Abstract
The inability to grow human noroviruses in cell culture has greatly impeded the studies of their pathogenesis and immunity. Vesiviruses, in the family Caliciviridae, grow efficiently in cell culture and encode a unique protein in the subgenomic region designated as leader of the capsid protein (LC). We hypothesized that LC might be associated with the efficient replication of vesiviruses in cell culture and promote the replication of human norovirus in cells. To test this hypothesis, a recombinant plasmid was engineered in which the LC region of feline calicivirus (FCV) was placed under the control of the cytomegalovirus promoter (pCI-LC) so that the LC protein could be provided in trans to replicating calicivirus genomes bearing a reporter gene. We constructed pNV-GFP, a recombinant plasmid containing a full-length NV genome with a green fluorescent protein (GFP) in the place of VP1. The transfection of pNV-GFP in MVA-T7-infected cells produced few GFP-positive cells detected by fluorescence microscopy and flow cytometry analysis. When pNV-GFP was cotransfected with pCI-LC in MVA-T7-infected cells, we observed an increase in the number of GFP-positive cells (ca. 3% of the whole-cell population). Using this cotransfection method with mutagenesis study, we identified potential cis-acting elements at the start of subgenomic RNA and the 3′ end of NV genome for the virus replication. We conclude that LC may be a viral factor which promotes the replication of NV in cells, which could provide a clue to growing the fastidious human noroviruses in cell culture.
Caliciviruses are plus-strand RNA viruses in the family Caliciviridae that consists of four genera: Norovirus, Sapovirus, Lagovirus, and Vesivirus. Viruses in the genera Norovirus and Sapovirus cause gastroenteritis in humans and animals. Recent studies estimate that noroviruses are responsible for more than 90% of nonbacterial gastroenteritis outbreaks (7) and as many as 23 million cases of gastroenteritis in the United States each year (21). Norwalk virus (NV) is a prototype strain of the noroviruses and was associated with an outbreak of gastroenteritis in Norwalk, OH, in 1968 (16). The viral genome of caliciviruses is 7 to 8 kb in length and organized into either two or three open reading frames (ORFs) that encode a polyprotein of approximately 1,800 amino acids (aa), the major capsid protein (VP1), and a minor capsid protein (VP2) (11, 14, 19, 33). The first known calicivirus, vesicular exanthema swine virus (Vesivirus), was initially recognized in 1932. Vesicular exanthema swine virus may have arisen in swine from feed containing marine mammals infected with San Miguel sea lion virus, which causes vesicular lesions and reproductive failure in sea lions (23). The genus Vesivirus includes feline caliciviruses (FCV), which is a major respiratory virus in cats and causes persistent respiratory infection (10). The vesiviruses have been efficiently isolated in cell culture and provided a surrogate system for studying replication of the fastidious human enteric caliciviruses. Interestingly, vesiviruses encode a unique protein in the subgenomic region designated leader of the capsid protein (LC), and the expression of LC is a part of the LC-VP1 precursor that is processed to each mature protein by viral proteinase (Pro) (27).
Although recent studies have presented evidence for the replication of human noroviruses in cells (1, 12, 29), studies of the replication of these viruses have been severely hampered by the lack of a reliable cell culture system (6). Among the noroviruses, only murine noroviruses (including MNV-1) (17) have been successfully propagated in cell culture (30). Murine noroviruses are present widely in laboratory mouse colonies without apparent clinical symptoms (13, 31). MNV-1 has a tissue tropism of macrophage-like cells in vivo and in vitro (30), but it is not clear if human noroviruses target such cells. In the present study, we hypothesized that LC might be associated with the efficient replication of vesiviruses in cell culture and as a consequence may promote human norovirus replication in cells. We constructed a recombinant plasmid containing cDNA encoding the LC from FCV under the control of the cytomegalovirus (CMV) promoter (pCI-LC) to test this hypothesis. Previously, we reported an NV replicon system (NV replicon-bearing cells) using a recombinant plasmid containing a full-length NV genome (pNV101) (8) with a neomycin resistance gene (Neor) in the place of VP1 (pNV-Neo) (3, 5). Using a similar strategy, we constructed pNV-GFP, where Neor was replaced by a gene encoding green fluorescent protein (GFP). We also constructed a similar plasmid with GFP inserted into the ORF2 of the infectious clone of FCV (pQ14), pQ-GFP, for comparative studies. Cotransfection studies with pNV101, pNV-GFP, pQ14, or pQ-GFP with pCI-LC showed that LC promoted the replication of both FCV and NV in cells. Furthermore, we found that this cotransfection method was useful in the identification of cis-acting elements for NV replication in cells, which provides a novel tool for studying RNA elements. Using this system, we identified RNA elements at the start of the subgenomic RNA and 3′ end of NV genome essential for virus replication.
MATERIALS AND METHODS
Cells and reagents.
BHK21, HEK293T, and Vero cells were maintained in Dulbecco minimal essential medium containing 10% fetal bovine serum and antibiotics (chlortetracycline [25 μg/ml], penicillin [250 U/ml], and streptomycin [250 μg/ml]). Crandell-Ress feline kidney (CRFK) cells were maintained in minimal essential medium containing 5% fetal bovine serum and antibiotics. Polyclonal antibodies to NV VP1 or FCV VP1 have been described previously (24). We also used a polyclonal antibody to FCV LC that was described previously (25). Monoclonal antibodies to NV VP1 were purchased from Chemicon (Temecula, CA).
Plasmid construction and generation of region-specific antisera.
Standard recombinant DNA methods were used for the construction of plasmids. The consensus full-length clone of NV, pNV101, has been described previously (8). We reported pNV-Neo, which is used for generating NV replicon-bearing cells based in BHK21 or Huh-7 cells (5). We generated a similar reporter plasmid based on NV101 with a gene encoding GFP, pNV-GFP, to study the replication of NV (Fig. 1). For the construction of pNV-GFP, the GFP gene was amplified from the pAcGFP1 vector (Clontech, Mountain View, CA) with the primers GFP-NV-F (aattggatccTATGGTG AGCAAGGGCGAGGA) and GFP-NV-R (aattaccggtTCAAGCTCGAGATCTGAGTC) (the italic bases in the primers indicate the start and stop codons of GFP). The PCR product was digested with BamHI and AgeI (the sites underlined in the primers) and cloned into the corresponding sites of pNV-Neo. The resulting construct contained the GFP gene engineered into the 5′-end region of the ORF2 so that the expressed product would contain the first 33 aa of the VP1 fused in frame with GFP (Fig. 1B). Similarly, we constructed pNV-RL (for Renilla luciferase) after the Renilla luciferase was amplified with primers NV-Bam-Luc-F (ataaggatccTATGGAAGACGCCAAAAAC) and NV-Age-Luc-R (aattaccggtTTACAATTTGGACTTTCCG), using pRL-CMV (Promega, Madison, WI) as a template. The viral polymerase (Pol) active site, GDD in pNV101, pNV-GFP, or pNV-RL, was deleted by using a site-directed mutagenesis kit (Stratagene, La Jolla, CA) and the primers N-ΔGDD-F (CATGTCATATTTCTCATTTTATGAGATTGTGTCAACTGACATAG) and N-ΔGDD-R (CTATGTCAGTTGACACAATCTCATAAAATGAGAAATATGACATG), generating the pNV101ΔGDD, pNV-GFPΔGDD, or pNV-RLΔGDD plasmids, respectively (underlined bases indicate the position of the deletion of nine bases encoding GDD between them). To examine transfection efficiency, we generated a plasmid expressing chimeric VP1(33aa)-GFP fused to the end of NV ORF1 by deleting a base (G) at position 5354 in pNV-GFP by mutagenesis. The primers used for this deletion were M5354DG-F (CTTCTGCCCGAATTCTAAATGAT GATGGCG) and M5354DG-R (CGCCATCATCATTTAGAATTCGGGCAGAAG) (underlined bases indicate the position of the deletion of G between them). Transfection of this plasmid yields expression of GFP, as a part of viral Pol-VP1(33aa)-GFP (unpublished observation). This plasmid was used to measure the transfection efficiency for the expression of NV ORF1 in MVA-T7-infected cells. We also generated pCI-GFP, which produces the expression of GFP under the CMV promoter control to measure overall transfection efficiency. The GFP gene was amplified by PCR using pAcGFP1 as a template with the primers GFP-Xho-F (aattctcgagATGGTGAGCAAGGGCGAGGA) and GFP-Not-R (aattgcggccgcTCAAGCTCGAGATCTGAGTC). The amplicon was cloned into pCI vector using the enzyme sites for XhoI and NotI.
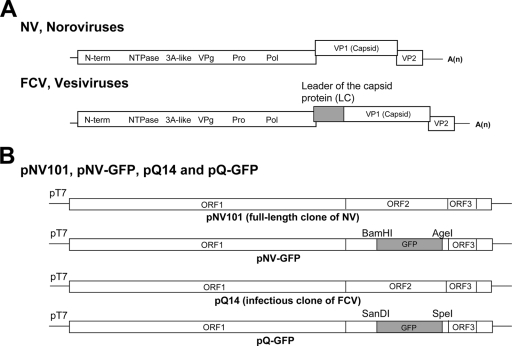
FIG. 1.
Genomic organization of NV, FCV, and recombinant plasmids based on the full-length genome of NV or FCV. (A) Schematic diagram of the genome organization of NV and FCV. FCV has a unique protein, LC, at the start of the subgenomic RNA. (B) Recombinant plasmids of pNV101 (a plasmid containing the full-length genome of NV under the T7 promoter): pNV-GFP, pQ14 (an infectious clone of FCV), and pQ-GFP. pNV-GFP was generated using unique sites of BamHI and AgeI in ORF2 of the NV genome, and GFP gene was cloned in place of VP1. pQ-GFP was generated using the unique restriction sites SanDI and SpeI in ORF2 of the FCV genome, and the GFP gene was cloned in the place of LC and VP1.
For comparative studies, we also generated pQ-GFP based on the infectious clone of FCV Urbana strain, pQ14 (24). We utilized the enzyme sites of SanDI and SpeI in the ORF2 of FCV to insert GFP. Briefly, the GFP gene was amplified from the pAcGFP1 vector (Clontech) with the primers GFP-FCV-F (aattgggacccCATGGTGAGCAAGGGCGAGGA) and GFP-FCV-R (aattactagtTCAAGCTCGAGATCTGAGTC). The PCR product was digested with SanDI and SpeI and cloned into the corresponding sites of the pQ14 plasmid. Also, the viral Pol active-site GDD in pQ14 or pQ-GFP was deleted by using the site-directed mutagenesis kit with the primers FCV-ΔGDD-F (CGACATGATGACTTATGGTGTTTACATGTTTC) and FCV-ΔGDD-R (GAAACATGTAAACACCATAAGTCATCATGTCG), generating pQ14ΔGDD and pQ-GFPΔGDD, respectively.
To examine the effects of LC from FCV on the replication of FCV or NV, we generated a plasmid containing the LC gene under the control of CMV promoter (pCI-LC). For the pCI-LC, the region encoding LC in FCV was amplified with the primers FCVLC-Xho-F (aattctcgagATGTGCTCAACCTGCGCTAACG) and FCVLC-Not-R (aattgcggccgcTTATCATTCCAATCTGAACAATGGCA) by PCR, using pQ14 as a template. The amplicon was cloned into pCI vector using the enzyme sites of XhoI and NotI. Similarly, we generated a plasmid containing the V gene of simian virus 5 (SV5) under the control of the CMV promoter (pCI-V). The gene encoding the V protein in SV5 (DA strain) was amplified with the primers SV5V-Nhe-F (aattgctagcATGGATCCCACTGATCTGAGC) and SV5V-Not-R (aattgcggccgcTTAAGTATCTCGTTCACATTCAG) by reverse transcription-PCR (RT-PCR), using viral RNA as a template. The RT-PCR was performed using the following conditions: 45°C for 30 min (for RT) and 95°C for 10 min, followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 50°C for 1 min, and elongation at 72°C for 1 min. The amplicon was cloned into pCI vector using the enzyme sites of NheI and NotI. Also, we generated a plasmid, pNVP1 to express NV VP1 in cells for a control. The region encoding NV VP1 was amplified by PCR with the primers NVP1-Xho-F (aattctcgagATGATGATGGCGTCTAAGG) and NVP1-Not-R (aattgcggccgcTTATCGGCGCA GACCAAGC) (the italic bases in the primers indicate the start and stop codons of VP1) using pNV101 as a template. The amplicon was cloned into pCI vector using the enzyme sites XhoI and NotI.
Transfection study.
All transfections were performed with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) with 2 μg of each plasmid per well in six-well plates (ca. 1 × 106 to 2 × 106 cells). Cells including BHK21, HEK293T, or Vero cells were infected with the modified vaccinia virus (Ankara strain) expressing T7 Pol (MVA-T7) (32) at a multiplicity of infection of 10 for 1 h before transfection. The plasmid pNV101, pNV-GFP, pQ14, pQ-GFP, pNV101ΔGDD, pNV-GFPΔGDD, pQ14ΔGDD, or pQ-GFPΔGDD was transfected alone or cotransfected with pCI-LC or pCI-V to study the effects of LC or V in the replication of FCV or NV. The plasmid NV-GFP or pNV-GFPΔGDD was also cotransfected with pCI-LC and pCI-V to study the combined effects of LC and V on NV replication. Similarly, each mutant plasmid of pNV-GFP (described below) was cotransfected with pCI-LC and pCI-V into MVA-T7-infected Vero cells to study cis-acting elements of NV. After transfection, the replication of FCV or NV was monitored by detecting the expression of VP1 or GFP (GFP-based constructs) with the various assays described below. In addition, plasmid pNV-RL or plasmid pNV-RLΔGDD was transfected alone or with pCI-LC, pCI-V, or pCI-LC plus pCI-V into MVA-T7-infected Vero cells. For the control of this transfection, we used pCI and pNV-GFP. At 20 h posttransfection, cell lysates were prepared for Renilla luciferase activity using the Renilla luciferase assay system (Promega).
Virus replication assay.
Virus replication was measured by various assays including immunofluorescence assay (IFA), Western blot analysis, enzyme-linked immunosorbent assay (ELISA), plaque-forming assay (for FCV), and flow cytometry analysis.
(i) Detection of NV or FCV VP1.
The expression of NV or FCV VP1 was measured by IFA using antibody against NV virus-like particles (VLPs) or FCV virions. Cells transfected with various plasmids were fixed with cold methanol at 18 to 24 h posttransfection for IFA. Monoclonal antibody to NV VLPs or polyclonal antibody (guinea pig serum) to FCV virions were added to the fixed monolayers, and the binding of antibodies was detected with fluorescein isothiocyanate-conjugated, affinity-purified goat antibodies to mouse or guinea pig immunoglobulin G (Sigma-Aldridge, St. Louis, MO) as described previously (4). NV VP1 was also detected by Western blot analysis using polyclonal antibody to the protein as described previously (5). We also used ELISA for detecting NV VP1 after the transfections. Briefly, 96-well plates (Nunc, Rochester, NY) were coated with monoclonal antibody to NV VP1 as a capture antibody. Reagents were added to triplicate wells in the following sequence: cell lysate samples; hyperimmune guinea pig antiserum specific for NV VP1; goat anti-guinea pig immunoglobulin G conjugated to horseradish peroxidase; and the substrate tetramethylbenzidine (Kirkegaard & Perry Laboratories, Gaithersburg, MD). The absorbance was measured at 610 nm. The cutoff value was calculated as an absorbance at 610 nm that was at three standard deviations above the absorbance in cell control wells (cells infected with MVA-T7).
(ii) Detection of NV genome.
To examine NV genome levels in the cells with various transfections, real-time quantitative RT-PCR (qRT-PCR) was performed by using a One-Step Platinum qRT-PCR kit (Invitrogen) according to the protocol established for the analysis of genogroup 1 norovirus samples (15). The primers COG1F (CGYTGGATGCGNTTYCATGA) and COG1R (CTTAGACGCCATCATCATTYAC) and the probe RING1(a)-TP (FAM-AGATYGCGATCYCCTGTCCA-TAMRA) were used for the real-time qRT-PCR, which targets genomic RNA (sequence between positions 5291 and 5375) (15). As a quantity control of cellular RNA levels, qRT-PCR for β-actin with the primers actin-F (GGCATCCACGAAACTACCTT) and actin-R (AGCACTGTGTTGGCGTACAG) and the probe actin-P (HEX-ATCATGAAGTGTGACGTGGACATCCG-TAMRA) was performed as described previously (3, 28). For qRT-PCR, the total RNA of cells (in six-well plates) was extracted with an RNeasy kit (Qiagen, Valencia, CA). The qRT-PCR amplification was performed in a Cepheid SmartCycler with the following parameters: 45°C for 30 min and 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 30 s, annealing at 50°C for 1 min, and elongation at 72°C for 30 s. The relative genome levels in cells with various transfection were calculated after the RNA levels were normalized with those of β-actin.
(iii) Expression of GFP.
After the transfection of plasmids encoding the GFP gene, GFP-positive cells were detected by fluorescence microscopy or enumerated by using flow cytometry analysis. After 16 to 20 h of transfection, the cells were treated with trypsin, fixed with 4% formaldehyde for 1 h, and then washed twice with phosphate-buffered saline by centrifugation and resuspended with phosphate-buffered saline. Flow cytometry analysis was performed on a population of 10,000 cells by using a FACSCalibur (BD Biosciences, Franklin Lakes, NJ).
Mutagenesis of potential cis-acting elements in pNV-GFP.
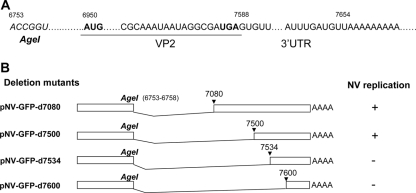
The potential promoter or cis-acting elements located at the start of subgenomic RNA or the 3′ end in NV genome was examined by substitution and deletion mutagenesis of pNV-GFP (see Fig. 6A and 7). First, we generated a series of substitute mutants at the start of the subgenomic RNA focusing on four untranslated bases, GUAA, using the primers listed in Table 1. The substitution mutants at the first base (G) in the subgenomic RNA to U, A, or C were designated as pNV-GFP-G5354U, pNV-GFP-G5354A, or pNV-GFP-G5354C, respectively. These mutations resulted in amino acid changes in the sequences of the virus Pol from valine to leucine (pNV-GFP-G5354U) or isoleucine (pNV-GFP-G5354A and pNV-GFP-G5354C). The second base (U) in the subgenomic RNA was also mutated to A, C, or G with the plasmids designated pNV-GFP-U5355A, pNV-GFP-U5354C, or pNV-GFP-U5354G, yielding amino acid changes from valine to glutamic acid, alanine, or glycine, respectively. The third base (A) was mutated to U, C, or G with the plasmids designated pNV-GFP-A5356U, pNV-GFP-A5356C, or pNV-GFP-A5356G (all were silent mutations). Mutation of the fourth base (A) to U, C, or G yielded plasmids designated pNV-GFP-A5357U, pNV-GFP-A5357C, or pNV-GFP-A5357G. These mutations changed the asparagine to tyrosine, histidine, or aspartic acid, respectively. Second, to examine the RNA elements at the 3′ end of NV genome for virus replication, we generated a series of deletion mutations targeting the regions between the stop codon of GFP and the end of genome (Fig. 7). Nine deletion mutants were generated between the bases after the GFP gene, and the deletions corresponded to bases 7080, 7089, 7480, 7500, 7534, 7600, 7610, 7630, and 7640 (Fig. 7). These mutations were generated by inserting an AgeI site at the desired bases using the mutagenesis kit and primers listed in Table 1. The plasmids containing the mutants were digested with AgeI, which removed the deletion fragments and then they were religated to generate each mutant plasmid. The presence of the mutation for all plasmids described above was confirmed by sequencing analysis.
FIG. 6.
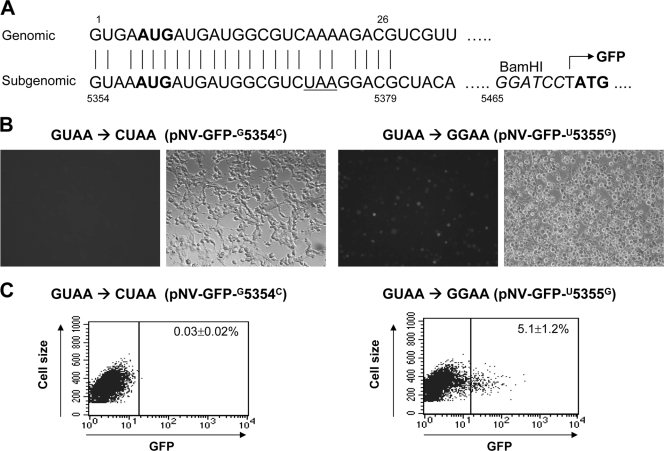
Mutagenesis study for the potential promoter activity at the start of subgenomic RNA of NV genome. (A) Conserved bases at the start of NV genome and subgenomic RNA. The first 26 conserved bases contain the start codons (boldface) for ORF1 and VP1 and the stop codon for ORF1 (underlined). In pNV-GFP, the GFP gene was inserted in place of VP1 using the BamHI site (positions 5465 to 5470). (B) GFP-positive cells observed under a fluorescent (left panel) or a light microscope (right panel) after Vero cells were cotransfected with pNV-GFP-G5354C or pNV-GFP-U5355G and pCI-LC plus pCI-V (after MVA-T7 infection). (C) Flow cytometry analysis of Vero cells were cotransfected with pNV-GFP-G5354C or pNV-GFP-U5355G and pCI-LC plus pCI-V (after MVA-T7 infection).
FIG. 7.
Mutagenesis study for cis-acting elements in 3′-end region of NV genome. (A) Schematic diagram of the 3′-end region of NV genome including ORF2, ORF3, and 3′UTR. The enzyme site of AgeI is located at positions 6753 to 6758 in ORF2, and there are 66 bases in the 3′UTR between the stop codon of VP2 (ORF3) and the poly(A) tail. (B) Summary of deletion mutants targeting ORF2, ORF3, and 3′UTR and GFP expression after cotransfection of each mutant, pCI-LC and pCI-V into MVA-T7-infected Vero cells.
TABLE 1.
Primers used in the mutagenesis study
| Primer | Designation | Sequence (5′ to 3′)a |
|---|---|---|
| M5354RC-F | G5354C | CTTCTGCCCGAATTCCTAAATGATGATGGCG |
| M5354RA-F | G5354A | CTTCTGCCCGAATTCATAAATGATGATGGCG |
| M5354RT-F | G5354U | CTTCTGCCCGAATTCTTAAATGATGATGGCG |
| M5355RA-F | U5355A | CTTCTGCCCGAATTCGAAAATGATGATGGCG |
| M5355RC-F | U5355C | CTTCTGCCCGAATTCGCAAATGATGATGGCG |
| M5355RG-F | U5355G | CTTCTGCCCGAATTCGGAAATGATGATGGCG |
| M5356RT-F | A5356U | CTTCTGCCCGAATTCGTTAATGATGATGGCGTC |
| M5356RC-F | A5356C | CTTCTGCCCGAATTCGTCAATGATGATGGCGTC |
| M5356RG-F | A5356G | CTTCTGCCCGAATTCGTGAATGATGATGGCGTC |
| M5357RT-F | A5357U | TTCTGCCCGAATTCGTATATGATGATGGCGTCTA |
| M5357RC-F | A5357C | TTCTGCCCGAATTCGTACATGATGATGGCGTCTA |
| M5357RG-F | A5357G | TTCTGCCCGAATTCGTAGATGATGATGGCGTCTA |
| M7080Age-F | D to 7080 | CAAAGCCAAAGGTATCACCGGTATTTGCAACTGCAAG |
| M7280Age-F | D to 7280 | CTGCTCCCGAGTCCACCGGTACCACATTGAGATCCG |
| M7480Age-F | D to 7489 | GCGGAGGCTCTCAATACCGGTTGGTTGACTCCACCC |
| M7534Age-F | D to 7534 | CTACACTGTCTTCTGTACCGGTTGGTTATTTCAATACAG |
| M7500Age-F | D to 7500 | TGGTTGACTCCAACCGGTTCAACAGCCTCTTCTAC |
| M7600Age-R | D to 7600 | CGCAAATAATAGGCACCGGTGTTGTAATATGAAATGTGG |
| M7610Age-F | D to 7610 | GTGGGCATCATATTCACCGGTTTAGGTTTAATTAGG |
| M7630Age-F | D to 7630 | CATCATATTCATTTAATTACCGGTAATTAGGTTTAATTTG |
| M7640Age-F | D to 7640 | CATTTAATTAGGTTACCGGTGGTTTAATTTGATG |
The sequences listed in the table represent the forward primers only. Underlined bases are mutated bases in the primers. Italic bases are AgeI recognition sites in the primers.
Statistical analysis.
All experiments, including flow cytometry, Renilla luciferase assay, IFA, and ELISA, were conducted with at least three independent measurements, and the effects of LC protein, V protein, and the combination of LC protein plus V protein on FCV or NV replication were analyzed by using the Student t test. The results were considered statistically significant when the P value was <0.05.
RESULTS
The expression of LC in cells promoted the replication of FCV.
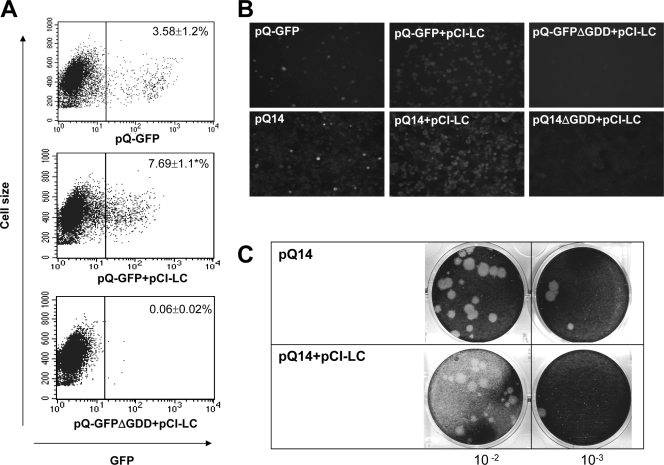
The expression of LC in various cell lines (BHK21, HEK293T, and Vero cells) after the transfection of pCI-LC with MVA-T7 infection showed that more than 50% of the cells were positive with LC as detected by IFA. Also, the transfection of pCI-GFP showed similar rates of cells expressing GFP in each cell type. We first examined the effects of LC in FCV replication using pQ14 and pQ-GFP, which served as the framework of the system for studying the replication of NV. The transfection of pQ14 or pQ-GFP into MVA-T7-infected BHK21, HEK293T, and Vero cells resulted in the cells expressing VP1 or GFP, respectively. When we enumerated cells expressing GFP after the transfection of pQ-GFP in Vero cells, <4% of the whole population were GFP positive (Fig. 2A). The overall proportions of cells expressing GFP or VP1 (detected by IFA) after transfection of pQ14 or pQ-GFP were similar (Fig. 2B). Cells transfected with pQ14 produced viable FCV, which indicated complete replication in these cell lines (Fig. 2). For example, the titers of viral progeny after transfections of pQ14 in Vero cells were between 103 and 104 PFU/ml (Fig. 2C). The transfection of RNA transcripts derived from pQ14 or pQ-GFP showed lower percentages (<1%) of cells expressing VP1 or GFP, respectively (unpublished observations).
FIG. 2.
Transfection of pQ14 or pQ-GFP with or without pCI-LC into MVA-T7-infected Vero cells. (A) Flow cytometry analysis of cells transfected with pQ-GFP alone (upper panel), cotransfected with pQ-GFP and pCI-LC (middle panel), or cotransfected with pQ-GFPΔGDD and pCI-LC (bottom panel) into MVA-T7-infected Vero cells. In all flow cytometry analyses, cells were collected after 18 to 20 h of transfection, and the figures represent the counts of 10,000 cells with cell size (y axis) and GFP expression (x axis). The numbers in the panel represent the averages and standard deviations of at least three independent experiments in all figures. An asterisk (*) indicates that the number of GFP-expressing cells by cotransfection with pQ-GFP and pCI-LC was significantly higher (P < 0.05) than the transfection with pQ-GFP alone. (B) In the upper panel, GFP-positive cells were observed under a fluorescence microscope after Vero cells were transfected with pQ-GFP alone (left panel), pQ-GFP plus pCI-LC (middle panel), or pQ-GFPΔGDD plus pCI-LC (right panel). In the bottom panel, IFA staining was performed with antibody against FCV VP1 after Vero cells were transfected with pQ14 alone (left panel), pQ14 plus pCI-LC (middle panel), or pQ14ΔGDD plus pCI-LC (right panel). (C) Plaque-forming assay of recovered progeny viruses after Vero cells were transfected with pQ14 alone (upper panel) or pQ14 plus pCI-LC (bottom panel).
When pQ14 or pQ-GFP was cotransfected with pCI-LC in Vero cells after MVA-T7 infection, the cells expressing VP1 or GFP increased to ∼2-fold over those with pQ14 or pQ-GFP transfection alone (Fig. 2A and B). The number of GFP expressing cells by the cotransfection with pCI-LC and pQ-GFP was significantly higher (P < 0.05) than the transfection with pQ-GFP alone. Although the cotransfection increased the number of GFP-positive cells, the overall intensity of GFP expression in each cell was reduced by the cotransfection. In Fig. 2A, the distribution of GFP-expressing cells leaned toward to the cutoff line by the cotransfection with pQ-GFP and pCI-LC (middle panel) compared to the transfection with pQ-GFP alone (upper panel). The virus titers after the transfection of pQ14 with or without pCI-LC in Vero cells were similar to each other, yielding approximately 2 × 103 PFU/ml (Fig. 3C). Transfection of pQ14ΔGDD or pQ-GFPΔGDD with or without pCI-LC did not produce VP1- or GFP-positive cells above the level of the control MVA-T7-infected cells (Fig. 2A and B).
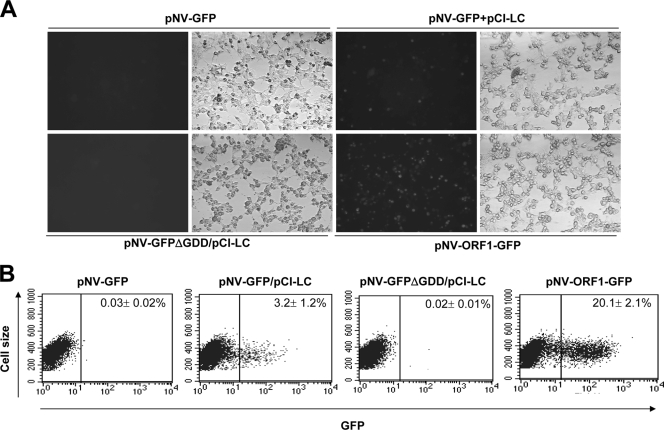
FIG. 3.
Transfection of pNV-GFP or pNV-GFPΔGDD with or without pCI-LC into MVA-T7-infected Vero cells. (A) GFP-positive cells observed under a fluorescent (left panel) or a light microscope (right panel) after cells were transfected with pNV-GFP with or without pCI-LC. Cotransfection of pNV-GFPΔGDD and pCI-LC served as a negative control, and transfection of pNV-ORF1-GFP serves as a measurement of transfection efficiency in the present study. (B) Flow cytometry analysis to enumerate GFP-positive cells after the transfection of pNV-GFP, pCI-GFP plus pCI-LC, pNV-GFPΔGDD plus pCI-LC, or pNV-ORF1-GFP into MVA-T7-infected Vero cells. The numbers in the panel represent the averages and standard deviations of at least three independent experiments in the figures.
In the presence of LC, the replication of NV was promoted in cells.
The transfection of pNV-GFP in BHK21, HEK293T, or Vero cells did not produce GFP-positive cells above the level of control MVA-T7-infected cells as measured by flow cytometry analysis (Fig. 3B), although under a fluorescence microscope, few GFP-positive cells (fewer than five cells) were observed in one well of a six-well plate. The transfection of pNV-ORF1-GFP, which served as a control for transfection efficiency in Vero cells infected with MVA-T7, produced ca. 20% GFP-positive cells by flow cytometry analysis (Fig. 3A and B). When pNV-GFP was cotransfected with pCI-LC into MVA-T7-infected Vero cells, we found that ca. 3% cells expressed GFP as detected by flow cytometry analysis (Fig. 3A and B). Cotransfection of the same plasmids in BHK21 or HEK293T cells also produced GFP-positive cells, but with a rate of ca. 1%. The transfection of pNV-GFPΔGDD with or without pCI-LC into MVA-T7-infected Vero, BHK21, and HEK293T cells did not yield GFP-positive cells by fluorescence microscopy or flow cytometry analysis (Fig. 3A and B). The transfection study with pNV-RL or pNV-RLΔGDD was consistent with the results of pNV-GFP or pNV-RLΔGDD, showing that while transfection of pNV-RL alone induced a slight increase in Renilla luciferase expression, the cotransfection with pCI-LC significantly increased (P < 0.01) the expression levels of Renilla luciferase (Table 2).
TABLE 2.
Expression of Renilla luciferase after transfection of pNV-RL or pNV-RLΔGDD alone or with pCI-LC, pCI-V, or pCI-LC+pCI-V into MVA-T7-infected Vero cells
| Plasmid | Fold induction in Renilla luciferase (mean ± SD)a |
|---|---|
| pCI | 1 |
| pNV-GFP | 0.95 ± 0.1 |
| pNV-RL | 1.5 ± 0.7 |
| pNV-RLΔGDD | 0.90 ± 0.3 |
| pNV-RL/pCI-LC | 7.1 ± 2.1** |
| pNV-RL/pCI-V | 4.8 ± 2.4* |
| pNV-RL/pCI-LC+pCI-V | 20.2 ± 8.7** |
| pNV-RLΔGDD/pCI-LC+pCI-V | 1.1 ± 0.2 |
Cell lysates were prepared after 20 h of transfection for the Renilla luciferase activity. Standard deviations were calculated with at least three independent measurements. *, P < 0.05; **, P < 0.01 (compared to the pNV-RL group).
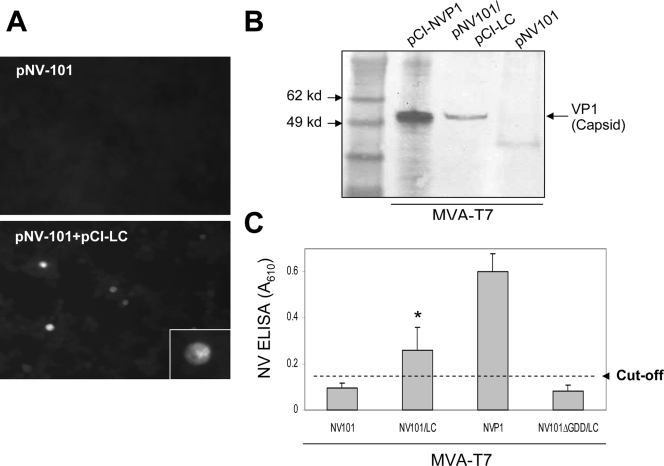
We next examined cotransfection of pNV101 and pCI-LC into Vero cells to evaluate the effect of LC on NV replication. The transfection of pNV101 alone into MVA-T7-infected cells showed no evidence of VP1 expression (by IFA, Western blot analysis, or ELISA; Fig. 4). However, when pNV101 was cotransfected with pCI-LC into MVA-T7-infected cells, we could observe cells expressing VP1 by IFA (Fig. 4A) and Western blot analysis (Fig. 4B) and ELISA (Fig. 4C). When the Pol motif GDD was deleted in pNV101 (pNV101ΔGDD), the transfection of this mutant plasmid with or without pCI-LC in cells did not produce any expression of VP1 (Fig. 4). The NV RNA levels after the transfection measured by real-time qRT-PCR showed that cotransfection of pNV101 or pNV-GFP with pCI-LC increased the RNA levels ∼2-fold (P < 0.05) over those of cells transfected with pNV101 or pNV-GFP alone (Table 3).
FIG. 4.
Transfection of pNV101 or pNV101ΔGDD with or without pCI-LC into MVA-T7-infected Vero cells. (A) IFA staining (with monoclonal antibody) detecting NV VP1 in cells transfected with pNV101 with or without pCI-LC (magnification, ×100). A cell in the box in the bottom panel shows the expression of NV VP1 in the cytoplasm with a higher magnification (×400). (B) Western blot analysis detecting NV VP1 in cells transfected with pNV101 with or without pCI-LC. Transfection of pCI-NVP1 serves as the positive control for VP1 expression in the transfected cells. (C) ELISA to detect NV VP1 in cell lysates after transfecting pNV101 with or without pCI-LC into MVA-T7-infected Vero cells. Transfection of pCI-NVP1 or cotransfection pNV-GFPΔGDD and pCI-LC serves as a positive or negative control, respectively, in the present study. The cutoff value was calculated as the average ±3 standard deviations of cell lysates prepared with the transfection of mock-medium into MVA-T7-infected cells. An asterisk (*) indicates that the level of VP1 expression by cotransfection with pCI-LC and pNV101 was significantly higher (P < 0.05) than the transfection with pNV101 alone.
TABLE 3.
NV RNA levels after transfection of pNV-GFP or pNV-GFP alone or with pCI-LC in MVA-T7-infected Vero cells detected by real-time qRT-PCR
| Plasmid | Fold induction in RNA levels (mean ± SD)a |
|---|---|
| pCI | NA |
| pNV-GFP | 1 |
| pNV-GFPΔGDD | 0.92 ± 0.3 |
| pNV-GFP/pCI-LC | 2.1 ± 0.3* |
| pNV-GFPΔGDD/pCI-LC | 1.1 ± 0.2 |
Total RNA was extracted from cell lysates after 20 h of transfection for the real-time qRT-PCR. NA, not available. *, P < 0.05 (compared to the pNV-GFP group).
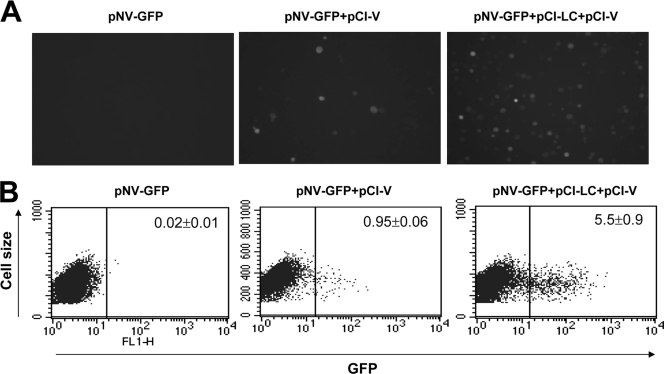
The expression of LC and V synergistically increased NV replication.
The V protein of SV5 downregulates the interferon (IFN) system by degrading STAT1 and enhances the replication of various viruses in vitro (34). First, we confirmed the expression of V protein after transfection of pCI-V in various cells (BHK21, HEK293T, and Vero cells) using a firefly luciferase reporter plasmid under the control of the IFN stimulating response element (pISRE-luc) (unpublished observations) as described previously (5). The cotransfection of pNV-GFP and pCI-V into MVA-T7-infected Vero cells also produced GFP-positive cells but with much less efficiency than pCI-LC (ca. 1% of total cells; Fig. 5). Interestingly, there were synergistic effects of LC and V in promoting NV replication. Cotransfection of pNV-GFP, pCI-LC, and pCI-V yielded ca. 5.5% of GFP-positive cells in Vero cells (Fig. 5). The cotransfection study with pNV-RL, pCI-LC, and pCI-V confirmed the synergistic effects of LC and V protein, since cotransfection significantly increased Renilla luciferase expression levels by cotransfection over that of LC or V protein alone (Table 2).
FIG. 5.
Transfection of pNV-GFP alone, pNV-GFP+pCI-V, or pNV-GFP+pCI-LC+pCI-V in MVA-T7-infected Vero cells. (A) GFP-positive cells observed under fluorescence after cells were transfected with pNV-GFP alone (left panel), pNV-GFP+pCI-V (middle panel), or pNV-GFP+pCI-LC+pCI-V (right panel). (B) Flow cytometry analysis to enumerate GFP-positive after cells were transfected with pNV-GFP alone (left panel), pNV-GFP+pCI-V (middle panel), or pNV-GFP+pCI-LC+pCI-V (right panel).
Mutagenesis studies.
We applied the cotransfection method with three plasmids—pNV-GFP, pCI-LC, and pCI-V—to the identification of a potential cis-acting element in the start of subgenomic region and 3′ end of the NV genome. Each pNV-GFP-based mutant plasmid was cotransfected with pCI-LC plus pCI-V, and GFP expression was observed under a fluorescence microscope and enumerated with flow cytometry analysis. Table 4 and Fig. 6 to 8 summarize the results of the cotransfection study. First, the noncoding bases (GUAA) at the start of subgenomic RNA were examined for their role in virus replication by mutagenesis study. The first base at the beginning of subgenomic RNA, G, was essential for the replication of NV in this system. When the first base, G, was mutated to U, A, or C (pNV-GFP-G5354U, pNV-GFP-G5354A, or pNV-GFP-G5354C), cotransfection of each plasmid with pCI-LC and pCI-V yielded few GFP-positive cells ranging from ca. 0.02 to 0.03% (Fig. 6B and Table 4). For the second base, U, while its mutation to A or C abolished the replication of NV in the system (0.02 to 0.03%), its mutation to G resulted in comparable numbers of GFP-positive cells (5.1%) to that of the parental pNV-GFP (Fig. 6C, Table 4, and Fig. 5). The third (A) or fourth base (A) did not appear to be essential for virus replication. The mutation of the third or fourth base to any bases yielded similar (or slightly lower, but without statistical significance) numbers of GFP-positive cells ranging from 4.1 to 4.5% to that of the parental pNV-GFP after cotransfection in Vero cells (Table 4).
TABLE 4.
Summary of mutagenesis study
| Plasmid | Bases | Amino acid change | Mean no. of GFP-positive cells ± SDa | NV replication |
|---|---|---|---|---|
| Wild type | GUAA | |||
| pNV-GFP-G5354C | CUAA | Val to Leu | 0.03 ± 0.02 | - |
| pNV-GFP-G5354A | AUAA | Val to Ile | 0.02 ± 0.01 | - |
| pNV-GFP-G5354U | UUAA | Val to Ile | 0.02 ± 0.02 | - |
| pNV-GFP-U5355A | GAAA | Val to Glu | 0.03 ± 0.01 | - |
| pNV-GFP-U5355C | GCAA | Val to Ala | 0.02 ± 0.01 | - |
| pNV-GFP-U5355G | GGAA | Val to Gly | 5.1 ± 1.2 | + |
| pNV-GFP-A5356U | GUUA | Val to Val | 4.3 ± 1.4 | + |
| pNV-GFP-A5356C | GUCA | Val to Val | 4.1 ± 0.8 | + |
| pNV-GFP-A5356G | GUGA | Val to Val | 4.5 ± 1.1 | + |
| pNV-GFP-A5357U | GUAU | Asn to Tyr | 4.2 ± 2.2 | + |
| pNV-GFP-A5357C | GUAC | Asn to His | 4.5 ± 1.3 | + |
| pNV-GFP-A5357G | GUAG | Asn to Asp | 4.4 ± 0.9 | + |
| pNV-GFP-Dto7080 | NA | NA | 5.4 ± 1.2 | + |
| pNV-GFP-Dto7089 | NA | NA | 5.1 ± 1.4 | + |
| pNV-GFP-Dto7480 | NA | NA | 4.9 ± 0.8 | + |
| pNV-GFP-Dto7500 | NA | NA | 4.7 ± 1.6 | + |
| pNV-GFP-Dto7534 | NA | NA | 0.02 ± 0.02 | - |
| pNV-GFP-Dto7600 | NA | NA | 0.02 ± 0.01 | - |
| pNV-GFP-Dto7610 | NA | NA | 0.03 ± 0.02 | - |
| pNV-GFP-Dto7630 | NA | NA | 0.02 ± 0.01 | - |
| pNV-GFP-Dto7640 | NA | NA | 0.03 ± 0.01 | - |
GFP-positive cells were measured as the expression of GFP as determined by flow cytometry analysis.
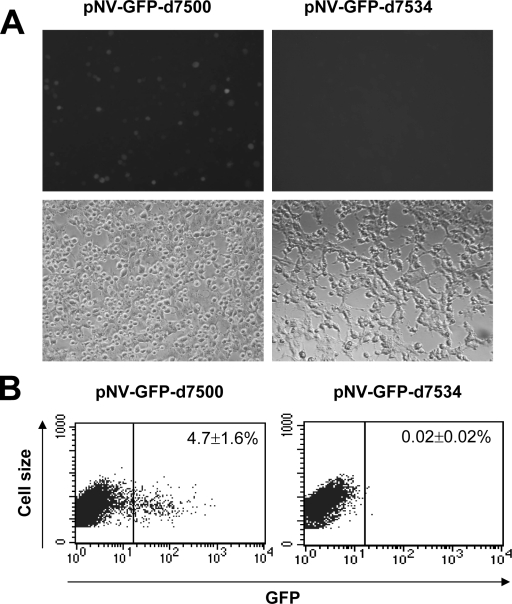
FIG. 8.
Cotransfection of each pNV-GFP mutant, pCI-LC, and pCI-V into MVA-T7-infected Vero cells. (A) GFP-positive cells observed under a fluorescence microscope after cells were transfected with pNV-GFP-d7500 or pNV-GFP-d7534 and pCI-LC plus pCI-V. The bottom panel is the corresponding area observed under a light microscope (after MVA-T7 infection). (B) Flow cytometry analysis to enumerate GFP-positive cells after Vero cells were transfected with pNV-GFP-d7500 or pNV-GFP-d7534 and pCI-LC plus pCI-V (after MVA-T7 infection).
We also examined the 3′ end of the NV genome for potential cis-acting elements by generating a series of deletion mutants of pNV-GFP. The designation of each plasmid—pNV-GFPD7080, pNV-GFPD7089, pNV-GFPD7480, pNV-GFPD7500, pNV-GFPD7534, pNV-GFPD7600, pNV-GFPD7610, pNV-GFPD7630, and pNV-GFPD7640—was based on the deletion that starts just after the stop codon of the GFP gene to the base numbered in the construct (Fig. 7). While cotransfection of pNV-GFPD7080, pNV-GFPD7089, pNV-GFPD7480, or pNV-GFPD7500 with pCI-LC and pCI-V in Vero cells produced GFP-positive cells at rates similar to that of the parental plasmid (pNV-GFP), the cotransfection of pNV-GFPD7534, pNV-GFPD7600, pNV-GFPD7610, pNV-GFPD7630, or pNV-GFPD7640 did not produce any GFP-positive cells (Table 4 and Fig. 7 and 8), These results indicated that the 3′ untranslated region (3′UTR) and the last part of the RNA sequences encoding ORF3 was essential, containing cis-acting elements for virus replication.
DISCUSSION
The inability to grow human noroviruses in cell culture greatly impedes studies of their pathogenesis and immunity. In this report, we report NV replicon plasmids with reporter genes, pNV-GFP or pNV-RL, for studying the replication of NV in cells. During the generation of NV replicon-bearing cells with pNV-Neo in our previous study, we noticed that only a few populations of cells (either BHK21 or Huh-7) could support NV replication when selected by the antibiotics. The initial study of transfecting pNV-GFP alone was consistent with the observation that only few of the transfected cells expressed GFP. The enumeration of GFP-positive cells by flow cytometry failed to yield significant numbers above the level of control cells. However, we were consistently able to observe some GFP-positive cells under a fluorescence microscope after the transfection of pNV-GFP regardless of cell types. In the case of hepatitis C virus (HCV), a phenomenon similar to that seen for NV was observed during the generation of replicon-bearing cells: only a few cells in the population of Huh-7 were susceptible to replication of HCV (20). Viral and cellular factors that promote the replication of HCV in cell culture have been identified. As viral factors, adaptive mutations in the HCV genome led to the susceptible population and efficiency of selecting replicon-harboring cells increased up to 1,000-fold in Huh-7 cells (2, 18). Cellular factors have also been identified for HCV replication: cells with mutations in the IFN system (the retinoid-inducible gene 1) were much more susceptible to HCV replication than the parental cells (2, 9). However, unlike HCV, we could not identify such adaptive mutations in the viral genome for NV replication in a similar study with NV replicon-bearing cells (5). To identify any viral factors which may promote the replication of NV, our attention was focused on the observation that vesiviruses grow efficiently in cell culture and encode a unique protein in the subgenomic region designated LC. Our hypothesis was that LC might be associated with the efficient replication of vesiviruses in cell culture and, if so, it might promote human norovirus replication in cells. When we enumerated cells expressing GFP after the transfection of pQ-GFP alone in various cells, including Vero, BHK21, or 293T cells, with MVA-T7, <4% of the cell population was GFP positive, suggesting that even FCV has some restrictions on replication within the cells. This was consistent with the transfection of pQ14, where virus replication was measured by the IFA staining with VP1 antibody. However, cotransfection of pQ14 or pQ-GFP with pCI-LC indicated that LC increased the cell population susceptible to FCV replication by ∼2-fold in these cell lines (Fig. 2A and B). Although the number of cells expressing GFP or VP1 increased by the cotransfection, virus titers after the transfection of pQ14 with or without pCI-LC were similar to each other (Fig. 2C). One possible explanation is that the intensity of GFP and VP1 expression was reduced by the cotransfection with pCI-LC in the cells (Fig. 2A and B), which may offset the titers. The overexpression of LC triggers morphological changes (cell rounding) and apoptosis of cells (unpublished observation). We also found that LC significantly reduced the expression of luciferase under the promoter of CMV after transfection (unpublished observation), suggesting that it might have an inhibitory effect on overall protein translation, probably due to the apoptosis. We are conducting further experiments to examine the role of LC in protein translation and apoptosis in correlation with the increased expression of FCV VP1 and GFP. When testing pCI-based plasmids containing LC from other FCV strains such as F9 (vaccine strain) or Ari (22) for the cotransfection study, we observed similar results, as they enhanced the expression of VP1 or GFP with FCV-based plasmids (unpublished observation).
When we performed the same experiments with the reporter plasmids of NV, we found that LC promoted the replication of NV as well. First, the transfection of pNV101 or pNV-GFP into MVA-T7-infected cells (BHK21, HEK293T, or Vero cells) produced few VP1- or GFP-positive cells. However, cotransfection of pNV101 or pNV-GFP and pCI-LC produced significantly increased numbers of VP1- or GFP-positive cells, respectively, in those cells. The percentage of GFP-positive cells after the cotransfection of pNV-GFP and pCI-LC was ca. 3% of the whole population (Fig. 3). Cotransfection of pNV-GFP and pCI-V also produced increased numbers of GFP-expressing cells, indicating the importance of innate immunity as a restriction factor for NV replication in cells. Furthermore, we found that there were synergistic effects of LC and V in promoting NV replication. We observed similar results with the full-length clone of NV, pNV101. Although transfection of pNV101 alone produced few VP1-positive cells, cotransfection of pNV101 and pCI-LC resulted in significant numbers of cells expressing VP1 as detected by various methods including IFA, Western blot analysis, and ELISA. Using IFA with monoclonal antibody against NV VP1, we could observe distinct localization of VP1 in the cells reminiscent of that of MNV-1-infected cells (Fig. 4A) (30), suggesting that VP1 may be the part of the replicase complexes and/or virions are assembled within the replicase complexes during the replication of NV in cells. Since the transfection of pQ14 resulted in the production of approximately 2 × 103 PFU of viable viruses/ml, it is possible that the cotransfection of pNV101 and pCI-LC produces viable viruses in cells. We are currently working on the identification of viable NV or NV particles in the supernatant and/or cell lysates of the cotransfection. To rule out the effects of MVA-T7 in NV replication in the cotransfection study, we used BHK21 cells stably expressing T7 Pol for the cotransfection study and found similar results. Cotransfection of pNV-GFP and pCI-LC into the cells yielded GFP-positive cells but with much less efficiency than the MVA-T7 system (unpublished observation).
Using the cotransfection approach, we could identify potential cis-acting elements for NV replication. This is a significant advance because this cell-based system provides a valuable tool to study RNA elements for the fastidious virus. In the present study, we focused on RNA elements at the beginning of the subgenomic RNA and 3′-end region, including 3′UTR and ORF3 for NV replication. Our first observation noted that the first base (G) at the beginning of subgenomic RNA was essential for virus replication. The mutant plasmids with substitution to other bases at the first base (G) did not produce any GFP-positive cells. It has been postulated that the first base G is the site of VPg linkage, and the mutations may abolish this activity. The second base (U) is also important in the virus replication, and the mutation to bases other than G abolished the replication, as confirmed through the cotransfection study (Table 4). However, we cannot rule out that the lack of GFP expression by pNV-GFP-G5354U, pNV-GFP-G5354A, pNV-GFP-G5354C, pNV-GFP-U5355A, or pNV-GFP-U5355C is due to defects in Pol activity because the mutations also altered the amino acids at the end of the enzyme (Table 4). The third and fourth bases were not essential for virus replication, and substitution mutations only slightly reduced GFP-positive cells in the cotransfection experiments. As for RNA bases essential for NV replication at the 3′ end of genome, we found that both the 3′UTR (66 bases) and the last stretch of bases encoding VP2 were essential for virus replication (Table 4 and Fig. 7 and 8). This finding was in accordance with our previous studies with FCV in which we also demonstrated that the ORF3 nucleotide sequence itself overlaps a cis-acting RNA signal at the genomic 3′ end (26). It is possible that the secondary RNA structures in the region may interact with viral replicase complexes for successful replication. We plan to generate additional mutants in the region based on the predicted RNA structure to examine its roles in NV replication.
In summary, we conclude that LC may be a viral factor that promotes the replication of NV in cells. Also, we found that inhibiting the STAT1 pathway promoted NV replication. At present, we are trying to produce cell lines constitutively expressing LC, V, or LC plus V to examine whether they support NV replication without cotransfection. It is not yet clear by what mechanism LC has an effect on the replication of FCV and NV. It is possible that LC may interfere with the IFN pathway or interact with viral components to promote virus replication. We are currently conducting experiments to identify the potential mechanisms focused on the role of LC in the IFN pathway and interaction with viral proteins. We also report a reliable NV replicon system with reporter genes to study the replication of NV. Identification of viral or cellular factors which promote NV replication could provide vital clues to growing the fastidious human noroviruses in cell culture.
Acknowledgments
This study was partly supported by NIH COBRE grant 2 P20 RR016443-07.
This paper is contribution no. 08-208-J from the Kansas Agricultural Experiment Station.
Footnotes
Published ahead of print on 16 July 2008.
REFERENCES
- 1.Asanaka, M., R. L. Atmar, V. Ruvolo, S. E. Crawford, F. H. Neill, and M. K. Estes. 2005. Replication and packaging of Norwalk virus RNA in cultured mammalian cells. Proc. Natl. Acad. Sci. USA 10210327-10332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blight, K. J., J. A. McKeating, J. Marcotrigiano, and C. M. Rice. 2003. Efficient replication of hepatitis C virus genotype 1a RNAs in cell culture. J. Virol. 773181-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, K. O., and D. W. George. 2007. Interferons and ribavirin effectively inhibit Norwalk virus replication in replicon-bearing cells. J. Virol. 8112111-12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, K. O., Y. Kim, K. Y. Green, and L. J. Saif. 2002. Cell-culture propagation of porcine enteric calicivirus mediated by intestinal contents is dependent on the cyclic AMP signaling pathway. Virology 304302-310. [DOI] [PubMed] [Google Scholar]
- 5.Chang, K. O., S. V. Sosnovtsev, G. Belliot, A. D. King, and K. Y. Green. 2006. Stable expression of a Norwalk virus RNA replicon in a human hepatoma cell line. Virology 353463-473. [DOI] [PubMed] [Google Scholar]
- 6.Duizer, E., K. J. Schwab, F. H. Neill, R. L. Atmar, M. P. Koopmans, and M. K. Estes. 2004. Laboratory efforts to cultivate noroviruses. J. Gen. Virol. 8579-87. [DOI] [PubMed] [Google Scholar]
- 7.Fankhauser, R. L., J. S. Noel, S. S. Monroe, T. Ando, and R. I. Glass. 1998. Molecular epidemiology of “Norwalk-like viruses” in outbreaks of gastroenteritis in the United States. J. Infect. Dis. 1781571-1578. [DOI] [PubMed] [Google Scholar]
- 8.Fernandez-Vega, V., S. V. Sosnovtsev, G. Belliot, A. D. King, T. Mitra, A. Gorbalenya, and K. Y. Green. 2004. Norwalk virus N-terminal nonstructural protein is associated with disassembly of the Golgi complex in transfected cells. J. Virol. 784827-4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foy, E., K. Li, R. Sumpter, Jr., Y. M. Loo, C. L. Johnson, C. Wang, P. M. Fish, M. Yoneyama, T. Fujita, S. M. Lemon, and M. Gale, Jr. 2005. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc. Natl. Acad. Sci. USA 1022986-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geissler, K., K. Schneider, G. Platzer, B. Truyen, O. R. Kaaden, and U. Truyen. 1997. Genetic and antigenic heterogeneity among feline calicivirus isolates from distinct disease manifestations. Virus Res. 48193-206. [DOI] [PubMed] [Google Scholar]
- 11.Green, K. Y. 2007. Caliciviridae: the noroviruses, p. 949-980. In D. M. Knipe (ed.), Fields virology, 5th ed., vol. 1. Lippincott/The Williams & Wilkins Co., Philadelphia, PA. [Google Scholar]
- 12.Guix, S., M. Asanaka, K. Katayama, S. E. Crawford, F. H. Neill, R. L. Atmar, and M. K. Estes. 2007. Norwalk virus RNA is infectious in mammalian cells. J. Virol. 8112238-12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu, C. C., L. K. Riley, H. M. Wills, and R. S. Livingston. 2006. Persistent infection with and serologic cross-reactivity of three novel murine noroviruses. Comp. Med. 56247-251. [PubMed] [Google Scholar]
- 14.Jiang, X., M. Wang, K. Wang, and M. K. Estes. 1993. Sequence and genomic organization of Norwalk virus. Virology 19551-61. [DOI] [PubMed] [Google Scholar]
- 15.Kageyama, T., S. Kojima, M. Shinohara, K. Uchida, S. Fukushi, F. B. Hoshino, N. Takeda, and K. Katayama. 2003. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 411548-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kapikian, A. Z., R. G. Wyatt, R. Dolin, T. S. Thornhill, A. R. Kalica, and R. M. Chanock. 1972. Visualization by immune electron microscopy of a 27-nm particle associated with acute infectious nonbacterial gastroenteritis. J. Virol. 101075-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karst, S. M., C. E. Wobus, M. Lay, J. Davidson, and H. W. t. Virgin. 2003. STAT1-dependent innate immunity to a Norwalk-like virus. Science 2991575-1578. [DOI] [PubMed] [Google Scholar]
- 18.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 754614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lambden, P. R., E. O. Caul, C. R. Ashley, and I. N. Clarke. 1993. Sequence and genome organization of a human small round-structured (Norwalk-like) virus. Science 259516-519. [DOI] [PubMed] [Google Scholar]
- 20.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285110-113. [DOI] [PubMed] [Google Scholar]
- 21.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ossiboff, R. J., A. Sheh, J. Shotton, P. A. Pesavento, and J. S. Parker. 2007. Feline caliciviruses (FCVs) isolated from cats with virulent systemic disease possess in vitro phenotypes distinct from those of other FCV isolates. J. Gen. Virol. 88506-517. [DOI] [PubMed] [Google Scholar]
- 23.Smith, A. W., D. E. Skilling, N. Cherry, J. H. Mead, and D. O. Matson. 1998. Calicivirus emergence from ocean reservoirs: zoonotic and interspecies movements. Emerg. Infect. Dis. 413-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sosnovtsev, S., and K. Y. Green. 1995. RNA transcripts derived from a cloned full-length copy of the feline calicivirus genome do not require VpG for infectivity. Virology 210383-390. [DOI] [PubMed] [Google Scholar]
- 25.Sosnovtsev, S. V., G. Belliot, K. O. Chang, and K. Y. Green. 2004. Induction of apoptosis in feline calicivirus-infected cells: implication of the p30 (3A-like) protein in caspase activation, abstr. P2H7. Seventh International Symposium on Positive Strand RNA Viruses, San Francisco, CA.
- 26.Sosnovtsev, S. V., G. Belliot, K. O. Chang, O. Onwudiwe, and K. Y. Green. 2005. Feline calicivirus VP2 is essential for the production of infectious virions. J. Virol. 794012-4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sosnovtsev, S. V., S. A. Sosnovtseva, and K. Y. Green. 1998. Cleavage of the feline calicivirus capsid precursor is mediated by a virus-encoded proteinase. J. Virol. 723051-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spann, K. M., K. C. Tran, B. Chi, R. L. Rabin, and P. L. Collins. 2004. Suppression of the induction of alpha, beta, and lambda interferons by the NS1 and NS2 proteins of human respiratory syncytial virus in human epithelial cells and macrophages. J. Virol. 784363-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Straub, T. M., K. Honer zu Bentrup, P. Orosz-Coghlan, A. Dohnalkova, B. K. Mayer, R. A. Bartholomew, C. O. Valdez, C. J. Bruckner-Lea, C. P. Gerba, M. Abbaszadegan, and C. A. Nickerson. 2007. In vitro cell culture infectivity assay for human noroviruses. Emerg. Infect. Dis. 13396-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wobus, C. E., S. M. Karst, L. B. Thackray, K. O. Chang, S. V. Sosnovtsev, G. Belliot, A. Krug, J. M. Mackenzie, K. Y. Green, and H. W. T. Virgin. 2004. Replication of norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol. 2e432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wobus, C. E., L. B. Thackray, and H. W. t. Virgin. 2006. Murine norovirus: a model system to study norovirus biology and pathogenesis. J. Virol. 805104-5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wyatt, L. S., B. Moss, and S. Rozenblatt. 1995. Replication-deficient vaccinia virus encoding bacteriophage T7 RNA polymerase for transient gene expression in mammalian cells. Virology 210202-205. [DOI] [PubMed] [Google Scholar]
- 33.Xi, J. N., D. Y. Graham, K. N. Wang, and M. K. Estes. 1990. Norwalk virus genome cloning and characterization. Science 2501580-1583. [DOI] [PubMed] [Google Scholar]
- 34.Young, D. F., L. Andrejeva, A. Livingstone, S. Goodbourn, R. A. Lamb, P. L. Collins, R. M. Elliott, and R. E. Randall. 2003. Virus replication in engineered human cells that do not respond to interferons. J. Virol. 772174-2181. [DOI] [PMC free article] [PubMed] [Google Scholar]