoplasm reticulum and Golgi complex during protein synthesis, leading to CD4 degradation (26, 29). In addition, two accessory proteins, Nef and Vpu, interact and remove CD4 from the plasma membrane (12, 16, 26, 29). Superinfection resistance mechanisms of other retroviruses, such as mouse mammary tumor virus and foamy viruses, are independent of the envelope (3, 9).
As a consequence of superinfection resistance, it is uncommon for a retrovirus to display superinfection. However, a number of superinfecting strains have been described, and these variant viruses have evolved a number of different mechanisms to evade superinfection resistance. Superinfection can occur when low-affinity interactions between the viral glycoprotein and receptor do not adequately mask or downregulate the receptor (9, 19). Superinfection can also occur when viruses utilize overlapping domains of the same receptor. For instance, cells infected with polytropic MuLV (P-MuLV) can be superinfected by xenotropic MuLV (X-MuLV) because these viruses utilize different domains of the surface protein Xpr1 (28). Type E avian sarcoma and leukosis virus can be superinfected by avian sarcoma and leukosis virus type B because these viruses utilize different isoforms of the same receptor (1).
Equine infectious anemia virus (EIAV) is a lentivirus that infects equine macrophages and endothelial cells in vivo (21, 25). Once adapted to tissue culture, EIAV has the ability to infect and persistently replicate in equine endothelial cells, fetal kidney cells, and fibroblasts as well as canine and feline fibroblast cell lines. The tissue culture cells become chronically infected with EIAV without apparent deleterious effects on the cells. Studies with the tissue culture strains EIAVMA-1 and EIAVSP19 have demonstrated that infected cultures of an equine dermis fibroblastic line (ED cells) exhibit superinfection resistance against further infection with wild-type strains of EIAV (13).
EIAVvMA-1c is a variant strain of EIAV that spontaneously arose by multiple passages of EIAVMA-1 in ED cells (13). EIAVvMA-1c superinfects equine fibroblasts, but not the other permissive cells, while retaining the same cellular tropism as the parental strain (13). EIAVvMA-1c induces large syncytia and rapid cell death of equine fibroblasts. Both phenotypes are atypical of an EIAV infection (13). We recently demonstrated that EIAVvMA-1c superinfection of equine fibroblasts is clathrin and low pH independent, whereas productive entry of wild-type virus is dependent on a low-pH, clathrin-mediated endocytosis event, indicating that EIAVvMA-1c can productively enter fibroblasts through a pathway distinct from that used by wild-type virus (4, 5). Here, we demonstrate that both wild-type EIAV and the superinfecting strain EIAVvMA-1c can utilize the equine lentiviral receptor 1 (ELR1) to enter cells in a low-pH and clathrin-dependent manner. In equine fibroblasts, we propose that EIAVvMA-1c has evolved the use of an unidentified receptor. Use of this second receptor is required for clathrin- and low pH-independent superinfection by EIAVvMA-1c.
MATERIALS AND METHODS
Cell lines and viral strains.
Equine cells were used to characterize the superinfection phenotype of EIAVMA-1 and EIAVvMA-1c. ED cells, an equine fibroblastic cell line derived from dermis cells (ATCC CCL57), and primary equine umbilical vein endothelial cells (eUVECs) were used for infection. Human 293T cells (8) were used for transfections and protein production. Cells were maintained in high-glucose Dulbecco's modified Eagle's medium with penicillin and streptomycin. Medium was supplemented with 15% fetal calf serum (FCS) for ED cells, 10% FCS for 293T cells, and 40% FCS for eUVECs.
Several tissue culture strains of EIAV were used in this study. EIAVMA-1 is an avirulent, tissue culture-adapted strain of EIAV (6). EIAVvMA-1c is a cytopathic, superinfecting strain of EIAV that was derived by serial passages of EIAVMA-1 in ED cells as previously described (13). EIAVSP19 was generated from the avirulent infectious molecular clone pSP19 (23) by transfection of ED cells. EIAVTh.1 is a macrophage-tropic strain obtained from the first viremic episode of a horse inoculated with a field isolate from Massachusetts (6). EIAVWSU5 is a strain from Washington State University that was derived from the Wyoming strain and continues to be passaged in equine fetal kidney cells and back-passaged through ponies to maintain virulence (22).
Generation of virus stocks and detection of EIAV replication.
Viral stocks were produced in ED cells. Supernatants were harvested from cells that were >90% positive for EIAV antigen as determined by EIAV antigen immunostaining. Supernatants were filtered through a 0.45-μm filter to remove cell debris, aliquoted, and frozen at −80°C until needed.
EIAV infection and replication were assessed by immunostaining infected cell populations for viral antigens as previously described (14). Virus titers were determined on ED cells by serial dilution of stocks followed by immunostaining of cells for viral antigens at 40 h after infection. Acetone-fixed cells were immunostained with polyclonal horse anti-EIAV antiserum (1:800) from a long-term-infected horse (WSU 2085; a kind gift from J. Lindsay Oaks). Primary antisera were incubated for 3 h at 37°C, followed by several washes with phosphate-buffered saline. Peroxidase-conjugated goat anti-horse immunoglobulin (1:800; Jackson Immunoresearch) was incubated for 1 h at 37°C. Peroxidase activity was detected using the substrate 3-amino-9-ethylcarbazole (Sigma). The EIAV antigen-positive cells within the infected cell monolayer were counted and titers were determined.
VSV-G-pseudotyped EIAV production.
Fifteen-cm plates of 293T cells were transfected with a total of 75 μg of DNA consisting of a vesicular stomatitis virus glycoprotein (VSV-G) expression construct, pONY3.1 (EIAV gag-pol expression plasmid) and pONYϕβgal (18) at a ratio of 1:2:3. The DNA was transfected into 80% confluent 15-cm dishes of 293T cells via a calcium-phosphate transfection. Supernatants were collected at 24, 36, 48, 60, and 72 h, passed through a 0.45-μm filter, and pelleted by a 16-h centrifugation step (7,000 rpm at 4°C in a Sorvall GSA rotor). The pellet was resuspended in 250 μl of Dulbecco's modified Eagle's medium to an approximate 200-fold concentration. Pseudotyped particles were aliquoted and stored at −80°C until use.
Flow cytometry.
ED cells, eUVECs, and chronically infected ED cells and eUVECs were scraped off the flask and washed with phosphate-buffered saline. The live cells were stained with either a control rabbit antiserum against human Src or anti-ELR1 rabbit antiserum (1:100) for 1 h at 4°C. Cells were washed and incubated with allophycocyanin-goat anti-rabbit antiserum (1:100; Jackson Immunoresearch) for 20 min on ice. Cells were washed three times and analyzed by flow cytometry using a FACScan cytometer. Live cells that were gated by forward scatter and side scatter were evaluated for FL-4 intensity.
Soluble SU expression and competition assay.
The DNA encoding the EIAVSP19 SU protein (ATG-Hpa1 site) was codon optimized for human expression (GenScript). We cloned the codon-optimized SU sequence into the pcDNA3.1/hygro eukaryotic expression vector (Invitrogen). The SU-expressing plasmid or an empty plasmid was transfected into 293T cells using GenePorter as per the manufacturer's instructions. The cell supernatant was collected 48 h after transfection, filtered through a 0.22-μm filter to remove any cellular debris, and frozen at −80°C until use.
SU competitions were performed by adding increasing amounts (1:300, 1:30, and 1:3) of the collected supernatant to media on ED cells, and SU was allowed to bind at 4°C for 20 min. The cells were infected with EIAVMA-1 or EIAVvMA-1c virus or VSV-G-pseudotyped EIAV particles containing a β-galactosidase (β-Gal) reporter gene at a multiplicity of infection (MOI) of 0.005. The EIAVMA-1- and EIAVvMA-1c-infected cells were fixed and immunostained for EIAV antigen 40 h after infection, and the VSV-G-transduced particles were formalin fixed 2 days after transduction and stained for β-Gal activity. The numbers of antigen- or β-gal-positive cells were counted and divided by the positive control (the number of infected or transduced cells in the absence of supernatant addition) to obtain a ratio (percentage of controls, shown in the figures). Supernatant from 293T cells transfected with an empty pcDNA plasmid was used as a negative control.
ELR1 ectodomain competition assay.
The ectodomain of ELR1 was cloned, expressed in Escherichia coli, and purified by B. Zhang at the University of Pittsburgh as previously described (31). Serial dilutions of ELR1 ectodomain (0.06, 0.3, and 1.5 μg/ml) were diluted in medium and incubated with viral particles at 4°C for 30 min. After ELR1 binding, either EIAVMA-1 or EIAVvMA-1c virus or VSV-G-pseudotyped EIAV/β-Gal particles were added to the cells at an MOI of 0.005 and incubated at 37°C. EIAVMA-1- and EIAVvMA-1c-infected cells were fixed and immunostained for EIAV antigen 40 h after infection, and the VSV-G-transduced particles were formalin fixed 2 days after transduction and stained for β-Gal. The number of infected or transduced cells was counted and divided by the positive control (the number of infected or transduced cells in the absence of ELR1 ectodomain addition) to obtain the ratio for the percentage of control, as shown in the figures.
ELR1 antibody competition.
Serial dilutions (1:1500, 1:300, and 1:60) of either control rabbit serum or anti-ELR1 rabbit serum (31) were added to the culture medium of either ED cells or eUVECs. Antibodies were allowed to bind for 30 min at 4°C. The cells were inoculated with either EIAVMA-1 or EIAVvMA-1c at an MOI of 0.005 and incubated at 37°C. Cells were fixed and stained for EIAV antigen 40 h after infection. The number of antigen-positive cells was counted and divided by the positive control results (the number of infected or transduced cells in the absence of antibodies) to obtain the ratio for the percentage of control, as shown in the figures.
ELR1 RNA interference knockdown.
One million ED cells were mock transfected or transfected with small interfering RNA (siRNA) against ELR1 (custom synthesized by Integrated DNA Technologies; target sequence, 5′-GUGGAACCUGGGACUAGCAGCACAG-3′) or control siRNA that was a fluorescently labeled nonspecific RNA inhibitor (Block-iT; Invitrogen). Cells were transfected using Amaxa protocols with a final siRNA concentration of 2 μM. Each transfected cell population was plated into 12 wells of a 48-well plate. Forty-eight hours after transfection, the cells were infected with EIAV or harvested for immunoblotting analysis.
ELR1 immunoblotting.
Proteins in cell lysates were separated on a 4 to 20% Tris-glycine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel (Invitrogen) and transferred onto nitrocellulose. ELR1 was detected by incubating the membranes with rabbit anti-ELR1 polyclonal sera (1:1,000) for 3 h and with secondary peroxidase-conjugated goat anti-rabbit antiserum (1:20,000; Sigma) for 1 h. Membranes were visualized by the chemiluminescence method according to the manufacturer's instructions (Pierce).
Cell killing superinfection assay.
To assess the ability of EIAVvMA-1c to superinfect and kill cells, ED cells were treated with a variety of agents that we have shown inhibit the ELR1-dependent pathway of EIAV entry. We used concentrations of agents that inhibited 90% of productive virus infectivity (4, 5). These included EIAV-neutralizing antiserum 2085 (1:60), ELR1 antibodies (1:60), EIAV SU protein (50 μl), ammonium chloride (30 mM), and chlorpromazine (20 μg/ml). The cells were infected with a low MOI of EIAVvMA-1c (0.05). Cell viability was evaluated by the ATPLite assay (Packard Biosciences) per the manufacturer's instructions 4 days after infection.
RESULTS
Chronically infected cells retain ELR1 on the cell surface.
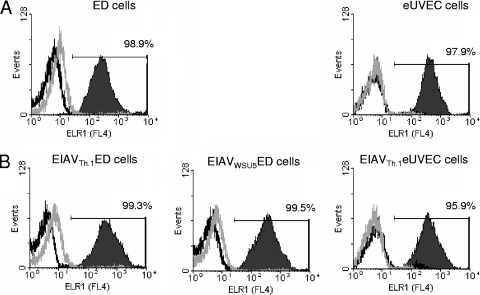
Wild-type strains of EIAV have been demonstrated to prevent superinfection (13). In order to determine if these strains inhibit superinfection by downregulating the cellular receptor from the cell surface, live cells were immunostained with a polyclonal antiserum directed against the ELR1 ectodomain and analyzed by flow cytometry. Surface staining of uninfected ED or eUVECs demonstrated that greater than 95% of the populations were positive for ELR1 surface expression (Fig. 1A). Chronically infected EIAVWSU5 ED, EIAVTh.1 ED, and EIAVTh.1 eUVECs that were more than 90% EIAV antigen positive (data not shown) were also positive for ELR1 (Fig. 1B). These findings indicate that ELR1 is not decreased on the surface of EIAV-infected cells.
FIG. 1.
ELR1 remains on the surface of EIAV-infected cells. ED cells and eUVECs (A) and chronically EIAV-infected cells (B) were immunostained for surface expression of ELR1 using rabbit polyclonal antisera against the ectodomain of ELR1. EIAV-infected populations were greater than 90% positive for EIAV antigens. Allophycocyanin-conjugated goat-anti-rabbit was used as a secondary antibody. Flow cytometry of live, stained cells was used to determine the percentage of the population that was positive for ELR1 (solid area). Secondary antisera alone (black line) as well as rabbit control sera (gray line) were used as negative controls. A representative flow experiment is shown. The experiments were performed three independent times.
The EIAV SU protein is sufficient to block entry of wild-type EIAV.
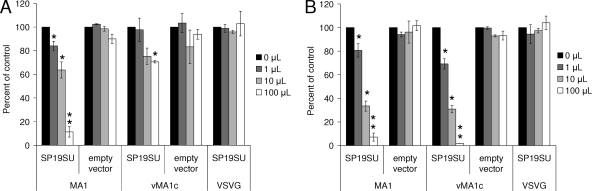
Previously we demonstrated that two unrelated strains of EIAV, EIAVMA-1 and EIAVSP19, exhibited superinfection resistance against each other (13). Because ELR1 is found on the surface of chronically infected cells, we next sought to determine if the superinfection resistance mechanism involved receptor masking by the EIAV Env protein. To determine if expression of the EIAV SU was sufficient for receptor interference, 293T cells were transfected with a codon-optimized EIAVSP19 SU expression construct (27), and the expressed SU protein was collected in the supernatant. As a control, supernatants were collected from 293T cells that had been transfected with an empty plasmid. Increasing quantities of supernatant were added to ED cells or eUVECs that were subsequently infected with EIAVMA-1 or EIAVvMA-1c virus. The SU protein inhibited entry of EIAVMA-1 in a dose-dependent manner with the highest quantity of competitor inhibiting more than 90% of EIAVMA-1 infectivity (Fig. 2A and B). The combined findings that ELR1 remains on the surface of EIAV-infected cells and that EIAV SU inhibits entry of EIAV in a dose-dependent manner strongly suggested that EIAV SU masks ELR1 on the surface of cells and is sufficient to mediate receptor interference for wild-type strains of EIAV.
FIG. 2.
EIAV SU blocks entry of wild-type strains of EIAV. ED cells (A) or eUVECs (B) were incubated with increasing amounts of 293T cell supernatants that had been either transfected with an empty vector or a codon-optimized EIAVSP19 SU expression vector. The cells were subsequently infected with EIAVMA-1 or EIAVvMA-1c or transduced with VSV-G-pseudotyped EIAV particles that express β-galactosidase. Cells were evaluated for infection or transduction at 40 h. Shown is the ratio of the number of infected or transduced cells in the presence of supernatant divided by the number of infected or transduced cells when no supernatant was added. Data represent the means and standard errors of the means from three separate experiments performed in triplicate. *, P < 0.05; **, P < 0.001.
In contrast, only the highest quantity of EIAVSP19 SU protein resulted in modest inhibition of entry of EIAVvMA-1c into ED cells, indicating that EIAVvMA-1c was less sensitive to inhibition by wild-type EIAV SU (Fig. 2A). However, EIAVvMA-1c infection of endothelial cells was as sensitive to increasing concentrations of SU as EIAVMA-1 (Fig. 2B). These findings suggest that EIAVSP19 SU-induced receptor masking is not sufficient to inhibit entry by EIAVvMA-1c in ED cells but is able to inhibit eUVEC infection by this virus.
EIAVvMA-1c uses ELR1 but utilizes a second receptor to infect ED cells.
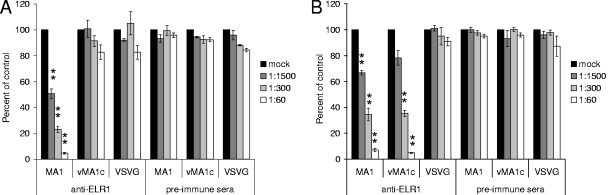
EIAVvMA-1c retains the same cell tropism as the parental strain, EIAVMA-1, and in addition can superinfect equine fibroblasts (13). Because the superinfection phenotype is limited to one cell type and superinfection resistance against EIAVvMA-1c is exhibited in other chronically infected cell populations (13), we hypothesized that EIAVvMA-1c enters cells using the ELR1 receptor and in addition has evolved the ability to enter equine fibroblast cells through other interactions specific for equine fibroblasts. To determine if ELR1 is necessary for EIAVvMA-1c entry into two equine cell populations, primary eUVECs and the ED cell line, we performed infections in the presence of increasing concentrations of polyclonal antibodies against ELR1. In eUVECs, EIAVMA-1 and EIAVvMA-1c entry levels were inhibited by anti-ELR1 antiserum in a dose-dependent manner, and no decrease was seen when preimmune serum was used as control (Fig. 3B). In a similar manner, EIAVMA-1 showed a significant dose-dependent inhibition of infection with increasing amounts of anti-ELR1 antibodies added to ED cells (Fig. 3A). To ensure that EIAVMA-1 infectivity was not being blocked nonspecifically by antibodies binding to the cells, the experiment was repeated with antitransferrin monoclonal antibody that recognizes equine transferrin. The transferrin antibody did not impede entry (data not shown), providing additional evidence that the competition seen with anti-ELR1 antiserum is specific. In these same studies, EIAVvMA-1c infection of ED cells or VSV-G-pseudotyped EIAV particle transduction was not blocked by anti-ELR1 antibodies.
FIG. 3.
EIAVvMA-1c can enter ED cells in an ELR1-independent manner. ED cells (A) or eUVECs (B) were incubated with preimmune serum or polyclonal anti-ELR1 antiserum at 4°C for 30 min. The cells were subsequently infected with EIAVMA-1 or EIAVvMA-1c or transduced with VSV-G-pseudotyped EIAV particles. Cells were stained 40 h after infection. Shown is the ratio of the number of infected or transduced cells in the presence of antiserum divided by the number of infected or transduced cells when no antiserum was added. Data represent the means and standard errors of the means from three separate experiments performed in triplicate. *, P < 0.05; **, P < 0.001.
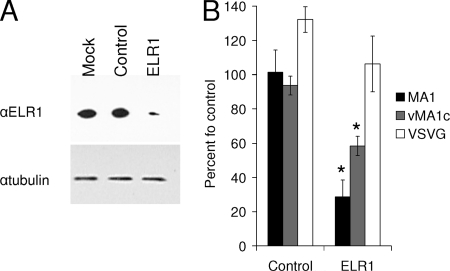
To further examine the necessity of ELR1 in EIAVvMA-1c entry, siRNA knockdown of ELR1 was performed. The greatest reduction of ELR1 protein was observed 48 h after transfection, although the knockdown was incomplete at all time points (Fig. 4A). siRNA-treated cells were infected with EIAV or transduced with VSV-G-pseudotyped EIAV at 48 h. EIAVMA-1 infection was reduced more than 70% when ELR1 was decreased (Fig. 4B). EIAVvMA-1c infection was inhibited by 40%, indicating that EIAVvMA-1c infection of ED cells was not as severely impacted by the reduction of ELR1 in the cell population (Fig. 4B). The results in both the anti-ELR1 antisera and siRNA studies suggest that EIAVvMA-1c entry into ED cells is not solely dependent upon ELR1.
FIG. 4.
siRNA knockdown of ELR1 decreases EIAV entry into ED cells. ED cells were transfected with a control siRNA or ELR1 siRNA. Forty-eight hours after transfection, cells were either harvested for immunoblotting or used in infection/transduction studies. Cell lysates were immunoblotted for ELR1 and a tubulin loading control (A). Cells were stained for EIAV antigens or β-Gal activity at 40 h after infection (88 h after transfection) (B). Data represent the means and standard errors of the means from three separate experiments performed in triplicate. *, P < 0.05.
The ELR1 ectodomain blocks both EIAVMA-1 and EIAVvMA-1c entry into ED cells.
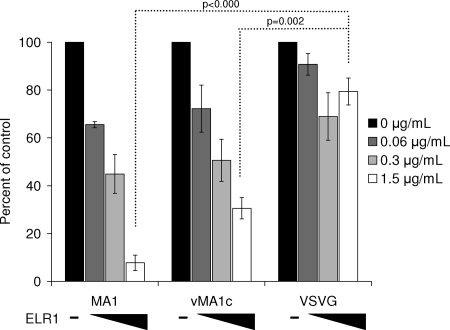
As EIAVvMA-1c retains the ability to use ELR1, we sought to determine if ELR1 ectodomain binding to EIAVvMA-1c particles interferes with virion binding to ED cells. Infectivity assays were performed in the presence of increasing quantities of ELR1 ectodomain. EIAVMA-1 infectivity was inhibited in a dose-dependent manner with increasing amounts of soluble ELR1, with greater than 90% inhibition at the highest dose (Fig. 5). Surprisingly, the infectivity of EIAVvMA-1c was also significantly inhibited by the ELR1 ectodomain; however, VSV-G-pseudotyped particles were unaffected. These data indicate that both EIAVMA-1 and EIAVvMA-1c have the ability to bind to the ELR1 ectodomain and upon binding the ectodomain is sufficient to block interaction of the virus with receptors on the cell. ELR1 ectodomain inhibition of EIAVvMA-1c infectivity may be due to direct competition for the receptor binding site on the virion or due to blocking the site through steric hindrance.
FIG. 5.
EIAVvMA-1c retains the ability to interact with ELR1, and this interaction inhibits entry. EIAVMA-1 or EIAVvMA-1c virions and VSV-G-pseudotyped EIAV particles (MOI, 0.005) were incubated with the soluble ectodomain of ELR1 for 30 min at 4°C. The viruses were evaluated for ED cell infection. Cells were stained 40 h after infection. Shown is the ratio of the number of infected or transduced cells in the presence of ELR1 ectodomain divided by the number of infected or transduced cells when no ectodomain was added. Data represent the means and standard errors of the means from three separate experiments performed in triplicate.
EIAVvMA-1c cell killing is independent of the ELR1 entry pathway.
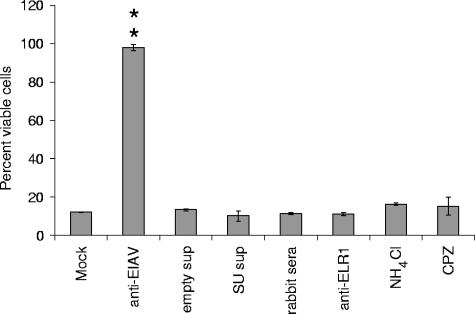
Our data suggest that EIAVvMA-1c can utilize a second receptor in equine fibroblasts and that this alternative receptor is involved in superinfection. To examine cell killing as a result of superinfection, we treated ED cells with several different agents that we have previously shown inhibit wild-type EIAV entry, which is ELR1 dependent. Ammonium chloride and chlorpromazine are agents that block endocytic events downstream of ELR1 binding (4, 5), whereas soluble EIAVSP19 SU and ELR1 antisera were shown earlier in this study to block wild-type EIAV entry. All these agents failed to prevent the cell killing caused by EIAVvMA-1c superinfection (Fig. 6). Only neutralizing antisera against EIAV was successful in preventing EIAVvMA-1c-mediated cell death, demonstrating EIAVvMA-1c superinfection does not require the ELR1 low-pH, clathrin-mediated pathway used by wild-type EIAV.
FIG. 6.
Interference with ELR1-EIAV interactions does not inhibit EIAVvMA-1c superinfection-dependent cell killing. ED cells were treated with inhibitors of ELR1-dependent entry at concentrations previously shown to inhibit 90% of EIAVMA-1 infectivity. The treated cells were infected with EIAVvMA-1c, and 4 days after infection the cells were assayed for viability. Data represent the means and standard errors of the means from three separate experiments performed in triplicate. Anti-EIAV, 1:60 dilution of equine anti-EIAV serum 2085; control sups, 100 μl of supernatant from 293T cells; WT EIAV SU sups, 100 μl of supernatant from codon-optimized SU-transfected 293T cells; normal rabbit serum, 1:60 dilution of normal rabbit serum; anti-ELR1, 1:60 of rabbit anti-ELR1 antiserum; NH4Cl, 30 mM ammonium chloride; CPZ, 20 μg/ml of chlorpromazine. **, P < 0.001.
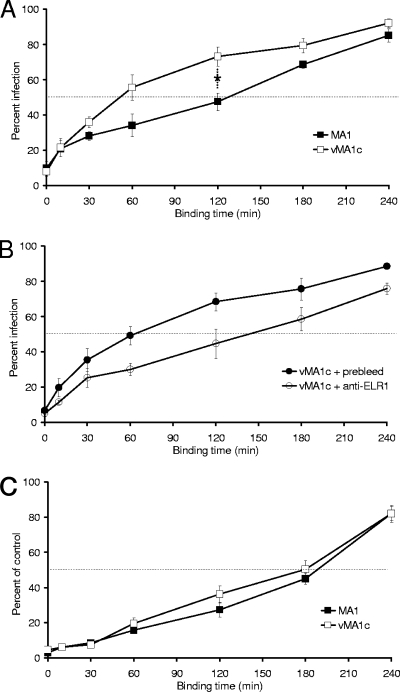
EIAVvMA-1c and EIAVMA-1 have similar binding kinetics when entry is dependent on ELR1 interactions.
In some superinfecting scenarios, a reduction of viral envelope-receptor affinity prevents efficient superinfection resistance, thereby allowing superinfection to occur (9, 19). One estimate of envelope-receptor affinity is the rapidity of virion binding to the cells. To examine the interaction of EIAVMA-1 and EIAVvMA-1c with their receptors, we examined their binding kinetics on both ED cells and eUVECs. Virus was incubated with ED cells for increasing periods of time, and then unbound virions were removed and medium was replaced with fresh medium. The cells were fixed and stained 40 h after infection to evaluate the rapidity of virus binding. EIAVvMA-1c bound to ED cells twice as fast as EIAVMA-1 (t1/2 = 60 min versus 120 min) (Fig. 7A). The experiment was repeated with EIAVvMA-1c in the presence of ELR1 antibodies to examine EIAVvMA-1c's ability to bind to ED cells without ELR1 interaction (Fig. 7B). EIAVvMA-1c binding kinetics in the presence of preimmune serum was not altered, but the anti-ELR1 antiserum altered the slope of the curve and increased the t1/2 to ∼125 min. The impact of ELR1 antiserum on the slope of the binding curve suggests that interactions between EIAVvMA-1c and the superinfecting receptor have a similar avidity as EIAVMA-1 does for ELR1. The binding of virus to eUVECs was also examined, as both EIAVMA-1 and EIAVvMA-1c binding and productive infection of eUVECs are dependent on ELR1. Binding of both viruses to eUVECs was slower than to ED cells (t1/2 = 3 h) and the two viruses had indistinguishable binding curves, suggesting that they have similar affinities for ELR1 (Fig. 7C).
FIG. 7.
EIAVvMA-1c has similar binding kinetics for ELR1 as EIAVMA-1. EIAVMA-1 or EIAVvMA-1c binding kinetics were evaluated in ED cells (A), in ED cells in the presence of preimmune serum or anti-ELR1 antiserum (diluted 1:60) (B), and eUVECs (C). Virus particles were added to cells and then removed at the time points indicated. Cells were stained 40 h after infection and compared to the number of infected cells when particles were not removed. Data represent the means and standard errors of the means from three separate experiments performed in triplicate. *, P < 0.05.
DISCUSSION
The mechanism of HIV superinfection resistance involves the downregulation of the cellular receptor from the surface of infected cells by CD4 interactions with the envelope protein as well as two accessory proteins (12, 29). The multipronged means of preventing superinfection suggest that the virus is under strong selective pressure to inhibit superinfection. Here, we demonstrate that the cellular receptor for EIAV ELR1 remains on the surface of chronically infected cells and the expression of EIAV SU protein is sufficient to prevent infection of the complex lentivirus EIAV in ED cells and eUVECs. It should be noted, however, that this study does not exclude the possibility that other EIAV-encoded proteins can contribute to superinfection resistance. Thus, we find that the receptor masking by EIAV SU protein is more reminiscent of superinfection resistance mechanisms used by simple retroviruses rather than those that have been characterized for the complex retroviruses.
ELR1 was recently identified as the cellular receptor for EIAVUK and EIAVWYO (30). Our study demonstrated that interference with the ELR1-virus interaction via ELR1 antibodies or soluble ELR1 ectodomain inhibits entry of two additional strains of virus, EIAVMA-1 and EIAVvMA-1c, into eUVECs. This further confirms ELR1 as the receptor for wild-type tissue culture-adapted strains.
All the experimental approaches used here to interfere with ELR1-dependent entry into ED cells reduced the entry of EIAVMA-1 into ED cells to a greater degree than the entry of EIAVvMA-1c. Addition of wild-type SU, anti-ELR1 antiserum, or siRNA against ELR1 effectively reduced or eliminated EIAVMA-1 entry into both eUVECs and ED cells. These same treatments impacted EIAVvMA-1c entry into eUVECs but were less effective or ineffective at inhibiting EIAVvMA-1c entry into ED cells. These findings along with our previously published findings that EIAVvMA-1c enters ED cells in a pH-independent, endosomal-independent manner strongly suggest that EIAVvMA-1c does not mediate superinfection of ED cells through the same ELR1 isoform that EIAVMA-1 utilizes for entry. Instead, we propose that a second, independent receptor is used by EIAVvMA-1c for superinfection of ED cells.
It is possible that alternatively spliced forms of ELR1 that are specific for equine fibroblasts and contain additional binding sites for EIAVvMA-1c may serve as the second, currently unidentified receptor. To date, the splicing patterns for ELR1 have not been characterized. As our siRNA experiment failed to remove all of ELR1 from the ED cells, we cannot exclude this possibility. Alternatively, the receptor used by EIAVvMA-1c to superinfect ED cells may be a protein that is entirely unrelated to ELR1.
Interestingly, EIAVvMA-1c infection of ED cells was inhibited by incubation of the ELR1 ectodomain with virions. This unexpected finding may be due to the ability of the soluble ectodomain of ELR1 to block EIAVvMA-1c infection of ED cells by sterically interfering with the virus's binding to its other receptor. Alternatively, the ELR1 binding site on the SU of EIAVvMA-1c may overlap with the new receptor binding site.
Some superinfecting strains of retroviruses display poor affinity for their receptors (19). Poor receptor affinity can prevent adequate receptor masking, permitting superinfection. However, we found both ED cells and eUVECs display high levels of surface ELR1, and the binding kinetics for EIAVMA-1 and EIAVvMA-1c in eUVECs were indistinguishable, indicating that SU changes present in EIAVvMA-1c do not result in a reduced affinity of the viral glycoprotein for ELR1. Because EIAVvMA-1c can superinfect cells previously infected with EIAVvMA-1c with no receptor interference observed, the EIAVvMA-1c interaction with its unknown receptor in equine fibroblasts would be predicted to have low affinity and/or avidity. However, the binding kinetics for EIAVvMA-1c with the second receptor were indistinguishable from EIAVMA-1 binding to ELR1, suggesting a strong interaction. An alternative possibility is that the second receptor is expressed in high abundance on ED cells such that EIAVvMA-1c cannot produce enough envelope protein to mask both ELR1 and the second receptor, thus enabling superinfection to occur.
There are 18 residue differences in the SU protein between EIAVMA-1 and EIAVvMA-1c. In addition, EIAVvMA-1c contains a premature stop codon that truncates the cytoplasmic tail of TM. Presumably the changes within the SU protein confer the new EIAVvMA-1c receptor binding site. Several of the mutations occur within regions of the SU that have been found to be highly variable among differing EIAV strains (2, 15, 24). Although the changes in the variable regions have been implicated in immune evasion, some of the changes may confer new receptor interactions. The EIAV SU residues necessary for ELR1 interaction have not been identified. Because ELR1 is the confirmed receptor for several diverse strains of EIAV, the ELR1 receptor binding site would presumably occur within conserved regions of the SU. Efforts are under way to identify the residue changes required for interaction with the EIAVvMA-1c receptor.
The ability of EIAVvMA-1c to interact with the additional ED cell-specific receptor must result from strong selective pressures because long-term tissue culture passage of EIAVMA-1 in ED cells predictably yields a superinfecting virus with genetic and phenotypic characteristics similar to EIAVvMA-1c. This evolution has been observed on numerous separate occasions. Selection for high viral titers and a quick replication rate must evolve a virus that can interact with an additional surface receptor on equine fibroblasts.
In conclusion, wild-type strains of EIAV can efficiently prevent superinfection by other wild-type strains of EIAV via expression of the SU protein. The SU protein masks ELR1 on the cell surface. EIAVvMA-1c infects ED cells through interactions with ELR1 and in addition has evolved with the use of an additional unidentified receptor. Interactions with the additional equine fibroblast-specific receptor allow superinfection of ED cells. EIAVvMA-1c's superinfection scenario is very similar to amphotropic MuLV and 10A1MuLV. 10A1MuLV can interact with an additional related receptor, enabling it to superinfect A-MuLV (17). We are interested in determining if this novel receptor for EIAVvMA-1c is related to ELR1 or if it has evolved to interact with a completely unrelated receptor. These studies are ongoing.
Acknowledgments
This work was supported by NIH 9R56 AI07326 (R.C.M.). M.A.B. was supported through the University of Iowa Training in Molecular Virology grant (NIH T32 A1007533).
Footnotes
Published ahead of print on July 30 2008.
REFERENCES
- 1.Adkins, H. B., S. C. Blacklow, and J. A. Young. 2001. Two functionally distinct forms of a retroviral receptor explain the nonreciprocal receptor interference among subgroups B, D, and E avian leukosis viruses. J. Virol. 753520-3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ball, J. M., K. E. Rushlow, C. J. Issel, and R. C. Montelaro. 1992. Detailed mapping of the antigenicity of the surface unit glycoprotein of equine infectious anemia virus by using synthetic peptide strategies. J. Virol. 66732-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bock, M., M. Heinkelein, D. Lindemann, and A. Rethwilm. 1998. Cells expressing the human foamy virus (HFV) accessory Bet protein are resistant to productive HFV superinfection. Virology 250194-204. [DOI] [PubMed] [Google Scholar]
- 4.Brindley, M. A., and W. Maury. 2005. Endocytosis and a low-pH step are required for productive entry of equine infectious anemia virus. J. Virol. 7914482-14488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brindley, M. A., and W. Maury. 2008. Equine infectious anemia virus entry occurs through clathrin-mediated endocytosis. J. Virol. 821628-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carpenter, S., and B. Chesebro. 1989. Change in host cell tropism associated with in vitro replication of equine infectious anemia virus. J. Virol. 632492-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delwart, E. L., and A. T. Panganiban. 1989. Role of reticuloendotheliosis virus envelope glycoprotein in superinfection interference. J. Virol. 63273-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DuBridge, R. B., P. Tang, H. C. Hsia, P. M. Leong, J. H. Miller, and M. P. Calos. 1987. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell. Biol. 7379-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dzuris, J. L., W. Zhu, D. Kapkov, T. V. Golovkina, and S. R. Ross. 1999. Expression of mouse mammary tumor virus envelope protein does not prevent superinfection in vivo or in vitro. Virology 263418-426. [DOI] [PubMed] [Google Scholar]
- 10.Heard, J. M., and O. Danos. 1991. An amino-terminal fragment of the Friend murine leukemia virus envelope glycoprotein binds the ecotropic receptor. J. Virol. 654026-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jobbagy, Z., S. Garfield, L. Baptiste, M. V. Eiden, and W. B. Anderson. 2000. Subcellular redistribution of Pit-2 Pi transporter/amphotropic leukemia virus (A-MuLV) receptor in A-MuLV-infected NIH 3T3 fibroblasts: involvement in superinfection interference. J. Virol. 742847-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindwasser, O. W., R. Chaudhuri, and J. S. Bonifacino. 2007. Mechanisms of CD4 downregulation by the Nef and Vpu proteins of primate immunodeficiency viruses. Curr. Mol. Med. 7171-184. [DOI] [PubMed] [Google Scholar]
- 13.Maury, W., P. J. Wright, and S. Bradley. 2003. Characterization of a cytolytic strain of equine infectious anemia virus. J. Virol. 772385-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maury, W. J., S. Carpenter, K. Graves, and B. Chesebro. 1994. Cellular and viral specificity of equine infectious anemia virus Tat transactivation. Virology 200632-642. [DOI] [PubMed] [Google Scholar]
- 15.Mealey, R. H., S. R. Leib, S. L. Pownder, and T. C. McGuire. 2004. Adaptive immunity is the primary force driving selection of equine infectious anemia virus envelope SU variants during acute infection. J. Virol. 789295-9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michel, N., I. Allespach, S. Venzke, O. T. Fackler, and O. T. Keppler. 2005. The Nef protein of human immunodeficiency virus establishes superinfection immunity by a dual strategy to downregulate cell-surface CCR5 and CD4. Curr. Biol. 15714-723. [DOI] [PubMed] [Google Scholar]
- 17.Miller, D. G., and A. D. Miller. 1994. A family of retroviruses that utilize related phosphate transporters for cell entry. J. Virol. 688270-8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitrophanous, K., S. Yoon, J. Rohll, D. Patil, F. Wilkes, V. Kim, S. Kingsman, A. Kingsman, and N. Mazarakis. 1999. Stable gene transfer to the nervous system using a non-primate lentiviral vector. Gene Ther. 61808-1818. [DOI] [PubMed] [Google Scholar]
- 19.Murphy, S. L., M. Chung-Landers, M. Honczarenko, and G. N. Gaulton. 2006. Linkage of reduced receptor affinity and superinfection to pathogenesis of TR1.3 murine leukemia virus. J. Virol. 804601-4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nethe, M., B. Berkhout, and A. C. van der Kuyl. 2005. Retroviral superinfection resistance. Retrovirology 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oaks, J. L., C. Ulibarri, and T. B. Crawford. 1999. Endothelial cell infection in vivo by equine infectious anaemia virus. J. Gen. Virol. 802393-2397. [DOI] [PubMed] [Google Scholar]
- 22.O'Rourke, K., L. E. Perryman, and T. C. McGuire. 1988. Antiviral, anti-glycoprotein and neutralizing antibodies in foals with equine infectious anaemia virus. J. Gen. Virol. 69667-674. [DOI] [PubMed] [Google Scholar]
- 23.Payne, S. L., J. Rausch, K. Rushlow, R. C. Montelaro, C. Issel, M. Flaherty, S. Perry, D. Sellon, and F. Fuller. 1994. Characterization of infectious molecular clones of equine infectious anaemia virus. J. Gen. Virol. 75425-429. [DOI] [PubMed] [Google Scholar]
- 24.Payne, S. L., K. Rushlow, B. R. Dhruva, C. J. Issel, and R. C. Montelaro. 1989. Localization of conserved and variable antigenic domains of equine infectious anemia virus envelope glycoproteins using recombinant env-encoded protein fragments produced in Escherichia coli. Virology 172609-615. [DOI] [PubMed] [Google Scholar]
- 25.Sellon, D. C., S. T. Perry, L. Coggins, and F. J. Fuller. 1992. Wild-type equine infectious anemia virus replicates in vivo predominantly in tissue macrophages, not in peripheral blood monocytes. J. Virol. 665906-5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stevenson, M., C. Meier, A. M. Mann, N. Chapman, and A. Wasiak. 1988. Envelope glycoprotein of HIV induces interference and cytolysis resistance in CD4+ cells: mechanism for persistence in AIDS. Cell 53483-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tallmadge, R. L., M. A. Brindley, J. Salmans, R. H. Mealey, W. Maury, and S. Carpenter. 2008. Development and characterization of an equine infectious anemia virus Env pseudotyped reporter virus. Clin. Vaccine Immunol. 151138-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Hoeven, N. S., and A. D. Miller. 2005. Use of different but overlapping determinants in a retrovirus receptor accounts for non-reciprocal interference between xenotropic and polytropic murine leukemia viruses. Retrovirology 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wildum, S., M. Schindler, J. Munch, and F. Kirchhoff. 2006. Contribution of Vpu, Env, and Nef to CD4 down-modulation and resistance of human immunodeficiency virus type 1-infected T cells to superinfection. J. Virol. 808047-8059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang, B., S. Jin, J. Jin, F. Li, and R. C. Montelaro. 2005. A tumor necrosis factor receptor family protein serves as a cellular receptor for the macrophage-tropic equine lentivirus. Proc. Natl. Acad. Sci. USA 1029918-9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang, B., C. Sun, S. Jin, M. Cascio, and R. C. Montelaro. 2008. Mapping of equine lentivirus receptor 1 residues critical for equine infectious anemia virus envelope binding. J. Virol. 821204-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]