Abstract
A quantitative proteome study using the stable isotope labeling with amino acids in cell culture technique was performed on bovine kidney cells after infection with the alphaherpesvirus pseudorabies virus (PrV), the etiological agent of Aujeszky's disease. To enhance yields of proteins to be identified, raw extracts were fractionated by affinity solid-phase extraction with a combination of a cibacron blue F3G-A and a heparin matrix and with a phosphoprotein-specific matrix. After two-dimensional gel electrophoresis in different pH ranges between pH 3 and pH 10, 2,600 proteins representing 565 genes were identified by mass spectrometry and screened for virus-induced changes in relative protein levels. Four hours after infection, significant quantitative variations were found for constituents of the nuclear lamina, representatives of the heterogeneous nuclear ribonucleoproteins, proteins involved in membrane trafficking and intracellular transport, a ribosomal protein, and heat shock protein 27. Several proteins were present in multiple charge variants that were differentially affected by infection with PrV. As a common pattern for all these proteins, a mass shift in favor of the more acidic isoforms was observed, suggesting the involvement of viral or cellular kinases.
Herpesviral genes are generally expressed in three kinetic classes (18, 19), which are regulated sequentially by a number of positive and negative feedback mechanisms exerted by virus-encoded proteins. However, herpesvirus infection also influences the expression of cellular genes, e.g., by host cell shutoff mechanisms that target the integrity of cellular mRNA and thus block the synthesis of cellular proteins. Herpes simplex virus type 1 (HSV-1), the prototypical alphaherpesvirus, expresses two shutoff proteins, ICP27 and pUL41. Whereas ICP27 interferes with mRNA splicing (16, 17), pUL41 degrades mRNA by virtue of its endoribonucleolytic activity (10, 29, 54), with specificity for mRNAs containing AU-rich elements (11). Homologs of both proteins are also present in pseudorabies virus (PrV), an alphaherpesvirus causing Aujeszky's disease. PrV, whose main host is the pig, infects numerous mammalian species except higher primates including humans. In contrast to HSV-1 ICP27, the PrV homolog pUL54 is dispensable for virus replication in cell culture (47, 49). Transcript analyses of PrV-infected rat (44), human (6), and porcine (12) cells demonstrated alterations in the abundance of individual cellular transcripts, which resulted in the depletion or accumulation of specific mRNAs. Of the 9,850 genes examined in one study (6), the number of significantly up- or downregulated genes increased from approximately 1,000 to over 2,400 between 6 and 9 h after infection. Evaluation of the functions annotated for highly regulated cellular genes shows that PrV infection influences numerous cellular pathways and that genes involved in protein and nucleic acid metabolism, signaling, transport, cell cycle control, adhesion, transcription, the stress response, and innate immunity are affected most frequently. However, alphaherpesvirus infection has an impact not only on the transcription of cellular genes but also on posttranslational protein metabolism. Alphaherpesviruses encode several gene products with enzymatic functions to alter proteins by posttranslational modification. These include the protein kinases pUL13 and pUS3 as well as pUL36, a large structural protein that mediates deubiquitination (21, 23). Examples for posttranslational modifications of cellular proteins induced by infection with HSV-1 are the phosphorylation of lamins A/C and B (35, 40), the ICP27-stimulated phosphorylation of heterogeneous nuclear ribonucleoprotein (hnRNP) K by casein kinase 2 (CK2) (25), and the block of histone deacetylase 1 by pUS3 and viral ICP0 (43). Infection with HSV-1 also induces the proteasome-dependent degradation of a number of proteins like CD83 (26), the ND10-related proteins PML and Sp100 (8), or the catalytic subunit of the DNA-dependent protein kinase (41). However, a systematic examination of the impact of alphaherpesvirus infection on the protein composition of the infected cell has not yet been performed. Thus, the objective of this study was to establish a quantitative protein expression profile of PrV-infected cultured cells, which includes posttranslationally modified isoforms of individual proteins.
MATERIALS AND METHODS
Cells and viruses.
Madin-Darby bovine kidney cells (31) were provided by the Collection of Cell Lines in Veterinary Medicine, Insel Riems, Germany. PrV strain Kaplan (22) was used.
Stable isotope labeling.
The original stable isotope labeling procedure (38) was adapted as follows. Dulbecco's modified Eagle (DME)/F12 medium (D-9785; Sigma-Aldrich, Taufkirchen, Germany) was supplemented with 5% dialyzed fetal calf serum and all missing amino acids (Sigma-Aldrich) except l-leucine. Medium was then divided and supplemented with conventional or deuterated l-leucine (l-leucine-5,5,5-D3 [99 atom% D]) (catalog number 486825; Sigma-Aldrich) to produce PROLeu-DME/F12 or DEULeu-DME/F12 medium, respectively. MDBK cells were passaged in parallel in both media at a 1:10 ratio every 3 days. After four passages, aliquots of the cell cultures were lysed, and proteins were separated by gel electrophoresis. The efficiency of the exchange of normal by deuterated leucine was controlled by mass spectrometry. The incorporation of l-leucine-5,5,5-D3 increases the mass of leucine-containing peptides by 3 mass units per leucine residue.
Infection.
Deuterium-labeled cells were used as mock-infected controls, and cells grown on the conventional amino acid source were infected. Cell batches passaged in the two media were seeded in 75-cm2 or 150-cm2 cell culture flasks and inoculated with virus stock corresponding to a multiplicity of infection of 10 (cells grown in PROLeu-DME/F12 medium) or mock inoculated (cells grown in DEULeu-DME/F12 medium) on ice for 1 h and then incubated at 37°C for 4 h.
Extraction of phosphoproteins.
Phosphoproteins of infected and mock-infected cells were purified with the PhosphoProtein purification kit (catalog number 37101; Qiagen, Hilden, Germany). In short, 1 × 107 cells were extracted with the buffer supplied with the kit, containing 0.24% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propansulfonate (CHAPS), and unsolubilized material was removed by centrifugation. The protein concentration in the clarified extract was determined with the BCA protein assay (Pierce, Rockford, IL), and equal amounts of proteins were mixed and applied onto the column supplied with the kit. The eluate containing mainly phosphoproteins and the flowthrough containing mainly the nonphosphorylated proteins were concentrated by ultrafiltration in Nanosep tubes equipped with a 10-kDa-cutoff membrane. Protein concentrations were assayed with the BCA protein assay (Pierce). Total eluates, usually containing approximately 200 μg of protein and aliquots of 1 mg of the concentrated flowthrough, were precipitated (2D clean-up kit, catalog number 80-6484-51; GE Healthcare, Braunschweig, Germany), and the pellets were dissolved in 200 μl of rehydration buffer (RHB) (7) with mild sonication and either used immediately or stored at −20°C. The resulting fractions are referred to as the phosphoprotein fraction and nonphosphoprotein fraction (flowthrough). Both fractions were tested for the phosphorylation status of the included proteins by Western blotting with antibodies directed against phosphoserine, phosphothreonine, or phosphotyrosine, and the efficiency of the separation for HeLa cells as given by the manufacturer (PhosphoProtein purification kit user manual) was confirmed for the MDBK cells used.
ASPE with cibacron blue F3G-A-Sepharose and heparin-Sepharose.
Two 150-cm2 flasks each of labeled and unlabeled cells corresponding to approximately 8 × 107 cells in total were harvested in 10 ml extraction buffer (10 mM Na2HPO4-KH2PO4, 150 mM NaCl, 1% CHAPS [pH 7.0]) supplemented with protease inhibitors (Complete Mini, 1 tablet/10 ml; Roche), extracted for 60 min on ice with occasional shaking, and centrifuged (15 min at 4°C at 4,000 × g) to remove unsoluble material. Extracts from unlabeled and isotope-labeled cells were mixed at a 1:1 protein ratio. HiTrap columns (1-ml column volume) (catalog numbers 17-0412-01 and 17-0406-01; GE Healthcare) were washed with 10 column volumes each of water, elution buffer (10 mM Na2HPO4-KH2PO4, 2 M NaCl, 1% CHAPS [pH 7.0]), and extraction buffer before the extract was applied with a peristaltic pump and recirculated for 30 min. The columns were washed with 10 column volumes of extraction buffer, and bound material was eluted with 5 ml elution buffer. Samples containing 1 mg of the eluates or the flowthrough were precipitated with trichloroacetic acid, and precipitates were resuspended in RHB and stored at −20°C. The resulting fractions are referred to as the cibacron fraction, the heparin fraction, and the affinity solid-phase extraction (ASPE) flowthrough in the text. Protein yields were approximately 11% in the cibacron fraction and 17% in the heparin fraction, and the rest was found in the flowthrough. Protein recoveries approximated 100%.
Two-dimensional (2D) gel electrophoresis.
Phosphoproteins were analyzed on 11-cm ReadyStrips (Bio-Rad, Munich, Germany) with a nonlinear pH range of 3 to 10, nonphosphoproteins were analyzed on 24-cm strips with a nonlinear pH range of 3 to 10, and all other fractions were analyzed on 24-cm strips with linear pH ranges of 3 to 6, 4 to 7, and 6 to 9. Precipitated samples were resuspended in RHB, briefly sonicated on ice, and extracted for 2 h at 20°C with intensive shaking. Undissolved material was removed by centrifugation (20°C at 10 min at 14,000 × g), and sample proteins were allowed to diffuse into ReadyStrips and focused in an IEF cell (Bio-Rad) according to the guidelines provided by the manufacturer. Focused strips were frozen at −80°C, thawed, and sequentially equilibrated in buffers containing dithiothreitol and iodoacetamide as recommended by the manufacturer. The second dimension was run on hand-cast full-size 12% acrylamide gels in a Dodeca cell electrophoresis chamber (Bio-Rad) with two 11-cm strips or one 24-cm strip loaded per gel. After the electrophoretic run, gels were fixed, stained overnight with colloidal Coomassie brilliant blue (36), and scanned.
Peptide mass fingerprint (PMF) analysis.
After evaluation of the gel scans with Delta2D software (version 3.4; Decodon, Greifswald, Germany), lists of protein spots to be picked were used to operate a Proteineer SPII Spotpicker (Bruker Daltonics, Bremen, Germany). Tryptic digestion (46) was carried out in 96-well V-bottom polypropylene microtiter plates for 3 h with 30 ng trypsin (catalog number V5111; Promega, Mannheim, Germany) per sample at 37°C. Fingerprint and up to four tandem mass (MS/MS) spectra per sample were registered on a Bruker Ultraflex I tandem time-of-flight instrument (Bruker) and processed by flexAnalysis 2.0 software (Bruker). For the quantitation of mass-tagged peptide peak pairs, it was crucial to choose the “SNAP” option as the peak detection algorithm in the flexAnalysis software, which is robust with respect to overlapping isotope patterns. A batch database search (MASCOT Server 2.0.0 software; Matrix Science Ltd., London, United Kingdom) (42) was launched by Biotools 2.2 (Bruker) using the bovine International Protein Index (IPI) database (www.ebi.ac.uk) (24) or an in-house database covering the PrV proteome as compiled from the Swiss-Prot database (www.expasy.org) (13). Carbamidomethylation was set as a fixed modification for cysteine residues, the significance level was set to 95%, and mass tolerance was set to 50 ppm for the fingerprint spectra. Proteins were considered as identified and selected for quantitative evaluation if significant molecular weight search (MOWSE) scores (39) were obtained with the fingerprint spectrum or in a combined search of the fingerprint and MS/MS spectra.
Quantitation and data processing.
Quantitation was carried out by in-house software (AMaDEuS) based on Visual Basic for applications (Microsoft, Redmond, WA) macros. For identified samples, peaks representing the protein with the highest MOWSE score were selected, and the masses of the expected isotope-labeled peaks within error margins of 15 ppm were calculated on the basis of the number of leucine residues present in the peptide and the mass shift of +3 Da per leucine residue. If the spectrum contained the mass of the expected deuterated peak, the peak pair was selected for quantitative evaluation, and the intensity ratio of the peaks was calculated. If more than four peak pairs were obtained from one spectrum, outliers were eliminated by the symmetrical truncation of intensity ratios. Truncation was limited to a maximum of three rounds or the number of rounds that would leave a minimum of three values. Truncated mean values and standard deviations of the intensity ratios of qualified peak pairs representing the identified protein were calculated. Relative standard deviations rarely exceeded 15%. The database query was then repeated with a fixed modification of +3 Da for leucine residues to ensure the identification of proteins that were strongly downregulated after infection. Quantitative evaluation was again carried out as described above, with the only difference being that a −3-Da shift was used to calculate the expected mass of the unlabeled peptide peak in the AMaDEuS software. Results of both queries and calculations were combined and compared. Data sets resulting from the same sample (protein spot) yielding higher MOWSE scores were preferred to those with lower scores. The final output for every sample was the truncated mean of the ratios (unlabeled over labeled, that is, infected over mock [IOM]) of qualified peak pairs, which will be abbreviated as IOM ratios. IOM ratios greater than 1.0 indicate higher relative levels, and IOM ratios below 1.0 indicate lower relative levels of the respective proteins after infection with PrV. The average number of qualified peptides in identified proteins was 3.3, and the maximum number was 18.
RESULTS
Experimental design.
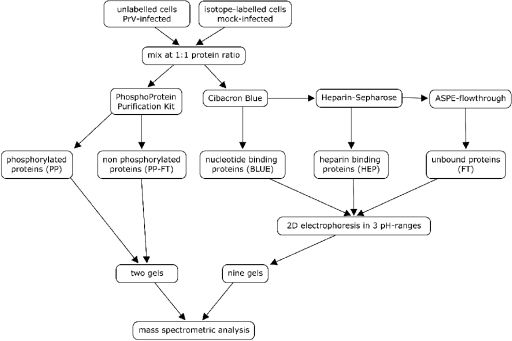
The strategy for the fractionation and analysis of raw cell extracts is depicted in Fig. 1. The design of the experiment aimed at two goals. First, the very complex protein mixture present in whole-cell extracts was prefractionated into well-defined fractions in order to facilitate further analysis (45); second, proteins with interesting features like phosphorylation as well as nucleotide-binding capacities (cibacron blue F3G-A-Sepharose) or DNA- and RNA-binding capacities (heparin-Sepharose) were enriched by affinity purification. Quantitation was carried out with the stable isotope labeling with amino acids in cell culture (SILAC) technique (38), which makes use of the high resolution of mass spectrometry to allow the differentiation and quantitation of isotopically modified peptides that are chemically identical. Thus, the major steps for quantitation by SILAC are the metabolic labeling of a sample with stable isotopes, mixing with an unlabeled reference (or vice versa), purification of individual proteins from the mix, digestion of the proteins to peptides, and calculation of intensity ratios of protein-specific peptides in a peptide mass fingerprint spectrum. Ratios of isotope-labeled peptides reflect the relative levels of the protein in sample and reference with high accuracy. In the experiments reported here, the intensity ratios of unlabeled over labeled peptide peaks were calculated to result in IOM ratios given in the text and figures. Cell cultures metabolically labeled with deuterated l-leucine and unlabeled cultures served as starting material. After infection with PrV or mock infection and incubation for 4 h, a mix of raw extracts from labeled (mock-infected) and unlabeled (PrV-infected) cells at 1:1 protein ratios was then used for the parallel extraction of phosphoproteins with a commercially available reagent kit and a two-step ASPE procedure with cibacron blue F3G-A-Sepharose and heparin-Sepharose. All five resulting fractions (phosphoproteins and nonphosphoproteins from the phosphoprotein-specific extraction and the heparin-binding proteins, the cibacron blue-binding proteins, and the flowthrough fraction from the ASPE) were then analyzed by large-format 2D gel electrophoresis in different pH ranges in order to maximize the yield of separated proteins. Isolated proteins were identified by PMF analysis using matrix-assisted laser desorption-ionization mass spectrometry with the MS/MS option and quantitated from the PMF spectra. Three repetitions of the experiment were performed to screen for proteins with significantly altered relative protein levels. Candidate proteins were then evaluated by additional experiments focusing on the fractions and gel regions of interest.
FIG. 1.
Flowchart of sample preparation and analysis. Note that all separation and analytical procedures (affinity extractions, 2D gel electrophoresis, tryptic digestion, and mass spectrometric analysis) were carried out with a 1:1 protein mixture of extracts of conventional and heavy-isotope-labeled cells. After extraction with three different affinity matrices and separation in different pH ranges, a total of 11 2D electrophoretic gels were analyzed for each experiment. FT, flowthrough.
The time point of 4 h after infection was chosen to monitor early effects of infection under conditions where cellular protein synthesis is already effectively blocked by viral shutoff mechanisms (data not shown), but the cell is still morphologically intact and not massively damaged by the release of progeny virions.
In the course of this study, over 4,000 proteins were identified, and 2,374 proteins were quantified from a minimum number of three qualified peptide pairs. Of the 1,490 spots quantified in infection experiments, IOM ratios of 109 samples (7.3%) representing 55 genes (Table 1) were beyond the empirical 1% and 99% quantiles of 0.63 and 1.63. Thus, approximately 30 of the 109 samples were expected to result from the statistical variance of the experiment itself, and approximately 79 samples were expected to result from virus-induced changes.
TABLE 1.
Proteins up- or downregulated after infection with PrV-Kad
| Genea | IPI accession number | Descriptionb | IOM | SDd |
|---|---|---|---|---|
| 540272 | IPI00695524 | Proliferation-associated 2G4 | 0.18 | 0.04 |
| 505686 | IPI00706431 | Ladinin 1 | 0.29 | 0.08 |
| 511512 | IPI00690613 | RNA binding protein 14 | 0.36 | 0.01 |
| 509771 | IPI00688922 | RNA binding motif protein 3 | 0.36 | 0.04 |
| 281165 | IPI00692328 | Filamin A | 0.40 | 0.45 |
| 527471 | IPI00725795 | hnRNP D (AU-rich element RNA binding protein 1) | 0.42 | 0.09 |
| 282419 | IPI00707334 | CK2 subunit alpha | 0.47 | 0.04 |
| 505242 | IPI00706906 | EBP50 | 0.48 | 0.06 |
| 404098 | IPI00695802 | Staphylococcal nuclease domain-containing protein 1 | 0.48 | 0.13 |
| 533874 | IPI00705755 | Proteasome subunit beta type 3 | 0.49 | 0.16 |
| 510041 | IPI00837986 | Proteasome activator subunit 1 | 0.49 | 0.28 |
| 511048 | IPI00726962 | RuvB-like 2 | 0.50 | 0.15 |
| 507345 | IPI00710727 | Transitional endoplasmic reticulum ATPase | 0.53 | 0.02 |
| 539060 | IPI00827112 | Serpine 1 mRNA binding protein 1 | 0.53 | 0.05 |
| 533851 | IPI00829551 | Enigma protein | 0.54 | 0.23 |
| 353121 | IPI00693691 | Macrophage-capping protein | 0.55 | 0.18 |
| 530409 | IPI00688006 | Prohibitin | 0.55 | 0.03 |
| 511475 | IPI00685691 | RuvB-like 1 | 0.57 | 0.09 |
| 327682 | IPI00700792 | Guanine-nucleotide-binding protein subunit beta-2-like 1 | 0.58 | 0.15 |
| 615447 | IPI00686420 | Nucleoside diphosphate kinase B | 0.59 | 0.21 |
| 281574 | IPI00694641 | Ezrin | 0.59 | 0.20 |
| 515646 | IPI00693645 | Chloride intracellular channel protein 1 | 0.59 | 0.19 |
| 513410 | IPI00691068 | hnRNP AB | 0.60 | 0.16 |
| 281615 | IPI00692627 | Retinal dehydrogenase 1 | 0.60 | 0.09 |
| 541202 | IPI00734138 | hnRNP H | 0.62 | 0.10 |
| 282485 | IPI00695563 | Sulfotransferase 1A1 | 0.62 | 0.09 |
| 535119 | IPI00689197 | Cytokine-induced apoptosis inhibitor 1 | 0.63 | 0.13 |
| 514355 | IPI00700509 | T-complex protein 1 subunit beta | 0.63 | 0.02 |
| 507197 | IPI00702566 | Serine hydroxymethyltransferase, mitochondrial precursor | 1.65 | 0.42 |
| 282689 | IPI00706002 | Annexin A2 | 1.67 | 0.01 |
| 539218 | IPI00716493 | Calumenin precursor | 1.68 | 0.23 |
| 533746 | IPI00698338 | Dihydropyrimidinase-related protein 2 | 1.73 | 0.4 |
| 513793 | IPI00715791 | eIF-3F | 1.73 | 0.29 |
| 505968 | IPI00704474 | Acyl-coenzyme A dehydrogenase, C-4 to C-12 straight chain | 1.73 | 0.23 |
| 506059 | IPI00699601 | Acyl-coenzyme A synthetase long-chain family member 6 | 1.75 | 0.02 |
| 540643 | IPI00701266 | Lamin B1 | 1.77 | 0.21 |
| 281544 | IPI00708322 | Tropomyosin alpha-1 chain | 1.79 | 0.27 |
| 512584 | IPI00711352 | Small nuclear ribonucleoprotein polypeptide A | 1.79 | 0.15 |
| 510201 | IPI00698102 | Serine/threonine kinase receptor-associated protein | 1.85 | 0.32 |
| ENSBTAG00000033117 | IPI00842846 | Histone H3 | 1.88 | 0.17 |
| 504912 | IPI00711479 | Target of Myb1 | 2.04 | 0.50 |
| 281660 | IPI00686760 | l-Caldesmon | 2.31 | 0.27 |
| 281181 | IPI00713814 | GAPDH | 2.32 | 0.3 |
| 732539 | IPI00697070 | Cytokine-induced protein 29 kDa | 2.64 | 0.66 |
| 539060 | IPI00827112 | Serpine 1 mRNA binding protein 1 | 2.67 | 0.66 |
| 507564 | IPI00691167 | hnRNPs A2 and B1 | 2.69 | 1.50 |
| 782669 | IPI00690667 | hnRNP A3 | 2.80 | 0.23 |
| 281961 | IPI00717623 | Osteoclast-stimulating factor 1 | 4.57 | 1.02 |
| 404144 | IPI00689750 | Lamin A/C | 0.72->10c | |
| 528135 | IPI00696554 | hnRNP K | 0.43-3.03c | |
| 506218 | IPI00697355 | SNX-9 | 0.53-1.99c | |
| 505850 | IPI00700182 | eIF-4B | 0.32-3.19c | |
| 286868 | IPI00703564 | 60S acidic ribosomal protein P0 | 0.61-2.32c | |
| 516099 | IPI00704836 | Heat shock 27-kDa protein 1 | 0.42->10c | |
| 516326 | IPI00713660 | Lamin B2 | 0.38-2.01c |
Gene identifiers referenced in the IPI database from the Entrez Gene (33) or the Ensembl (12) database are given.
Descriptions given in the IPI data set or the referenced gene in the Entrez Gene database.
See the text for details.
In three rounds of screening. One hundred nine of the 1,490 quantified protein spots representing 55 genes showed IOM ratios beyond the empirical cutoff values of 1.63 and 0.63.
Performance of the ASPE.
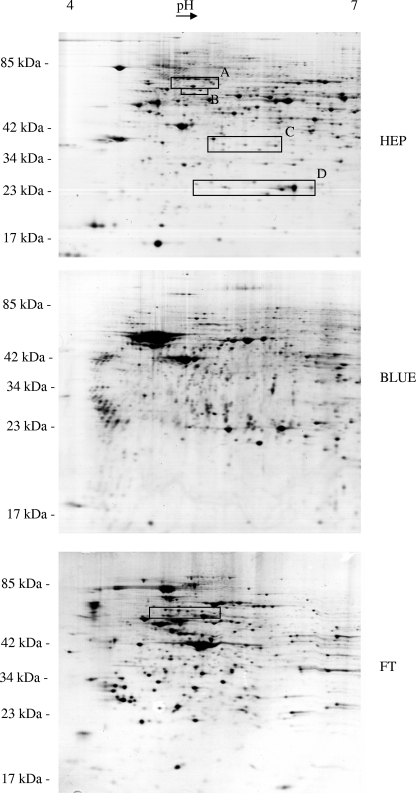
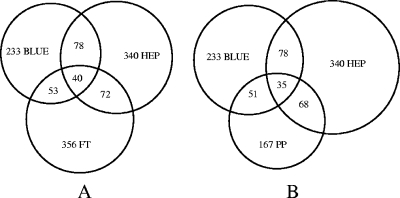
Representative gels from the three fractions of the ASPE at the pH range of 4 to 7 show very distinct protein spot patterns (Fig. 2). The distribution of identified proteins over the ASPE fractions is shown in Fig. 3A (9). Overlaps among the three fractions were low and are not necessarily attributable to the unsatisfactory performance of the ASPE, since different isoforms of the same protein may distribute into different fractions on the basis of their biochemical properties. Yields of identified proteins in the range of pH 3 to 6 (12%) were lower than those in the ranges of pH 4 to 7 (51%) and pH 6 to 9 (37%). Protein compositions of the cibacron and heparin fractions of the ASPE differ markedly from the composition of the phosphoprotein fraction (Fig. 3B), indicating that phosphoprotein extraction and the ASPE procedure are largely complementary, and a combination of both significantly enhances the yield of identified proteins from a single sample. The specificity of the ASPE procedure was estimated by statistical evaluation (http://gostat.wehi.edu.au) (3) of the Gene Ontology (GO) annotations (“molecular function” branch at www.geneontology.org) (2) characterizing the proteins of each fraction. In both affinity fractions (cibacron and heparin), the GO accession numbers with the highest frequencies were GO:0000166 (“nucleotide binding”) (78/233 in the cibacron fraction and 98/340 in the heparin fraction) and GO:0017076 (“purine nucleotide binding”) (59/233 in the cibacron fraction and 82/340 in the heparin fraction). In the heparin fraction, GO:0008135 (“translation factor activity”) and GO:0003743 (“translation initiation factor activity”) followed, whereas in the cibacron fraction, GO:0003676 (“nucleic acid binding”) and GO:0017111 (“nucleoside-triphosphatase activity”) were the next most numerous. All mentioned annotations were significantly overrepresented in the affinity fractions compared to the raw extract on the basis of P values of 0.05. Thus, both affinity fractions contained a high abundance of proteins with molecular functions in accordance with the properties of cibacron blue F3G-A (nucleotide binding) and heparin (DNA binding, with affinity for translation factors).
FIG. 2.
Scans of 2D electrophoretic gels representing the two affinity-purified fractions (BLUE, cibacron fraction; HEP, heparin fraction) and the flowthrough (FT) of the ASPE separation procedure. Differing protein patterns reflect an efficient separation into three well-defined protein fractions. Boxes indicate gel regions that were analyzed in more detail (Fig. 6 and 7) for modifications of eIF-4B and SNX-9 (box A), lamin B2 (box B), 60S acidic ribosomal protein P0 (box C), Hsp27 (box D), and hnRNP K (box in panel FT).
FIG. 3.
Graphic representation of separation results. Overlaps were calculated on the basis of the number of entries of identified proteins found in the IPI database. Shown are overlaps between the different fractions of the ASPE (BLUE, cibacron fraction; HEP, heparin fraction; FT, flowthrough) (A) and overlaps between the cibacron, heparin, and phosphoprotein (PP) fractions (B) demonstrating a high selectivity of the ASPE procedure (A) and a good complementarity of the ASPE with phosphoprotein extraction (B).
Determination of a cutoff value.
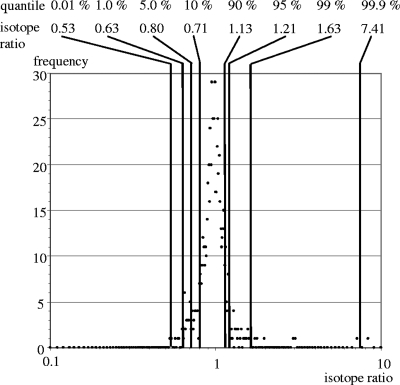
In a pilot experiment, a mix of unlabeled and labeled cell extracts, both originating from mock-infected cells, was analyzed. The purpose of this experiment was to determine the precision and the variation of our quantitation. As expected, most of the isotope ratios were close to 1.0, and the frequency distribution of isotope ratios was very narrow (Fig. 4) so that empirical 1% and 99% quantiles were determined for isotope ratios of 0.63 and 1.63 and were used as cutoff values in the following infection experiments.
FIG. 4.
Distribution of isotope ratios of proteins identified in a control experiment with labeled and unlabeled mock-infected cells. Isotope ratios were calculated as described in Materials and Methods. The frequency of isotope ratios was registered in steps of 0.01. As expected, the distribution centers around 1.0. Quantiles are given in percentages, and the corresponding isotope ratios are shown as plain numbers.
Infection experiments.
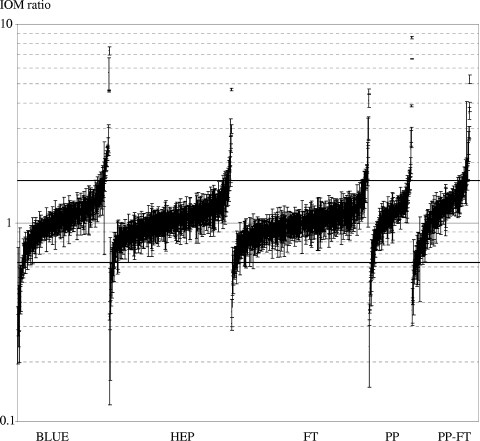
In three independent infection experiments, quantitative protein profiles were determined for PrV-infected MDBK cells. Figure 5 shows that the distribution of IOM ratios in the five protein fractions is much broader than the distribution of isotope ratios in the control experiment (Fig. 4), indicating that infection leads to numerous changes in the quantitative composition of the MDBK proteome. Up- and downregulated proteins were about equal in all fractions, which was surprising, since under the experimental conditions used, the biosynthesis of cellular proteins is severely inhibited due to PrV-mediated host cell shutoff (4, 20), and a significant bias to IOM ratios smaller than 1.0 had been expected. For a number of proteins that were identified in multiple isoforms, differential or even inverse effects were found for different charge variants (Fig. 6 and 7). In subsequent experiments, proteins that showed relative protein levels beyond the calculated cutoff values or showed interesting differential regulations were reanalyzed. All qualitative and quantitative data reported in the text or in figures rely on a minimum of three experiments.
FIG. 5.
Distribution of relative cellular protein levels 4 h after infection with PrV-Ka. Error bars indicate the standard deviations of the isotope ratios of the peptides used for the calculation of the IOM ratio. PP, phosphoprotein fraction; PP-FT, flowthrough of the PhosphoProtein purification kit (nonphosphoproteins); BLUE, cibacron fraction; HEP, heparin fraction; FT, ASPE flowthrough. Cutoff values of 0.63 and 1.63 are given as horizontal lines.
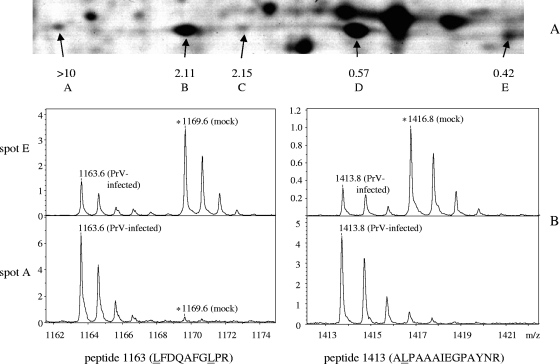
FIG. 6.
(A) Detailed view from a 2D electrophoretic gel of a heparin fraction analyzed in the pH range 4 to 7 (box D in Fig. 2). Five protein spots were identified as being Hsp27, and the relative protein levels, given in numbers, were calculated as described in the text. Details of mass spectra originating from spot A (most acidic isoform) and spot E (least acidic isoform) show two peptide peaks containing one (1,413 Da) or two (1,163 Da) leucine residues. IOM ratios were calculated from intensity ratios of peak pairs with a mass distance corresponding to the number of leucine residues present, in this case 3 Da (1,413 Da) and 6 Da (1,163 Da). The respective deuterium-labeled peaks at 1,416 Da and 1,169 Da representing peptides from mock-infected cells are marked with asterisks. Both labeled and unlabeled peptides occur in their natural isotope patterns, giving rise to multiple peaks with descending intensities in 1-Da distances. Note that reliable relative quantitation was restricted to ratios between 1:10 and 10:1 so that IOM ratios of the most acidic spots of Hsp27 and lamin C (Fig. 7A) could be assessed only as “greater than 10”.
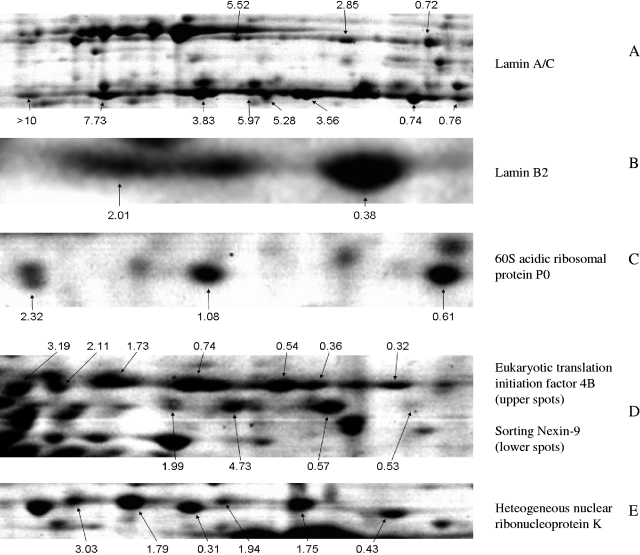
FIG. 7.
Proteins that were differentially regulated in different isoforms. Relative abundances of the respective isoforms after infection are given in plain numbers.
Stress response proteins.
Heat shock protein 27 was found in five spots, two of which (spots B and D) (Fig. 6A) represent over 90% of the total protein. Relative protein levels of 2.11 and 0.57 in spots B and D, respectively, indicate that a considerable fraction of available Hsp27 had undergone modification at 4 h after infection. Spot A was not detectable by Coomassie staining of gels from mock-infected cell extracts (not shown) and contains only very little material from mock-infected reference cells used in the SILAC procedure (see spectra in Fig. 6B). Other stress-related proteins that had been identified and quantified (alpha crystalline B chain; heat shock 70-kDa proteins 1B, 5, and 9B; heat shock cognate 71-kDa protein; and heat shock protein 90-alpha) did not show significant variations.
Lamins.
Lamin A/C was found mainly in the phosphoprotein fraction but was also present in the nonphosphoprotein and heparin fractions of the ASPE in two strings of poorly resolved protein spots representing the unprocessed lamin A and the approximately 20-kDa-smaller proteolytic cleavage product, lamin C (Fig. 7A). For both proteins, a shift to more acidic isoforms was observed after infection. As found for the most acidic modification of Hsp27, the protein spot representing the most acidic variant of lamin C contained hardly any material from mock-infected cells. Lamin B2 was found in one major and one minor more acidic spot (Fig. 7B). Considering the relative protein levels and the distribution of the total mass between the two spots, gains and losses are not nearly balanced so that under the condition that no other lamin B2 isoforms have been missed, a total loss of protein occurred after infection. Lamin B1 was also identified in three protein spots with quantitative variations within the isoforms being less pronounced than those for lamin B2, although for lamin B1 increasing abundances were also found with increasing acidity (IOM ratios from the most acidic spot to the least acidic spot were 1.77, 1.08, and 0.80).
Proteins involved in translation.
Although numerous ribosomal proteins and translation factors were identified and quantified, only one ribosomal protein (60S acidic ribosomal protein P0) (Fig. 7C) and two translation initiation factors showed a significant (eukaryotic initiation factor 4, subunit B [eIF-4B]) (Fig. 7D) or a moderate response (eIF-3F) (Table 1) to PrV infection. eIF-4B was present in seven charge variants most likely representing different phosphorylation levels.
hnRNPs.
hnRNPs were highly enriched in the cibacron fraction with the exception of hnRNP K, which reliably separated into the flowthrough of the ASPE (Fig. 2). A number of hnRNP representatives like hnRNP A3 (IOM of 2.80 ± 0.23), hnRNP D (IOM of 0.42 ± 0.09), and hnRNP A2/B1 (IOM of 2.69 ± 1.50) showed significant variation after infection, while others (hnRNP A1 and hnRNP F) remained constant or were only slightly changed (IOM of 0.62 ± 0.1 for hnRNP H and IOM of 0.60 ± 0.16 for hnRNP A/B). Of the six spots that were identified as being hnRNP K (Fig. 7E), two migrated with a slightly smaller apparent molecular weight and decreased in abundance after infection, whereas relative levels of the slightly larger proteins increased, with gains being more pronounced for the more acidic isoforms.
Proteins related to intracellular transport and the cytoskeleton.
Sorting nexin 9 (SNX-9) (Fig. 7D), enriched in the heparin fraction, appeared in four distinct spots, which followed the observed shift to more acidic isoforms. However, in this case, the mass gain was not highest for the most acidic form. A functionally related protein, ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) was found with decreased relative levels (IOM of 0.48 ± 0.06) in the heparin fraction. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which, apart from its function in the glycolytic pathway, is involved in intracellular membrane trafficking (55, 57), was present in the cibacron fraction and found to be markedly upregulated (IOM of 2.32 ± 0.30). Relative levels of the other glycolytic enzymes that were identified and quantified (fructose-bisphosphate aldolase, triosephosphate isomerase, phosphoglycerate kinase, enolase, and pyruvate kinase) did not differ significantly between infected and noninfected cells.
DISCUSSION
The aim of this study was to systematically screen for alterations in the amounts of cellular proteins during infection with PrV. To increase the number of cellular proteins that could be identified by 2D gel electrophoresis followed by mass spectrometry, a simple prefractionation scheme was established, which yielded three fractions with well-defined compositions. Overlaps between fractions were low, and the fractions contained a high abundance of proteins with the expected biochemical properties and molecular functions. The parallel extraction of phosphoproteins was shown to be highly efficient, as both procedures yielded largely complementary protein fractions. Prefractionation was not detrimental for the subsequent mass spectrometric quantitation using the SILAC technique. The high precision of the mass spectrometric quantitation allowed us to set cutoff values close to 1.0, which provided the high sensitivity of the procedure for the detection of regulated protein spots.
Overall, the cellular proteome appeared to be very stable. The relative levels of the vast majority of proteins were unaffected by infection with PrV, and the calculated IOM ratios were close to 1.0 despite the known detrimental effects of PrV infection on the stability of mRNA and cell protein synthesis due to viral host cell shutoff functions (29, 49). Thus, the delayed character of PrV-induced host cell shutoff indicated by microarray studies in porcine (12), rat (44), and human (6) cells was confirmed on the protein level for the bovine cells used here. From time course studies with unfractioned material and analysis with 2D electrophoresis in the pH range of 3 to 10 (data not shown), there was no indication for a global decrease of protein levels up to 8 h after infection with PrV, indicating that the degradation of cellular mRNA by viral pUL41 and other mechanisms of host cell shutoff seem to exclusively target the RNA metabolism but do not interfere with the physiological steady-state levels of most proteins, at least not at early times after infection.
Nevertheless, the IOM ratios of a considerable number of protein spots did change significantly in either direction. Moreover, in several cases, e.g., Hsp27, lamin A/C, lamin B2, hnRNP K, SNX9, eIF-4B, or 60S acidic ribosomal protein P0, inverse modulations of different charge variants of the same protein were observed.
Stress response proteins.
The most acidic of the five isoforms of Hsp27 was detected almost exclusively in infected cells, indicating that PrV infection caused a sharp rise in these isoforms. Most probably, the charge variants are the result of differential phosphorylation, which was previously described for human (37, 48) and bovine (28) Hsp27. A similar shift to higher phosphorylated forms of Hsp27 has been observed after treatment of bovine cells with cadmium (28), indicating that this reaction is not specific for infections with herpesviruses but rather reflects phosphorylation of Hsp27 during cellular stress (27). However, other stress-related proteins that were identified and quantified showed no significant changes in relative protein levels (data not shown). Hsp27 is a multifunctional protein (1) with strong antiapoptotic properties. It is unclear so far if the changes in levels of Hsp27 are related to the pUS3-mediated suppression of apoptosis (14), which is observed after infection with PrV.
Lamins.
The observed shift to more acidic isoforms of lamin A/C parallels the reported changes of 2D gel electrophoretic patterns of lamin A/C caused by the viral kinase pUS3 after infection with HSV-1 (35). In an ongoing study with a US3-deleted PrV mutant, the impact of pUS3(PrV) on the phosphorylation pattern of lamin A/C will be addressed. Infection with HSV-1 is also accompanied by an intracellular rearrangement (5) and loss (50) of B-type lamins, which were identified and quantified from the heparin fraction. A shift to more acidic isoforms was observed for both, which corresponds to the previously reported phosphorylation of B-type lamins following HSV-1 infection (40). The hyperphosphorylation of the lamins precedes the disintegration of the nuclear lamina in many physiological processes, which is presumably a prerequisite for the transfer of PrV capsids from the nucleus into the cytoplasm for secondary envelopment (34).
Proteins related to translation.
After infection with HSV-1, the synthesis of several ribosomal proteins and their assembly into ribosomes continue in spite of a general inhibition of cellular protein synthesis and a concomitant loss of the mRNA encoding the respective ribosomal proteins (15, 52). With the exception of the redistribution seen within the isoforms of the 60S acidic ribosomal protein P0, we also found stable levels of ribosomal proteins after PrV infection. Likewise, significant changes in relative levels of translation factors were not observed, with the exception of eIF-4B and the F subunit of eIF-3.
Proteins related to intracellular transport and the cytoskeleton.
So far, none of the three proteins related to intracellular transport and the cytoskeleton (SNX9, EBP50, and GAPDH), which we found to be significantly altered in abundance, have been correlated with any step during herpesvirus replication. However, they exhibit interesting features, which may have functions related to herpesvirus infections. Members of the SNX family of proteins contain a Phox domain that mediates binding to membrane-anchored phospholipids. They are involved in intracellular membrane trafficking (51), which is important for the replication of enveloped viruses. SNX9 shares a relative promiscuity with other SNXs with respect to the different phosphatidylinositols that are bound by the Phox domain, the presence of an SH3 domain, and the presence of a BAR dimerization domain. SNX9 is activated by phosphorylation (30), which correlates with the observed shift to more negatively charged variants, which we observed after infection (Fig. 7D). After phosphorylation, SNX9 is translocated to the plasma membrane, where it can interact with transmembrane proteins, but also recruits dynamin to the plasma membrane (30). It is also required for effective clathrin-mediated endocytosis (53). By virtue of its BAR domain, it may be involved in the stabilization but also in the sensoring of strongly curved membranes (30), which occur during budding.
EBP50 is a peripheral membrane protein that resides at the plasma membrane and supports functions of the ezrin-radixin-moesin proteins in connecting the actin cytoskeleton to the plasma membrane. Cytomorphological alterations of PrV infection have been attributed to actin stress fiber breakdown mediated by the pUS3 kinase (58), which may occur via the phosphorylation of EBP50.
Apart from its function in glycolysis, GAPDH plays a major role in the early steps of the secretory pathway during the transport of cargo from the endoplasmic reticulum to the Golgi apparatus (55). As was exemplarily demonstrated with the G glycoprotein of vesicular stomatitis virus, the transport function of GAPDH was independent of its enzymatic activity but depended on the phosphorylation of GAPDH mediated by the cellular Src protein (56, 57). In the context of PrV infection, GAPDH may play a role in the transport of viral glycoproteins to late secretory compartments like the trans-Golgi network, where the PrV particles acquire their final envelope in a budding process (33).
hnRNPs.
Four types of hnRNP were significantly (hnRNPs A3, A2/B1, D, and K) and two were moderately (hnRNPs H and A/B) modulated after infection with PrV. Since infection with alphaherpesviruses is accompanied by drastic changes in RNA metabolism, which is, at least in part, caused by herpesviral shutoff mechanisms, the modulation of hnRNPs, which are involved in RNA processing and turnover, was not unexpected. The host shutoff protein pUL41 of HSV-1 acts as an endoribonuclease, preferentially degrading mRNA containing AU-rich elements (11). In this context, the loss of hnRNP D, which is also known as AU-rich element binding protein 1 (AUF-1), might be important for the function of pUL41.
Another viral protein that is involved in HSV-1-mediated host cell shutoff, ICP27, interacts with hnRNPs K and CK2 (59), which phosphorylates hnRNP K in an ICP27-dependent manner (25). The impact of this phosphorylation, which is probably reflected by the observed shift to more acidic variants of hnRNP K, in the progress of the infection is still unclear but might be of importance across the herpesvirus family, since a similar interaction between hnRNP K and CK2 and the ORF57 protein has been found in Kaposi's sarcoma-associated herpesvirus-infected cells (32).
The proteomic screen described here resulted in the identification of a number of cellular proteins that are potentially of significance for herpesvirus infection. The fact that several of them have previously been linked to the replication cycle of alphaherpesviruses (lamin A/C, the B-type lamins, and hnRNP K) serves to validate our experimental system. The newly established protocol can be easily adapted to study virus-host cell interactions of any virus that can be propagated in cultured cells of a species with a sequenced and annotated genome. It can be further refined by the application of additional affinity matrices or the use of narrow-range isoelectric focusing strips to improve yields of less abundant proteins. The demonstration that most of the observed changes affected the distribution of protein in different posttranslationally modified isoforms rather than absolute expression levels underlines the importance of analysis at the protein level for a comprehensive understanding of the molecular effects of virus infections on the cellular metabolism.
Acknowledgments
This study was in part supported by the Deutsche Forschungsgemeinschaft (DFG Me 854/8).
We thank Barbara Bettin for expert technical assistance.
Footnotes
Published ahead of print on 23 July 2008.
REFERENCES
- 1.Arrigo, A. P. 2007. The cellular “networking” of mammalian Hsp27 and its functions in the control of protein folding, redox state and apoptosis. Adv. Exp. Med. Biol. 59414-26. [DOI] [PubMed] [Google Scholar]
- 2.Ashburner, M., C. A. Ball, J. A. Blake, D. Botstein, H. Butler, J. M. Cherry, A. P. Davis, K. Dolinski, S. S. Dwight, J. T. Eppig, M. A. Harris, D. P. Hill, L. Issel-Tarver, A. Kasarskis, S. Lewis, J. C. Matese, J. E. Richardson, M. Ringwald, G. M. Rubin, G. Sherlock, et al. 2000. Gene Ontology: tool for the unification of biology. Nat. Genet. 2525-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beissbarth, T., and T. P. Speed. 2004. GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics 201464-1465. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Porat, T., T. Rakusanova, and A. S. Kaplan. 1971. Early functions of the genome of herpesvirus. II. Inhibition of the formation of cell-specific polysomes. Virology 46890-899. [DOI] [PubMed] [Google Scholar]
- 5.Bjerke, S. L., and R. J. Roller. 2006. Roles for herpes simplex virus type 1 UL34 and US3 proteins in disrupting the nuclear lamina during herpes simplex virus type 1 egress. Virology 347261-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanchard, Y., N. Le Meur, M. Le Cunff, P. Blanchard, J. Leger, and A. Jestin. 2006. Cellular gene expression survey of pseudorabies virus (PRV) infected human embryonic kidney cells (HEK-293). Vet. Res. 37705-723. [DOI] [PubMed] [Google Scholar]
- 7.Buttner, K., J. Bernhardt, C. Scharf, R. Schmid, U. Mader, C. Eymann, H. Antelmann, A. Volker, U. Volker, and M. Hecker. 2001. A comprehensive two-dimensional map of cytosolic proteins of Bacillus subtilis. Electrophoresis 222908-2935. [DOI] [PubMed] [Google Scholar]
- 8.Chelbi-Alix, M. K., and H. de Thé. 1999. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene 18935-941. [DOI] [PubMed] [Google Scholar]
- 9.Chow, S., and P. Rodgers. 2005. Constructing area—proportional Venn and Euler diagrams with three circles. Euler Diagrams Workshop, Paris, France.
- 10.Elgadi, M. M., C. E. Hayes, and J. R. Smiley. 1999. The herpes simplex virus vhs protein induces endoribonucleolytic cleavage of target RNAs in cell extracts. J. Virol. 737153-7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esclatine, A., B. Taddeo, L. Evans, and B. Roizman. 2004. The herpes simplex virus 1 UL41 gene-dependent destabilization of cellular RNAs is selective and may be sequence-specific. Proc. Natl. Acad. Sci. USA 1013603-3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flori, L., C. Rogel-Gaillard, M. Cochet, G. Lemonnier, K. Hugot, P. Chardon, S. Robin, and F. Lefevre. 2008. Transcriptomic analysis of the dialogue between pseudorabies virus and porcine epithelial cells during infection. BMC Genomics 9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gasteiger, E., A. Gattiker, C. Hoogland, I. Ivanyi, R. D. Appel, and A. Bairoch. 2003. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 313784-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geenen, K., H. W. Favoreel, L. Olsen, L. W. Enquist, and H. J. Nauwynck. 2005. The pseudorabies virus US3 protein kinase possesses anti-apoptotic activity that protects cells from apoptosis during infection and after treatment with sorbitol or staurosporine. Virology 331144-150. [DOI] [PubMed] [Google Scholar]
- 15.Greco, A., A. M. Laurent, and J. J. Madjar. 1997. Repression of beta-actin synthesis and persistence of ribosomal protein synthesis after infection of HeLa cells by herpes simplex virus type 1 infection are under translational control. Mol. Gen. Genet. 256320-327. [DOI] [PubMed] [Google Scholar]
- 16.Hardwicke, M. A., and R. M. Sandri-Goldin. 1994. The herpes simplex virus regulatory protein ICP27 contributes to the decrease in cellular mRNA levels during infection. J. Virol. 684797-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardy, W. R., and R. M. Sandri-Goldin. 1994. Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. J. Virol. 687790-7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Honess, R. W., and B. Roizman. 1974. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J. Virol. 148-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Honess, R. W., and B. Roizman. 1975. Regulation of herpesvirus macromolecular synthesis: sequential transition of polypeptide synthesis requires functional viral polypeptides. Proc. Natl. Acad. Sci. USA 721276-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ihara, S., L. Feldman, S. Watanabe, and T. Ben Porat. 1983. Characterization of the immediate-early functions of pseudorabies virus. Virology 131437-454. [DOI] [PubMed] [Google Scholar]
- 21.Jarosinski, K., L. Kattenhorn, B. Kaufer, H. Ploegh, and N. Osterrieder. 2007. A herpesvirus ubiquitin-specific protease is critical for efficient T cell lymphoma formation. Proc. Natl. Acad. Sci. USA 10420025-20030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaplan, A. S., and A. E. Vatter. 1959. A comparison of herpes simplex and pseudorabies viruses. Virology 7394-407. [DOI] [PubMed] [Google Scholar]
- 23.Kattenhorn, L. M., G. A. Korbel, B. M. Kessler, E. Spooner, and H. L. Ploegh. 2005. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol. Cell 19547-557. [DOI] [PubMed] [Google Scholar]
- 24.Kersey, P. J., J. Duarte, A. Williams, Y. Karavidopoulou, E. Birney, and R. Apweiler. 2004. The International Protein Index: an integrated database for proteomics experiments. Proteomics 41985-1988. [DOI] [PubMed] [Google Scholar]
- 25.Koffa, M. D., J. Kean, G. Zachos, S. A. Rice, and J. B. Clements. 2003. CK2 protein kinase is stimulated and redistributed by functional herpes simplex virus ICP27 protein. J. Virol. 774315-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kummer, M., N. M. Turza, P. Muhl-Zurbes, M. Lechmann, C. Boutell, R. S. Coffin, R. D. Everett, A. Steinkasserer, and A. T. Prechtel. 2007. Herpes simplex virus type 1 induces CD83 degradation in mature dendritic cells with immediate-early kinetics via the cellular proteasome. J. Virol. 816326-6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Landry, J., H. Lambert, M. Zhou, J. N. Lavoie, E. Hickey, L. A. Weber, and C. W. Anderson. 1992. Human HSP27 is phosphorylated at serines 78 and 82 by heat shock and mitogen-activated kinases that recognize the same amino acid motif as S6 kinase II. J. Biol. Chem. 267794-803. [PubMed] [Google Scholar]
- 28.Leal, R. B., T. Posser, A. P. Rigon, C. S. Oliveira, C. A. Goncalves, D. P. Gelain, and P. R. Dunkley. 2007. Cadmium stimulates MAPKs and Hsp27 phosphorylation in bovine adrenal chromaffin cells. Toxicology 23434-43. [DOI] [PubMed] [Google Scholar]
- 29.Lin, H. W., Y. Y. Chang, M. L. Wong, J. W. Lin, and T. J. Chang. 2004. Functional analysis of virion host shutoff protein of pseudorabies virus. Virology 324412-418. [DOI] [PubMed] [Google Scholar]
- 30.Lundmark, R., and S. R. Carlsson. 2004. Regulated membrane recruitment of dynamin-2 mediated by sorting nexin 9. J. Biol. Chem. 27942694-42702. [DOI] [PubMed] [Google Scholar]
- 31.Madin, S. H., and N. B. Darby, Jr. 1958. Established kidney cell lines of normal adult bovine and ovine origin. Proc. Soc. Exp. Biol. Med. 98574-576. [DOI] [PubMed] [Google Scholar]
- 32.Malik, P., and J. B. Clements. 2004. Protein kinase CK2 phosphorylation regulates the interaction of Kaposi's sarcoma-associated herpesvirus regulatory protein ORF57 with its multifunctional partner hnRNP K. Nucleic Acids Res. 325553-5569. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 761537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mettenleiter, T. C., B. G. Klupp, and H. Granzow. 2006. Herpesvirus assembly: a tale of two membranes. Curr. Opin. Microbiol. 9423-429. [DOI] [PubMed] [Google Scholar]
- 35.Mou, F., T. Forest, and J. D. Baines. 2007. US3 of herpes simplex virus type 1 encodes a promiscuous protein kinase that phosphorylates and alters localization of lamin A/C in infected cells. J. Virol. 816459-6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neuhoff, V., N. Arold, D. Taube, and W. Ehrhardt. 1988. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie brilliant blue G-250 and R-250. Electrophoresis 9255-262. [DOI] [PubMed] [Google Scholar]
- 37.Olsen, J. V., B. Blagoev, F. Gnad, B. Macek, C. Kumar, P. Mortensen, and M. Mann. 2006. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127635-648. [DOI] [PubMed] [Google Scholar]
- 38.Ong, S. E., B. Blagoev, I. Kratchmarova, D. B. Kristensen, H. Steen, A. Pandey, and M. Mann. 2002. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1376-386. [DOI] [PubMed] [Google Scholar]
- 39.Pappin, D. J., P. Hojrup, and A. J. Bleasby. 1993. Rapid identification of proteins by peptide-mass fingerprinting. Curr. Biol. 3327-332. [DOI] [PubMed] [Google Scholar]
- 40.Park, R., and J. D. Baines. 2006. Herpes simplex virus type 1 infection induces activation and recruitment of protein kinase C to the nuclear membrane and increased phosphorylation of lamin B. J. Virol. 80494-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parkinson, J., S. P. Lees-Miller, and R. D. Everett. 1999. Herpes simplex virus type 1 immediate-early protein vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J. Virol. 73650-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perkins, D. N., D. J. Pappin, D. M. Creasy, and J. S. Cottrell. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 203551-3567. [DOI] [PubMed] [Google Scholar]
- 43.Poon, A. P., H. Gu, and B. Roizman. 2006. ICP0 and the US3 protein kinase of herpes simplex virus 1 independently block histone deacetylation to enable gene expression. Proc. Natl. Acad. Sci. USA 1039993-9998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ray, N., and L. W. Enquist. 2004. Transcriptional response of a common permissive cell type to infection by two diverse alphaherpesviruses. J. Virol. 783489-3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Righetti, P. G., A. Castagna, P. Antonioli, and E. Boschetti. 2005. Prefractionation techniques in proteome analysis: the mining tools of the third millennium. Electrophoresis 26297-319. [DOI] [PubMed] [Google Scholar]
- 46.Rosenfeld, J., J. Capdevielle, J. C. Guillemot, and P. Ferrara. 1992. In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal. Biochem. 203173-179. [DOI] [PubMed] [Google Scholar]
- 47.Sacks, W. R., C. C. Greene, D. P. Aschman, and P. A. Schaffer. 1985. Herpes simplex virus type 1 ICP27 is an essential regulatory protein. J. Virol. 55796-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sarto, C., C. Valsecchi, F. Magni, L. Tremolada, C. Arizzi, N. Cordani, S. Casellato, G. Doro, P. Favini, R. A. Perego, F. Raimondo, S. Ferrero, P. Mocarelli, and M. Galli-Kienle. 2004. Expression of heat shock protein 27 in human renal cell carcinoma. Proteomics 42252-2260. [DOI] [PubMed] [Google Scholar]
- 49.Schwartz, J. A., E. E. Brittle, A. E. Reynolds, L. W. Enquist, and S. J. Silverstein. 2006. UL54-null pseudorabies virus is attenuated in mice but productively infects cells in culture. J. Virol. 80769-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scott, E. S., and P. O'Hare. 2001. Fate of the inner nuclear membrane protein lamin B receptor and nuclear lamins in herpes simplex virus type 1 infection. J. Virol. 758818-8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seet, L. F., and W. Hong. 2006. The Phox (PX) domain proteins and membrane traffic. Biochim. Biophys. Acta 1761878-896. [DOI] [PubMed] [Google Scholar]
- 52.Simonin, D., J. J. Diaz, T. Masse, and J. J. Madjar. 1997. Persistence of ribosomal protein synthesis after infection of HeLa cells by herpes simplex virus type 1. J. Gen. Virol. 78435-443. [DOI] [PubMed] [Google Scholar]
- 53.Soulet, F., D. Yarar, M. Leonard, and S. L. Schmid. 2005. SNX9 regulates dynamin assembly and is required for efficient clathrin-mediated endocytosis. Mol. Biol. Cell 162058-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taddeo, B., and B. Roizman. 2006. The virion host shutoff protein (UL41) of herpes simplex virus 1 is an endoribonuclease with a substrate specificity similar to that of RNase A. J. Virol. 809341-9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tisdale, E. J. 2001. Glyceraldehyde-3-phosphate dehydrogenase is required for vesicular transport in the early secretory pathway. J. Biol. Chem. 2762480-2486. [DOI] [PubMed] [Google Scholar]
- 56.Tisdale, E. J., and C. R. Artalejo. 2007. A GAPDH mutant defective in Src-dependent tyrosine phosphorylation impedes Rab2-mediated events. Traffic 8733-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tisdale, E. J., C. Kelly, and C. R. Artalejo. 2004. Glyceraldehyde-3-phosphate dehydrogenase interacts with Rab2 and plays an essential role in endoplasmic reticulum to Golgi transport exclusive of its glycolytic activity. J. Biol. Chem. 27954046-54052. [DOI] [PubMed] [Google Scholar]
- 58.Van Minnebruggen, G., H. W. Favoreel, L. Jacobs, and H. J. Nauwynck. 2003. Pseudorabies virus US3 protein kinase mediates actin stress fiber breakdown. J. Virol. 779074-9080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wadd, S., H. Bryant, O. Filhol, J. E. Scott, T. Y. Hsieh, R. D. Everett, and J. B. Clements. 1999. The multifunctional herpes simplex virus IE63 protein interacts with heterogeneous ribonucleoprotein K and with casein kinase 2. J. Biol. Chem. 27428991-28998. [DOI] [PubMed] [Google Scholar]