Abstract
The env gene of gammaretroviruses encodes a glycoprotein conserved among diverse retroviruses, except for the domains involved in receptor binding. Here we show that pairs of gammaretrovirus envelope proteins (from Friend virus and GALV or xenotropic viruses) assemble into heteromers when coexpressed. This assembly results in a strong inhibition of infectivity. An unrelated envelope protein does not assemble in heteromers with the gammaretrovirus glycoproteins tested and does not affect their infectivity, demonstrating the specificity of the mechanism we describe. We propose that the numerous copies of endogenous retroviral env genes conserved within mammalian genomes act as restriction factors against infectious retroviruses.
Retroviruses are a group of relatively simple viruses that share the same general organization and contain three major genes (gag, pol, and env) that are homologous among members of a given genus (reviewed in reference 19). The env gene encodes an envelope glycoprotein that is expressed at the surface of the viral particles and is responsible for the tropism of the virus through its interaction with a specific cellular receptor. This protein is a prime target of the host immune system, as it is the major antigen for the generation of neutralizing antibodies, and as a consequence of immune selection of escape mutants, it evolves particularly quickly. Despite this high evolution rate, its general structure has been conserved, consisting of a leader peptide followed by the surface subunit (SU) and transmembrane subunit (TM), which are processed from the full-length precursor in the Golgi apparatus (reviewed in reference 17). Among gammaretroviruses, these two subunits are remarkably well conserved, with most variations occurring in the N-terminal region of the SU, the receptor binding domain, which determines the specificity of cellular receptor usage (1, 2, 13, 20), and at the C-terminal end of the TM in the cytoplasmic tail, which is responsible for the intracellular trafficking of the envelope (Env) protein and its interaction with the Gag structural protein (15). The other regions of gammaretrovirus Env proteins are less divergent and correspond to domains important in the structure of the protein (SU-TM association and assembly into trimers that takes place during its synthesis) and in its conformational change upon interaction with the receptor, leading to the fusion of the viral and cellular membranes (reviewed in reference 11).
Here we demonstrate that the Env proteins encoded by several gammaretroviruses are sufficiently conserved to associate as heteromers when coexpressed in mammalian cells and that this association renders the Env proteins nonfunctional, resulting in a strong decrease in the viral titer. This mechanism provides a possible function for the endogenous retrovirus (ERV) env genes that are conserved and expressed in mammals (reviewed in references 3, 4, and 7). We propose that endogenous Env proteins act as restriction factors that inhibit infection by exogenous retroviruses through association with their envelope.
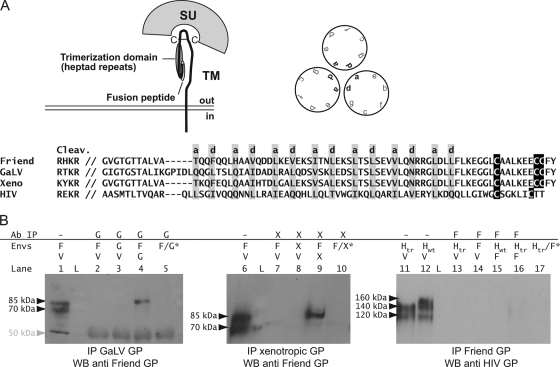
Except for the receptor binding domain, the env gene of gammaretroviruses is well conserved, with very few amino acid modifications in the ectodomain of the TM subunit, which is organized as a pair of α-helixes involved in the trimeric assembly of the protein within the cells (Fig. 1A) (8, 10). We thus asked whether this high homology may enable different but related Env proteins to interact with each other when expressed within the same cell. To test this hypothesis, we performed immunoprecipitation experiments on Cos cells transiently transfected with two gammaretrovirus Env proteins (from Friend virus and gibbon ape leukemia virus [GALV] and from Friend virus and a mouse endogenous xenotropic provirus [from C57BL/6 mice, chromosome 2; coordinates in Ensembl, 156182828 to 156184762]). Two days following transfection, the cells were lysed under stringent conditions (radioimmunoprecipitation assay buffer), and the lysates were immunoprecipitated using antibodies against GALV or xenotropic-virus proteins. The immunoprecipitates were then tested for the presence of Friend virus Env by Western blotting. As shown in Fig. 1B, we observed in both cases the presence of the Friend virus glycoprotein in the precipitated fraction. The different controls indicate that the signal we observed is not due to cross-recognition of the two retroviral Env proteins by the antibodies during the immunoprecipitation (Fig. 1B, lanes 2 and 7) or hybridization step (lanes 3 and 8). We also did not detect cross-immunoprecipitation of the Friend virus Env protein by the anti-GALV Env antibody (or by the anti-xenotropic-virus Env antibody) when cells were singly transfected with each expression vector and mixed before the lysis step (lanes 5 and 10). This demonstrates that the signal we detected is not an artifact due to aggregation of the Env proteins during the lysis step.
FIG. 1.
(A) Organization of the envelope protein of retroviruses. The trimerization is mediated by the heptad repeats localized in TM (interaction between residues a and d of the repeats), in a region particularly conserved among the envelope proteins encoded by distinct gammaretroviruses. A sequence alignment is shown for the glycoproteins from Friend virus, GALV, HIV, and the xenotropic virus (from the cleavage site between SU and TM to the conserved cysteine residues in TM). HIV does not belong to gammaretroviruses, and its glycoprotein is far less conserved in this domain; in its case, the alignment is structural (a and d residues) and is based on reference 3. (B) Detection by coimmunoprecipitation of heteromers formed by distinct retroviral glycoproteins when coexpressed within cells. Cos cells were cotransfected with expression vectors for two retroviral glycoproteins and lysed 2 days later. The cell lysates were subjected to immunoprecipitation using antiserum against one of the transfected glycoproteins (left, anti-GALV Env rabbit serum generated using a recombinant protein corresponding to SU; middle, rat monoclonal antibody 83A25, which reacts with the xenotropic-virus envelope proteins but not the Friend virus glycoprotein [9]; right, anti-Rauscher leukemia virus gp70 goat serum, which reacts with the Friend virus glycoprotein [obtained from the National Cancer Institute, Frederick, MD]) and protein G-Sepharose; the precipitated fractions were then tested for the presence of the second Env protein by Western blotting (for the Friend virus Env we used a rat antiserum generated against a recombinant protein corresponding to the 5′ half of SU; for the HIV glycoprotein we used rabbit polyclonal antibody ADP422 [MRC AIDS Directed Programme]). For each sample, the glycoproteins that were transfected are indicated above the gels, together with the antibodies used for the immunoprecipitation (F, Friend virus; G, GALV; V, vesicular stomatitis virus G protein, an unrelated glycoprotein used to normalize the total amount of transfected DNA in the cells; L, molecular weight ladder). *, cells were singly transfected with an expression vector for either the Friend virus or the GALV glycoprotein (lane 5), for either the Friend virus or the xenotropic-virus glycoprotein (lane 10), or for either the Friend virus or the truncated HIV glycoprotein (lane 17) and mixed 48 h later, just before the lysis step preceding the immunoprecipitation. Lanes 1, 6, 11, and 12 correspond to samples collected before the immunoprecipitation step (total cell lysates) and were used as positive controls for the Western blot (the proportion of total cell lysate versus immunoprecipitation product is the same for the three gels, allowing quantitative comparison of the intensity of the signals obtained). The approximate size of the main products is indicated next to the gels. The expected masses (in kDa) for the glycoproteins are as follows (precursor/processed SU): F, 85/70; Hwt, 160/120; Htr, 140/120. In the left panel, the background band at 50 kDa is due to cross-reactivity with the heavy chain of the immunoglobulins used for the immunoprecipitation.
Finally, we checked the specificity of these interactions by determining whether we could coimmunoprecipitate an unrelated Env protein, the human immunodeficiency virus (HIV) glycoprotein, with an antiserum specific for the Friend virus Env when the two proteins are coexpressed. This experiment was performed with two variants of the HIV glycoprotein (described in reference 16): the native full-length protein (Hwt) and a mutant truncated in its cytoplasmic tail (Htr), which we used for the experiments described below. As illustrated in Fig. 1B (right), we detected no (or very little) HIV protein in the immunoprecipitates, indicating that the signal obtained in the presence of the two gammaretrovirus proteins was not due to partial lysis of the cells that would have resulted in the persistence, during immunoprecipitation, of small membrane vesicles harboring a mixture of two types of homotrimers, each made of one of the two Env proteins.
Last, it is noteworthy that the Friend virus Env protein we detected by Western blotting in the case of the Friend virus-xenotropic-virus and Friend virus-GALV heteromers migrated with an apparent molecular mass of approximately 85 kDa, which is the size of the major band observed in the cell lysates (Fig. 1B, lanes 1 and 6) and corresponds most probably to the unprocessed envelope precursor. This does not necessarily mean that the heteromers cannot be processed, since the lysis conditions we used were probably too harsh to preserve the interaction between the SU and TM of the mature forms of the Env proteins. Together, our immunoprecipitation results indicate that different gammaretrovirus Env proteins can interact and form heteromers when they are coexpressed within mammalian cells. This association is specific, since an unrelated lentiviral glycoprotein is not coprecipitated under the same conditions.
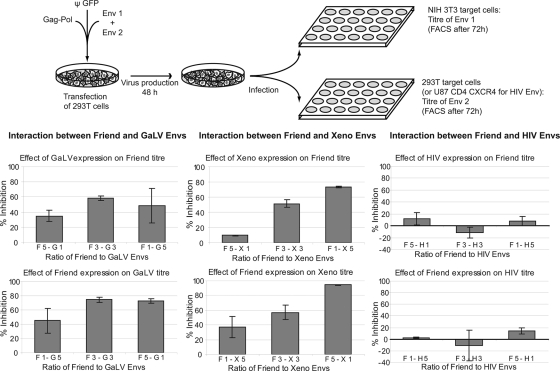
We then investigated whether these interactions could have functional consequences. We took advantage of the different tropism of the retroviral Env proteins tested in the immunoprecipitation experiments described above (reviewed in reference 14). We carried out a single infection cycle assay (scheme in Fig. 2), using nonreplicative retroviral pseudotypes generated by transient transfection of 293T cells with four plasmids: the expression vector for the Gag-Pol polyprotein, the green fluorescent protein-marked retroviral RNA expression vector, and expression vectors for the two Env proteins (or β-galactosidase in the case of the single Env pseudotypes) at three ratios. Since the two glycoproteins tested possess distinct tropisms, we could then measure the titer of each of them by assaying the virus-containing supernatants on two different target cells: murine NIH 3T3 cells for the Friend virus glycoprotein (since these cells cannot be infected by GALV or the xenotropic virus) and human 293T cells for the GALV and xenotropic-virus Env proteins (because these cells are resistant to infection by the Friend virus). As shown in Fig. 2 (left and middle), we observed a strong decrease (up to 80%) in the titer of the pseudotypes when two gammaretrovirus glycoproteins are coexpressed. The decrease is less when the “interfering” Env is expressed at a low level. However, in the case of the Friend virus and GALV proteins, the extent of the decrease does not correlate linearly with the ratios of the two proteins. This may be at least partially due to the cytotoxic effects caused by a high level of expression of the GALV Env protein on the cells (data not shown). In contrast and as actually expected, the extent of the decrease in titer observed when the Friend virus and xenotropic-virus proteins are coexpressed matches the ratios of expression of the two Env proteins, with an inhibition of infection ranging from 10 to 90%. These values are in the range of the theoretical maximal inhibition that can be calculated from the probability of formation of functional homotrimers at the different ratios we used (42.1%, 87.5%, and 99.5% inhibition). We then performed the same experiments using the Friend virus and HIV proteins (more precisely, the truncated variant of the latter, to allow its incorporation into the murine leukemia virus [MLV]-derived retroviral pseudotypes), replacing the 293T cells with U87 CD4 CXCR4 cells as a target to measure the HIV titer (Fig. 2, right). In contrast to the observations described above (using the Friend virus and the GALV or xenotropic-virus Env proteins), we found no reproducible effect of the coexpression of the HIV and Friend virus Env proteins on their respective titers. Since these two glycoproteins do not assemble into heteromers, as determined by immunoprecipitation experiments, this observation supports the proposal that heteromerization and the decrease in infection have a cause-effect relationship. Altogether, these experiments show that coexpression of two gammaretrovirus Env proteins specifically causes a decrease in the titer of the mixed pseudotypes, as measured for each of them on appropriate target cells.
FIG. 2.
Effect of coexpression of distinct retroviral glycoproteins on the titers of their respective retroviral pseudotypes. Retroviral pseudotypes were generated by a four-plasmid transfection of 293T cells, and the viral particles were collected 48 h later for titration (top; also, see the text for details). In addition to the retroviral proteins and marked viral vector, the cells (seeded in 6-cm dishes) were transfected with a total of 0.6 μg of Env1 and/or Env2 expression vectors for the retroviral Env proteins or of lacZ control plasmid (Env1 + Env2, Env1 + lacZ, lacZ + Env2), at three ratios: 0.1 μg + 0.5 μg, 0.3 μg + 0.3 μg and 0.5 μg + 0.1 μg. The decrease in infectivity observed when a second glycoprotein is expressed, compared to the control vector (encoding β-galactosidase), is represented on the histograms, with the standard deviation indicated (F, Friend virus; G, GALV; X, xenotropic virus; numbers refer to the amount of each Env protein). The experiment was performed with three pairs of glycoproteins, as indicated (in the Friend virus-HIV interaction, the HIV glycoprotein is the truncated variant so as to enable its incorporation on MLV-derived particles), the titers of which can be measured independently using appropriate target cells (see the text). The viral titers (105 IU per ml) for each glycoprotein expressed alone at the three doses (0.1, 0.3, and 0.5 μg DNA) were as follows: Friend virus envelope, 1.93, 4.00, and 4.61; GALV envelope, 2.01, 7.81, and 5.15; xenotropic-virus envelope, 3.91, 8.98, and 10.71; Htr, 0.51, 0.98, 0.74.
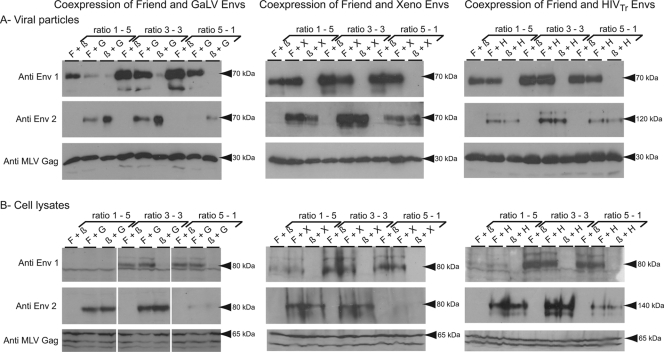
To examine the mechanism of this decrease in titer, we analyzed the protein content of the viral particles produced in these experiments. The viral particles from the supernatants of transfected cells were concentrated by ultracentrifugation before analysis by Western blotting. As shown in Fig. 3 (left), coexpression of the Friend virus and GALV Env proteins leads to the production of viral particles containing a smaller amount of each protein, whereas the same Western blot analysis performed on the transfected cell lysates does not show any decrease in the intracellular content of each of them. We are thus led to conclude that the decrease in infectivity is related to the lower level of incorporation of both Env proteins on the particles. This defect could be a consequence of the assembly of the two glycoproteins into heteromers leading either to misfolding or to inappropriate subcellular localization, thus preventing their proper incorporation on the particles. Rather surprisingly, the same analyses performed after coexpression of the Friend virus and xenotropic-virus glycoproteins gave opposite results, the content of the viral particles being unaffected by the expression of the interfering Env protein (Fig. 3, middle). However, the Friend virus and xenotropic-virus proteins are much more closely related to one another than to the GALV protein, and it is possible that the heteromers they form are more stable and can thus be efficiently incorporated on viral particles, even though they are not functional for virus entry and infection. Finally, the coexpression of the Friend virus and HIV proteins affected neither their incorporation on the viral particles nor their intracellular expression level, in agreement with their inability to assemble as heteromers and with the absence of any decrease in titer when they are coexpressed.
FIG. 3.
Effects of cotransfection with two retroviral glycoproteins on their respective expression. Retroviral pseudotypes containing one or two retroviral Env proteins were generated as described for Fig. 2, using the same ratios, and the level of expression of each Env protein was analyzed by Western blotting. (A) Analysis was performed on viral particles obtained from the cell supernatants (collected 48 h posttransfection) by ultracentrifugation on a 20% sucrose cushion. (B) The virus-producing cells were lysed 48 h after transfection for analysis of the retroviral glycoproteins in whole-cell extracts. Env1, Friend virus envelope; Env2, GALV envelope (left), xenotropic-virus envelope (middle), or HIV envelope (right), using rat polyclonal antiserum for Friend virus and rabbit polyclonal antiserum for GALV (see the legend to Fig. 1), using mouse polyclonal antiserum generated against a recombinant protein corresponding to the 5′ part of SU for the xenotropic-virus glycoprotein, and using rabbit polyclonal antiserum for HIV. The anti-MLV Gag antibody (rat anticapsid monoclonal antibody obtained from the American Type Culture Collection [CRL-1912]) was used as a loading and transfection control. The approximate sizes of the main products are given next to the gels. The expected sizes of the proteins (in kDa) are as follows (precursor/processed): Friend virus envelope, 85/70; GALV envelope, 85/70; xenotropic-virus envelope, 85/70; Hwt, 160/120; Htr, 140/120; MLV Gag, 65/30.
In this study we have shown that distinct gammaretrovirus Env proteins can physically associate as heteromers when they are coexpressed in mammalian cells. This association does not seem to alter the expression of the glycoproteins, but it can be correlated with a decrease in the titer measured for each Env protein. Interestingly, such heterotypic interactions have been previously suspected, with the finding that the coexpression of the native 4070A amphotropic envelope and a mutated feline leukemia virus A (FeLV-A) glycoprotein can modify the tropism of the resulting viral particles, enabling them to use a new receptor, probably because the interaction modifies the shape of the receptor binding domain of the 4070A envelope (6). In that study, it was also found that the 4070A Env had a positive effect on the mutant FeLV-A protein, rescuing its cellular localization and helping its incorporation on the viral particles. In our experiments, we found that coexpression had either no effect (in the case of the Friend virus and xenotropic-virus Env) or a negative effect (in the case of the Friend virus and GALV Env) on the incorporation on the particles. However, in the study by Bupp et al., the mutant FeLV-A was generated by inserting a short peptide into the SU subunit, thus probably dramatically altering its structure and destabilizing the Env protein, which could be partially rescued via its association with the 4070A Env (6). In our case, both proteins are native and presumably optimal with regard to expression and stability, and heteromerization can probably only lead to a decrease in the stability of the protein complex, explaining the opposite effects on the incorporation on the viral particles detected between the two studies.
We were able to demonstrate heterotypic interactions between the Env proteins isolated from infectious retroviruses but also between the glycoproteins encoded by an exogenous retrovirus and an endogenous one. If the observation of the interactions between the Friend virus and GALV proteins is not directly relevant to retrovirus biology (the former is of murine origin whereas the latter was isolated from an ape), the pair constituted by the Friend virus and the xenotropic retrovirus Env proteins may reflect a natural situation, since the Friend retrovirus naturally infects mice and most mouse strains possess in their genome one or several copies of xenotropic endogenous retrovirus env genes (reviewed in reference 4). It is thus likely that the two envelope proteins can be coexpressed within the same host. Indeed, this observation suggests a possible role that may account for the conservation of xenotropic retroviruses—by definition unable to infect murine tissues—in the mouse genome. Even if their Env protein cannot prevent the initial stage of infection by exogenous mouse retroviruses (such as the Friend virus) as typical restriction factors do, its expression by infected cells may reduce the production of infectious particles and thus help the immune system to clear or at least contain the infection by lowering the viral load. In fact, this model is consistent with a previous study in which a locus containing an expressed xenotropic-virus endogenous envelope was genetically linked to resistance against a polytropic recombinant Friend retrovirus in the C57BL/10 mouse strain (5). Such a role has also already been postulated for the Fv4 gene, a bona fide mouse restriction factor expressing a mutated nonfunctional retroviral Env protein which acts mainly by interacting with the ecotropic receptor and prevents its expression at the membrane of the cells, thus making it unavailable for infectious retroviruses (12, 18). However, it was also suggested that the Fv4 protein can also act at a later stage in the infection cycle, via a different mechanism, independently of receptor binding, by decreasing the amount of infectious viral particles produced by the infected cells in vivo (12, 18). Our study gives credence to this proposal by providing a molecular scenario through which it could happen. We further demonstrate that the envelope protein does not need to be mutated (as is Fv4) but that indeed any endogenous retroviral envelope might play a restriction factor-like role when confronted with a cognate infectious retrovirus. This mechanism provides a new insight into the complicated interplay between endogenous and infectious retroviruses, with the first being a reservoir of sequences that can be at the same time used for the generation of new, more efficient viruses via recombination (as observed in mice or in cats [reviewed in reference 4]) but also recruited as restriction factors able to prevent or limit the infection of the host.
Acknowledgments
We thank Valerie Bosch for the expression vectors encoding the wild-type and truncated forms of HIV glycoproteins, Benjamin Chain for the U87 CD4 CXCR4 cell line, François-Loïc Cosset for the expression vector encoding GALV glycoprotein, Thierry Heidmann for the expression vector encoding the Friend virus envelope protein, and Yasuhiro Takeuchi for critical reading of the manuscript and helpful discussions.
We have no competing interests.
Footnotes
Published ahead of print on 30 July 2008.
REFERENCES
- 1.Battini, J. L., O. Danos, and J. M. Heard. 1995. Receptor-binding domain of murine leukemia virus envelope glycoproteins. J. Virol. 69713-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Battini, J. L., J. M. Heard, and O. Danos. 1992. Receptor choice determinants in the envelope glycoproteins of amphotropic, xenotropic, and polytropic murine leukemia viruses. J. Virol. 661468-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benit, L., P. Dessen, and T. Heidmann. 2001. Identification, phylogeny, and evolution of retroviral elements based on their envelope genes. J. Virol. 7511709-11719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boeke, J. D., and J. P. Stoye. 1997. Retrotransposons, endogenous retroviruses, and the evolution of retroelements, p. 343-435. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, New York, NY. [PubMed]
- 5.Buller, R. S., K. Wehrly, J. L. Portis, and B. Chesebro. 1990. Host genes conferring resistance to a central nervous system disease induced by a polytropic recombinant Friend murine retrovirus. J. Virol. 64493-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bupp, K., A. Sarangi, and M. J. Roth. 2006. Selection of feline leukemia virus envelope proteins from a library by functional association with a murine leukemia virus envelope. Virology 351340-348. [DOI] [PubMed] [Google Scholar]
- 7.de Parseval, N., and T. Heidmann. 2005. Human endogenous retroviruses: from infectious elements to human genes. Cytogenet. Genome Res. 110318-332. [DOI] [PubMed] [Google Scholar]
- 8.Einfeld, D., and E. Hunter. 1988. Oligomeric structure of a prototype retrovirus glycoprotein. Proc. Natl. Acad. Sci. USA 858688-8692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans, L. H., R. P. Morrison, F. G. Malik, J. Portis, and W. J. Britt. 1990. A neutralizable epitope common to the envelope glycoproteins of ecotropic, polytropic, xenotropic, and amphotropic murine leukemia viruses. J. Virol. 646176-6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fass, D., S. C. Harrison, and P. S. Kim. 1996. Retrovirus envelope domain at 1.7 angstrom resolution. Nat. Struct. Biol. 3465-469. [DOI] [PubMed] [Google Scholar]
- 11.Hunter, E. 1997. Viral entry and receptors, p. 71-119. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, New York, NY. [PubMed]
- 12.Ikeda, H., and T. Odaka. 1983. Cellular expression of murine leukemia virus gp70-related antigen on thymocytes of uninfected mice correlates with Fv-4 gene-controlled resistance to Friend leukemia virus infection. Virology 128127-139. [DOI] [PubMed] [Google Scholar]
- 13.Kim, F. J., N. Manel, E. N. Garrido, C. Valle, M. Sitbon, and J. L. Battini. 2004. HTLV-1 and -2 envelope SU subdomains and critical determinants in receptor binding. Retrovirology 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Overbaugh, J., A. D. Miller, and M. V. Eiden. 2001. Receptors and entry cofactors for retroviruses include single and multiple transmembrane-spanning proteins as well as newly described glycophosphatidylinositol-anchored and secreted proteins. Microbiol. Mol. Biol. Rev. 65371-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sandrin, V., D. Muriaux, J. L. Darlix, and F. L. Cosset. 2004. Intracellular trafficking of Gag and Env proteins and their interactions modulate pseudotyping of retroviruses. J. Virol. 787153-7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schnierle, B. S., J. Stitz, V. Bosch, F. Nocken, H. Merget-Millitzer, M. Engelstadter, R. Kurth, B. Groner, and K. Cichutek. 1997. Pseudotyping of murine leukemia virus with the envelope glycoproteins of HIV generates a retroviral vector with specificity of infection for CD4-expressing cells. Proc. Natl. Acad. Sci. USA 948640-8645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly and processing of viral proteins, p. 263-334. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, New York, NY. [PubMed]
- 18.Taylor, G. M., Y. Gao, and D. A. Sanders. 2001. Fv-4: identification of the defect in Env and the mechanism of resistance to ecotropic murine leukemia virus. J. Virol. 7511244-11248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vogt, V. M. 1997. Historical introduction to the general properties of retroviruses, p. 1-25. In J. M. Coffin, C. L. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, New York, NY. [PubMed]
- 20.Zhang, Y., J. C. Rassa, M. E. deObaldia, L. M. Albritton, and S. R. Ross. 2003. Identification of the receptor binding domain of the mouse mammary tumor virus envelope protein. J. Virol. 7710468-10478. [DOI] [PMC free article] [PubMed] [Google Scholar]