Abstract
Actively replicating endogenous retroviruses entered the human genome millions of years ago and became a stable part of the inherited genetic material. They subsequently acquired multiple mutations, leading to the assumption that these viruses no longer replicate. However, certain human tumor cell lines have been shown to release endogenous retroviral particles. Here we show that RNA from human endogenous retrovirus K (HERV-K) (HML-2), a relatively recent entrant into the human genome, can be found in very high titers in the plasma of patients with lymphomas and breast cancer as measured by either reverse transcriptase PCR or nucleic acid sequence-based amplification. Further, these titers drop dramatically with cancer treatment. We also demonstrate the presence of reverse transcriptase and viral RNA in plasma fractions that contain both immature and correctly processed HERV-K (HML-2) Gag and envelope proteins. Finally, using immunoelectron microscopy, we show the presence of HERV-K (HML-2) virus-like particles in the plasma of lymphoma patients. Taken together, these findings demonstrate that elements of the endogenous retrovirus HERV-K (HML-2) can be found in the blood of modern-day humans with certain cancers.
Over the course of millions of years, actively replicating retroviruses entered into the human genome and ultimately became a stable part of the inherited genetic material (3, 35, 43, 45). These viruses are termed human endogenous retroviruses (HERVs), and endogenous retroviral elements make up approximately 8 percent of the human genome (3, 34, 45, 48). HERVs exist in the genome in a proviral form and consist of three genes (gag, pol, and env) flanked by two long terminal repeats (3, 44). After entering the genome, HERVs subsequently acquired multiple mutations and deletions, leading to the assumption that none of them are competent to replicate. In fact, almost all of the proviruses thus far characterized in the human genome appear to be nonfunctional and fixed. However, some have recently been found to contain polymorphisms in different humans, suggesting that they have integrated into the genome relatively recently in human history and may still be evolving (4, 7, 8, 38, 40, 41, 55).
The endogenous retrovirus HERV-K (HML-2) is the most recent entrant into the human genome, having entered 200,000 to 5 million years ago (4, 55), as well as being the most transcriptionally active (29, 47, 49, 51, 54, 57, 59). In the past 2 years, one group has created an infectious clone of HERV-K (HML-2) (23), and another has shown that virus can be generated using several transcomplementary plasmids (32). Thus, at least in its reanimated form with mutations corrected to reintroduce open reading frames, HERV-K (HML-2) can be shown to replicate. Debate exists as to whether HERV-K (HML-2) is still capable of replication in modern humans.
HERV-K (HML-2) has been linked to oncogenesis. Indeed, this virus first came to the notice of biologists due to its similarity to mouse mammary tumor virus (MMTV), a virus that causes mammary tumors in mice, replicates in lymphocytes, and has an exogenous phase and an endogenous phase (9, 37). Consistent with the similarity of HERV-K (HML-2) to MMTV, HERV-K (HML-2) env has been found to be overexpressed in breast cancer tissue, where it also exhibits novel, alternatively spliced forms (24, 56, 57). The overexpression of HERV-K gag has additionally been seen in the peripheral blood cells of leukemia patients (21). It has also been shown that HERV-K (HML-2) is capable of forming viral particles in megakaryocytes from patients with essential thrombocythemia and in cell lines from melanoma, breast cancer, and teratocarcinoma, although it has not been demonstrated that these particles are infectious (10, 16, 36, 39, 42, 49).
The mechanisms by which HERV-K (HML-2) might prove to be oncogenic, if it is at all, have only recently begun to be elucidated. In this regard, HERV-K (HML-2), which has two forms, type 1 and type 2, can encode at least two putative oncoproteins (1, 2, 15, 20). Type 1 encodes the newly identified Np9 oncoprotein, whereas type 2 encodes an accessory protein, Rec, which is necessary to export unspliced RNA from the nucleus to the cytoplasm like its counterparts human immunodeficiency virus type 1 (HIV-1) Rev and human T-cell leukemia virus type 1 Rex (38, 58). Both Np9 and Rec have been shown to be capable of cellular transformation under certain circumstances, and Rec can induce carcinoma in situ in mice (1, 2, 6, 11, 15, 20, 25, 38).
We recently made the observation that HERV-K (HML-2) RNA can be found in the plasma of HIV-1-infected patients (18, 19). In view of this and the factors cited above, we investigated whether HERV-K (HML-2) RNA might also be present in the blood of patients with either lymphoma or breast cancer. We report that these patients indeed have extremely high titers of viral RNA in their blood, and these titers fall precipitously when patients are treated for lymphoma. We further demonstrate that individuals with lymphoma who have high titers of HERV-K (HML-2) RNA also have reverse transcriptase activity and viral Gag and Env proteins in the same plasma fractions in which viral RNA is found. Finally, using electron microscopy (EM) and immunogold staining we visualized, for the first time, the presence of HERV-K (HML-2)-like particles in the plasma of people (lymphoma patients). These findings demonstrate that HERV-K (HML-2) viral elements can be found circulating in the blood of humans with lymphoma and breast cancer.
MATERIALS AND METHODS
Study subjects.
Following informed consent, plasma samples were obtained from human subjects following protocols approved by the Institutional Review Boards of the University of Michigan and the North Shore University Hospital. Subjects included 18 healthy individuals and patients with rheumatoid arthritis, breast cancer, HIV infection with diffuse large B-cell lymphoma, non-HIV-associated diffuse large B-cell lymphoma, and HIV infection with Hodgkin lymphoma.
Viral RNA extraction.
For reverse transcriptase PCR (RT-PCR), plasma was treated with 200 U RNase-free DNase (Roche Laboratories) for 2 h. RNA was extracted from 140 μl of plasma using the QIAamp viral RNA minikit following the manufacturer's instructions (Qiagen, Valencia, CA). Absence of DNA contamination was confirmed by PCR. For nucleic acid sequence-based amplification (NASBA), RNA was extracted using the NucliSENS easyMAG extraction method (Biomerieux, France) (17).
Synthesis of RNA transcripts in vitro and real-time RT-PCR.
Synthesis of RNA transcripts for calibration and real-time RT-PCR were performed as described previously (18). Primers were designed to amplify and quantify the HERV-K gag (KgagRTF, 5′-AGC AGG TCA GGT GCC TGT AAC ATT-3′; KgagRTR, 5′-TGG TGC CGT AGG ATT AAG TCT CCT-3′) and the HERV-K env tm (KenvTMF, 5′-GCT GTA GCA GGA GTT GCA TTG-3′; KenvTMR, 5′-TAA TCG ATG TAC TTC CAA TGG TC-3′).
NASBA.
NASBA amplification was performed as described previously (17), using primers specific to the HERV-K (HML-2) env (Ktype1F, 5′-AGA AAA GGG CCT CCA CGG AGA TG-3′; Ktype1R, 5′-AAT TCT AAT ACG ACT CAC TAT AGG GAG AAG GCT CTC CCT AGG CAA ATA GGA-3′; Ktype2F, 5′-AGA CAC CGC AAT CGA GCA CCG TTG A-3′; and Ktype2R, 5′-AAT TCT AAT ACG ACT CAC TAT AGG GAG AAG GAT CAA GGC TGC AAG CAG CAT ACT C-3′) with molecular beacons specific for type 1 and type 2 viruses labeled with the fluorophore FAM (6-carboxyfluorescein) or ROX (6-carboxy-X-rhodamine) at the 5′ end and a quencher (Dabsyl) at the 3′ end. Probe sequences were Ktype1P (5′-FAM-CGA TCG ACG GAG ATG GTA ACA CCA GTC ACA TGG ACG ATC G-3′) and Ktype2P (5′-ROX-CGA TCG AAG TTG CCA TCC ACC AAG AAG GCA GAC GAT CG-ROX-3′).
Iodixanol gradients and viral particle purification.
One milliliter of each plasma sample was diluted in 10 ml phosphate-buffered saline (PBS) and centrifuged at 3,000 rpm for 10 min. Supernatants were overlaid on 20% iodixanol cushions (Optiprep density gradient medium; Sigma, St. Louis, MO) and centrifuged at 45,000 × g for 2 h. Pellets were resuspended in 500 μl PBS and overlaid in 50% iodixanol solution in 0.85% NaCl. A self-gradient was achieved through ultracentrifugation in a vertical fixed-angle rotor at 350,000 × g for 6 h. Fractions of 350 μl were collected, and their density was calculated by measuring the absorbance at 340 nm and also reading the refraction index.
RT assays.
The RT activity was measured in 5 μl of each fraction using the EnzCheck RT assay kit (Invitrogen) as described by the manufacturer. Serial dilutions of murine leukemia virus RT (Stratagene) were used as calibrators.
Western blotting.
Proteins were precipitated from the fractions with chloroform-methanol, separated on 10% sodium dodecyl sulfate-polyacrylamide gels, and blotted onto nitrocellulose membranes. The membranes were blocked in 10% milk for 1 h and incubated with primary anti-HERV-K Env (HERM-1811-5) or anti-HERV-K Gag (HERM-1841-5) monoclonal antibody (Austral Biologicals) in blocking solution. As a control for nonspecific cross-reactivity, blots were incubated with mouse serum. Membranes were washed five times in PBS containing 1% Tween. The bound primary antibody was then detected with horseradish peroxidase-conjugated goat anti-mouse secondary antibody using the Super Signal West Pico system (Pierce Chemical Co., Rockford, IL).
EM.
Plasma fractions obtained by iodixanol gradients were diluted in PBS, and particles were pelleted at 250,000 × g. Negative staining, conventional EM, and immunogold labeling of particles were performed as described previously (30). Briefly, particles were absorbed to 300-mesh nickel grids and blocked in Tris-buffered saline-1% bovine serum albumin. Particles were incubated with primary mouse monoclonal anti-HERV-K Env (HERM-1811-5, diluted 1:10 in blocking solution) (Austral Biologicals) for 1 h. In addition, another set of grids was incubated with purified mouse immunoglobulin G (IgG) for 1 h to control for cross-reactivity of mouse serum with plasma proteins. Grids were washed five times and incubated with gold-labeled anti-mouse secondary antibody for 1 h. In the experiments done with 10-nm gold particles, the secondary antibodies came from Sigma (St. Louis, MO), and in experiments done with 5-nm gold particles, the secondary antibodies came from BB International (Madison, WI). Particles were fixed in 2% glutaraldehyde, negative stained in 2% uranyl acetate, and visualized with a Philips CM-100 (Ann Arbor, MI) or a CM-10 (Siena) transmission electron microscope operating at 80 kV.
RESULTS
We recently made the observation that HERV-K (HML-2) RNA, but not that of the related HERV, HERV-H, can be found in the plasma of HIV-1-positive patients using a quantitative real-time RT-PCR method to amplify the gag and pol transcripts of HERV-K (HML-2) (18, 19). In these experiments, plasma was treated with DNase prior to extraction of RNA in order to try to eliminate contaminating DNA that could cause false-positive RT-PCR results. Using degenerate primers that amplify all 10 HERV-K families (HML-1 to HML-10) present in the human genome, we found that expression of only HML-2 and HML-3 was increased in HIV-1 patients. RNA from the viruses of the HERV-H family, of which there are at least 1,000 copies in the human genome as opposed to ∼50 copies of HERV-K (HML-2), was not detected in plasma (18, 19). The finding that plasma HERV-K RNA titers are substantially elevated in HIV-1-infected individuals has now been confirmed by at least two other groups (26, 32). As HERV-K (HML-2) bears considerable resemblance to MMTV, which causes mammary tumors in rodents and replicates in lymphocytes (9, 37), we investigated whether titers of HERV-K (HML-2) are also elevated in the plasma of humans with breast cancer or lymphoma. As measured by the same quantitative real-time RT-PCR assay for HERV-K (HML-2) gag RNA, control individuals generally had little to no evidence of gag in their plasma (102 copies/ml on average), whereas patients with either HIV- or non-HIV-associated lymphomas had very high titers (ranging up to 1010 copies of RNA per ml, with an average of 107 copies/ml), and patients with breast cancer also had high titers of gag RNA in their plasma (Fig. 1A).
FIG. 1.
HERV-K (HML-2) RNA titers in the plasma of patients and healthy individuals. (A) Viral RNA was isolated from plasma samples, and the HERV-K (HML-2) gag RNA load was measured by real-time RT-PCR. (B and C) The env RNA loads of type 1 (B) and type 2 (C) HERV-K (HML-2) strains were quantified by NASBA. Bars indicate the log median HERV-K (HML-2) RNA viral load. A statistically significant difference was observed between the mean HERV-K (HML-2) RNA load of patients with lymphoma (t test, P < 0.0001) or breast cancer (t test, P < 0.0001) and that of healthy individuals.
To further prove that high titers of HERV-K (HML-2) viral RNA could be found in the plasma of lymphoma and breast cancer patients, we employed a second, independent RNA amplification method, NASBA, which does not require thermocycling and is not interfered with by contaminating DNA that might be present in plasma (see Fig. S1 in the supplemental material). In addition, we used this technique to amplify viral env RNA, rather than gag as was used in the RT-PCRs. Using this methodology, we were able to detect the two different forms of HERV-K (HML-2), type 1 and type 2, which are distinguished by a 292-bp fragment in env that is present in type 2 but deleted in type 1. As can be seen in Fig. 1B, very high titers of type 1 env RNA were again seen in breast cancer and lymphoma patients but not in normal controls. A similar pattern was seen with type 2 HERV-K (HML-2), except that here the titers were considerably lower, or nonexistent, in Hodgkin lymphoma (Fig. 1C). Therefore, not only are there very high titers of HERV-K (HML-2) RNA in the plasma of lymphoma patients, as opposed to healthy people, there is specificity as to the type of HERV-K (HML-2) found in individuals with distinct types of lymphoma. In lymphoma and breast cancer patients, sequencing of HERV-K (HML-2) RNAs confirmed the transcriptional activation of env sequences from at least 16 different type 1 and 18 different type 2 polymorphic HERV-K (HML-2) viruses (data not shown). Strikingly, when patients were treated and their lymphoma went into remission, titers of HERV-K (HML-2) became undetectable in most cases (Fig. 2). In two patients who received incomplete treatment or obtained only a partial remission, titers dropped but still remained well above the normal range.
FIG. 2.
HERV-K (HML-2) plasma titers in HIV-associated lymphoma patients at time of diagnosis and remission. Plasma was available pre- and posttreatment for nine patients (four with diffuse large B-cell lymphoma, three with Hodgkin lymphoma, one with Burkitt lymphoma, and one with marginal zone lymphoma). The plasma HERV-K (HML-2) gag viral load was measured by real-time RT-PCR. The P value was calculated using the t test and comparing the mean HERV-K (HML-2) RNA loads at diagnosis and upon remission. Bars indicate the log median HERV-K (HML-2) RNA viral loads pre- and posttreatment. One patient with diffuse large B-cell lymphoma was treated with foscarnet alone, which resulted in dramatic shrinkage of the tumor. Four other patients with diffuse large B-cell lymphoma were treated with cycles of cyclophosphamide, adriamycin, vincristine, and prednisone. The one patient with Burkitt lymphoma was treated with cycles of cyclophosphamide, vincristine, prednisone, and intrathecal methotrexate. The single patient with marginal zone lymphoma was treated with rituxan plus cyclophosphamide, adriamycin, vincristine, and prednisone. The patients with Hodgkin disease were treated with cycles of adriamycin, bleomycin, velban, prednisone, and dacarbazine. In two patients, viral loads did not become undetectable following treatment: one patient with Hodgkin lymphoma completed only three of six cycles of chemotherapy, and the single patient with marginal zone lymphoma had only a partial remission. The other seven patients completed therapy and achieved complete and sustained remissions.
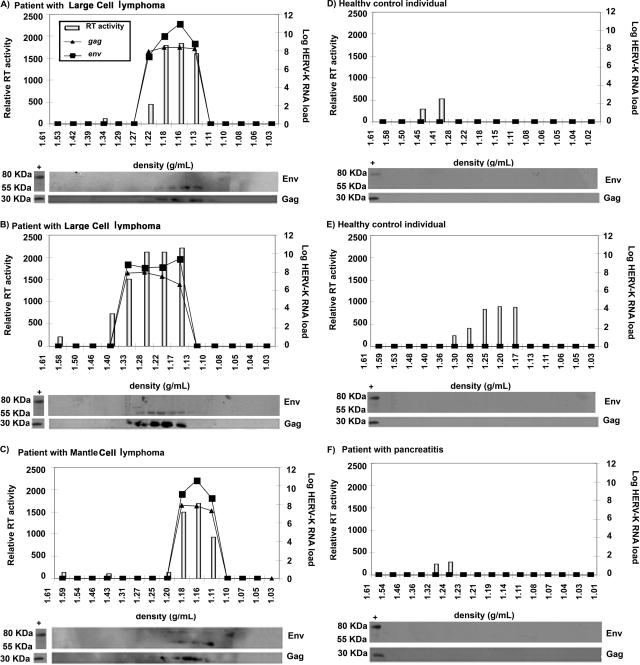
Having obtained evidence using two independent RNA amplification methods (NASBA and RT-PCR) that HERV-K (HML-2) elements are likely found in the plasma of patients with lymphoma, we examined this issue further using additional parameters. We studied the plasma of three HIV-negative lymphoma patients with high titers of HERV-K (HML-2) RNA, as detected by RT-PCR, and three control individuals without lymphoma (two healthy young people and one lymphoma-free individual with pancreatitis) who lacked detectable HERV-K (HML-2) RNA titers. Plasma specimens from these individuals were first subjected to iodixanol gradient ultracentrifugation in order to fractionate the plasma by density. We then looked for viral gag and env RNA using RT-PCR, for RT activity using a standard enzymatic assay, and for Gag and Env proteins using Western blotting in the different fractions obtained from the iodixanol gradients. Protein extract from the HERV-K (HML-2) particle-producing cell line NCCIT was used as a control for Western blotting, since both HERV-K (HML-2) Gag and Env proteins are produced by these cells (10). As can be seen in Fig. 3, no viral RNA or viral protein, and very little RT activity, was found in the three control individuals who had no detectable RNA in plasma by RT-PCR. In contrast, in the three lymphoma patients, RT activity, viral gag and env RNAs, and correctly processed viral Gag and Env proteins were all seen, in each case in the fractions that were at the appropriate density (1.15 to 1.17 g/ml) for a putative HERV-K (HML-2) virus (22, 23, 50). The sizes of the Gag (30 kDa) and Env (55 kDa) proteins detected by Western blotting in all three lymphoma patients correspond to the processed Gag protein and the Env surface (SU) subunit, indicating that these proteins were correctly processed (10, 12, 13, 22, 23, 34). This observation suggested that some HERV-K (HML-2) particles found in the blood might be mature virions. Further, as cleavage of the Env and Gag proteins by the viral protease is required for virion maturation (50), the presence of processed Gag and Env indicates that the viral protease is likely functional in this setting. One lymphoma patient (Fig. 3C) showed not only the 30-kDa band, corresponding to the processed Gag protein, and the 55-kDa band, representing the processed Env SU subunit, but also an 80-kDa band corresponding to the unprocessed Env (10, 12, 23). This suggests that immature and mature HERV-K virions may coexist in the blood of this patient.
FIG. 3.
HERV-K RNA titers, proteins, and RT activity in fractions from plasma samples of healthy individuals and non-HIV lymphoma patients. Plasma samples from three patients with lymphoma (large-cell lymphoma [A and B] and mantle cell lymphoma [C]) and three control individuals (healthy individuals [D and E] and a patient with pancreatitis [F]) were fractionated by density in iodixanol gradients. The density of each fraction is given on the x axis. The RT activity in each fraction, represented by the light bars, is given on the y axis in relative fluorescent units. The HERV-K (HML-2) gag and env RNA titers (lines) were assessed by real-time RT-PCR. HERV-K (HML-2) viral proteins were detected by Western blotting of protein extracts from each fraction. Cell lysates from the HERV-K (HML-2) particle-producing cell line NCCIT were used as a positive control (first lane of the Western blot in each panel). The band representing processed Env is found at 55 kDa and that corresponding to the unprocessed Env at 80 kDa.
It is generally very difficult to detect pathogenic viral particles in the blood of patients by EM; for example, this has never been done successfully with HIV-1-infected patients. However, a few RNA viruses found in very high titers in the blood, such as hepatitis C virus, can be seen directly in the blood by EM (30). As some of our lymphoma patients had HERV-K (HML-2) RNA titers of as high as 1010 copies/ml, we searched for evidence of viral particles in the fractionated plasma of lymphoma patients. Indeed, we were able to detect viral particles in the fraction (density, ∼1.16 g/ml) containing HERV-K (HML-2) viral RNA and proteins (Fig. 4A to C), and prominent spikes characteristic of a retroviral envelope can be seen in some particles (for example, in Fig. 4A). We then performed immuno-EM using an antibody against the HERV-K (HML-2) Env SU protein (Fig. 4D to M). In spite of the difficulty of preserving virions from envelope shearing as a result of ultracentrifugation in gradients such as sucrose (30), using iodixanol gradients we were able to detect clusters of gold-labeled viral particles with an anti-HERV-K (HML-2) Env mouse monoclonal antibody. Some particles showed condensed cores and spikes symmetrically distributed around the viral membrane, suggesting mature viruses. No similar clustering of gold particles was seen in viral preparations probed with purified control mouse IgG antibody and detected with gold-labeled secondary antibody (Fig. 4N to O). These EM findings, confirmed in two independent laboratories (Ann Arbor, MI, and Siena, Italy), demonstrate that the viral particles, some mature and some immature, seen in the blood of the lymphoma patients are indeed HERV-K (HML-2). Thus, multiple lines of evidence obtained in our laboratories suggest the presence of HERV-K (HML-2) elements in the plasma of modern-day human beings with cancer.
FIG. 4.
HERV-K (HML-2) particles are found in the plasma of lymphoma patients. Viral particles found in the plasma fraction at a density of 1.16 g/ml were visualized by EM. (A to C) Particles were found in all lymphoma patients. High magnification of one particle (A) shows mature morphology with a condensed core and prominent spikes. Two particles (B and C) show Env spikes, but the core is incompletely condensed, indicating that these virions are likely still not completely mature. (D to M) Immuno-EM images of particles labeled with an antibody specific to the HERV-K Env and a secondary antibody linked to 5 (D to K) or 10-nm gold particles (L and M). All viral particles detected have a size of between 90 and 100 nm, as expected for HERV-K (HML-2). Some particles (G, I, J, K, and L) showed condensed cores and spikes symmetrically distributed around the viral membrane, suggesting mature viruses. (N and O) No clustering of gold particles was observed in the negative control in which the same preparations were labeled with purified mouse IgG antibody and detected with gold-labeled (5-nm particles) secondary antibody.
DISCUSSION
Virus-like particles generated by mammalian tissues have often been found to have their genesis in endogenous retroviral proviruses. Mouse tissues generate such endogenous retrovirus-derived virus-like particles under both pathological and normal conditions, including in the developing embryo (31, 46, 60). In addition to mice, the presence of endogenous retroviruses and retroviral particles has been linked to cancer in several other animals, including humans. Recently, Tarlinton et al. isolated a new retrovirus from koala bears, termed KoRV. This virus was isolated from koalas that developed lymphomas. The authors showed that KoRV is a new endogenous koala retrovirus currently in transition between an exogenous and endogenous phase. Furthermore, KoRV is found in high titers in the blood of certain koalas as demonstrated by real-time RT-PCR (52, 53). Interestingly, in these koalas there is a strong association between plasma viral load, as assessed by RNA levels, and the development of leukemia/lymphoma (51). HERV-K (HML-2) virus-like particles have previously been found in cell lines from human teratocarcinoma, melanoma, and breast cancer (10, 42, 49). Here we demonstrate, for the first time, that HERV-K (HML-2) RNA can be found in the blood of human patients with lymphoma, as well as in patients with breast cancer. In addition, these patients have remarkably high plasma titers of HERV-K (HML-2) RNAs, and these titers drop dramatically with successful treatment of lymphoma. By analogy to MMTV, it is likely that mammary or lymphocytic tumor cells are the main source of production of HERV-K (HML-2) particles, the number of which appears to be drastically decreased in the plasma after the tumor burden is reduced by chemotherapy. However, ongoing experiments in our laboratory aim to clarify that this is indeed the case.
It must be cautioned that what we are detecting as HERV-K (HML-2) plasma RNA could conceivably be viral DNA that is released in response to the increased cellular proliferation and turnover seen with malignancy (27, 28). Several factors suggest that this is not likely the case, however. First, we detected no amplification of HERV-K (HML-2) RNA when RT was not added into the reaction mixtures (reference 19 and data not shown). Second, the RT-PCRs are performed in the presence of DNase, and, perhaps more significantly, we obtained very similar results with both RT-PCR and NASBA, the latter being a method of amplifying RNA that does not require thermocycling and is not interfered with by the presence of free DNA. In addition, even intentional contamination of NASBA reactions did not lead to amplification of DNA (see Fig. S1 in the supplemental material). Thus, while it is impossible to completely rule out DNA contamination, these considerations, as well as the presence of viral particles and proteins in the plasma of these patients, suggest that it is much more likely to be HERV-K (HML-2) RNA than DNA that we are detecting in these studies.
In addition to HERV-K (HML-2) RNA, we have also presented evidence suggesting the presence of other viral elements in the plasma of lymphoma patients. First, when plasma is separated over iodixanol gradients, the fractions corresponding to the appropriate density for a retrovirus contain RT activity, HERV-K (HML-2) gag and env RNA, and Gag and Env proteins, whereas no RT activity, viral RNA, or viral proteins are seen in the plasma of control individuals. Further, likely because the titers of virus-like particles are so high, we were able to visualize HERV-K (HML-2)-like particles in the plasma of the lymphoma patients. Immunogold staining demonstrated that these particles are indeed quite likely to be HERV-K (HML-2), as suggested by the Western blots, RT-PCR, and NASBA results. To our knowledge, this is the first demonstration that, similar to the case for the supernatants of human malignant cell lines, HERV-K (HML-2) particles can be found in the sera of actual cancer patients. Although the presence of these viral elements correlates with disease, whether HERV-K (HML-2) plays an actively pro-oncogenic role remains to be elucidated. However, should these viral elements in the plasma ultimately prove to be from truly infectious viral particles (see discussion below), targeting HERV-K (HML-2) with antiretroviral compounds might ultimately emerge as a therapeutic strategy in patients with lymphoma or breast cancer. Therefore, our findings have the potential to affect both the understanding of viral oncogenesis and therapies for important malignancies.
Whether or not replicating HERV-K (HML-2) plays a role in the pathogenesis of breast cancer or lymphoma, it appears that, consistent with the koala data (52, 53), HERV-K (HML-2) viral loads may prove to be an important new biomarker in these diseases. First, whereas some biomarkers in clinical use show changes over a range of a single log unit, the titers of HERV-K (HML-2) RNA in patients versus controls are markedly different: normal individuals have titers of 102 copies/ml on average, whereas patients with lymphoma can have titers of up to 1010 copies/ml. In addition, while the specificity of these findings will require significantly greater investigation before conclusions are reached, it does appear that there are differences within cancer groupings that are not simply based on overall titer. For example, patients with Hodgkin disease have very high titers of HERV-K (HML-2) type 1 but have negligible titers of HERV-K (HML-2) type 2 (Fig. 1B and C). Finally, we find that when patients are successfully treated for lymphoma (Fig. 2), the titers of HERV-K (HML-2) RNA in the blood return to low or even undetectable levels. Thus, HERV-K (HML-2) titers have the potential to be developed into a badly needed biomarker for this important cancer. However, it must be emphasized that further work must be done before the true clinical utility of HERV-K (HML-2) as a biomarker is established.
Endogenous retroviral elements make up approximately 8 percent of the human genome (33, 45), and they are generally considered to have lost the ability to replicate due to having acquired multiple mutations. Further, there is a paucity of complete functional elements [those that encode functional copies of all HERV-K (HML-2) genes contiguously] to be found in the human genome. Two groups have recently shown that reanimated versions of HERV-K (HML-2), made from cloned constructs in which mutations have been corrected, are able to replicate (23, 34). These elegant experiments demonstrate that HERV-K (HML-2) could indeed replicate in the past, but they do not directly address whether HERV-K (HML-2) can still replicate in modern humans. Interestingly, at least one full-length HERV-K with intact open reading frames, HERV-K 113, has been found to be present in about 15 to 30 percent of individuals who have been tested, and genetic analysis reveals that it is a very recent addition to the human genome (14, 55). This led the authors to suggest that HERV-K 113 is a candidate for active replication in modern humans, but three groups have now produced evidence that HERV-K113 is by itself likely defective, at least in cell culture assays (5, 13, 22). Belshaw and colleagues have shown that HERV-K (HML-2) has been under continuous purifying selection, a finding that they interpret to suggest that proliferation of this family has been almost entirely due to germ line reinfection (7, 8). Conservation of the env gene was further thought to support this idea, as it would suggest a need for ongoing reinfection in the life cycle of these viral elements. Finally, at least eight elements from the HERV-K (HML-2) family appear to be polymorphic with respect to their presence in the human population, indicating that they have inserted into the human genome subsequent to the last common ancestor of humans and chimpanzees (7, 8, 38, 40, 41). Therefore, while the majority of investigators today believe that HERV-K (HML-2) is no longer capable of replication in modern humans, some evidence to the contrary does exist. Our finding of HERV-K (HML-2) RNA, RT activity, processed viral proteins, and what appear to be mature viral particles in the blood of cancer patients raises the issue of whether active HERV-K (HML-2) replication might take place in these patients under some circumstances. However, proof to that effect will require transmission of the modern virus in the laboratory.
Supplementary Material
Acknowledgments
We thank Rino Rappuoli, John Donnelly, John Moran, Steve Goff, and Joseph Pagano for their thoughtful comments on the work; Donna Gschwend for manuscript preparation; Dotty Sorenson for help with the electron microscopy studies; and Judith Estes and Kathryn Jacobi for patient recruitment.
This work was supported primarily by a generous grant to M.H.K. from the Concerned Parents for AIDS Research (05-5089), with additional funding coming from a gift from students at Roslyn High School, Roslyn, NY. D.F.H. was supported by the Fashion Footwear Charitable Foundation of New York/QVC Presents Shoes on Sale. S.D.G. was supported by a grant from the Michigan Institute for Clinical and Health Research. D.M.M. was supported by grant R01 AI062248 from the National Institutes of Health and is the recipient of a Burroughs Wellcome Fund Clinical Scientist Award in Translational Research.
Footnotes
Published ahead of print on 16 July 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Armbruester, V., M. Sauter, E. Krautkraemer, E. Meese, A. Kleiman, B. Best, K. Roemer, and N. Mueller-Lantzsch. 2002. A novel gene from the human endogenous retrovirus K expressed in transformed cells. Clin. Cancer Res. 81800-1807. [PubMed] [Google Scholar]
- 2.Armbruester, V., M. Sauter, K. Roemer, B. Best, S. Hahn, A. Nty, A. Schmid, S. Philipp, A. Mueller, and N. Mueller-Lantzsch. 2004. Np9 protein of human endogenous retrovirus K interacts with ligand of numb protein X. J. Virol. 7810310-10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannert, N., and R. Kurth. 2004. Retroelements and the human genome: new perspectives on an old relation. Proc. Natl. Acad. Sci. USA 10114572-14579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbulescu, M., G. Turner, M. Seaman, A. Deinard, K. Kidd, and J. Lenz. 1999. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr. Biol. 9861-868. [DOI] [PubMed] [Google Scholar]
- 5.Beimforde, N., K. Hanke, I. Ammar, R. Kurth, and N. Bannert. 2008. Molecular cloning and functional characterization of the human endogenous retrovirus K113. Virology 371216-225. [DOI] [PubMed] [Google Scholar]
- 6.Belancio, V. P., D. J. Hedges, and P. Deininger. 2008. Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health. Genome Res. 18343-358. [DOI] [PubMed] [Google Scholar]
- 7.Belshaw, R., A. L. A. Dawson, J. Woolven-Allen, J. Redding, A. Burt, and M. Tristem. 2005. Genomewide screening reveals high levels of insertional polymorphism in the human endogenous retrovirus family HERV-K(HML2): implications for present-day activity. J. Virol. 7912507-12514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belshaw, R., V. Pereira, A. Katzourakis, G. Talbot, J. Paces, A. Burt, and M. Tristem. 2004. Long-term reinfection of the human genome by endogenous retroviruses. Proc. Natl. Acad. Sci. USA 1014894-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhadra, S., M. M. Lozano, and J. P. Dudley. 2005. Conversion of mouse mammary tumor virus to a lymphomagenic virus. J. Virol. 7912592-12596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bieda, K., A. Hoffmann, and K. Boller. 2001. Phenotypic heterogeneity of human endogenous retrovirus particles produced by teratocarcinoma cell lines. J. Gen. Virol. 82591-596. [DOI] [PubMed] [Google Scholar]
- 11.Boese, A., M. Sauter, U. Galli, B. Best, H. Herbst, J. Mayer, E. Kremmer, K. Roemer, and N. Mueller-Lantzsch. 2000. Human endogenous retrovirus protein cORF supports cell transformation and associated with the promyelocytic leukemia zinc finger protein. Oncogene 194328-4336. [DOI] [PubMed] [Google Scholar]
- 12.Boller, K., H. Konig, M. Sauter, N. Mueller-Lantzsch, R. Löwer, J. Löwer, and R. Kurth. 1993. Evidence that HERV-K is the endogenous retrovirus sequence that codes for the human teratocarcinoma-derived retrovirus HTDV. Virology 196349-353. [DOI] [PubMed] [Google Scholar]
- 13.Boller, K., K. Schönfeld, S. Lischer, N. Fischer, A. Hoffmann, R. Kurth, and R. R. Tönjes. 2008. Human endogenous retrovirus HERV-K113 is capable of producing intact viral particles. J. Gen. Virol. 89567-572. [DOI] [PubMed] [Google Scholar]
- 14.Burmeister, T., A. D. Ebert, W. Pritze, C. Loddenkemper, S. Schwartz, and E. Thiel. 2004. Insertional polymorphisms of endogenous HERV-K113 and HERV-K115 retroviruses in breast cancer patients and age-matched controls. AIDS Res. Hum. Retroviruses 201223-1229. [DOI] [PubMed] [Google Scholar]
- 15.Büscher, K., S. Hahn, M. Hofmann, U. Trefzer, M. Ozel, W. Sterry, J. Löwer, R. Löwer, R. Kurth, and J. Denner. 2006. Expression of the human endogenous retrovirus-K transmembrane envelope, Rec and Np9 proteins in melanoma cell lines. Melanoma Res. 16223-234. [DOI] [PubMed] [Google Scholar]
- 16.Büscher, K., U. Trefzer, M. Hofmann, W. Sterry, R. Kurth, and J. Denner. 2005. Expression of human endogenous retrovirus K in melanomas and melanoma cell lines. Cancer Res. 654172-4180. [DOI] [PubMed] [Google Scholar]
- 17.Caliendo, A. M., J. Ingersoll, A. M. Fox-Canale, S. Pargman, T. Bythwood, M. K. Hayden, J. W. Bremer, and N. S. Lurain. 2007. Evaluation of real-time PCR laboratory-developed tests using analyte-specific reagents for cytomegalovirus quantification. J. Clin. Microbiol. 451723-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Contreras-Galindo, R., M. Gonzalez, S. Almodovar, S. Gonzalez-Ramirez, E. Lorenzo, and Y. Yamamura. 2006. A new real-time-RT-PCR for quantitation of human endogenous retroviruses type K (HERV-K) RNA load in plasma samples: increased HERV-K RNA titers in HIV-1 patients with HAART non-suppressive regimens. J. Virol. Methods 13651-57. [DOI] [PubMed] [Google Scholar]
- 19.Contreras-Galindo, R., M. H. Kaplan, D. M. Markovitz, E. Lorenzo, and Y. Yamamura. 2006. Detection of HERV-K (HML-2) viral RNA in plasma of HIV type 1-infected individuals. AIDS Res. Hum. Retroviruses 22979-984. [DOI] [PubMed] [Google Scholar]
- 20.Denne, M., M. Sauter, V. Armbruester, J. D. Licht, K. Roemer, and N. Mueller-Lantzsch. 2007. Physical and functional interactions of human endogenous retrovirus proteins Np9 and rec with the promyelocytic leukemia zinc finger protein. J. Virol. 815607-5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Depil, S., C. Roche, P. Dussart, and L. Prin. 2002. Expression of a human endogenous retrovirus, HERV-K, in the blood cells of leukemia patients. Leukemia 16254-259. [DOI] [PubMed] [Google Scholar]
- 22.Dewannieux, M., S. Blaise, and T. Heidmann. 2005. Identification of a functional envelope protein from the HERV-K family of human endogenous retroviruses. J. Virol. 7915573-15577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dewannieux, M., F. Harper, A. Richaud, C. Letzelter, D. Ribet, G. Pierron, and T. Heidmann. 2006. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res. 161548-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ejthadi, H. D., J. H. Martin, J. Junying, D. A. Roden, M. Lahiri, P. Warren, P. G. Murray, and P. N. Nelson. 2005. A novel multiplex RT-PCR system detects human endogenous retrovirus-K in breast cancer. Arch. Virol. 150177-184. [DOI] [PubMed] [Google Scholar]
- 25.Galli, U. M., M. Sauter, B. Lecher, S. Maurer, H. Herbst, K. Roemer, and N. Mueller-Lantzsch. 2005. Human endogenous retrovirus rec interferes with germ cell development in mice and may cause carcinoma in situ, the predecessor lesion of germ cell tumors. Oncogene 243223-3228. [DOI] [PubMed] [Google Scholar]
- 26.Garrison, K. E., R. B. Jones, D. A. Meiklejohn, N. Anwar, L. C. Ndhlovu, J. M. Chapman, A. L. Erickson, A. Agrawal, G. Spotts, F. M. Hecht, S. Rakoff-Nahoum, J. Lenz, M. A. Ostrowski, and D. F. Nixon. 2007. T cell responses to human endogenous retroviruses in HIV-1 infection. PloS Pathog. 3e165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giacona, M. B., G. C. Ruben, K. A. Iczkowski, T. B. Roos, D. M. Porter, and G. D. Sorenson. 1998. Cell-free DNA in human blood plasma: length measurements in patients with pancreatic cancer and healthy controls. Pancreas 1789-97. [DOI] [PubMed] [Google Scholar]
- 28.Jahr, S., H. Hentze, S. Englisch, D. Hardt, F. O. Fackelmayer, R. D. Hesch, and R. Knippers. 2001. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 611659-1665. [PubMed] [Google Scholar]
- 29.Johnston, J., C. Silva, J. Holden, K. Warren, A. Clark, and C. Power. 2001. Monocyte activation and differentiation augment human endogenous retrovirus expression: implications for inflammatory brain diseases. Ann. Neurol. 50434-442. [DOI] [PubMed] [Google Scholar]
- 30.Kaito, M., S. Watanabe, K. Tsukiyama-Kohara, K. Yamaguchi, Y. Kobayashi, M. Konishi, M. Yokoi, S. Ishida, S. Suzuki, and M. Kohara. 1994. Hepatitis C virus particle detected by immunoelectron microscopic study. J. Gen. Virol. 751755-1760. [DOI] [PubMed] [Google Scholar]
- 31.Kigami, D., N. Minami, H. Takayama, and H. Imai. 2003. MuERV-L is one of the earliest transcribed genes in mouse one-cell embryos. Biol. Reprod. 68651-654. [DOI] [PubMed] [Google Scholar]
- 32.Laderoute, M. P., A. Giulivi, L. Larocque, D. Bellfoy, Y. Hou, H. X. Wu, K. Fowke, J. Wu, and F. Mitoma. 2007. The replicative activity of human endogenous retrovirus K102 (HERV-K102) with HIV viremia. AIDS 212417-2424. [DOI] [PubMed] [Google Scholar]
- 33.Lander, E. S., R. H. Waterston, J. Sulston, and F. S. Collins. 2001. Initial sequencing and analysis of the human genome. Nature 409860-921. [DOI] [PubMed] [Google Scholar]
- 34.Lee, Y., and P. D. Bieneasz. 2007. Reconstitution of an infectious human endogenous retrovirus. PLoS Pathog. 3e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Löwer, R., J. Löwer, and R. Kurth. 1996. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc. Natl. Acad. Sci. USA 935177-5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Löwer, R., K. Boller, B. Hasenmaier, C. Korbmacher, N. Mueller-Lantzsch, J. Löwer, and R. Kurth. 1993. Identification of human endogenous retroviruses with complex mRNA expression and particle formation. Proc. Natl. Acad. Sci. USA 904480-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsuzawa, A., H. Nakano, Y. Yoshimoto, and K. Sayama. 1995. Biology of mouse mammary tumor virus (MMTV). Cancer Lett. 903-11. [DOI] [PubMed] [Google Scholar]
- 38.Mayer, J., S. Ehlhardt, M. Seifert, M. Sauter, N. Mueller-Lantzsch, Y. Mehraein, K. D. Zang, and E. Meese. 2004. 2004. Human endogenous retrovirus HERV-K(HML-2) proviruses with Rec protein coding capacity and transcriptional activity. Virology 322190-198. [DOI] [PubMed] [Google Scholar]
- 39.Morgan, D., and I. Brodsky. 2004. Human endogenous retrovirus (HERV-K) particles in megakaryocytes cultured from essential thrombocythemia peripheral blood stem cells. Exp. Hematol. 32520-525. [DOI] [PubMed] [Google Scholar]
- 40.Moyes, D., A. Martin, S. Sawcer, N. Temperton, J. Worthington, D. Griffiths, and P. Venables. 2005. The distribution of the endogenous retroviruses HERV-K113 and HERV-K115 in health and disease. Genomics 86337-341. [DOI] [PubMed] [Google Scholar]
- 41.Moyes, D., D. J. Griffiths, and P. J. Venables. 2007. Insertional polymorphisms: a new lease of life for endogenous retroviruses in human disease. Trends Genet. 23326-333. [DOI] [PubMed] [Google Scholar]
- 42.Muster, T., A. Waltenberger, A. Grassauer, S. Hirschl, P. Caucig, I. Romirer, D. Födinger, H. Seppele, O. Schanab, C. Magin-Lachmann, R. Löwer, B. Jansen, H. Pehamberger, and K. Wolff. 2003. An endogenous retrovirus derived from human melanoma cells. Cancer Res. 638735-8741. [PubMed] [Google Scholar]
- 43.Nelson, P., P. Carnegie, J. Martin, E. Davari, P. Hooley, D. Roden, S. Rowland-Jones, P. Warren, J. Astley, and P. Murray. 2003. Demystified human endogenous retroviruses. Mol. Pathol. 5611-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ono, M., T, Yasunaga. T. Miyata, and H. Ushikubo. 1986. Nucleotide sequence of human endogenous retrovirus genome related to the mouse mammary tumor virus genome. J. Virol. 60589-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paces, J., A. Pavlicek, and V. Paces. 2002. HERVd: the Human Endogenous Retroviruses Database. Nucleic Acids Res. 30205-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ribet, D., S. Louvet-Vallee, F. Harper, N. de Parseval, M. Dewannieux, O. Heidmann, G. Pierron, B. Maro, and T. Heidmann. 2008. Murine endogenous retrovirus MuERV-L is the progenitor of the “orphan” epsilon viruslike particles of the early mouse embryo. J. Virol. 821622-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruda, V. M., S. B. Akopov, D. O. Trubetskoy, N. L. Manuylov, A. S. Vetchinova, L. L. Zavalova, L. G. Nikolaev, and E. D. Sverdlov. 2004. Tissue specificity of enhancer and promoter activities of a HERV-K(HML-2) LTR. Virus Res. 10411-16. [DOI] [PubMed] [Google Scholar]
- 48.Ryan, F. P. 2004. Human endogenous retroviruses in health and disease: a symbiotic perspective. J. R. Soc. Med. 97560-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seifarth, W., C. Baust, A. Murr, H. Skladny, F. Krieg-Schneider, J. Blusch, T. Werner, R. Hehlmann, and C. Leib-Mosch. 1998. Proviral structure, chromosomal location, and expression of HERV-K-T47D, a novel human endogenous retrovirus derived from T47D particles. J. Virol. 728384-8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simpson, G. R., C. Patience, R. Löwer, R. R. Tönjes, H. D. Moore, R. A. Weiss, and M. T. Boyd. 1996. Endogenous D-type (HERV-K) related sequences are packaged into retroviral particles in the placenta and possess open reading frames for reverse transcriptase. Virology. 222451-456. [DOI] [PubMed] [Google Scholar]
- 51.Sugimoto, J., N. Matsuura, Y. Kinjo, N. Takasu, T. Oda, and Y. Jinno. 2001. Transcriptionally active HERV-K genes: identification, isolation, and chromosomal mapping. Genomics 72137-144. [DOI] [PubMed] [Google Scholar]
- 52.Tarlinton, R., J. Meers, and P. R. Young. 2006. Retroviral invasion of the koala genome. Nature 44279-81. [DOI] [PubMed] [Google Scholar]
- 53.Tarlinton, R., J. Meers, J. Hanger, and P. Young. 2005. Real-time reverse transcriptase PCR for the endogenous koala retrovirus reveals an association between plasma viral load and neoplastic disease in koalas. J. Gen. Virol. 86783-787. [DOI] [PubMed] [Google Scholar]
- 54.Tönjes, R. R., R. Löwer, K. Boller, J. Denner, B. Hasenmaier, H. Kirsch, H. König, C. Korbmacher, C. Limbach, R. Lugert, R. C. Phelps, J. Scherer, K. Thelen, J. Löwer, and R. Kurth. 1996. HERV-K: the biologically most active human endogenous retrovirus family. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 13(Suppl. 1)S261-S267. [DOI] [PubMed] [Google Scholar]
- 55.Turner, G., M. Barbulescu, M. Su, M. Jensen-Seaman, K. Kidd, and J. Lenz. 2001. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr. Biol. 111531-1535. [DOI] [PubMed] [Google Scholar]
- 56.Wang-Johanning, F., A. Frost, B. Jian, L. Epp, D. Lu, and G. Johanning. 2003. Quantitation of HERV-K env gene expression and splicing in human breast cancer. Oncogene 221528-1535. [DOI] [PubMed] [Google Scholar]
- 57.Wang-Johanning, F., A. R. Frost, G. L. Johanning, M. B. Khazaeli, A. F. LoBuglio, D. R. Shaw, and T. V. Strong. 2001. Expression of human endogenous retrovirus K envelope transcripts in human breast cancer. Clin. Cancer Res. 71553-1560. [PubMed] [Google Scholar]
- 58.Yang, J., H. P. Bogerd, S. Peng, H. Wiegand, R. Truant, and B. R. Cullen. 1999. An ancient family of human endogenous retroviruses encodes a functional homolog of the HIV-1 Rev. protein. Proc. Natl. Acad. Sci. USA 9613404-13408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yi, J. M., H. M. Kim, and H. S. Kim. 2001. Molecular cloning and phylogenetic analysis of the human endogenous retrovirus HERV-K long terminal repeat elements in various cancer cells. Mol. Cells 12137-141. [PubMed] [Google Scholar]
- 60.Yotsuyanagi, Y., and D. Szollosi. 1980. Embryo development and intracisternal particles in the mouse. Biol. Cell 39201-204. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.