Abstract
APOBEC3G limits the replication of human immunodeficiency virus type 1, other retroviruses, and retrotransposons. It localizes predominantly to the cytoplasm of cells, which is consistent with a model wherein cytosolic APOBEC3G packages into assembling virions, where it exerts its antiviral effect by deaminating viral cDNA cytosines during reverse transcription. To define the domains of APOBEC3G that determine cytoplasmic localization, comparisons were made with APOBEC3B, which is predominantly nuclear. APOBEC3G/APOBEC3B chimeric proteins mapped a primary subcellular localization determinant to a region within the first 60 residues of each protein. A panel of 25 APOBEC3G mutants, each with a residue replaced by the corresponding amino acid of APOBEC3B, revealed that several positions within this region were particularly important, with Y19D showing the largest effect. The mislocalization phenotype of these mutants was only apparent in the context of the amino-terminal half of APOBEC3G and not the full-length protein, suggesting the existence of an additional localization determinant. Indeed, a panel of five single amino acid substitutions within the region from amino acids 113 to 128 had little effect by themselves but, in combination with Y19D, two substitutions—F126S and W127A—caused full-length APOBEC3G to redistribute throughout the cell. The critical localization-determining residues were predicted to cluster on a common solvent-exposed surface, suggesting a model in which these two regions of APOBEC3G combine to mediate an intermolecular interaction that controls subcellular localization.
The human genome encodes seven apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3 proteins (abbreviated as APOBEC3 or A3) (35). These proteins are capable of defending cells against a variety of genetic pathogens such as endogenous retrotransposons and exogenous retroviruses (for reviews, see references 3, 16, 20, 27, 32, and 48). The most-studied A3 target is human immunodeficiency virus type 1 (HIV-1). A3 proteins, in particular APOBEC3F (A3F) and APOBEC3G (A3G), are able to limit the replication of Vif-deficient HIV-1. They do this by gaining access to viral particles as they bud from infected cells. The A3s hitchhike within the virus particle to a cell that will be infected. There, the A3s use their DNA cytosine deaminase enzymatic activity to convert cytosine bases in the viral genome to uracils during reverse transcription, thereby blocking infection.
Members of the A3 family possess DNA cytosine deaminase activity, but they can be differentiated on the basis of other activities (see, for example, references 5, 7, 11, 21, 31, 34, 39, 52, 56, and 61). For instance, some A3s such as A3F and A3G are potent inhibitors of HIV-1 replication, while others such as APOBEC3B (A3B) have modest anti-HIV activity (see, for example, references 5, 22, 52, and 60). The subcellular distribution of A3 proteins is also a distinguishing feature (1, 2, 7, 8, 10, 12, 25, 26, 28, 37, 41-43, 45, 47, 52, 54, 55). For instance, A3G is predominantly cytoplasmic, whereas A3B is primarily nuclear. These differences in subcellular localization presumably reflect differences in the sets of proteins and/or nucleic acids with which the A3s interact, and they also suggest that the A3s perform distinct functions within the cell.
The A3 proteins are evolutionarily related to the mRNA editor APOBEC1 and the antibody gene DNA deaminase AID. AID is present in all vertebrates, but A3s are found only in mammals, suggesting that an ancestral AID gene duplicated and diverged to give rise to the present day A3 genes (19, 30, 35, 40, 50, 53). It was therefore reasonable to hypothesize that the A3s and AID share conserved properties in addition to DNA cytosine deaminase activity. For instance, both AID and APOBEC1 are nucleocytoplasmic shuttling proteins that are exported from the nucleus by the CRM1-dependent nuclear export pathway (9, 15, 33, 44, 58, 59). A3G also has a putative leucine-rich CRM1 nuclear export sequence spanning residues 369 to 379 (Fig. 1). However, localization studies indicated that A3G is not subject to CRM1-dependent nuclear export (1). Rather, its cytoplasmic localization was ascribed to a 16-residue peptide, dubbed a cytoplasmic retention signal (CRS), spanning residues 113 to 128 (2).
FIG. 1.
Alignment of A3B, A3F, and A3G amino acid sequences, highlighting regions of concentrated identity. Blue and green shading indicates regions of concentrated identity between A3G and A3F and between A3B and A3F, respectively. Gray-shaded residues are identical between at least two of the proteins. The crossing arrows indicate the junctions of chimeric proteins. Dashed boxes outline the putative CRS and NES of A3G and the putative NLS of A3B. The histidine, glutamic acid, and cysteine residues of the two zinc-coordinating motifs are indicated in boldface. The amino acids deleted in A3G1-369 are underlined. Additional details can be found in the text.
In our present studies, we inhibited CRM1 and deleted the leucine-rich region of A3G to independently confirm some of this prior work. However, our analyses of chimeric A3G-A3B proteins and A3G amino acid substitution mutants suggested a more complex molecular explanation for A3G's cytoplasmic localization, namely, that residues within amino acids 1 to 60 and amino acids 113 to 128 cooperate to determine APOBEC3G's cytoplasmic localization. A structural model of the amino (N)-terminal half of A3G supported these data by indicating that critical residues within this region are likely to be solvent exposed and therefore available for macromolecular interactions.
MATERIALS AND METHODS
DNA constructs.
The pEGFP-N3 (Clontech)-based plasmids encoding carboxy (C)-terminally green fluorescent protein (GFP)-tagged full-length AID, A3B, and A3G were described previously (AID [49]; A3B and A3G [52]).
PCR was used to create the truncated A3B and A3G/GFP fusion constructs. The primers were as follows: A3G1-369 (5′-NNN NGA GCT CAG GTA CCA CCA TGA AGC CTC ACT TCA GAA AC [RSH431] and 5′-NNN NGT CGA CTT GGC TGT GCT CAT CTA GTC C), A3B-NTD (5′-NNN NGA GCT CGG TAC CAC CAT GAA TCC ACA GAT CAG AAA T [RSH567] and 5′-NNN NGT CGA CCA TCC TTC CCA GGT ATC TGA GAA TCT CCT TTA G), A3B-CTD (5′-NNN NGT CGA CCA TCC TTC ACA GGT ATC TGA GAA TCT CCT TTA G and 5′-NNN NGT CGA CCA TCC TTC CGT TTC CCT GAT TCT GGA G [RSH580]), A3G-NTD (RSH431 and 5′-NNN NGT CGA CCG AGT GTC TGA GAA TCT CCC CC), and A3G-CTD (5′-NNN GAG CTC AGG TAC CAC CAT GGA TCC ACC CAC ATT CAC TTT C and 5′-NNN GTC GAC TCC GTT TTC CTG ATT CTG GAG AAT [RSH498]). PCR products were digested with SacI and SalI and ligated into similarly digested pEGFP-N3.
The A3B/G chimeric constructs were created using overlapping PCR. The primers used were A3B1-60G60-384 (RSH567, 5′-CTG GGT GGT ACT TAA GTT CGG AAT ACA CCT GGC CTC GAA AGA C, 5′-TCC GAA CTT AAG TAC CAC CCA G, and RSH498), A3G1-59B61-382 (RSH431, 5′-GCG TGG TAC TGA GGC TTG AAA TAC ACC TGG CCT CGA AAG AC, 5′-TTC AAG CCT CAG TAC CAC GC, and RSH580), A3B1-190G195-384 (RSH567, 5′-GTG GGT GGA TCC ATC GAG TGT CTG AGA ATC TCC TTT AGC G, 5′-CAC TCG ATG GAT CCA CCC AC, and RSH498), and A3G1-194B191-382 (RSH431, 5′-GTG TCT GGA TCC ATC AGG TAT CTG AGA ATC TCC CCC AGC A, 5′-TAC CTG ATG GAT CCA GAC AC, and RSH580). PCR products were digested with SacI and SalI and ligated into a similarly digested pEGFP-N3.
The GFP-tagged truncated and chimeric constructs were subcloned into C-terminal hemagglutinin (HA)-tagged mammalian and bacterial expression vectors. The A3 coding region was cut out of the pEGFP-N3-based plasmids by using KpnI and SalI and ligated into KpnI/XhoI-digested pcDNA3.1-HA and pTrc99-HA (52).
Amino acid substitution mutants were generated by using site-directed mutagenesis (QuikChange, Stratagene; primer sequences available on request). A3G double mutants were created by site-directed mutagenesis using single mutants as PCR templates. pEGFP-N3-A3G-NTD mutants were subcloned as KpnI/RsrII fragments into similarly digested full-length A3G expression constructs pEGFP-N3-A3G and pcDNA3.1-A3G-HA, reported previously (52). All constructs were confirmed by restriction digestion and DNA sequencing.
Cell culture.
Human embryonic kidney 293T and HeLa cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum and 25 U of penicillin/ml and 25 μg of streptomycin/ml at 37°C and 5% CO2. All transfections used FuGENE (Roche Applied Science) or TransIT-LT1 (Mirus Bio Corp.) according to the manufacturers' protocols.
Microscopy.
HeLa or 293T cells were seeded on eight-well LabTek chambered cover glasses (Nunc). After 24 h of incubation, the cells were transfected with the indicated pEGFP-N3-based constructs. After an additional 24 h of incubation, images of the live cells were captured on a Zeiss Axiovert 200 microscope. Images were cropped, and their brightness and contrast were adjusted linearly using ImageJ software (http://rsb.info.nih.gov/ij/). For leptomycin B (LMB) experiments, cells were treated for 2 h with 20 ng of LMB (LC Laboratories)/ml or mock treated with an equivalent dilution of ethanol (the vehicle).
For immunofluorescence experiments, HeLa cells were seeded onto sterilized cover glasses set in culture dishes. After 24 h of incubation, the cells were transfected with the indicated pcDNA3.1-HA-based constructs. After an additional 24 h of incubation, cells were fixed with 4% formaldehyde, permeabilized in 0.2% Triton X-100, incubated first with anti-HA antibody (Covance) and then with fluorescein-conjugated anti-mouse secondary antibody (Jackson Immunoresearch), and imaged as described above.
Escherichia coli mutation assays.
An E. coli-based rifampin-resistance assay was used to quantify the intrinsic DNA cytosine deaminase activity of A3B, A3G, and chimeric and mutant derivatives. This assay has been described previously (see, for example, references 29 and 31).
Immunoblotting.
293T cells were plated in six-well plates. After 24 h of incubation, the cells were transfected with APOBEC expression plasmid (or appropriate control plasmids). At 36 h later, the cells were harvested, and total proteins were extracted in a buffer containing 50 mM Tris (pH 7.5), 150 mM NaCl, 1% Nonidet P-40, 0.1% sodium dodecyl sulfate, 0.5% sodium deoxycholate, and a protease inhibitor cocktail (Roche). The extracts were clarified by centrifugation for 10 min at 20,800 × g at 4°C. The extracted proteins were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to a polyvinylidene difluoride membrane (Millipore), and probed with monoclonal anti-HA (Covance) or monoclonal anti-GFP (Clontech) antibody. Primary antibodies were detected by incubation with horseradish peroxidase-coupled anti-mouse immunoglobulin G (Bio-Rad) or anti-rabbit immunoglobulin G (Bio-Rad), followed by chemiluminescence (Roche Applied Science). Ponceau S (Sigma) staining of the polyvinylidene difluoride membrane following immunoblotting served as a protein loading control.
Amino acid alignments.
The amino acid sequences of A3B, A3F, and A3G were aligned by using CLUSTAL W and edited manually (Fig. 1). The protein sequences correspond to GenBank accession numbers NP_004891, NP_660341, and NP_068594, respectively.
Structural modeling of A3Gntd.
The primary amino acid sequences of A3G-NTD (residues 1 to 196) and A3G-CTD (residues 197 to 384) were aligned by using the homology modeling module of the InsightII program (Accelrys) (see Fig. S1 in the supplemental material). These amino acid sequences are 35% identical and 52% similar (according to the bl2seq program [National Center for Biotechnology Information]). The secondary structure elements of A3G-NTD were predicted by projecting the actual secondary structure of A3G-CTD (PDB 2jyw[13]) over the A3G-NTD/A3G-CTD primary amino acid sequence alignment. Because of amino acid conservation in the corresponding regions, the secondary structure elements of A3G-NTD were predicted to correspond directly with the actual elements in the A3G-CTD (see Fig. S1 in the supplemental material). Finally, this information was used to construct a three-dimensional model of the A3G-NTD (InsightII program; Accelrys). Images of the model were created by using MacPyMol (DeLano Scientific).
RESULTS
A predicted nuclear export signal is not required for A3G's cytoplasmic localization.
A3G residues 369 to 379 QDLSGRLRAIL are similar to the canonical leucine-rich nuclear export motif (LXLX2-3LX2-3L) that enables proteins to be bound by CRM1 and shuttled to the cytoplasm (e.g., the nuclear export signal [NES] of AID: LRDAFRTLGL [9, 23, 33, 44, 46, 51]). We analyzed the localization of A3G-GFP and AID-GFP in the presence or absence of the CRM1 inhibitor LMB (57). LMB caused AID-GFP to accumulate in nuclei of transiently transfected HeLa cells, as shown previously (Fig. 2A) (1, 9, 33, 44). In contrast, LMB treatment of cells expressing A3G-GFP had no effect (Fig. 2A) (1). To support this finding, we deleted the entire putative NES of A3G and observed that this construct, encoding A3G1-369 fused to GFP, was still predominantly cytoplasmic (Fig. 2B). These data corroborated prior studies that used LMB and substitution mutants to show that A3G is not being shuttled by CRM1 out of the nucleus (1).
FIG. 2.

Predicted nucleocytoplasmic shuttling signals in A3G and A3B are not required for their subcellular distributions. Images of representative live HeLa cells transiently transfected with the indicated A3-GFP expression plasmids are shown. Scale bar, 10 μm. (A) LMB caused AID-GFP but not A3G-GFP to accumulate in the nucleus. Minus (−) LMB indicates mock-treated cells. (B) Localization of A3G-GFP and the A3G1-369-GFP mutant lacking the putative C-terminal NES. (C) Localization of wild-type A3B and a mutant A3B with four alanines replacing residues in the putative NLS (A3B4Ala).
A putative NLS in A3B is not required for its nuclear localization.
To contrast with A3G, we also tested the hypothesis that A3B, a nuclear resident, is actively imported into the nucleus. In support of this hypothesis, A3B has a putative nuclear localization signal (NLS) at amino acids 206 to 212 (PLVLRRR; Fig. 1), and it has been reported to be a nucleocytoplasmic shuttling protein (7). This amino acid motif is similar to a functional NLS in the simian virus 40 large T antigen (PKKKRKVE) (24, 36). We performed site-directed mutagenesis, replacing the proline and arginines in this motif with alanines. All of these A3B mutants remained nuclear, including a quadruple mutant with the NLS motif changed at four positions to ALVLAAA (Fig. 2C and data not shown). This result demonstrated that the putative NLS is not required for A3B's nuclear localization in HeLa cells and that A3B may be transported to or retained in the nucleus by other means. We did note that there was a slight increase in cytoplasmic fluorescence with the quadruple mutant, but the bulk of the fluorescence clearly remained nuclear.
The N-terminal halves of A3G and A3B recapitulate the localization of the full-length proteins.
A3G and A3B are termed “double-domain” A3s because each has two zinc-binding domains. The genes of the double-domain proteins are likely to have arisen from the duplication of an ancestral “single-domain” AID-like APOBEC3 gene (19, 30, 35, 53). We therefore examined the localization of the N- and C-terminal halves of A3B and A3G by themselves to map the determinants of the localization of the full-length protein. Previous structural studies on the C-terminal domain (CTD) informed the selection of the dividing line between the domains, which was chosen so as to not disrupt secondary structure and to ensure that the CTD remained catalytically intact (Fig. 1 and see Fig. S1 in the supplemental material) (13, 14).
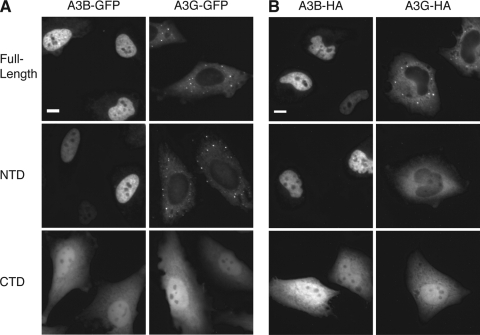
The N-terminal domain (NTD) of A3G (A3G1-196-GFP) localized predominantly to the cytoplasm, and the NTD of A3B (A3B1-192-GFP) localized predominantly to the nucleus, much like their respective full-length parental proteins (Fig. 3A). In contrast, their CTDs, A3G197-384-GFP and A3B193-382-GFP, were distributed throughout the cell in a manner indistinguishable from that of GFP alone. Similar data were obtained with HA-tagged proteins in fixed cells (Fig. 3B). Thus, the N-terminal halves of the proteins clearly contained their primary localization determinants. These data are largely consistent with prior deletion studies (2, 7).
FIG. 3.
The N-terminal halves of A3B and A3G recapitulate the localization patterns of the full-length proteins. Representative images of HeLa cells transfected with the indicated A3-GFP or A3-HA expression plasmids are shown. The NTD of A3B and A3G consists of amino acids 1 to 192 and amino acids 1 to 196, respectively. The CTD of A3B and A3G consists of amino acids 193 to 382 and amino acids 197 to 384, respectively. (A) Live cells expressing the indicated GFP-fusion proteins. (B) Fixed cells expressing the indicated HA-fusion proteins. Scale bar, 10 μm.
As an additional test that the N-terminal half of A3G and A3B contained these proteins' major subcellular localization determinants, we constructed full-length chimeric proteins combining one domain of A3B with one domain of A3G. We created a chimera combining the NTD of A3B with the CTD of A3G (A3B1-190G195-384) and one that fused the NTD of A3G with the CTD of A3B (A3G1-194B191-382). The chimera crossover point was selected to maintain overall structural integrity, occurring within a region of high similarity that is predicted to link the N- and C-terminal domains of these proteins (i.e., after α5 in the A3G-NTD model structure; Fig. 1 and see Fig. S1 in the supplemental material). Similar crossover points were previously used to generate functional A3G/F chimeras and an A3G-A3A chimera (4, 28, 29). Like A3B, the chimera consisting of the NTD of A3B and the CTD of A3G localized predominantly to the nucleus (Fig. 4A and B). In contrast, like A3G, the chimera fusing the NTD of A3G and the CTD of A3B localized predominantly to the cytoplasm (although we noted that this chimera exhibited a higher degree of nuclear fluorescence than wild-type A3G). Virtually identical results were obtained with GFP-tagged proteins in live cells and HA-tagged proteins and immunofluorescence in fixed cells (Fig. 4A and B). These data support the conclusion that the N-terminal halves of A3B and A3G provide the primary determinants of their subcellular localization.
FIG. 4.
The first 60 amino acids of A3B and A3G possess essential subcellular localization determinants. Representative images of HeLa cells transfected with the indicated chimeric-APOBEC3 expression plasmids are shown. The GFP-tagged constructs were visualized in live cells, and the HA-tagged proteins were visualized in fixed cells. (A) Cells expressing A3B or A3G. (B) Cells showing the localization of chimeras that fuse the NTD of A3B with the CTD of A3G and vice versa. (C) Cells showing the localization of chimeras that swap the first 60 amino acids of A3B for those of A3G and vice versa. Scale bar, 10 μm.
The first 60 amino acids of A3G and A3B harbor key subcellular localization determinants.
We next wanted to more finely map the region within the N-terminal half of A3B and A3G that determines their subcellular distributions. Our strategy to do this was guided by A3B, A3F, and A3G amino acid alignments (Fig. 1). We and others have previously shown that A3F, like A3G, is cytoplasmic (6, 37, 52, 55). We also noted that, although the full-length proteins distribute to opposite compartments of the cell, the amino-terminal halves of A3B and A3F are largely identical (Fig. 1; residues 65 to 191 of A3B and residues 66 to 192 of A3F are 93% identical). In contrast, residues 1 to 65 of A3F and residues 1 to 64 of A3B share only 56% identity, whereas A3F and A3G are virtually identical over the same region (59 of the first 60 residues; shaded blue in Fig. 1) (39). These correlations strongly implied that a key determinant of the subcellular localization of both A3G and A3B would reside within the first 60 amino acids.
We therefore constructed chimeras that replaced the first 60 amino acids of A3G with the corresponding residues or A3B and vice versa (Fig. 1). These chimeric proteins are also likely to be structurally sound, because the fusion junctions were predicted to lie in a flexible loop between the β2 strand and α1 helix (see Fig. 1 and Fig. S1 in the supplemental material) (13). To verify their structural integrity, we performed E. coli-based mutation assays. All of the chimeras exhibited intrinsic DNA deaminase activity equal to or greater than that of the proteins from which they were derived (data not shown). We next assessed the localization patterns of these chimeras. Like A3G, the A3G1-59B61-382 chimera exhibited a predominantly cytoplasmic localization in the majority of cells (Fig. 4C). In contrast, the A3B1-60G60-384 chimera localized predominantly to the nucleus in most cells. Similar results were again obtained with GFP- and HA-tagged proteins (Fig. 4C). We noted that these results were less clear-cut than those for the chimeras crossing over at the midway point. Nevertheless, the first 59 residues of A3G conferred an A3G-like localization pattern to the rest of A3B. Moreover, replacing the first 59 residues of A3G with those of A3B caused A3G to adopt an A3B-like localization pattern. These results supported the conclusion that the first 60 amino acids of A3G harbor a major subcellular localization determinant.
Single amino acid substitutions can disrupt the subcellular distribution of A3G.
To more finely map the region of A3G responsible for its cytoplasmic localization, we constructed a panel of amino acid substitution mutants within this critical 60-amino-acid N-terminal region. A3G and A3B differ at 27 residues within their first 63 amino acids (Fig. 1). At 25 of these differing positions, we generated a mutant with the A3G residue replaced by the corresponding A3B residue. For instance, we created an A3G mutant with lysine 2 replaced by asparagine 2 of A3B (A3G-K2N). We did not generate two possible mutants because they were deemed conservative (M9/V9, I53/V54).
We first examined the subcellular distribution of the single amino acid substitution mutants, initially as A3G-NTD-GFP derivatives. The localization of most of the mutants was predominantly cytoplasmic and indistinguishable from the wild-type protein (e.g., A3G-NTD-S18Y-GFP in Fig. 5A; see also Fig. S2 in the supplemental material for representative fields of cells expressing each mutant). However, several mutants, such as Y19D and Y22E, exhibited a clearly disrupted localization pattern (Fig. 5A). For instance, 86% of the Y19D-expressing cells and 64% of the Y22E-expressing cells had a cell-wide or predominantly nuclear fluorescence localization pattern (Fig. 5B). Several other mutants, such as the R24E, S28Y, T32Y, and S60F mutants, also mislocalized (Fig. 5B). As a control, we characterized the expression level of the A3G-NTD-GFP mutants by immunoblotting (Fig. 5C). Some of the mutations resulted in an apparent decrease in steady-state protein expression level or a decrease in protein solubility. Immunoblotting also revealed the presence of cross-reacting bands of higher mobility that could correspond to fragments of the A3G-NTD-GFP proteins. However, there was no apparent correlation between mislocalization and expression level or fragmentation. These data therefore defined single amino acids required for A3G-NTD localization and confirmed that the first 60 amino acids form a region critical for determining the protein's subcellular distribution.
FIG. 5.
Single amino acid substitutions within the first 60 amino acids of A3G disrupt cytoplasmic localization. (A) Representative images of live HeLa cells transiently transfected with the indicated A3G-NTD-GFP or derivative expression plasmids (see Fig. S2 in the supplemental material for a full set of representative fields). Scale bar, 10 μm. (B) Quantification of the extent of disrupted localization for the mutants. Individual cells expressing the indicated proteins were scored and grouped into three categories based on their overall pattern of cellular fluorescence: those with more apparent nuclear than cytoplasmic fluorescence (C < N), equivalent nuclear and cytoplasmic fluorescence (C = N), or more apparent cytoplasmic than nuclear fluorescence (C > N). The percentage of cells falling into each category is indicated for each mutant. Cells from two to four independent experiments were scored, and the tallies were combined. The total number of cells (N) scored for each construct is indicated. (C) Anti-GFP immunoblot showing the expression of the indicated A3G-NTD-GFP proteins. An arrowhead indicates the band corresponding to A3G-NTD-GFP and mutant derivatives. Ponceau S staining of the membrane served as a protein loading control.
Two regions of A3G cooperate to determine cytoplasmic localization.
Despite the fact that A3G-NTD-Y19D or -Y22E localized aberrantly, none of these substitutions by themselves significantly altered the cytoplasmic localization of the full-length protein (Fig. 6 and see Fig. S3 in the supplemental material) (also data not shown). While this article was in revision, another study was published that suggested that A3G residues 113 to 128 were important for cytoplasmic localization (2). We therefore hypothesized that residues within amino acids 1 to 60 and amino acids 113 to 128 might cooperatively determine A3G's subcellular distribution. To test this hypothesis, we combined Y19D with single amino acid substitutions within the F126 to D130 region, which is predicted to be solvent exposed (see below and Fig. 7). Specifically, we analyzed the subcellular distribution of A3G-F126S, -W127A, -D128K, -P129R, and -D130K alone and in combination with Y19D. Like the single amino acid substitutions within the first 60 amino acids, none of these mutations by themselves significantly disrupted full-length A3G's cytoplasmic localization (Fig. 6A). However, when combined with Y19D, two of these substitutions, F126S and W127A, caused full-length A3G to distribute cell-wide (Fig. 6B). These data supported the hypothesis that two distinct regions of A3G cooperate to determine cytoplasmic localization.
FIG. 6.
Two regions within the N-terminal half of A3G cooperatively determine the cytoplasmic localization of the full-length protein. Representative images of live HeLa cells transfected with the indicated A3-GFP expression plasmids. Scale bar, 10 μm. Representative cells showing the localization of the indicated full-length A3G single mutants (A) or full-length A3G double mutants (B) are shown.
FIG. 7.
Predicted three-dimensional structure of the N-terminal half of A3G highlighting residues that influence cytoplasmic localization. A3G residues 1 to 65 are colored light green. The conserved zinc-coordinating residues H65, E67, C97, and C100 are colored cyan and labeled (Zn2+). Residues within the first 60 amino acids whose mutation results in a mislocalization of A3G-NTD-GFP are highlighted in red, orange, and yellow according to the degree of mislocalization (>50%, 25 to 50%, or 10 to 25%, respectively). The previously reported CRS (2) is tan, and critical residues within this motif are indicated (F126 and W127). Surface and ribbon representations of the model structure are shown with secondary structures numbered according to the A3G-CTD structure from the N to the C terminus as β1-β2/2′ (not visible here)-α1-β3-α2-β4-α3-β5-α4-α5 (Fig. S1 in the supplemental material) (13). Two residues with a significantly altered localization pattern [L49W and ins(R42)] were not predicted to be on the same surface, but it is probable that these mutations perturb the protein's structure.
Residues critical for cytoplasmic localization are predicted to cluster on a common solvent-exposed surface.
We recently used nuclear magnetic resonance spectroscopy to obtain a high-resolution structure of the CTD of A3G (13). This structural information and sequence homology between the N- and C-terminal domains were used to generate a three-dimensional model of the N-terminal half of A3G (Fig. 7 and Fig. S1 in the supplemental material; see also Materials and Methods). Interestingly, the predicted A3G-NTD structure indicated that most of the critical localization-determining residues, including Y19, Y22, F126, and W127, cluster on a common solvent exposed surface. The proximity of the two critical regions on the protein's surface strongly suggests that they cooperate to mediate an interaction that governs A3G's cytoplasmic localization.
DISCUSSION
Many prior studies have noted that A3G localizes to the cytoplasm of cells in a diffuse cytosolic manner and in some cells in punctate bodies thought to be mRNA-processing centers (P bodies) or other RNA-containing structures (1, 2, 7, 8, 10, 12, 17, 18, 26, 28, 37, 38, 41-43, 45, 47, 52, 54, 55). However, a clear molecular explanation for this property has been elusive. We confirmed here prior work showing that, unlike its homolog AID, A3G is not subject to CRM1-dependent nuclear export and therefore that it is unlikely to be a nucleocytoplasmic shuttling protein (1). We additionally used A3G deletion, chimera, and single amino acid substitution derivatives to show that the first 60 residues strongly influence the protein's cytoplasmic localization. Several N-terminal amino acids, including aromatic residues Y19 and Y22, appeared particularly important.
Several lines of evidence clearly showed that the first 60 amino acids of A3G contribute to cytoplasmic localization. First, N-terminal amino acid conservation between cytoplasmic A3G and A3F, but not nuclear A3B, implicated this region in such a role. Second, replacing the first 60 amino acids of A3G with the corresponding A3B residues caused the chimeric protein to redistribute to the nucleus. Third, replacing the first 60 residues of A3B with this portion of A3G caused the resulting chimera to become predominantly cytoplasmic. Fourth, single amino acid substitutions in this region of A3G caused the N-terminal half of the protein, which is normally cytoplasmic, to redistribute throughout the cell. Fifth, the critical amino acids so identified clustered on a predicted solvent-exposed surface with the potential to mediate an interaction required for cytoplasmic localization. These data therefore combined to demonstrate that this N-terminal portion of A3G provides key determinants of cytoplasmic localization.
However, there was also evidence that this 60-residue region alone is not solely responsible. In particular, the single amino acid substitutions that disrupted the localization of the N-terminal half of the protein failed to mislocalize full-length A3G. A recent study from the Smith group indicated that A3G residues 113 to 128 may also be involved in cytoplasmic localization (2). Our data showed that, like the Y19 region, this region alone is not sufficient for cytoplasmic localization of full-length A3G because mutation of five of the predicted solvent-exposed residues within this region had no effect on the protein's cellular distribution. However, two double mutants that combined substitutions within the two critical regions, Y19D-F126S and Y19D-W127A, caused full-length A3G to distribute cell-wide. These data therefore indicated that these two regions cooperate to determine the cytoplasmic localization of full-length A3G. It was further interesting that these regions were predicted to be juxtaposed on the same solvent-accessible surface (Fig. 7). Taken together with prior studies demonstrating the propensity for A3G to bind RNA and to form ribonucleoprotein complexes (17, 18, 26, 34, 35, 38), it is likely that the surface defined here mediates at least one of these important macromolecular interactions and thereby controls A3G's subcellular distribution.
Supplementary Material
Acknowledgments
We thank G. Haché, B. Hoium, M. Liu, P. Gross, and N. Martemyanova for expert technical assistance; C. Rada and M. Neuberger for the pEGFP-N3-AID plasmid; M. Titus for microscopy facilities; the University of Minnesota Cancer Center Flow Cytometry Core Facility and A. Schumacher, G. Haché, D. MacDuff, M. Huseby, and L. Breshears for thoughtful discussions; and an anonymous reviewer for helpful feedback.
M.D.S. was supported in part by a University of Minnesota Cancer Biology Training Grant (CA009138) and a 3M Science and Technology Graduate Fellowship. H.M. was supported in part by NIH grant AI073167. R.S.H. was supported in part by a Searle Scholarship, a University of Minnesota McKnight Land Grant Assistant Professorship, and NIH grants AI064046 and GM080437.
Footnotes
Published ahead of print on 30 July 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Bennett, R. P., E. Diner, M. P. Sowden, J. A. Lees, J. E. Wedekind, and H. C. Smith. 2006. APOBEC-1 and AID are nucleo-cytoplasmic trafficking proteins but APOBEC3G cannot traffic. Biochem. Biophys. Res. Commun. 350214-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett, R. P., V. Presnyak, J. E. Wedekind, and H. C. Smith. 2008. Nuclear exclusion of the HIV-1 host defense factor APOBEC3G requires a novel cytoplasmic retention signal and is not dependent on RNA binding. J. Biol. Chem. 2837320-7327. [DOI] [PubMed] [Google Scholar]
- 3.Bieniasz, P. D. 2004. Intrinsic immunity: a front-line defense against viral attack. Nat. Immunol. 51109-1115. [DOI] [PubMed] [Google Scholar]
- 4.Bishop, K. N., R. K. Holmes, and M. H. Malim. 2006. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J. Virol. 808450-8458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bishop, K. N., R. K. Holmes, A. M. Sheehy, N. O. Davidson, S. J. Cho, and M. H. Malim. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 141392-1396. [DOI] [PubMed] [Google Scholar]
- 6.Bogerd, H. P., B. P. Doehle, H. L. Wiegand, and B. R. Cullen. 2004. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc. Natl. Acad. Sci. USA 1013770-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bogerd, H. P., H. L. Wiegand, A. E. Hulme, J. L. Garcia-Perez, K. S. O'Shea, J. V. Moran, and B. R. Cullen. 2006. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc. Natl. Acad. Sci. USA 1038780-8785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonvin, M., F. Achermann, I. Greeve, D. Stroka, A. Keogh, D. Inderbitzin, D. Candinas, P. Sommer, S. Wain-Hobson, J. P. Vartanian, and J. Greeve. 2006. Interferon-inducible expression of APOBEC3 editing enzymes in human hepatocytes and inhibition of hepatitis B virus replication. Hepatology 431364-1374. [DOI] [PubMed] [Google Scholar]
- 9.Brar, S. S., M. Watson, and M. Diaz. 2004. Activation-induced cytosine deaminase (AID) is actively exported out of the nucleus but retained by the induction of DNA breaks. J. Biol. Chem. 27926395-26401. [DOI] [PubMed] [Google Scholar]
- 10.Burnett, A., and P. Spearman. 2007. APOBEC3G multimers are recruited to the plasma membrane for packaging into human immunodeficiency virus type 1 virus-like particles in an RNA-dependent process requiring the NC basic linker. J. Virol. 815000-5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chelico, L., P. Pham, P. Calabrese, and M. F. Goodman. 2006. APOBEC3G DNA deaminase acts processively 3′→5′ on single-stranded DNA. Nat. Struct. Mol. Biol. 13392-399. [DOI] [PubMed] [Google Scholar]
- 12.Chen, H., C. E. Lilley, Q. Yu, D. V. Lee, J. Chou, I. Narvaiza, N. R. Landau, and M. D. Weitzman. 2006. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr. Biol. 16480-485. [DOI] [PubMed] [Google Scholar]
- 13.Chen, K. M., E. Harjes, P. J. Gross, A. Fahmy, Y. Lu, K. Shindo, R. S. Harris, and H. Matsuo. 2008. Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G. Nature 452116-119. [DOI] [PubMed] [Google Scholar]
- 14.Chen, K. M., N. Martemyanova, Y. Lu, K. Shindo, H. Matsuo, and R. S. Harris. 2007. Extensive mutagenesis experiments corroborate a structural model for the DNA deaminase domain of APOBEC3G. FEBS Lett. 5814761-4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chester, A., A. Somasekaram, M. Tzimina, A. Jarmuz, J. Gisbourne, R. O'Keefe, J. Scott, and N. Navaratnam. 2003. The apolipoprotein B mRNA editing complex performs a multifunctional cycle and suppresses nonsense-mediated decay. EMBO J. 223971-3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiu, Y. L., and W. C. Greene. 2008. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu. Rev. Immunol. 26317-353. [DOI] [PubMed] [Google Scholar]
- 17.Chiu, Y. L., V. B. Soros, J. F. Kreisberg, K. Stopak, W. Yonemoto, and W. C. Greene. 2005. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature 435108-114. [DOI] [PubMed] [Google Scholar]
- 18.Chiu, Y. L., H. E. Witkowska, S. C. Hall, M. Santiago, V. B. Soros, C. Esnault, T. Heidmann, and W. C. Greene. 2006. High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc. Natl. Acad. Sci. USA 10315588-15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Conticello, S. G., C. J. Thomas, S. K. Petersen-Mahrt, and M. S. Neuberger. 2005. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol. Biol. Evol. 22367-377. [DOI] [PubMed] [Google Scholar]
- 20.Cullen, B. R. 2006. Role and mechanism of action of the APOBEC3 family of antiretroviral resistance factors. J. Virol. 801067-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dang, Y., X. Wang, W. J. Esselman, and Y. H. Zheng. 2006. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J. Virol. 8010522-10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doehle, B. P., A. Schafer, and B. R. Cullen. 2005. Human APOBEC3B is a potent inhibitor of HIV-1 infectivity and is resistant to HIV-1 Vif. Virology 339281-288. [DOI] [PubMed] [Google Scholar]
- 23.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 901051-1060. [DOI] [PubMed] [Google Scholar]
- 24.Fried, H., and U. Kutay. 2003. Nucleocytoplasmic transport: taking an inventory. Cell Mol. Life Sci. 601659-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gallois-Montbrun, S., R. K. Holmes, C. M. Swanson, M. Fernandez-Ocana, H. L. Byers, M. A. Ward, and M. H. Malim. 2008. Comparison of cellular ribonucleoprotein complexes associated with the APOBEC3F and APOBEC3G antiviral proteins. J. Virol. 825636-5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gallois-Montbrun, S., B. Kramer, C. M. Swanson, H. Byers, S. Lynham, M. Ward, and M. H. Malim. 2007. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J. Virol. 812165-2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goff, S. P. 2004. Genetic control of retrovirus susceptibility in mammalian cells. Annu. Rev. Genet. 3861-85. [DOI] [PubMed] [Google Scholar]
- 28.Goila-Gaur, R., M. A. Khan, E. Miyagi, S. Kao, and K. Strebel. 2007. Targeting APOBEC3A to the viral nucleoprotein complex confers antiviral activity. Retrovirology 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haché, G., M. T. Liddament, and R. S. Harris. 2005. The retroviral hypermutation specificity of APOBEC3F and APOBEC3G is governed by the C-terminal DNA cytosine deaminase domain. J. Biol. Chem. 28010920-10924. [DOI] [PubMed] [Google Scholar]
- 30.Harris, R. S., and M. T. Liddament. 2004. Retroviral restriction by APOBEC proteins. Nat. Rev. Immunol. 4868-877. [DOI] [PubMed] [Google Scholar]
- 31.Harris, R. S., S. K. Petersen-Mahrt, and M. S. Neuberger. 2002. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol. Cell 101247-1253. [DOI] [PubMed] [Google Scholar]
- 32.Holmes, R. K., M. H. Malim, and K. N. Bishop. 2007. APOBEC-mediated viral restriction: not simply editing? Trends Biochem. Sci. 32118-128. [DOI] [PubMed] [Google Scholar]
- 33.Ito, S., H. Nagaoka, R. Shinkura, N. Begum, M. Muramatsu, M. Nakata, and T. Honjo. 2004. Activation-induced cytidine deaminase shuttles between nucleus and cytoplasm like apolipoprotein B mRNA editing catalytic polypeptide 1. Proc. Natl. Acad. Sci. USA 1011975-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iwatani, Y., H. Takeuchi, K. Strebel, and J. G. Levin. 2006. Biochemical activities of highly purified, catalytically active human APOBEC3G: correlation with antiviral effect. J. Virol. 805992-6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jarmuz, A., A. Chester, J. Bayliss, J. Gisbourne, I. Dunham, J. Scott, and N. Navaratnam. 2002. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79285-296. [DOI] [PubMed] [Google Scholar]
- 36.Kalderon, D., W. D. Richardson, A. F. Markham, and A. E. Smith. 1984. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature 31133-38. [DOI] [PubMed] [Google Scholar]
- 37.Kinomoto, M., T. Kanno, M. Shimura, Y. Ishizaka, A. Kojima, T. Kurata, T. Sata, and K. Tokunaga. 2007. All APOBEC3 family proteins differentially inhibit LINE-1 retrotransposition. Nucleic Acids Res. 352955-2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kozak, S. L., M. Marin, K. M. Rose, C. Bystrom, and D. Kabat. 2006. The anti-HIV-1 editing enzyme APOBEC3G binds HIV-1 RNA and messenger RNAs that shuttle between polysomes and stress granules. J. Biol. Chem. 28129105-29119. [DOI] [PubMed] [Google Scholar]
- 39.Liddament, M. T., W. L. Brown, A. J. Schumacher, and R. S. Harris. 2004. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr. Biol. 141385-1391. [DOI] [PubMed] [Google Scholar]
- 40.MacDuff, D. A., and R. S. Harris. 2006. Directed DNA deamination by AID/APOBEC3 in immunity. Curr. Biol. 16R186-R189. [DOI] [PubMed] [Google Scholar]
- 41.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 42499-103. [DOI] [PubMed] [Google Scholar]
- 42.Marin, M., S. Golem, K. M. Rose, S. L. Kozak, and D. Kabat. 2008. Human immunodeficiency virus type 1 Vif functionally interacts with diverse APOBEC3 cytidine deaminases and moves with them between cytoplasmic sites of mRNA metabolism. J. Virol. 82987-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marin, M., K. M. Rose, S. L. Kozak, and D. Kabat. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 91398-1403. [DOI] [PubMed] [Google Scholar]
- 44.McBride, K. M., V. Barreto, A. R. Ramiro, P. Stavropoulos, and M. C. Nussenzweig. 2004. Somatic hypermutation is limited by CRM1-dependent nuclear export of activation-induced deaminase. J. Exp. Med. 1991235-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muckenfuss, H., M. Hamdorf, U. Held, M. Perkovic, J. Lower, K. Cichutek, E. Flory, G. G. Schumann, and C. Munk. 2006. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J. Biol. Chem. 28122161-22172. [DOI] [PubMed] [Google Scholar]
- 46.Neville, M., F. Stutz, L. Lee, L. I. Davis, and M. Rosbash. 1997. The importin-beta family member Crm1p bridges the interaction between Rev. and the nuclear pore complex during nuclear export. Curr. Biol. 7767-775. [DOI] [PubMed] [Google Scholar]
- 47.Niewiadomska, A. M., C. Tian, L. Tan, T. Wang, P. T. Sarkis, and X. F. Yu. 2007. Differential inhibition of long interspersed element 1 by APOBEC3 does not correlate with high-molecular-mass-complex formation or P-body association. J. Virol. 819577-9583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perez, O., and T. J. Hope. 2006. Cellular restriction factors affecting the early stages of HIV replication. Curr. HIV/AIDS Rep. 320-25. [DOI] [PubMed] [Google Scholar]
- 49.Rada, C., J. M. Jarvis, and C. Milstein. 2002. AID-GFP chimeric protein increases hypermutation of Ig genes with no evidence of nuclear localization. Proc. Natl. Acad. Sci. USA 997003-7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rogozin, I. B., M. K. Basu, I. K. Jordan, Y. I. Pavlov, and E. V. Koonin. 2005. APOBEC4, a new member of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases predicted by computational analysis. Cell Cycle 41281-1285. [DOI] [PubMed] [Google Scholar]
- 51.Stade, K., C. S. Ford, C. Guthrie, and K. Weis. 1997. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 901041-1050. [DOI] [PubMed] [Google Scholar]
- 52.Stenglein, M. D., and R. S. Harris. 2006. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J. Biol. Chem. 28116837-16841. [DOI] [PubMed] [Google Scholar]
- 53.Wedekind, J. E., G. S. Dance, M. P. Sowden, and H. C. Smith. 2003. Messenger RNA editing in mammals: new members of the APOBEC family seeking roles in the family business. Trends Genet. 19207-216. [DOI] [PubMed] [Google Scholar]
- 54.Wichroski, M. J., K. Ichiyama, and T. M. Rana. 2005. Analysis of HIV-1 viral infectivity factor-mediated proteasome-dependent depletion of APOBEC3G: correlating function and subcellular localization. J. Biol. Chem. 2808387-8396. [DOI] [PubMed] [Google Scholar]
- 55.Wichroski, M. J., G. B. Robb, and T. M. Rana. 2006. Human retroviral host restriction factors APOBEC3G and APOBEC3F localize to mRNA processing bodies. PLoS Pathog. 2e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wiegand, H. L., B. P. Doehle, H. P. Bogerd, and B. R. Cullen. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 232451-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wolff, B., J. J. Sanglier, and Y. Wang. 1997. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem. Biol. 4139-147. [DOI] [PubMed] [Google Scholar]
- 58.Yang, Y., M. P. Sowden, Y. Yang, and H. C. Smith. 2001. Intracellular trafficking determinants in APOBEC-1, the catalytic subunit for cytidine to uridine editing of apolipoprotein B mRNA. Exp. Cell Res. 267153-164. [DOI] [PubMed] [Google Scholar]
- 59.Yang, Y., Y. Yang, and H. C. Smith. 1997. Multiple protein domains determine the cell type-specific nuclear distribution of the catalytic subunit required for apolipoprotein B mRNA editing. Proc. Natl. Acad. Sci. USA 9413075-13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu, Q., D. Chen, R. Konig, R. Mariani, D. Unutmaz, and N. R. Landau. 2004. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J. Biol. Chem. 27953379-53386. [DOI] [PubMed] [Google Scholar]
- 61.Zheng, Y. H., D. Irwin, T. Kurosu, K. Tokunaga, T. Sata, and B. M. Peterlin. 2004. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J. Virol. 786073-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.