Abstract
The generally accepted paradigm of transcription by regulated recruitment defines sequence-specific transcription factors and coactivators as separate categories that are distinguished by their abilities to bind DNA autonomously. The C2H2 zinc finger protein Zac1, with an established role in canonical DNA binding, also acts as a coactivator. Commensurate with this function, p73, which is related to p53, is here shown to recruit Zac1, together with the coactivators p300 and PCAF, to the p21Cip1 promoter during the differentiation of embryonic stem cells into neurons. In the absence of autonomous DNA binding, Zac1's zinc fingers stabilize the association of PCAF with p300, suggesting its scaffolding function. Furthermore, Zac1 regulates the affinities of PCAF substrates as well as the catalytic activities of PCAF to induce a selective switch in favor of histone H4 acetylation and thereby the efficient transcription of p21Cip1. These results are consistent with an authentic coactivator function of Zac1's C2H2 zinc finger DNA-binding domain and suggest coactivation by sequence-specific transcription factors as a new facet of transcriptional control.
The transcriptional activation of eukaryotic genes involves the regulated assembly of multiprotein complexes on enhancers and promoters (9, 22, 23). The specificity of this process is decisively controlled by sequence-specific DNA-binding transcription factors (henceforth termed “sequence-specific factors”) which play a key role in interpreting and transmitting the information contained in the primary DNA sequence to the factors and cofactors that, in turn, mediate the synthesis of RNA transcripts from the DNA template.
Sequence-specific factors typically contain a specific DNA-binding domain that directly contacts DNA, a multimerization domain that allows the formation of homo- or heteromultimers, and a transcription activation domain. The binding of sequence-specific factors to regulatory DNA elements, through which they are tethered to their correct location, is a prerequisite for gene regulation. This allows the positioning of the activation domain in the vicinity of the initiation complex and the subsequent recruitment of an active transcription complex (30). The sequential order of these events underlies the concept of gene activation via regulated recruitment (29).
Sequence-specific factors that do not directly contact the basal transcription machinery often bind to different classes of interconnecting coregulators (for a minimal classification see reference 22).
Among these, primary coactivators bind directly to sequence-specific factors and often contain relevant enzymatic activities that are necessary for changing chromatin structure from a quiescent state to one that permits active gene transcription; prototypical examples are the coactivators p300/CBP and PCAF, which are endowed with potent histone acetyltransferase (HAT) activities (21). In contrast, secondary coactivators dock onto sequence-specific factors and form a scaffold, thus facilitating the recruitment of other proteins containing relevant enzymatic activities (19). Generally, coregulators exhibit great variability in their enzymatic and scaffolding properties; however, they invariably depend on “locator” proteins—sequence-specific factors—to accomplish their roles (23, 29, 30).
The zinc finger protein Zac1 is a member of a new subfamily of sequence-specific factors which play a critical role in cell proliferation and tumorigenesis (16, 38). Zac1's antiproliferative actions in mesenchymal and neural progenitor/stem cells indicates its importance for cell fate and lineage decisions (36, 37). Previous studies demonstrate that either monomeric or dimeric Zac1 binding to GC-rich palindromic elements or to GC-rich direct- and reverse-repeat elements (4, 16, 38) underlies the activation of the H19, Lit1, peroxisome proliferator-activated receptor gamma, and CK14 target genes (1, 3, 16, 39).
Quite unexpectedly, Zac1 was isolated in a yeast screen for mammalian coregulator proteins that bind to the carboxyl-terminal activation domain of the nuclear receptor coactivator SRC-2 (18). Consistent with its putative role as a coactivator, Zac1 additionally associated with the ligand-binding domains of the androgen, estrogen, glucocorticoid, and thyroid hormone receptors and potently enhanced the transcriptional activities of these nuclear receptors. Also supporting the notion that Zac1 may have a broader role in coactivation, the protein was shown to increase the activity of the tumor suppressor p53 gene after binding to its activation domain (17).
Together, these data raise the fundamental question as to whether, and how, a sequence-specific factor can additionally function as a coactivator. This study reveals that Zac1 is recruited, together with the coactivators p300 and PCAF, by the sequence-specific factor p73 and that it selectively regulates the activity of PCAF. Consequently, Zac1 can refine p73-dependent transcription of the p21Cip1 gene during the early neuronal differentiation of embryonic stem cells.
MATERIALS AND METHODS
Cell cultures, plasmids, and transfections.
Saos-2, SK-N-MC, and U2OS (ATCC HTB-85, HTB-10, and HTB-96, respectively) cells were cultivated in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. The embryonic stem cell line 46C was routinely propagated without feeders in Lif-supplemented medium containing 10% fetal calf serum (PAN Biotech) and differentiated as described previously (42).
Plasmids or small interfering RNA (siRNA) oligonucleotides were transfected by electroporation, Lipofectamine 2000, or RNAiMAX reagent (Invitrogen) using 2 × 106 to 3 × 106 Saos-2, U2OS, or 46C cells. Luciferase values were normalized to β-galactosidase values (15).
For the dose-response reporter assays, the following concentrations of Zac1 or ZAC1 were used: 10 ng, 25 ng, or 50 ng Zac1 or 0.1 μg, 0.25 μg, or 0.5 μg ZAC1 for the reporter assays depicted in Fig. 1B, C, D, and F and 10 ng, 25 ng, 50 ng, or 100 ng Zac1 or 0.1 μg, 0.25 μg, 0.5 μg, or 1 μg ZAC1 for the reporter assays depicted in Fig. 3A, C, D, and G and Fig. 4C and F. The concentrations of the Zac1 zinc finger (ZΔZF), linker (ZΔL), proline-rich (ZΔPR), linker-proline-rich (ZΔLPR), and carboxyl-terminal (ZΔC) domain deletion mutants and the zinc finger (Z-ZF), linker-proline-rich (Z-LPR), and carboxyl-terminal (Z-C) domains were adjusted to the expression levels of Zac1, estimated by immunoblot (IB) analysis.
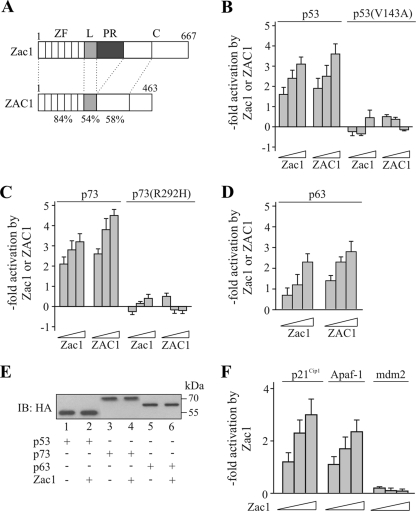
FIG. 1.
Zac1 coactivation of p73. (A) Scheme of mouse Zac1 and human ZAC1 proteins, depicting zinc finger (ZF), linker (L), proline-rich (PR), and carboxyl-terminal (C) domains. Amino acid identity (%) between the corresponding domains of mice and humans is indicated. (B to D) Reporter assays. Increasing concentrations of cotransfected Zac1 or ZAC1 enhanced p53-dependent (B), p73-dependent (C), or p63-dependent (D) PG12PYLuc reporter activity in Saos-2 cells. In contrast, Zac1 or ZAC1 did not enhance reporter activity in the presence of DNA-binding-defective p53(V143A) (B) or p73(R292H) (C). (E) IB analysis. Hemagglutinin (HA)-tagged p53 (25 ng) or p73 or p63 (50 ng each) was transfected in the absence (−) or presence (+) of Zac1 (50 ng) into Saos-2 cells. WCE were immunoblotted and tested with an anti-HA antibody. The expression levels of p53, p63, and p73 are unaltered in the presence of Zac1. (F) Reporter assays. Increasing concentrations of cotransfected Zac1 enhanced the p73-dependent promoter activities of p21Cip1 and Apaf-1, whereas the p73-dependent activity of mdm2 was not regulated.
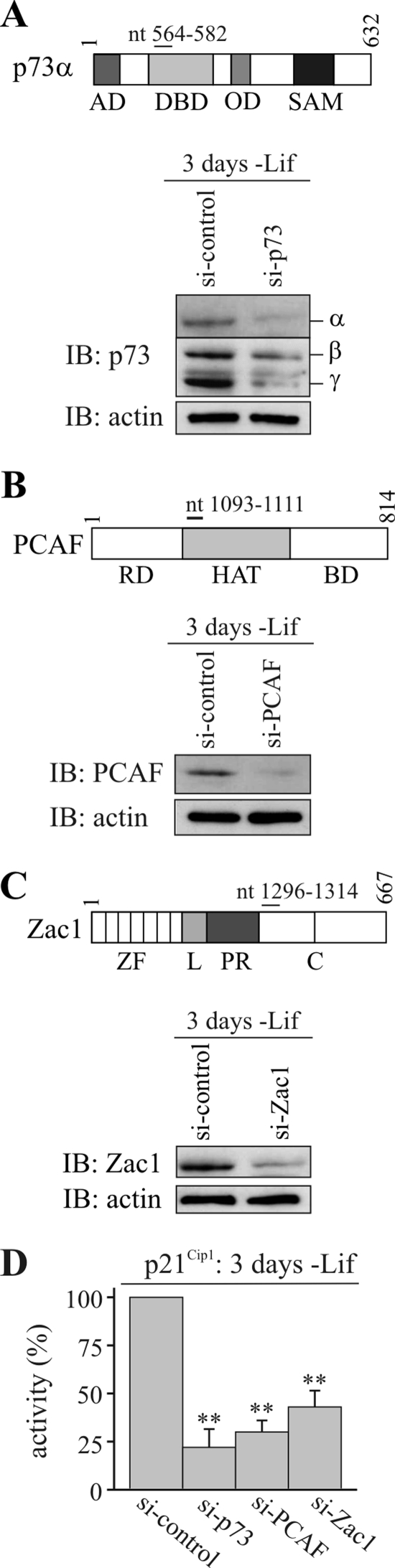
FIG. 10.
Knockdown of p73, PCAF, and Zac1 in embryonic stem cells. (A) Scheme depicting mouse p73α and the nucleotides (nt) against which the siRNA oligonucleotides were directed. The IB shows p73 expression in control or p73 siRNA-transfected mES cells. The IBs were retested with an antiactin antibody to verify the equal loading of WCE. The cells were transfected 1 day after Lif withdrawal and harvested 2 days later. Isoforms of p73 (α, β, and γ) are indicated. AD, activation domain; DBD, DNA-binding domain; OD, oligomerization domain; SAM, sterile alpha motif. (B) Scheme depicting mouse PCAF and the nucleotides (nt) against which the siRNA oligonucleotides were directed. The IB shows PCAF expression in control or PCAF siRNA-transfected mES cells. RD, amino-terminal regulatory domain; BD, carboxyl-terminal bromo domain. (C) Scheme depicting mouse Zac1 and the nucleotides (nt) against which the siRNA oligonucleo-tides were directed. The IB shows Zac1 expression in control or Zac1 siRNA-transfected mES cells. (D) qRT-PCR analysis. Downregulation of p21Cip1 mRNA expression on the third day of Lif withdrawal and after exposure to the above siRNA treatments. P values were calculated by Student's t test for differences between control versus factor-specific siRNA (*, P < 0.05; **, P < 0.01).
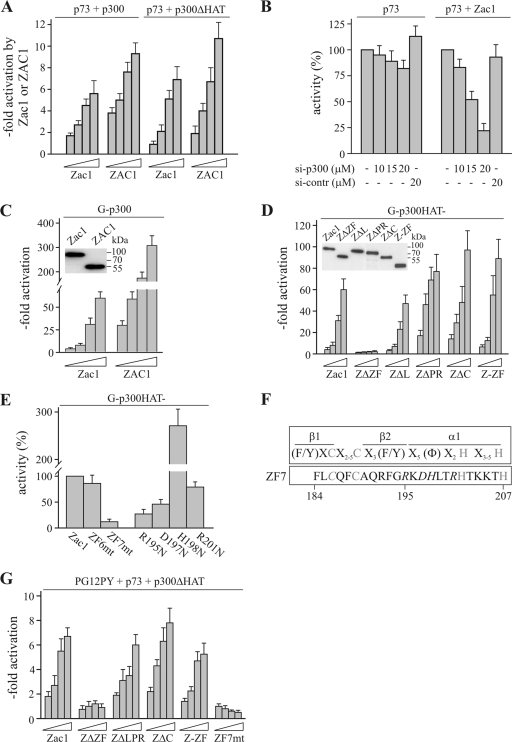
FIG. 3.
Zac1 zinc fingers mediate coactivation. (A) Reporter assays. p73-dependent PG12PYLuc reporter activity in the presence of p300 or HAT activity-deficient p300 (p300ΔHAT) was further strongly enhanced by the cotransfection of increasing concentrations of Zac1 or ZAC1 into Saos-2 cells. (B) Reporter assays and knockdown of p300 by siRNA treatment. p73 and Zac1 (50 ng each) and the reporter PG12PYLuc were cotransfected into Saos-2 cells in the absence (−) or presence of the indicated concentrations of p300 (si-p300) or control (si-contr) siRNA. Activities in the absence of siRNA were set to 100%. The knockdown of p300 weakly reduced p73 activity but strongly attenuated Zac1-dependent enhancement. (C) Reporter assays. Increasing concentrations of Zac1 or ZAC1 potently activated GAL4 reporter activity in the presence of a Gal4-p300 fusion protein (G-p300) in U2OS cells. The inset shows the adjusted expression of amino-terminal Flag-tagged Zac1 (50 ng) and ZAC1 following transfection into U2OS cells. IBs were tested with an anti-Flag antibody. (D) Reporter assays. A Gal4-p300 fusion construct encoding an inactive HAT domain (G-p300HAT-) was cotransfected as above with increasing concentrations of Zac1, ZΔZF, ZΔL, ZΔPR, ZΔC, or Z-ZF. Zinc finger-deleted Zac1 failed to activate G-p300HAT-, while the absence of the linker, proline-rich, or C-terminal domains, or the presence of the isolated zinc fingers, preserved activation. The inset shows the adjusted expression of amino-terminal hemagglutinin-tagged Zac1 (25 ng) and of the foregoing constructs upon transfection into U2OS cells. IBs were tested with an anti-HA antibody. (E) Reporter assays. G-p300HAT- was transfected as above with Zac1 or Zac1 containing broken zinc finger 6 or 7 (ZF6mt or ZF7mt) into U2OS cells. The activity of Zac1 was set to 100%. ZF7-mutated Zac1 strongly reduced the activation of p300. Moreover, Zac1 proteins containing single point mutations of key residues in the predicted α helix of ZF7 (see below) showed either impaired or enhanced activation, which is consistent with a regulatory role of ZF7. Each Zac1 construct was tested at 50 ng. (F) Scheme of a prototypical zinc finger depicting the localization of the cysteine and histidine residues (shown in light gray), the β-sheets, and the α-helix. The amino acid sequence of ZF7 is aligned below. Mutated residues are shown in italic letters. (G) Reporter assays. p73 was cotransfected with p300ΔHAT and the reporter PG12PYLuc into Saos-2 cells. Coactivation was investigated for increasing amounts of Zac1, ZΔZF, ZΔLPR, ZΔC, Z-ZF, or ZF7mt. In accord with the data from the heterologous Gal-p300 system (C to E), p73 coactivation depends on Zac1's zinc-finger region and an intact ZF7.
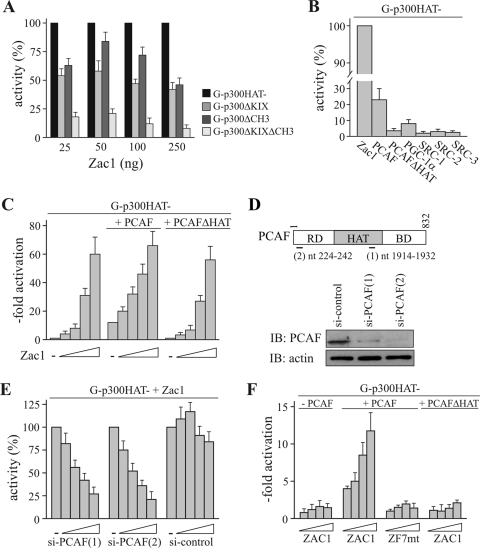
FIG. 4.
Zac1 binding to p300 enhances the association of PCAF. (A) Reporter assays. The HAT activity-deficient Gal4-p300 fusion construct (G-p300HAT-) was cotransfected with the reporter GAL4 and the indicated concentrations of Zac1 into U2OS cells. Activities were set to 100% and compared to those of G-p300ΔKIX, G-p300ΔCH3, or G-p300ΔKIXΔCH3, all of which harbor single or joint deletions of the KIX and CH3 domains. Zac1 activates through binding to either p300 docking site. (B) Reporter assays. G-p300HAT- was cotransfected with Zac1 (0.1 μg) into U2OS cells, and GAL4 reporter activity was set to 100%. Coactivators were tested at increasing concentrations in the absence of Zac1 and plotted with reference to maximal activations in the presence of Zac1. PCAF, but not HAT activity-deficient PCAF, could activate p300, while the other coactivators were ineffective. (C) Reporter assays. G-p300HAT- was cotransfected with the reporter GAL4 and increasing concentrations of Zac1 into U2OS cells. PCAF strongly enhanced Zac1 activation at low concentrations, indicating that PCAF is engaged in Zac1 coactivation. PCAF's HAT activity is critical, as evidenced by the failure of PCAFΔHAT to coactivate. (D) IB analysis of PCAF knockdown. Scheme depicting human PCAF and the nucleotides (nt) against which the siRNA oligonucleotides are directed. The IB shows PCAF expression in control or PCAF siRNA-treated U2OS cells (si-control and si-PCAF) 48 h after transfection. The IBs were retested with an antiactin antibody to verify the equal loading of WCE. RD, amino-terminal regulatory domain; BD, carboxyl-terminal bromo domain. (E) Reporter assays and knockdown of PCAF by siRNA treatments. G-p300HAT- was cotransfected with Zac1 (50 ng) into U2OS cells, and the activation of the GAL4 reporter was set to 100%. Activities following the cotransfection of increasing concentrations of two different PCAF [si-PCAF(1) and (2)] or one control (si-control) siRNA were normalized against Zac1 activation. (F) Reporter assays. G-p300HAT- was cotransfected with the GAL4 reporter and increasing concentrations of ZAC1 or ZF7-mutated ZAC1 (ZF7mt) into PCAF-negative human neuroblastoma cells. PCAF restored ZAC1 coactivation of G-p300HAT, while HAT activity-deficient PCAF (PCAFΔHAT) or ZF7mt was ineffective.
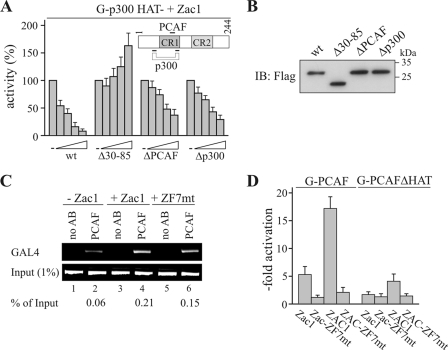
E1A12S (wild type [wt]) was transfected at a concentration of 2.5 ng, 5 ng, 10 ng, or 25 ng. The concentrations of Δ30-85, ΔPCAF (Δ55-60), or Δp300 (Δ64-67) were adjusted to the expression levels of E1A12S, estimated by IB analysis (Fig. 5A and B).
FIG. 5.
E1A prevents the enhanced association of PCAF, while a broken ZF7 blocks activation. (A) Reporter assays. G-p300HAT- was cotransfected with Zac1 (0.1 μg) into U2OS cells, and GAL4 reporter activity was set to 100%. Activities for the cotransfection of increasing concentrations of the adenovirus E1A12S (wt) or E1A deletion constructs were normalized to maximal activation by Zac1. E1A constructs contained deletions of either the common p300 and PCAF binding sites (Δ30-85) or of binding sites specific for PCAF (ΔPCAF) or p300 (Δp300). The inset shows E1A with PCAF and/or p300 binding sites indicated. CR1, conserved region 1; CR2, conserved region 2. (B) IB analysis. Expression of adjusted amounts of amino-terminal Flag-tagged adenovirus E1A12S (wt; 50 ng) or of the above deletion constructs in transfected U2OS cells. IBs were tested with an anti-Flag antibody. (C) ChIP assays. G-p300HAT- and the reporter GAL4 were cotransfected with Zac1 or ZF7-mutated (ZF7mt) Zac1 (0.1 μg each) into U2OS cells. ChIP experiments were conducted with an anti-PCAF antibody (AB) and a primer pair bracketing the minimal promoter of the GAL4 reporter. Zac1 strongly enhanced the association of PCAF (% of input), while ZF7-mutated Zac1 resulted in a reduction by one-third. (D) Reporter assays. Fusion constructs of Gal4 and PCAF or HAT activity-deficient PCAF (G-PCAF or G-PCAFΔHAT) were cotransfected with Zac1 or ZF7-mutated (ZF7mt) Zac1 (0.1 μg each), ZAC1 or ZF7-mutated ZAC1 (1 μg each), and the reporter GAL4 into U2OS cells. Data show that activation depends on an intact ZF7 and PCAF's HAT function.
The siRNA sequences are listed at http://www.mpipsykl.mpg.de/pages/13.07.pdf. In all the knockdown experiments (IB or chromatin immunoprecipitation [ChIP] analysis), PCAF, p73, Zac1, or control siRNA were used at 20 μM. For the dose-response experiments, PCAF siRNA concentrations were 10 μM, 15 μM, 20 μM, and 25 μM.
A series of cytomegalovirus-driven pRK (16, 32) or pcDNA (Invitrogen) vectors were used for cDNA expression. Members of the p53 family were p53, p73α, and p63α. Reporter plasmids contained multimerized p53 (PG12PYLuc [10]) or GAL4 DNA-binding sites upstream of the luciferase gene. The promoter plasmids were pGL3b, Apaf-1 (−871/+208) (24), p21Cip1(WWP-Luc) (11), and pGl2.hmdm2Luc (43). Further details on the generation of the constructs used in this study are available upon request.
The reporter plasmids (PG12PYLuc, p21Cip1, Apaf-1, mdm2, and GAL4) were used at 0.5 μg each. For the reporter assays, p53, p53(V143A), p73, p73(R292H), and p63 were transfected at 50 ng each. The concentrations of the Gal4 fusion constructs were 50 ng (G-p300, G-p300ΔKIX, G-p300ΔCH3, G-p300ΔKIXΔCH3, G-PCAF, or G-PCAFΔHAT) or 200 ng (G-p300HAT-). The concentrations of the cotransfected coactivator plasmids were 1 μg p300 or p300ΔHAT, 2 μg PCAF or PCAFΔHAT, 2.5 μg PGC-1α, and 0.5 μg each SRC-1 to -3.
Real-time quantitative PCR analysis.
RNA was isolated using the NucleoSpin RNA II kit (Macherey-Nagel). One microgram of total RNA was reverse transcribed using oligo(dT)18 and SuperScript II reverse transcriptase (Invitrogen). An analysis was performed with Absolute Blue QPCR Sybr green mix (ABgene) and the MJ Mini Opticon light cycler (Bio-Rad). The primer sequences used are listed at the URL mentioned above.
IP, IB, and generation of antibodies.
IP were performed as previously described (15). For the IB analysis, whole-cell extracts (WCE) were fractionated by polyacrylamide gel electrophoresis, immunoblotted, and tested with the indicated antibodies (16). Transfections for the IB and IP experiments were performed with 4 × 106 to 5 × 106 cells.
To detect the proteins translated after transfection of the cDNA expression vectors, 10 or 20 μg of WCE was immunoblotted. To detect endogenous PCAF, p73, Zac1, or cyclin kinase inhibitors, the IBs were performed with 100 μg WCE.
For the generation of polyclonal antisera against PCAF, cDNA fragments encoding the amino-terminal regulatory region (amino acids [aa] 1 to 352) or part of the carboxyl-terminal region (aa 452 to 717) were subcloned into pGEX-2TK. The rabbits were immunized four times with 100 μg of the fusion protein at 10-day intervals, and antisera were collected after 75 days. The specificity of the antibodies (1:1,000 each) was tested by immunoblotting of mock- or PCAF-transfected SK-N-MC cells and by preabsorbing PCAF antibodies on glutathione S transferase (GST)-PCAF-n-glutathione-Sepharose, or GST-PCAF-c-glutathione-Sepharose, respectively. The signals were specific for PCAF-transfected cells. No signals were detected with either the preimmune or preabsorbed PCAF sera (data not shown).
A p73 antibody against the amino-terminal activation domain (aa 1 to 72) was generated, preabsorbed, and characterized (data not shown).
The characteristics of all antibodies are provided at the URL mentioned above.
In vitro HAT assays.
GST-PCAF, GST-p73, and in vitro-translated p300ΔHAT were preincubated with an equimolar amount of GST-Zac1 or GST-Zac-ZF7mt and processed as previously described (15). Biotinylated N-terminal H4 (aa 2 to 24) or H3 (aa 1 to 21) peptides were purchased from Millipore.
GST pull-down assays and in vitro translations.
GST-PCAF, GST-Zac1, GST-ZF7mt, GST-p73, or derivatives were grown in DH5α, purified, and quantified as described previously (16). The GST pull-down experiments and in vitro translations were performed as described previously (15).
ChIP assays.
ChIP experiments were conducted with the Magna ChIP G kit (Millipore) according to the manufacturer's recommendations and as described previously (3). The ChIP primers for real-time PCR analysis and antibodies are listed at the URL mentioned above.
Statistical analysis.
The results show the means and standard deviations of the results from at least three independent experiments. The HAT assay and siRNA knockdown experiment results come from three to five independent experiments. Additional analyses were performed by Student's t test.
RESULTS
Zac1 coactivates and interacts with p73.
Mouse Zac1 and human ZAC1 proteins share virtually identical DNA-binding domains and show high conservation for the amino-terminal parts of their carboxyl termini; however, they differ by the presence of the central proline-rich activation domain, which is specific to mice (16) (Fig. 1A). The cotransfection of p53 and increasing concentrations of Zac1/ZAC1 consistently enhanced p53-dependent reporter activity in the p53-negative osteosarcoma cell line Saos-2. In contrast, Zac1 did not enhance reporter activity in the presence of a DNA-binding-defective form of p53 (Fig. 1B). Similar results were obtained for p73, while the maximal coactivation of p63 was about half of that seen with p53 (Fig. 1C and D). Importantly, the cotransfection of Zac1 did not alter the expression of p53, p63, or p73, indicating a direct transcriptional effect (Fig. 1E).
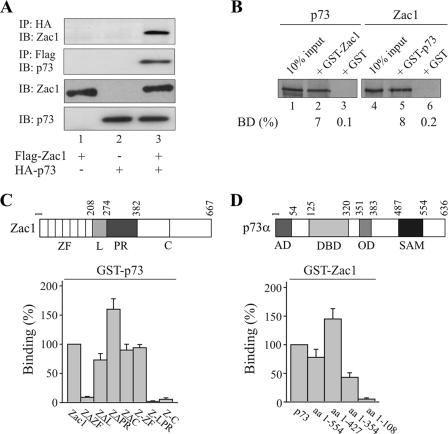
Given the shared role of Zac1 and p73 in neurodevelopment and differentiation (35-37), further experiments focused on the Zac1 coactivation of p73 and its relevance to the fine-tuning of p73 target genes. Zac1 strongly enhanced the p73-dependent activation of the p21Cip1 and Apaf-1 promoter plasmids but did not influence the activation of another p73 target, mdm2 (Fig. 1F). These findings resemble previous results on the context- and promoter-dependent coactivation of nuclear receptors by Zac1 (18). One possible explanation for this is that Zac1 can be recruited only as part of a promoter-specific coactivator complex and not by p73 itself. However, Zac1 was clearly detected in the presence of p73, and vice versa, when Zac1 and p73 were coimmunoprecipitated (Fig. 2A). Additional GST pull-down assays supported a direct interaction between Zac1 and p73; equivalent amounts of both fusion proteins (Fig. 2B; data not shown) efficiently retained Zac1 or p73. The deletion of the zinc finger domain of Zac1 (ZΔZF) strongly attenuated this interaction, whereas binding was preserved even in the absence of the linker (ZΔL), proline-rich (ZΔPR), and carboxyl-terminal (ZΔC) domains (Fig. 2C). Consistent with this, while the isolated transactivation (Z-LPR) and carboxyl-terminal (Z-C) domains were ineffective, the zinc fingers (Z-ZF) alone were sufficient for p73 binding. In a reverse experiment (Fig. 2D), the carboxyl terminus of p73 did not interact with Zac1, whereas the deletion of the oligomerization domain and the adjacent DNA-binding domain halved and eliminated binding, respectively. In contrast to p53, Zac1 did not interact with the amino-terminal activation domain of p73 (Fig. 2D) (17), raising the possibility that Zac1 modulates the DNA-binding activity of p73. However, there was no evidence for an increase in p73 occupancy on the endogenous p21Cip1 promoter in the presence of increasing amounts of Zac1 (see below; data not shown).
FIG. 2.
Zac1 and p73 interact through their DNA-binding domains. (A) Coimmunoprecipitation experiments. Flag-Zac1 (0.5 μg) and hemagglutinin (HA)-p73 (1 μg) were transfected alone or in combination into Saos-2 cells, and WCE (1 mg) were immunoprecipitated with anti-HA or -Flag antibodies. Immunoprecipitates were immunoblotted and tested with the indicated antibodies. +, presence; −, absence. (B to D) GST pull-down assays. Adjusted amounts of GST-Zac1, GST-p73, or GST alone were incubated with equal amounts of in vitro-translated Zac1 or p73 (B). The fraction of the input (100%) bound by each GST protein [BD (%)] is indicated. (C) Equal amounts of in vitro-translated Zac1 proteins, containing deletions of the zinc finger (ZΔZF), linker (ZΔL), and proline-rich (ZΔPR) or the C-terminal (ZΔC) domains, were each incubated with GST-p73 or GST alone. Similarly, equal amounts of the isolated zinc finger (Z-ZF), linker proline-rich (Z-LPR), or C-terminal (Z-C) domains were each incubated with GST-p73 or GST alone. Binding (%) by Zac1 was set to 100%. (D) Equal amounts of in vitro-translated p73 proteins containing successive deletions of the carboxyl terminus were incubated with GST-Zac1 or GST alone. Binding (%) by p73 was set to 100%. AD, activation domain; DBD, DNA-binding domain; OD, oligomerization domain; SAM, sterile alpha motif.
Collectively, these data indicate that Zac1 coactivates various members of the p53 gene family, albeit to different degrees; the coactivation of p73 results from a reciprocal interaction between the DNA-binding domains of p73 and Zac1.
p300 mediates Zac1 coactivation independent of its HAT function.
As previously shown, the C2H2 zinc fingers of Zac1 link canonical DNA binding to the coordinated regulation of p300 HAT activities (15). We therefore asked whether a related mechanism might also underlie Zac1 coactivation. In accord with earlier studies (44), cotransfected p300 resulted in an approximate threefold increase in p73 activity (data not shown), an effect that was enhanced further by Zac1/ZAC1 (Fig. 3A). Intriguingly, this was also the case for a HAT activity-deficient p300 protein, reinforcing the view that p300 can bridge sequence-specific factors, including p73, to other factors containing activational properties (19, 44). In contrast to p300, cotransfection with PCAF, another known coactivator of p73 (46), did not increase coactivation by Zac1 (data not shown). To dissect the relative importance of p300 versus Zac1 for the activation of p73, we depleted Saos-2 cells of endogenous p300 by siRNA treatment (15). This experiment revealed a major role for p300 in the Zac1-mediated coactivation of p73, the coactivation provided by p300 alone being minor (Fig. 3B).
One possible explanation for this behavior is that both sequence-specific factors jointly recruit p300, enabling a synergistic regulation of transcription. Therefore, binding to p73 could allow the localization of Zac1 to DNA even in the absence of a high-affinity Zac1 DNA-binding site. Alternatively, Zac1 might directly regulate p300 in the absence of autonomous DNA binding, a behavior that would be contrary to its established role as a sequence-specific factor. To distinguish between these two possibilities, we investigated a chimeric protein consisting of p300 and the yeast Gal4 DNA-binding domain. Strikingly, increasing concentrations of Zac1/ZAC1 potently induced GAL4 reporter activity (Zac1, ∼60-fold; ZAC1, ∼300-fold) in a manner that showed strict dependence on the p300 chimera (Fig. 3C; data not shown). Since Zac1 is not DNA bound in this model—the reporter plasmid does not contain Zac1 DNA-binding sites, and Zac1 does not interact with the Gal4 DNA-binding domain (16)—these results indicate a new role for Zac1 in regulating p300 activity. These findings were replicated in kidney (LLC-PK1) and cervical (HeLa) epithelial cell lines, demonstrating that the activating effects of Zac1 are not confined to a specific cell type (data not shown). Since Zac1 coactivation deviates from the principle that sequence-specific factors depend strictly on binding to DNA for the manifestation of their functions, we were interested in examining to what extent this type of regulation is specific to Zac1. To this end, we tested a representative number of unrelated eukaryotic sequence-specific factors that are known to interact with p300. Among these, only the estrogen and glucocorticoid receptors showed weak activities (data not shown). Furthermore, six viral activators known to interact with p300 conferred no activation.
In sum, independently of its HAT function, p300 is essential for Zac1 coactivation. When anchored directly to DNA, p300 enables Zac1 activation in the absence of autonomous DNA binding.
Zac1 zinc fingers underlie coactivation via p300.
To gain insight into the mechanism(s) of coactivation, we expressed adjusted concentrations of various Zac1 constructs (Fig. 3D, inset) together with a HAT activity-deficient p300 chimera (G-p300HAT-). Confirming that the HAT function of p300 is dispensable, the cotransfection of Zac1 or ZAC1 preserved activation (Fig. 3D; data not shown). While the deletion of either the activation domains or the entire carboxyl terminus of Zac1 did not alter this activity, the deletion of the zinc finger domain abolished activation; this was also the case when an additional localization signal was added to ensure nuclear localization (16). In fact, the expression of only the isolated zinc finger domains of Zac1/ZAC1 was sufficient to drive the potent activation of p300 (Fig. 3D; data not shown).
Since previous studies have shown that zinc fingers 6 and 7 (ZF6 and ZF7) are essential for DNA binding and interaction with p300 (15, 16), we investigated their importance for coactivation. Two zinc finger mutants were introduced, ZF6mt and ZF7mt, in which the first cysteine residue was replaced by alanine to destroy the zinc-dependent tetrahedral coordination of the zinc finger structure (broken zinc finger) (16). As shown in Fig. 3E, Zac1 which contained only a broken ZF7 (ZF7mt) failed to display coactivation properties. In accord, the mutation of key residues within the presumed α-helix of ZF7 resulted in various degrees of impaired (R195N, D197N, and R201N) or enhanced (H198N) activation (see Fig. 3F for topography). Since all of these constructs were similarly expressed and nuclear localized (16), these results suggest that, apart from the requirement of an intact ZF7, individual protein-protein contacts are critical to coactivation.
To ensure that the results obtained with the p300 chimera reflected Zac1 coactivation of p73, we retested the activity of Zac1 without (i) the central activation domain or (ii) the entire carboxyl terminus to activate a p73 reporter plasmid. These Zac1 deletion mutants coactivated p73 in a manner similar to Zac1, whereas coactivation was abrogated in the presence of a broken ZF7 (Fig. 3G). Moreover, the zinc finger domain alone was sufficient for p73 coactivation. Together, these results suggest a new role for ZF7 in the control of coactivation in a mode that is independent of the HAT function of p300.
Zac1 binding to either p300 docking site enhances association with PCAF.
Given that DNA-bound Zac1 coordinately binds to the amino-terminal (KIX) and the carboxyl-terminal (CH3) docking sites of p300 (15), we asked whether this also applies to coactivation. Fusion proteins of Gal4 and p300 lacking either the KIX or the CH3 domain halved Zac1 activation, which was abolished when both docking sites were deleted (Fig. 4A). This suggests that the KIX and CH3 sites operate individually to transmit Zac1 activation. Consistent with this idea, Zac1 potently activated the isolated amino- or carboxyl-terminal domain of p300 when fused to the Gal4 DNA-binding domain; the effects of ZF7 mutations closely mirrored those observed for the intact p300 chimera (data not shown).
The results presented so far are consistent with the “bridging” model of p300, in which the response to Zac1 leads to the recruitment and interactions of other coactivators with the amino and/or carboxyl terminus of p300, thereby allowing coactivation. While SRCs (which interact with both p300 and Zac1 [18; data not shown]) and PGC-1α (which interacts solely with p300; data not shown) revealed no effects, cotransfected PCAF, but not HAT activity-deficient PCAF, consistently activated p300 in the absence of Zac1; this suggests that the availability of endogenous PCAF might be a critical determinant of Zac1 coactivation (Fig. 4B). In support of this view, cotransfected PCAF strongly enhanced p300 activation at low concentrations of Zac1 in a HAT-dependent manner, indicating that PCAF is engaged in, and is a likely candidate for, the Zac1-dependent activation of p300 (Fig. 4C).
In a reverse experiment, the knockdown of PCAF by treatment with two different siRNAs largely abolished Zac1-mediated activation (Fig. 4D and 4E). This was also seen when a second unrelated cell line (LLC-PK1) was used, supporting the idea that Zac1 coactivation occurs through identical mechanisms in different cell types (data not shown). To assess whether PCAF alone is sufficient for Zac1 activation, the Gal4-p300 fusion construct was cotransfected into a PCAF-deficient human neuroblastoma cell line (SK-N-MC). As expected, ZAC1 failed to elicit any activation, but this was efficiently restored by exogenous PCAF (Fig. 4F). Importantly, rescue by PCAF depended on its HAT function and ZF7-intact ZAC1. Thus, PCAF seems to be necessary and sufficient to transmit ZAC1 coactivation via p300.
The coactivator PCAF was originally isolated as a p300/CBP-associated factor whose transient binding is thought to be stabilized by sequence-specific factors in an adenoviral E1A oncoprotein-competitive manner (41). In fact, E1A efficiently prevented the Zac1-dependent activation of p300, whereas the deletion of E1A's common binding site for p300 and for PCAF (Δ30-85) reversed inhibition (Fig. 5A). Furthermore, the selective deletion of either the PCAF (ΔPCAF) or the p300 (Δp300) binding sites alleviated inhibition (see the inset in Fig. 5A for the binding scheme of E1A). Concentrations of the various E1A constructs were adjusted to ensure similar expression levels and enable comparison of activities (Fig. 5B). Moreover, additional experiments with LLC-PK1 cells yielded essentially the same results, thus confirming E1A's disruptive effects on Zac1 coactivation (data not shown).
To examine whether Zac1 binding could, in turn, enhance the association of PCAF with p300, we conducted ChIP experiments with a PCAF-specific antibody (data not shown) in the absence or presence of Zac1. As shown in Fig. 5C, PCAF was strongly enriched at the GAL4 promoter in the presence of Zac1, while ZF7-mutated Zac1 resulted in slightly reduced promoter occupancy (∼30%). Given the complete absence of activation by mutated ZF7, Zac1's role in bridging p300 to PCAF appears to be essential, but not sufficient, to confer coactivation. This suggests that an intact structure of ZF7 is indispensable to the formation of a productive Zac1-p300-PCAF coactivator complex and additionally supports the view that, besides its role in binding, ZF7 has a regulatory function. To evaluate the last point, we fused PCAF, or HAT activity-deficient PCAF, to the yeast Gal4 DNA-binding domain and tested these chimeras with Zac1/ZAC1. Activation was strictly dependent on an intact HAT domain and an intact ZF7 (Fig. 5D).
Taken together, these results suggest that Zac1 enhances the association of PCAF with p300 in a manner that is subject to competition by E1A. Additionally, Zac1 controls PCAF's HAT function in a ZF7-dependent fashion.
Zac1 selectively modulates PCAF activities.
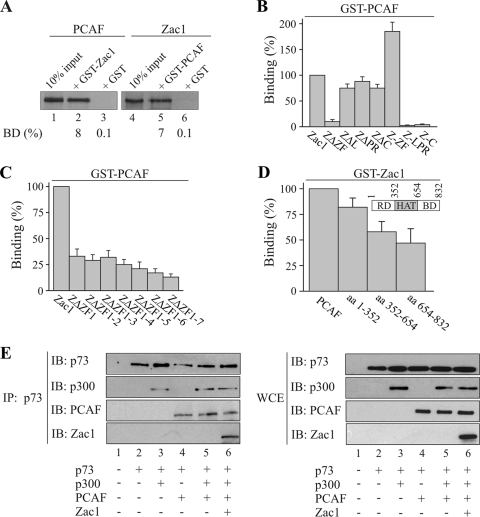
To determine the mechanistic basis through which Zac1 controls PCAF activity, we used GST pull-down assays to map their interacting regions. Equivalent amounts of each fusion protein (Fig. 6A; data not shown) efficiently retained Zac1 or PCAF. The deletion of the zinc finger domain abolished this interaction, whereas either the absence of the linker, proline-rich, or carboxyl-terminal domain or the presence of the isolated zinc fingers preserved binding (Fig. 6B). Moreover, to narrow down the contribution of each zinc finger to PCAF binding, mutants with successive deletions of the zinc finger domain were tested. The absence of ZF1 strongly diminished binding and was followed by a continuous, albeit faint, decline in the case of further truncations (Fig. 6C). Given previous evidence for unaltered DNA binding in the presence of ZF1-deleted Zac1 (16), it seems that this mutation is unlikely to interfere with the folding of adjacent fingers but rather suggests a direct interaction between PCAF and ZF1. In contrast to the accentuated role of ZF1 in binding, Zac1 bound more evenly across the different domains of the PCAF protein (Fig. 6D), suggesting that various contacts, in association with p73, could contribute to the stabilization of the PCAF-p300 complex.
FIG. 6.
Zac1 binds PCAF in a complex with p300 and p73. (A to D) GST pull-down assays. (A) Adjusted amounts of GST-Zac1, GST-PCAF, or GST alone were incubated with equal amounts of in vitro translated Zac1 or PCAF. The fraction of the input (100%) bound by each GST protein [BD (%)] is indicated. (B) Equal amounts of in vitro-translated Zac1 proteins containing deletions of the zinc-finger (ZΔZF), linker (ZΔΖF), proline-rich (ZΔPR), or C-terminal (ZΔC) domain were each incubated with GST-PCAF or GST alone. Additionally, equal amounts of the isolated zinc-finger (Z-ZF), linker-proline-rich (Z-LPR), or C-terminal (Z-C) domains were each incubated with GST-PCAF or GST alone. Binding (%) by Zac1 is set to 100%. Zac1's zinc finger domain appears necessary and sufficient for binding to PCAF. (C) Equal amounts of in vitro-translated Zac1 proteins containing progressive deletions of individual zinc fingers were incubated each with GST-PCAF or GST alone. Binding (%) by Zac1 is set to 100%. ZF1 is strongly involved in PCAF binding. (D) Equal amounts of in vitro-translated PCAF proteins containing the amino-terminal regulatory domain (RD), the central HAT domain, or the carboxyl-terminal bromo domain (BD) were incubated with GST-Zac1 or GST alone. The inset schematically depicts the localization of these domains. The binding (%) of PCAF was set to 100%. Zac1 equally binds across different domains of PCAF. (E) Coimmunoprecipitation assays. p73 (0.5 μg), p300 (5 μg), PCAF (1 μg), and Zac1 (0.25 μg) were transfected into Saos-2 cells, as shown in the scheme. Left panel, IP were performed with the anti-p73 antibody using 1 mg cell lysate. Immunoprecipitates were blotted and tested with the indicated antibodies. p73 binds Zac1 in a complex with p300 and PCAF. Right panel, WCE of the same transfections were immunoblotted and tested with the indicated antibodies.
In view of the reciprocal interactions between Zac1, PCAF, p300, and p73, we investigated whether these proteins can form a composite Zac1-p300-PCAF coactivator complex in vivo. We transfected p73 with p300 or PCAF alone or simultaneously, or both coactivators together in the presence of Zac1, and tested p73 immunoprecipitates for the presence of each factor. As shown in Fig. 6E, p73 interacted with p300 or PCAF alone or simultaneously, which is consistent with previous reports (45, 46). Moreover, transfected Zac1 was detectable in p73 immunoprecipitates, in addition to p300 and PCAF, indicating that these proteins can coexist as a single p73-bound complex in vivo. The coexpression of the different p73-binding partners did not change their individual expression levels (Fig. 6E, right panel).
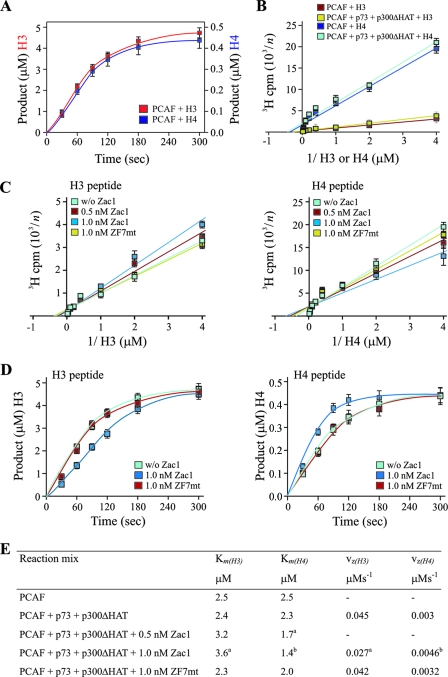
Assuming the existence of a composite p73 coactivator complex, and to exclude any other potential effectors in vivo besides Zac1, that could confound the analysis of PCAF HAT activity, we conducted in vitro HAT assays with the histone peptides H3 and H4 as substrates (15). A kinetic analysis for saturating concentrations of peptides revealed a strong preference of PCAF for histone H3 (Fig. 7A [note the different ordinates]). Although this has been commonly explained by a lower affinity for histone H4 (20, 28), the deduced Km values in this study did not support such differences [Km(H4), 2.5 μM; Km(H3), 2.5 μM] and instead pointed to a higher catalytic activity in the case of histone H3 (Fig. 7B and E). Virtually identical results were obtained when PCAF was incubated together with equimolar concentrations of p300ΔHAT and p73 (Fig. 7B and E) or Zac1 (data not shown).
FIG. 7.
Zac1 regulates PCAF's substrate affinities and catalytic activity. (A to D) PCAF in vitro HAT assays. (A) Kinetics of acetylation reactions for saturating concentrations of histone H3 or H4 peptides. The linear range of HAT enzymatic reactions was determined between 30 and 80 s. (B) Saturation analysis for various concentrations of H3 or H4 peptides in the absence or presence of p73 and p300ΔHAT, which do not alter PCAF's substrate affinities. (C) Affinities for H3 or H4 peptides in the presence of p73 and p300ΔHAT and in the absence (w/o) or presence of the indicated concentrations of Zac1 or ZF7-mutated Zac1 (ZF7mt). Zac1 significantly increases and decreases the affinity for histone H4 and H3, respectively. In contrast, ZF7-mutated Zac1 does not alter PCAF's substrate affinities. The y axis data in panels B and C are presented reciprocally, with the numbers on the y axis corresponding to n (e.g., a y axis value of 2 represents 500 cpm). (D) HAT reaction progress curves for H3 and H4 peptides in the presence of p73 and p300ΔHAT and in the absence (w/o) or presence of the indicated concentrations of Zac1 or ZF7-mutated Zac1 (ZF7mt). Zac1 significantly increases and decreases histone H4 and H3 catalytic activities, respectively. In contrast, ZF7-mutated Zac1 does not alter PCAF's catalytic activity. (E) Overview of deduced substrate affinities (Km) and maximal vz (Student's t test; a, P < 0.05; b, P < 0.01).
Interestingly, a different picture emerged for the coincubation of equimolar concentrations of PCAF, p300ΔHAT, and p73 in the presence of Zac1, which decreased and increased the affinities of histones H3 and H4, respectively. This reciprocal change in PCAF substrate specificity required an intact ZF7, corroborating its regulatory role (Fig. 7C and E).
Apart from an improvement in substrate recognition, Zac1 could enhance PCAF function through an increase in its enzymatic activity. Therefore, we determined the HAT reaction progress curves, in the absence or presence of Zac1, to analyze the catalytic activity of PCAF. Remarkably, equal concentrations of Zac1 decreased the vz (rate of formation) by 60% in the case of histone H3 and increased this measure by 60% in the case of H4 (Fig. 7D and E). In contrast, ZF7-mutated Zac1 showed no effects, reinforcing the concept of its critical role in controlling PCAF functions.
In sum, ZF1 interacts broadly with PCAF, consistent with the role of Zac1's DNA-binding domain in bridging PCAF to p300 and in enabling the formation of a composite Zac1-p300-PCAF coactivator complex. Thus, ZF7's interaction with p300 also appears to be necessary for the induction of a selective change in the substrate affinities and catalytic activities of PCAF.
Neuronal differentiation of embryonic stem cells induces the expression of both Zac1 and p73.
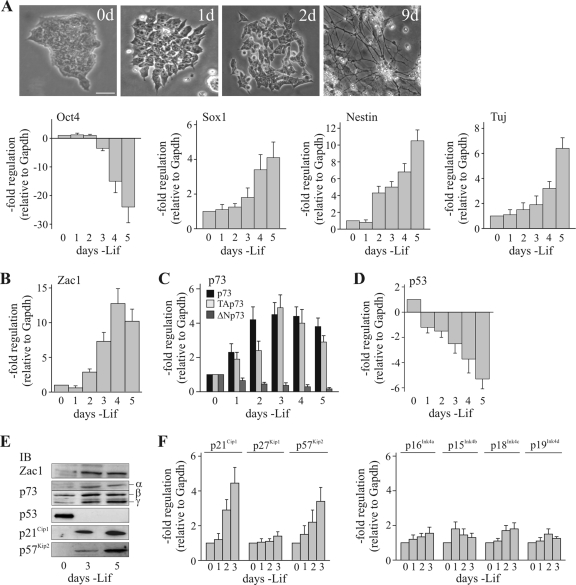
To elucidate the biological relevance of Zac1-dependent regulation of PCAF, we focused on the early neuronal differentiation of mouse embryonic stem (mES) cells. Following the withdrawal of leukemia inhibitory factor (Lif), mES colonies flattened and spread within 2 days, giving rise to single cells that progressively differentiated into cells with neuronal morphology (Fig. 8A, top panels). As evidenced by quantitative reverse transcription-PCR (qRT-PCR) analysis, a rapid downregulation of the pluripotency marker Oct4 was accompanied by an upregulation of the early neuroectoderm marker Sox1 and the progenitor cell marker Nestin, followed by the late neuronal marker Tuj1 (Fig. 8A, bottom panels). Interestingly, within 2 to 3 days of Lif withdrawal, Zac1 and p73 were strongly upregulated (Fig. 8B and C), and respective increases and decreases in the expression of the transactivation-competent (TAp73) and -deficient (ΔNp73) p73 isoforms were observed. In contrast, the expression of p53 rapidly declined upon Lif withdrawal (Fig. 8D); p63 was not expressed at any time point (data not shown). The changes in gene expression following Lif withdrawal showed a strong correlation with measurements of respective protein levels (Fig. 8E). Within 2 days of the removal of Lif, the proliferation of mES cells was reduced and there was a clear increase in the number of cells in the G1 phase on the third day (with Lif, 15.5% in the G1 phase and 63.4% in the S phase; without Lif, 25.8% in the G1 phase and 49.8% in the S phase). Time-shifted with respect to the onset of p73 induction, the cyclin kinase inhibitors and direct target p21Cip1 and p57Kip2 genes (5, 35) were upregulated at both the protein and mRNA levels (Fig. 8E and F), while the levels of p27Kip1 and members of the INK4 family remained unchanged (Fig. 8F). This finding raised the question of whether the coinduction of Zac1 and p73 is a general or lineage-specific feature of mES cell differentiation. Indeed, the induction of neuronal differentiation with retinoid acid potently upregulated Zac1 but repressed p73 (data not shown); in contrast, neither Zac1 nor p73 was upregulated during the induction of endodermal differentiation with bone morphogenetic protein 4 (BMP4)—the latter only induces p63 (data not shown). Therefore, the coinduction of Zac1 and p73 appears specific to Lif withdrawal-induced neuronal differentiation.
FIG. 8.
Neuronal differentiation of embryonic stem cells induces the expression of both Zac1 and p73. (A) Light microscopy of mES cells and qRT-PCR analysis of embryonic stem cell markers. Top panels, mES cells differentiate progressively into neurons following Lif withdrawal for the indicated number of days (d). Bottom panels, the expression of the pluripotency marker Oct4 is rapidly downregulated in mES cells upon Lif withdrawal. In contrast, the expression of the early neuroectoderm marker Sox1 and the progenitor marker Nestin were increased and followed by the increased expression of the late neuronal marker Tuj. (B to D) qRT-PCR analysis. The expression of Zac1 (B), entire p73, TAp73, or ΔNp73 (C), and p53 (D) was measured following Lif withdrawal for the indicated number of days. Zac1 and TAp73 are coinduced by Lif withdrawal, while p53 is downregulated. (E) IB analysis. WCE from mES cells grown for the indicated number of days in the absence of Lif were immunoblotted and tested with the indicated antibodies. Isoforms of p73 (α, β, and γ) are indicated. The coinduction of TAp73 and Zac1 mRNAs translates into their increased protein expression and the upregulation of the cyclin kinase inhibitor proteins p21Cip1 and p57Kip2. (F) qRT-PCR analysis. Expression of the Cip/Kip (p21Cip1, p27Kip1, and p57Kip2) and INK4 (p16Ink4a, p15Ink4b, p18Ink4c, and p19Ink4d) families following Lif withdrawal for the indicated number of days. The coinduction of TAp73 and Zac1 coincides with the upregulation of the direct p73 target p21Cip1 and p57Kip2 genes.
p73 recruits Zac1 together with PCAF and p300 to the p21Cip1 promoter.
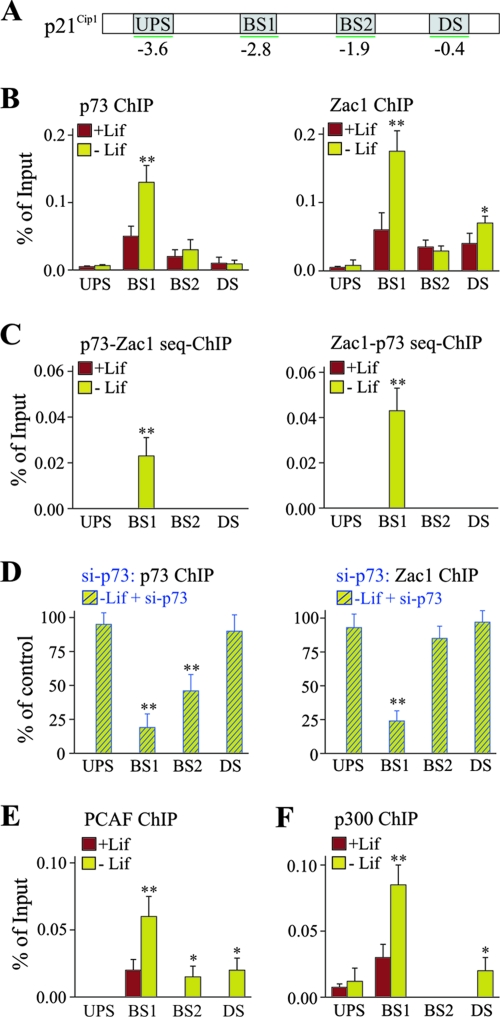
The p21Cip1 promoter contains two established p53 DNA-binding sites that potentially allow association with p73 (Fig. 9A). Indeed, as evidenced by ChIP analyses in undifferentiated cells, p73 bound preferentially to the distal site (BS1), compared to the proximal (BS2) site; this binding was barely detectable at unrelated upstream and downstream promoter regions (Fig. 9B). The withdrawal of Lif, which induced differentiation, caused a marked increase in binding that was restricted to the distal site. In undifferentiated cells, Zac1 associated preferentially with the distal site and the GC-rich proximal promoter; however, Zac1 occupancy increased strongly at the distal site only upon differentiation (Fig. 9B). Sequential ChIP experiments, in reciprocal orders with antibodies against p73 and Zac1, indicated that the respective proteins could co-occupy the distal site (Fig. 9C). Notably, despite the potent upregulation of Zac1, retinoid acid treatment caused a clear decrease in p73 and Zac1 occupancy at the distal site, suggesting that Zac1 binding depends on prior occupancy by p73 (data not shown). Indeed, p73 siRNA treatment strongly reduced Zac1 binding at the distal site 3 days following the withdrawal of Lif (Fig. 9D and 10A).
FIG. 9.
p73 recruits Zac1 together with PCAF and p300 to the p21Cip1 promoter. (A) Scheme of the p21Cip1 promoter. The locations of the upstream (UPS) and downstream (DS) promoter regions and the distal (BS1) and proximal (BS2) DNA-binding sites of p73 are shown. (B) ChIP analysis of p73 or Zac1 occupancy at the p21Cip1 promoter. ChIP assays were conducted in the presence (+Lif) or absence (-Lif) of Lif for 3 days. In the presence of Lif, p73 and Zac1 mainly colocalize to the distal p73 DNA-binding site and are selectively accumulated at this site upon Lif withdrawal. (C) Sequential ChIP analysis of p73 or Zac1 occupancy at the p21Cip1 promoter indicates that p73 and Zac1 co-occupy the distal p73 DNA-binding site. (D) ChIP for p73 or Zac1 occupancy at the p21Cip1 promoter following p73 or control siRNA transfections on day 1 of Lif withdrawal. The cells were harvested for the ChIP experiments 2 days later. The values for the control siRNA treatments were set to 100%. The knockdown of p73 strongly reduces the binding of p73 and Zac1 to the distal p73 DNA-binding site. (E to F) ChIP analysis of (E) PCAF or (F) p300 occupancy at the p21Cip1 promoter. (B to F) P values were calculated by Student's t test for differences with versus without Lif or control versus factor-specific siRNA. *, P < 0.05; **, P < 0.01.
Next, we investigated the recruitment of PCAF and p300 following Lif withdrawal. PCAF associated in undifferentiated cells with the distal p73 DNA-binding site solely; this binding showed a robust increase upon differentiation (Fig. 9E), although some weak associations could also be detected at the proximal site and the downstream region. Sequential ChIP experiments, with antibodies against p73 and PCAF, suggested that both proteins could co-occupy the distal site (data not shown). A similar picture also emerged for p300 which, in undifferentiated cells, was bound predominantly to the distal p73 DNA-binding site (Fig. 9F); this binding increased strongly upon Lif withdrawal. In agreement with the above results, sequential ChIP experiments for p73 and p300 or p300 and PCAF indicated their co-occupancies at the distal DNA-binding site of p73 (data not shown).
In sum, neuronal differentiation increases p73 occupancy at the distal DNA-binding site of the p21Cip1 promoter; this, in turn, enhanced the recruitment of the coactivators Zac1, PCAF, and p300.
Zac1 induces a selective switch of PCAF activity in vivo.
The p73-dependent recruitment of Zac1, PCAF, and p300 to the p21Cip1 promoter allowed us to investigate the impact of Zac1 on PCAF function in vivo and to analyze the roles of the different partners of the p73 coactivator complex by applying siRNA treatments. Cells were grown for 1 day without Lif before being transfected with control or factor-specific siRNA. IB analysis after an additional 2 days of culture revealed the efficient downregulation of p73, PCAF, and Zac1 proteins (Fig. 10A to C). In turn, the expression of the p21Cip1 gene was significantly attenuated in response to either treatment, with p73 siRNA eliciting the strongest effect (Fig. 10D).
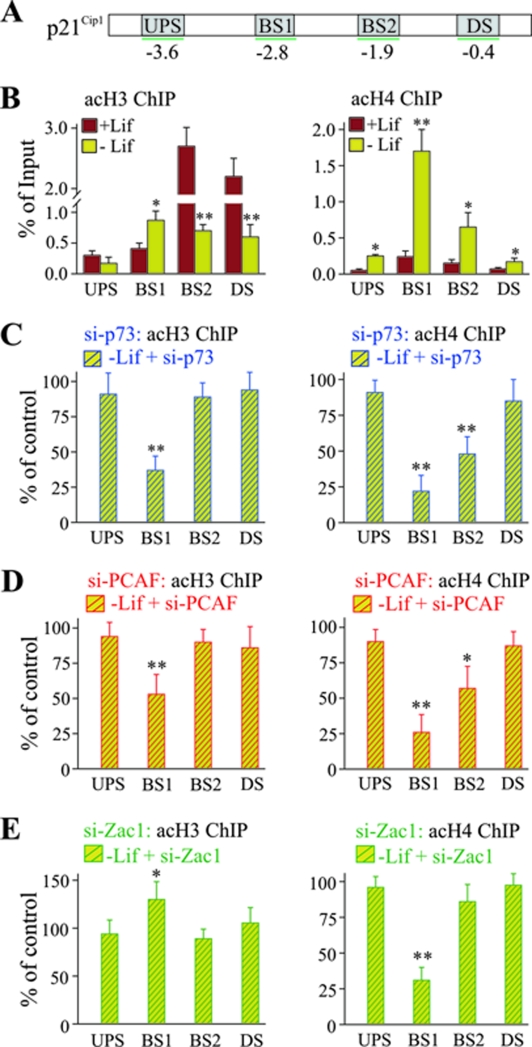
A ChIP promoter scan in undifferentiated cells revealed high levels of acetylated histone H3 at the proximal p73 DNA-binding site and downstream promoter region, whereas acetylated histone H4 was barely detectable at any site (Fig. 11B). However, high levels of acetylated histone H3 seemed not to translate into high transcription as suggested by a strong decrease of acetylated histone H3 following Lif withdrawal at all sites other than at the distal p73 DNA-binding site, which showed a twofold enrichment. In contrast, differentiation caused strong increases in acetylated histone H4 at the distal (sevenfold) and proximal (fourfold) p73 DNA-binding sites (Fig. 11B).
FIG. 11.
Zac1 induces a selective switch of PCAF activity in vivo. (A) Scheme of the p21Cip1 promoter, as described in the text. (B) ChIP analysis of pan-acetylated histones H3 or H4 at the p21Cip1 promoter. ChIP assays were conducted in the presence (+Lif) or absence (-Lif) of Lif for 3 days. Lif withdrawal causes an overall decrease of histone H3 acetylation. In contrast, histone H4 acetylation strongly increases at the distal p73 DNA-binding site. (C to E) ChIP analysis (as described in the text) following control or factor-specific siRNA treatments for p73 (C), PCAF (D), or Zac1 (E) on day 1 of Lif withdrawal. The cells were harvested for ChIP experiments 2 days later. The values for control siRNA treatments were set to 100%. The knockdown of p73 concomitantly reduced histone H3 and H4 acetylation at the distal p73 DNA-binding site (C), whereas the knockdown of PCAF preferentially affected histone H4 acetylation (D). In contrast, the knockdown of Zac1 significantly increased histone H3 acetylation, while histone H4 acetylation was strongly reduced (E). (B to E) P values were calculated by Student's t test for differences with versus without Lif or control versus factor-specific siRNA. *, P < 0.05; **, P < 0.01. UPS, upstream promoter region; DS, downstream promoter region; BS1, distal DNA-binding site; BS2, proximal DNA-binding site; acH3, acetylated H3; acH4, acetylated H4.
To assess the roles of p73, PCAF, and Zac1 in switching histone acetylation at the p21Cip1 promoter, we next determined the acetylation status of histones H3 and H4 by treating with siRNA for each factor or appropriate control siRNA. As anticipated, the knockdown of p73 largely prevented the switch in histone H3 and H4 acetylation, subsequently resulting in attenuated promoter acetylation (Fig. 11C). Similarly, the knockdown of PCAF interfered with the characteristic switch in acetylation at the distal p73 DNA-binding site (Fig. 11D), consistent with the concept that corecruited p300 cannot replace PCAF's HAT function. Interestingly, the knockdown of Zac1 selectively impaired histone H4 acetylation. Whereas histone H3 acetylation at the distal p73 DNA-binding site appeared to be even stronger than in the presence of Zac1, the concurrent increase in histone H4 acetylation was strongly reduced (Fig. 11E).
Together, these data suggest that p73 concomitantly recruits Zac1, PCAF, and p300 to the p21Cip1 promoter; once present, Zac1 can induce a selective switch of PCAF function, directing the transcription of p21Cip1 during early neuronal development.
DISCUSSION
The generally accepted paradigm of transcription by regulated recruitment considers sequence-specific factors and coactivators as separate categories that are distinguished on the basis of their abilities to autonomously bind to DNA. The results from this study are consistent with an authentic coactivator function of Zac1 and reveal that coactivation by sequence-specific factors could represent a hitherto unappreciated and, most likely, general facet of transcriptional control.
We show here that p73 recruits Zac1, together with the coactivators p300 and PCAF, to its direct target p21Cip1 gene during early neuronal differentiation. Furthermore, Zac1 mediates coactivation by (i) acting as a scaffold to enhance the transient association of PCAF with p300 and (ii) selectively increasing PCAF substrate affinity and catalytic activity for histone H4. Therefore, contrary to the well-established role of C2H2 zinc fingers in canonical DNA binding, Zac1 zinc fingers are now shown to be critical to protein-protein interaction and coactivation.
Zac1 coactivation requires ZF7.
Transcription via regulated recruitment implies that “even [when] expressed at high concentrations, an activating region that is not tethered to DNA cannot activate because it cannot recruit” (29). In lieu of direct DNA binding, some viral and cellular transcription activation domains associate with DNA-binding domains in trans via protein-protein interactions to control gene expression (7, 13). Zac1 coactivation does not require its activation domain in trans, neither in the context of a p300 chimera nor in the context of p73; moreover, while the isolated zinc finger domain, which does not contain any inherent activation potency (16), can coactivate on its own. This is a distinguishing mechanistic feature of Zac1; it highlights the explicit role of Zac1's zinc fingers in coactivation and the essentiality of an intact ZF7 for its regulation of PCAF functions.
Zac1 directs PCAF functions.
While PCAF exists in a stable complex comprising more than 20 polypeptides, p300/CBP is not a stoichiometric component of the complex (41). Interactions between PCAF and p300/CBP on promoters are facilitated by sequence-specific factors and/or intermediary proteins, as shown here for Zac1. Protein-protein interactions within the carboxyl-terminal domain of PCAF are responsible for complex formation, while interactions within the amino-terminal half appear transiently and seem to be tightly regulated. Indeed, the amino- and carboxyl-terminal regions of p300/CBP that flank the KIX and CH3 docking sites are well-known to interact with the amino terminus of PCAF (19). The results of this study suggest that Zac1 binding to either docking site facilitates the association of PCAF and, furthermore, enables the regulation of PCAF's HAT function. Intriguingly, the binding of ZF7-mutated Zac1 to either docking site largely preserves the enhanced association while abolishing PCAF's HAT activity, suggesting that these two functions are differentially dependent on an intact zinc finger structure.
In contrast to the general assumption, histones H3 and H4 varied little in their affinities in this study (20, 28); accordingly, the measured differences in acetylation rates most likely reflect differences in PCAF's catalytic activity. In fact, a comparison between the sequences of histones H3 and H4 failed to reveal any residues that would be likely to have a major impact on the enzyme's active site (28). This suggests that the weak acetylation of H4 peptides results from their respective acetyl-accepting lysines that are suboptimally positioned, in the lysine binding pocket, for efficient acetyl transfer. On the contrary, the incorporation of PCAF into multiprotein complexes is thought to enhance the in vivo affinity of PCAF for histone H4 and to facilitate contacts for optimal acetyl transfer under certain, albeit poorly understood, conditions; this results in a significant increase in kcat (28, 31). In support of this view, Zac1 is here shown to increase the rate of acetylation of histone H4 versus H3 due to changes in PCAF's substrate affinities and catalytic activities. In this context, it is important to note that previous studies showed that association with other factors leads to a context-dependent acetylation pattern for the PCAF-related acetyltransferase Gcn5 (14). Among the factors that associate with Gcn5 are the adaptor proteins Ada2, which potentiates Gcn5 catalytic activity, and Ada3, which facilitates nucleosomal acetylation and an expanded lysine specificity (2). Furthermore, loss-of-function mutations of the drosophila adaptor proteins Ada2a and Ada2b showed that they can provide HAT complexes with functional diversity in genome-wide and gene-specific regulation (8, 27). Clearly, many of the mechanistic details of how histone acetyltransferases are regulated by interacting subunits/factors and of how specific complexes are utilized in particular situations remain to be clarified (31).
Early neuronal differentiation induces Zac1 and p73 on top of p21Cip1.
Zac1 and p73 were robustly and specifically induced in mES cells after the withdrawal of Lif, an established model to mimic the permissive, or default, pathway of neuronal differentiation (25). Pluripotent cells of embryonic origin exhibit a short G1 phase that is driven by precocious and unrestrained cyclin expression (12, 26, 34). Although differentiation is coupled with the upregulation of p21Cip1 and p27Kip1 (40), many of the mechanisms and factors underlying cell cycle maturation in early neurons are largely unknown. While a role for p53 has been proposed in these processes (26), our data show that Lif withdrawal results in a strong downregulation of p53 mRNA and protein expression. However, we observed a marked switch in p73 isoform expression, as evidenced by a steady decrease in activation-deficient ΔNp73 as opposed to a strong increase in activation-proficient TAp73. Since ΔNp73 can promote immortalization and resistance to spontaneous senescence (35), its expression in undifferentiated mES cells could help sustain self-renewal capacity. In either case, the upregulation of p73 was found to be tightly coupled to a strong increase in the expression of p21Cip1 and p57Kip2 genes, two known direct target genes (5).
Zac1 induces a switch in PCAF function in vivo.
In undifferentiated mES cells, the p21Cip1 promoter was marked by high levels of acetylated histone H3 which possibly serve to keep p21Cip1 poised for differentiation cues (33). However, following the coinduction of p73 and Zac1, high levels of acetylated histone H4 localized to the p73 DNA-binding site, enabling efficient transcription.
The knockdown of Zac1 caused a selective loss of H4 acetylation at the distal DNA binding site of p73 while maintaining or even causing a slight increase in H3 acetylation, and without permitting efficient transcription. These data suggest that, in vivo, Zac1 promotes a selective switch of PCAF's HAT function in favor of histone H4 acetylation. Additional modifications of the p73 coactivator complex in vivo, compared to the use of recombinant proteins for the in vitro HAT assays, might explain the differences in the regulation of PCAF function by Zac1. More importantly, Zac1-mediated histone H4 acetylation in vitro reflects measurements based on an incubation time of approximately 1 min, compared to a period of 2 to 3 days in the Lif-withdrawn mES cells, arguing for the accumulation of histone H4 acetylation.
The transition from undifferentiated, pluripotent ES cells to committed or differentiated cells presumably involves the silencing of those parts of the genome that are not required for the newly forming lineage. Accordingly, the PCAF-mediated acetylation of histone H4 could secure the transcription of the p21Cip1 gene to advance progressive neuronal differentiation.
Multitasking zinc fingers could facilitate the coordination of biological programs.
Lewin (23) stated: “We may regard coactivators as transcription factors whose specificity is conferred by the ability to bind to DNA-binding transcription factors instead of directly to DNA.” The present study supports this concept through its revelation that certain factors could have evolved along a continuum of refined control mechanisms to accommodate both sequence-specific DNA binding and coactivation. In the case of Zac1, both functions operate through the same C2H2 zinc finger, illustrating how one of the simplest folds found in nature may be utilized to assemble complex, yet distinct, signaling properties. Evidently, C2H2 protein-protein interactions are more (i) abundant than previously thought, (ii) plastic than their DNA-binding counterparts, and (iii) variable and complex in their interaction surfaces (6). As a result, a coherent program of cell cycle maturation and early neuronal differentiation may be more efficiently coordinated through coupling of their “hard-wiring” (Zac1 sequence-specific DNA binding) and their “soft-wiring” ability to integrate developmental pathways (via Zac1 coactivation).
Acknowledgments
We thank members of the Molecular Neuroendocrinology group and Osborne Almeida for discussions on the manuscript.
Funding was provided by the Deutsche Forschungsgemeinschaft (SP 386/5-1 to D.S.) and the European Union (Crescendo LSHM-CT-2005-018652).
A.H. and D.S. conceived the study, designed the experimental strategy, and analyzed the data. D.S. wrote the paper, performed the cotransfection studies, and constructed part of the plasmids. A.H. prepared and performed the pull-down assays, RT-PCR experiments, ChIP assays, siRNA treatments, IBs and coimmunoprecipitations, in vitro HAT assays, and transfection studies.
Footnotes
Published ahead of print on 28 July 2008.
REFERENCES
- 1.Arima, T., T. Kamikihara, T. Hayashida, K. Kato, T. Inoue, Y. Shirayoshi, M. Oshimura, H. Soejima, T. Mukai, and N. Wake. 2005. Zac, Lit1 (Kcnq1Ot1) and P57(Kip2) (Cdkn1C) are in an imprinted gene network that may play a role in Beckwith-Wiedemann syndrome. Nucleic Acids Res. 332650-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balasubramanian, R., M. G. Pray-Grant, W. Selleck, P. A. Grant, and S. Tan. 2002. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J. Biol. Chem. 2777989-7995. [DOI] [PubMed] [Google Scholar]
- 3.Barz, T., A. Hoffmann, M. Panhuysen, and D. Spengler. 2006. Peroxisome proliferator-activated receptor gamma is a Zac target gene mediating Zac antiproliferation. Cancer Res. 6611975-11982. [DOI] [PubMed] [Google Scholar]
- 4.Bilanges, B., A. Varrault, A. Mazumdar, C. Pantaloni, A. Hoffmann, J. Bockaert, D. Spengler, and L. Journot. 2001. Alternative splicing of the imprinted candidate tumor suppressor gene ZAC regulates its antiproliferative and DNA binding activities. Oncogene 201246-1253. [DOI] [PubMed] [Google Scholar]
- 5.Blint, E., A. C. Phillips, S. Kozlov, C. L. Stewart, and K. H. Vousden. 2002. Induction of p57(KIP2) expression by p73beta. Proc. Natl. Acad. Sci. USA 993529-3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brayer, K. J., and D. J. Segal. 2008. Keep your fingers off my DNA: protein-protein interactions mediated by C2H2 zinc finger domains. Cell Biochem. Biophys. 50111-131. [DOI] [PubMed] [Google Scholar]
- 7.Brunnberg, S., K. Pettersson, E. Rydin, J. Matthews, A. Hanberg, and I. Pongratz. 2003. The basic helix-loop-helix-PAS protein ARNT functions as a potent coactivator of estrogen receptor-dependent transcription. Proc. Natl. Acad. Sci. USA 1006517-6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciurciu, A., O. Komonyi, T. Pankotai, and I. M. Boros. 2006. The Drosophila histone acetyltransferase Gcn5 and transcriptional adaptor Ada2a are involved in nucleosomal histone H4 acetylation. Mol. Cell. Biol. 269413-9423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Courey, A. J. 2008. Mechanisms in transcriptional regulation, 1st ed. Blackwell Publishing Professional, Oxford, United Kingdom.
- 10.el-Deiry, W. S., S. E. Kern, J. A. Pietenpol, K. W. Kinzler, and B. Vogelstein. 1992. Definition of a consensus binding site for p53. Nat. Genet. 145-49. [DOI] [PubMed] [Google Scholar]
- 11.el-Deiry, W. S., T. Tokino, V. E. Velculescu, D. B. Levy, R. Parsons, J. M. Trent, D. Lin, W. E. Mercer, K. W. Kinzler, and B. Vogelstein. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75817-825. [DOI] [PubMed] [Google Scholar]
- 12.Faast, R., J. White, P. Cartwright, L. Crocker, B. Sarcevic, and S. Dalton. 2004. Cdk6-cyclin D3 activity in murine ES cells is resistant to inhibition by p16(INK4a). Oncogene 23491-502. [DOI] [PubMed] [Google Scholar]
- 13.Flint, J., and T. Shenk. 1997. Viral transactivating proteins. Annu. Rev. Genet. 31177-212. [DOI] [PubMed] [Google Scholar]
- 14.Grant, P. A., A. Eberharter, S. John, R. G. Cook, B. M. Turner, and J. L. Workman. 1999. Expanded lysine acetylation specificity of Gcn5 in native complexes. J. Biol. Chem. 2745895-5900. [DOI] [PubMed] [Google Scholar]
- 15.Hoffmann, A., T. Barz, and D. Spengler. 2006. Multitasking C2H2 zinc fingers link Zac DNA binding to coordinated regulation of p300-histone acetyltransferase activity. Mol. Cell. Biol. 265544-5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffmann, A., E. Ciani, J. Boeckardt, F. Holsboer, L. Journot, and D. Spengler. 2003. Transcriptional activities of the zinc finger protein Zac are differentially controlled by DNA binding. Mol. Cell. Biol. 23988-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang, S. M., A. H. Schonthal, and M. R. Stallcup. 2001. Enhancement of p53-dependent gene activation by the transcriptional coactivator Zac1. Oncogene 202134-2143. [DOI] [PubMed] [Google Scholar]
- 18.Huang, S. M., and M. R. Stallcup. 2000. Mouse Zac1, a transcriptional coactivator and repressor for nuclear receptors. Mol. Cell. Biol. 201855-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korzus, E., J. Torchia, D. W. Rose, L. Xu, R. Kurokawa, E. M. McInerney, T. M. Mullen, C. K. Glass, and M. G. Rosenfeld. 1998. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science 279703-707. [DOI] [PubMed] [Google Scholar]
- 20.Lau, O. D., A. D. Courtney, A. Vassilev, L. A. Marzilli, R. J. Cotter, Y. Nakatani, and P. A. Cole. 2000. p300/CBP-associated factor histone acetyltransferase processing of a peptide substrate. Kinetic analysis of the catalytic mechanism. J. Biol. Chem. 27521953-21959. [DOI] [PubMed] [Google Scholar]
- 21.Lee, K. K., and J. L. Workman. 2007. Histone acetyltransferase complexes: one size doesn't fit all. Nat. Rev. Mol. Cell Biol. 8284-295. [DOI] [PubMed] [Google Scholar]
- 22.Lemon, B., and R. Tjian. 2000. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 142551-2569. [DOI] [PubMed] [Google Scholar]
- 23.Lewin, B. 2007. Activating transcription, p. 640-666. In B. Lewin (ed.), Genes IX, 1st ed. Jones and Bartlett Publishers International, London, United Kingdom.
- 24.Moroni, M. C., E. S. Hickman, E. Lazzerini Denchi, G. Caprara, E. Colli, F. Cecconi, H. Muller, and K. Helin. 2001. Apaf-1 is a transcriptional target for E2F and p53. Nat. Cell Biol. 3552-558. [DOI] [PubMed] [Google Scholar]
- 25.Munoz-Sanjuan, I., and A. H. Brivanlou. 2002. Neural induction, the default model and embryonic stem cells. Nat. Rev. Neurosci. 3271-280. [DOI] [PubMed] [Google Scholar]
- 26.Orford, K. W., and D. T. Scadden. 2008. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat. Rev. Genet. 9115-128. [DOI] [PubMed] [Google Scholar]
- 27.Pankotai, T., O. Komonyi, L. Bodai, Z. Ujfaludi, S. Muratoglu, A. Ciurciu, L. Tora, J. Szabad, and I. Boros. 2005. The homologous Drosophila transcriptional adaptors ADA2a and ADA2b are both required for normal development but have different functions. Mol. Cell. Biol. 258215-8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poux, A. N., and R. Marmorstein. 2003. Molecular basis for Gcn5/PCAF histone acetyltransferase selectivity for histone and nonhistone substrates. Biochemistry 4214366-14374. [DOI] [PubMed] [Google Scholar]
- 29.Ptashne, M. 2003. Regulated recruitment and cooperativity in the design of biological regulatory systems. Philos. Transact. A 3611223-1234. [DOI] [PubMed] [Google Scholar]
- 30.Ptashne, M., and A. Gann. 1997. Transcriptional activation by recruitment. Nature 386569-577. [DOI] [PubMed] [Google Scholar]
- 31.Shahbazian, M. D., and M. Grunstein. 2007. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 7675-100. [DOI] [PubMed] [Google Scholar]
- 32.Spengler, D., M. Villalba, A. Hoffmann, C. Pantaloni, S. Houssami, J. Bockaert, and L. Journot. 1997. Regulation of apoptosis and cell cycle arrest by Zac1, a novel zinc finger protein expressed in the pituitary gland and the brain. EMBO J. 162814-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spivakov, M., and A. G. Fisher. 2007. Epigenetic signatures of stem-cell identity. Nat. Rev. Genet. 8263-271. [DOI] [PubMed] [Google Scholar]
- 34.Stead, E., J. White, R. Faast, S. Conn, S. Goldstone, J. Rathjen, U. Dhingra, P. Rathjen, D. Walker, and S. Dalton. 2002. Pluripotent cell division cycles are driven by ectopic Cdk2, cyclin A/E and E2F activities. Oncogene 218320-8333. [DOI] [PubMed] [Google Scholar]
- 35.Stiewe, T. 2007. The p53 family in differentiation and tumorigenesis. Nat. Rev. Cancer 7165-168. [DOI] [PubMed] [Google Scholar]
- 36.Valente, T., and C. Auladell. 2001. Expression pattern of Zac1 mouse gene, a new zinc-finger protein that regulates apoptosis and cellular cycle arrest, in both adult brain and along development. Mech. Dev. 108207-211. [DOI] [PubMed] [Google Scholar]
- 37.Valente, T., F. Junyent, and C. Auladell. 2005. Zac1 is expressed in progenitor/stem cells of the neuroectoderm and mesoderm during embryogenesis: differential phenotype of the Zac1-expressing cells during development. Dev. Dyn. 233667-679. [DOI] [PubMed] [Google Scholar]
- 38.Varrault, A., E. Ciani, F. Apiou, B. Bilanges, A. Hoffmann, C. Pantaloni, J. Bockaert, D. Spengler, and L. Journot. 1998. hZAC encodes a zinc finger protein with antiproliferative properties and maps to a chromosomal region frequently lost in cancer. Proc. Natl. Acad. Sci. USA 958835-8840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varrault, A., C. Gueydan, A. Delalbre, A. Bellmann, S. Houssami, C. Aknin, D. Severac, L. Chotard, M. Kahli, D. A. Le, P. Pavlidis, and L. Journot. 2006. Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Dev. Cell 11711-722. [DOI] [PubMed] [Google Scholar]
- 40.White, J., E. Stead, R. Faast, S. Conn, P. Cartwright, and S. Dalton. 2005. Developmental activation of the Rb-E2F pathway and establishment of cell cycle-regulated cyclin-dependent kinase activity during embryonic stem cell differentiation. Mol. Biol. Cell 162018-2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang, X. J., V. V. Ogryzko, J. Nishikawa, B. H. Howard, and Y. Nakatani. 1996. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature 382319-324. [DOI] [PubMed] [Google Scholar]
- 42.Ying, Q. L., M. Stavridis, D. Griffiths, M. Li, and A. Smith. 2003. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol. 21183-186. [DOI] [PubMed] [Google Scholar]
- 43.Zauberman, A., Y. Barak, N. Ragimov, N. Levy, and M. Oren. 1993. Sequence-specific DNA binding by p53: identification of target sites and lack of binding to p53-MDM2 complexes. EMBO J. 122799-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeng, X., H. Lee, Q. Zhang, and H. Lu. 2001. p300 does not require its acetylase activity to stimulate p73 function. J. Biol. Chem. 27648-52. [DOI] [PubMed] [Google Scholar]
- 45.Zeng, X., X. Li, A. Miller, Z. Yuan, W. Yuan, R. P. Kwok, R. Goodman, and H. Lu. 2000. The N-terminal domain of p73 interacts with the CH1 domain of p300/CREB binding protein and mediates transcriptional activation and apoptosis. Mol. Cell. Biol. 201299-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao, L. Y., Y. Liu, N. R. Bertos, X. J. Yang, and D. Liao. 2003. PCAF is a coactivator for p73-mediated transactivation. Oncogene 228316-8329. [DOI] [PubMed] [Google Scholar]