Abstract
Drosophila Enabled (Ena) was initially identified as a dominant genetic suppressor of mutations in the Abelson tyrosine kinase and, more recently, as a member of the Ena/human vasodilator-stimulated phosphoprotein (VASP) family of proteins. We have used genetic, biochemical, and cell biological approaches to demonstrate the functional relationship between Ena and human VASP. In addition, we have defined the roles of Ena domains identified as essential for its activity in vivo. We have demonstrated that VASP rescues the embryonic lethality associated with loss of Ena function in Drosophila and have shown that Ena, like VASP, is associated with actin filaments and focal adhesions when expressed in cultured cells. To define sequences that are central to Ena function, we have characterized the molecular lesions present in two lethal ena mutant alleles that affected the Ena/VASP homology domain 1 (EVH1) and EVH2. A missense mutation that resulted in an amino acid substitution in the EVH1 domain eliminated in vitro binding of Ena to the cytoskeletal protein zyxin, a previously reported binding partner of VASP. A nonsense mutation that resulted in a C-terminally truncated Ena protein lacking the EVH2 domain failed to form multimeric complexes and exhibited reduced binding to zyxin and the Abelson Src homology 3 domain. Our analysis demonstrates that Ena and VASP are functionally homologous and defines the conserved EVH1 and EVH2 domains as central to the physiological activity of Ena.
INTRODUCTION
Changes in cytoskeleton assembly and cell adhesion in response to extracellular cues are critical in regulating diverse cellular functions (Damsky, 1993; Rubinfeld et al., 1993; Su et al., 1993; Montell, 1994). Translation of these extracellular signals into cytoskeletal changes are mediated by signal transduction pathways (Tanaka and Sabry, 1995). In Drosophila, the Abelson (Abl) tyrosine kinase and its substrate Enabled (Ena) are thought to play a role in regulating cytoskeletal changes during axonogenesis in the CNS of the developing embryo (Gertler et al., 1989, 1995; Bennett and Hoffmann, 1992; Hill et al., 1995). This hypothesis has been bolstered by the recent identification of Drosophila Ena as a member of the cytoskeletal-associated Ena/human vasodilator-stimulated phosphoprotein (VASP) family of proteins (Gertler et al., 1996). Members of this family in addition to Drosophila Ena include VASP, murine Ena (Mena), and murine Ena/VASP-like (EVL) protein. Mena and VASP have been localized to the actin cytoskeleton and are in vitro ligands for the focal adhesion-associated protein zyxin and the actin-binding protein profilin (Reinhard et al., 1992, 1995a; Haffner et al., 1995; Gertler et al., 1996). Drosophila Ena also binds profilin in vitro (our unpublished results). Mena and VASP interact with Act A, a protein from Listeria monocytogenes required for the recruitment of host actin filaments around the intracellular bacteria, and VASP and Mena may serve as links between ActA and profilin. Consequently, these proteins may enhance Act A-mediated F-actin recruitment that is used as a means of intracellular movement by the bacteria (Chakraborty et al., 1995; Pistor et al., 1995; Pollard, 1995; Gertler et al., 1996). Additional support for a direct role in cytoskeleton assembly comes from the observation that overexpression of neuronally enriched forms of Mena in cultured cells results in actin-rich membrane projections (Gertler et al., 1996) and that targeting of the Mena- and VASP-binding protein zyxin to the plasma membrane causes the elaboration of actin-rich projections (Golsteyn et al., 1997).
The Ena/VASP family of proteins is characterized by a common overall structural domain organization consisting of conserved N- and C-terminal domains separated by a less-conserved central proline-rich region (Gertler et al., 1996). This N-terminal 113-amino-acid domain, or the Ena/VASP homology domain 1 (EVH1), is 58% identical between Drosophila Ena and human VASP. The EVH1 domain mediates VASP and Mena binding to the focal adhesion-associated protein zyxin as well as Listeria Act A (Gertler et al., 1996). The EVH1 domain is also similar to the WP1 domain found in Wiskott–Aldrich syndrome protein (Symons et al., 1996). Wiskott–Aldrich syndrome is characterized by cytoskeletal abnormalities in T cells and platelets (Derry et al., 1995; Kolluri et al., 1995; Kwan et al., 1995; Villa et al., 1995; Wengler et al., 1995; Zhu et al., 1995). The central proline-rich regions of the Ena, Mena, and VASP proteins, which vary greatly in length, are important for binding to both Src homology 3 (SH3) domains and the actin-binding protein profilin (Gertler et al., 1995, 1996; Reinhard et al., 1995a) (our unpublished results). The C-terminal 35 amino acids of these proteins, or the EVH2 domain, is 31% identical between Drosophila Ena and human VASP and consists of a series of conserved charged repeats with a spacing predicted to form an extended highly charged α helix (Haffner et al., 1995). Homology to this region is also seen in human expressed sequence tag r74407 and the mouse cDNA NDPP-1 (Sazuka et al., 1992). Several functions have been proposed for the EVH2 domain, including subcellular localization and mediation of multimer formation (Haffner et al., 1995; Gertler et al., 1996), although conclusive evidence for these proposed functions has not been reported.
In addition to a role in cytoskeleton assembly, Ena/VASP proteins are constituents of signal transduction pathways. Ena has been linked genetically to the Abl tyrosine kinase signaling pathway by identification of ena mutations that act as dominant genetic suppressors of a loss of Abl tyrosine kinase function (Gertler et al., 1990). Heterozygous loss of function mutations in ena rescue the lethality and defects in axonal architecture in embryos with mutations in Abl and other components of Abl-mediated signaling (Gertler et al., 1989, 1993; Hill et al., 1995). Drosophila that lack Ena function die during embryogenesis and also display defects in the axonal architecture of the nervous system. These include a reduction in the integrity of axon bundles and some axon misrouting consistent with a role for Ena in regulating the cytoskeleton (Gertler et al., 1990 1995). Ena is a specific substrate for Abl and also specifically interacts with the Abl SH3 domain (Gertler et al., 1995).
Ena’s homology to a family of proteins clearly implicated in cytoskeletal dynamics led us to investigate possible conserved functions between Ena and VASP. We report that VASP can partially substitute for Ena function in Drosophila. In addition, we show that Ena and VASP are colocalized at focal adhesions and with actin filaments. We have characterized two lethal mutations and show that they result in defects in the EVH1 and EVH2 domains of Drosophila Ena. Furthermore, we demonstrate that these mutations disrupt conserved functions of Ena/VASP proteins, namely in vitro zyxin binding and multimerization. We further demonstrate that Ena and VASP are colocalized to focal adhesions with zyxin, providing indirect evidence that these proteins may interact in the intact cell. The evidence for conservation of function between Drosophila Ena and human VASP suggests that Ena may provide a regulated link between Abl-mediated signal transduction and cytoskeletal dynamics in the developing Drosophila CNS.
MATERIALS AND METHODS
Transfections and Western Blot Analysis
DNA encoding six histidine residues, the FLAG tag, or the hemaglutinin (Ha) tag was added to the 3′ end of the coding sequence of the ena cDNA by PCR. Drosophila S2 cells were transiently transfected with VASP, Abl, Ena, or tagged Ena cDNAs in the pPac-PL expression vector. Cells were harvested after 60 h and lysed in immunoprecipitation buffer (0.5% Triton X-100, 50 mM Tris, pH 8.0, 150 mM NaCl, 5 mM EDTA, 1 mM Na3VO4, 1 mM Pefabloc (Boehringer Mannheim, Indianapolis, IN), 1 μg/ml each pepstatin, leupeptin, and aprotinin) or His buffer (0.5% Triton X-100, 20 mM NaPO4, pH 7.8, 500 mM NaCl, 1 mM Na3VO4, 1 mM Pefabloc, 1 μg/ml each pepstatin, leupeptin, and aprotinin). After lysis, cell debris was pelleted at 12,000 × g for 20 min. Immunoprecipitations were carried out as previously described using anti-Ena (Gertler et al., 1995), anti-VASP (Halbrugge et al., 1990), anti-FLAG M2, or anti-Ha 12CA5 antibodies. His-tagged Ena was purified by incubating lysates with 35 μl of Ni-nitrilotriacetic acid (NTA)-agarose (Qiagen, Chatsworth, CA) for 1 h at 4°C. Nonspecifically bound proteins were eluted with His buffer plus 100 mM imidazole. Proteins were resolved on 7.5% SDS-polyacrylamide gels, transferred to a polyvinylidene difluoride membrane, and blotted with anti-Ena, anti-VASP, anti-Ha 12CA5, anti-FLAG M2 (Molecular Probes, Eugene, OR), or anti-phosphotyrosine mAb 4G10 (Upstate Biotechnology, Lake Placid, NY) antibodies.
Sequencing of Mutant ena Alleles
Total RNA was prepared from Drosophila pupae carrying heterozygous wild-type and mutant ena210 or ena23 alleles using Tri reagent (Molecular Research Center, Cincinnati, OH). Reverse transcription PCR was carried out by priming first-strand synthesis with an Ena oligonucleotide that hybridizes to the last 20 nucleotides in the Ena open reading frame. The Ena coding sequences were subsequently amplified with three sets of primers that amplified the 5′ 600 nucleotides, middle 600 nucleotides, and 3′ 700 nucleotides, respectively. PCR products were subcloned into the TA cloning vector system (Invitrogen, San Diego, CA), and at least eight independent subcloned PCR products were sequenced for each of the PCR products. As expected, half of the subcloned PCR products carried the ena210 or ena23 mutation, and half were wild type.
Site-directed Mutagenesis
DNA was purified by standard techniques (Maniatis et al., 1982). Site-directed mutagenesis of Ena was performed by the method of Deng and Nickoloff (1992). Oligonucleotides containing single-base changes that changed Ala-98 to Val and Lys-636 to a stop codon were incorporated into a full-length Ena cDNA. The entire mutagenized DNA fragment was sequenced to confirm the absence of any other mutations that may have been inadvertently generated during the procedure.
Preparation and Purification of Recombinant Ena and Fusion Proteins and In Vitro Synthesis of Proteins
The His-tagged Ena construct was subcloned into the baculovirus vector pVL1393 (Invitrogen). Spodoptera frugiperda SF9 cells were cotransfected with 2.5 μg of pVL1393-Ena plus 200 ng of Baculogold viral DNA (Pharmingen, San Diego, CA) to recover recombinant virus. High-titer virus stocks were generated and used to infect 2 × 108 SF9 cells. At 48 h after infection, cells were harvested and lysed in 20 ml of 0.5% Triton X-100, 20 mM NaPO4 (pH 7.8), 500 mM NaCl, 1 mM Na3VO4, 1 mM Pefabloc, and 1 μg/ml each pepstatin, leupeptin, and aprotinin. After lysis, cell debris was pelleted at 12,000 × g for 20 min, and the lysate was incubated with 2 ml of NTA-agarose (Qiagen) for 1 h at 4°C. Nonspecifically bound proteins were eluted with His buffer plus 100 mM imidazole and Ena protein was eluted with 20 mM piperazine-N,N′-bis(2-ethanesulfonic acid), 500 mM NaCl, and 300 mM imidazole and stored at 4°C.
A PCR fragment encoding the Drosophila Abl SH3 domain (amino acids 82–325) was subcloned into the BamHI site of the pGEX2TK vector (Pharmacia, Piscataway, NJ). The chicken zyxin translated region encoding amino acids 1–542 was subcloned into the EcoRI site of a modified pGEX expression vector. The chicken zyxin LIM domain (amino acids 349–542) and the chicken zyxin proline-rich region (amino acids 1–348) were also subcloned into the EcoRI site of a modified pGEX vector (Schmeichel and Beckerle, 1994). GST fusion proteins were expressed in Escherichia coli DH5α and prepared as previously described (Gertler et al., 1995). Ena protein and Ena A97V mutant protein were produced in vitro using the Single Tube Protein System (Novagen, Madison, WI) after subcloning into the pCITE vectors. All proteins were quantified by Coomassie blue staining or Western blot.
Solution- and Filter-binding Assays
For solution-binding assays, transfected S2 cell lysates were incubated with equivalent amounts of GST-SH3 or GST-Zyxin fusion proteins immobilized on glutathione-Sepharose (Pharmacia) for 1 h at 4°C. Beads were washed once with immunoprecipitation buffer and boiled in SDS sample buffer. Bound proteins were analyzed by SDS-PAGE, followed by Western blotting with anti-Ena or anti-VASP antibodies. Ena and VASP overlays were carried out essentially as previously described (Reinhard et al., 1995a). Fifty micrograms of total protein from human platelets and 100 ng of purified porcine platelet zyxin were separated by SDS-PAGE and transferred to nitrocellulose. The blot was overlaid with 1 μg/ml Ena purified from baculovirus-infected cells, and bound Ena was detected by the anti-Ena antibody followed by 125I-protein A.
Yeast Two-Hybrid System Screen
A cDNA encoding the C-terminal 243 amino acids of Ena was fused to the sequence encoding the GAL4 DNA binding domain in the pAS1-CYH2 vector to create pAS-EnaC. The yeast strain Saccharomyces cerevisiae Y190, which contains the reporter genes HIS3 and LacZ, was cotransformed with pAS-EnaC and the Drosophila larval library pAct (a gift of Dr. Stephen J. Ellidge, Baylor College of Medicine, Houston, TX) in which cDNAs are fused to the sequence encoding the GAL4 activation domain. Transformants (2.05 × 107) were screened for activation of the reporter genes by spreading the cells on medium lacking histidine but supplemented with 30 mM 3-aminotriazole. Colonies that grew in the absence of histidine appeared within 7 d after plating and were assayed for β-galactosidase production by a membrane transfer assay. Plasmid DNA from the positive clones was isolated and sequenced.
Genetic Stocks and Germ Line Transformation
Drosophila stocks containing ena point mutations and chromosomal deletions removing the ena gene have been previously described (Gertler et al., 1990, 1995). Flies were cultured on a Drosophila cornmeal-yeast extract medium. To construct the transposons used in this study, Ena or VASP coding sequences were subcloned into the pUAST vector, which contains a minimal promoter preceded by five GAL4 binding sites (Brand and Perrimon, 1993). The plasmid DNA was injected into white mutant Drosophila embryos, and P element transposase was supplied by coinjection of plasmid pPi25.7wc. Transformants were identified by pigmentation in the eyes provided by the white gene in the transposon, and the chromosomal location of the transposons was determined genetically. Stocks were generated containing both the transgene and heterozygous enaGC5 mutations.
ena Mutant Rescue Crosses
Chromosomes containing inversions that eliminate the ena gene product (enaGC1 and enaGC5) were used in this analysis (Gertler et al., 1990, 1995). Ubiquitous expression of the Ena transgenes was driven by the GAL4-e22c enhancer trap (generously provided by N. Perrimon, Harvard University, Boston, MA). For rescue experiments, enaGC5/Cyo, UAS-Ena, or UAS-VASP virgin females were crossed with enaGC1, GAL4-e22c/Cyo males. Progeny were scored for the presence of rescued Cy+ enaGC1/enaGC5 flies. Animals containing two ena mutant chromosomes were identified as Cy+. For each cross, the percentage of adult survival was calculated by dividing the number of Cy+ adult flies recovered by the expected number of Cy+ animals. The expected number of Cy+ animals was assumed to be equal to one-half the number of CyO siblings scored. The entire progeny were scored from each cross. Rescue with the other transgenes was normalized with respect to the rescue seen with the wild-type Ena transposon. The values presented are averages from two or three independent crosses. The absolute amount of rescue seen with the Ena transposons varies from cross to cross depending on temperature and how crowded the crosses were.
Immunofluorescence Microscopy of Transfected Cells
Ptk2 cells (CCL56; American Type Culture Collection, Manassas, VA) were grown on coverslips in MEM (Life Technologies, Gaithersburg, MD) supplemented with 1% glutamine, 1% nonessential amino acids, and 10% FCS. Cells were transfected by the calcium phosphate method with pCMV/Ena which consists of the Ena cDNA in the pRc/cytomegalovirus (CMV) expression vector (Invitrogen), or pVSV-VASP which consists of human VASP N-terminally tagged with an epitope of VSV glycoprotein G in the expression vector pcDNA3 (Invitrogen). Sixteen hours after addition of the precipitate, the cells were washed once in PBS, and fresh medium was added. Human fibroblasts were grown on coverslips in DMEM (Life Technologies)/10%. Cells were transfected by Fugene 6 transfection reagent (Boehringer Mannheim, Indianapolis, IN) with pCMV/Ena or the respective constructs into which the Ena23 or Ena210 mutations had been introduced.
After 42–53 h, cells were washed with PBS, fixed in 3.7% formaldehyde, permeabilized by 0.2% Triton X-100, and incubated with rabbit antiserum raised against amino acids 55–235 of Ena (diluted 1:6 in PBS, 11 μg/ml) and, for the detection of VSV-G-tagged VASP, with mAb P5D4 (Sigma, St. Louis, MO), diluted 1:5000 in PBS. Zyxin was detected with the mAb 164D4 raised against human zyxin. In PtK2 cells, primary antibodies were detected by TRITC-labeled donkey anti-rabbit antibodies (Ena staining) and DTAF-labeled goat anti-mouse antibodies (Dianova, Hamburg, Germany). Actin was detected with fluorescein-labelled phalloidin (Molecular Probes, Eugene, OR). In human fibroblasts, primary antibodies were detected by Oregon Green–labeled goat anti-rabbit antibodies (Ena staining) and Cy3-labeled donkey anti-mouse antibodies (zyxin staining). The specimens were examined with a Leitz (Wetzlar, Germany) Aristoplan microscope equipped with epifluorescence. Photographs were taken with Kodak Ektachrome Elite 400 film (Eastman Kodak, Rochester, NY).
RESULTS
VASP Can Functionally Substitute for a Complete Loss of Ena Protein in Drosophila
Ena and VASP share sequence identity in two regions of the protein thought to be important for function (Figure 1A). We wondered whether these similarities could be sufficient to permit VASP to partially compensate for a lack of Ena protein during Drosophila development. To test this, we generated stable transgenic Drosophila expressing human VASP or Drosophila Ena. Five VASP and three Ena transgenic lines that were independently derived were tested for their ability to rescue the lethality of ena null mutants. When expressed ubiquitously via the UAS/GAL4 binary expression system (Brand and Perrimon, 1993), VASP partially rescued ena mutant lethality, allowing 25–85% of the ena mutant progeny to survive to adulthood compared with 79–100% rescue by the Ena transgene (Table 1). As expected, no ena mutant flies from these crosses eclosed in the absence of expression of an Ena or VASP transgene. All the flies rescued with the VASP transgene were visibly normal and had comparable fertility and survival 4 wk after eclosion to flies that were rescued with a Drosophila Ena transgene. The range in rescue seen with the different transgenes is likely due to differences in expression levels or patterns as a result of the different insertion sites of the transgenes. This result suggests that Drosophila Ena and human VASP have overlapping in vivo functions and affect similar cellular processes.
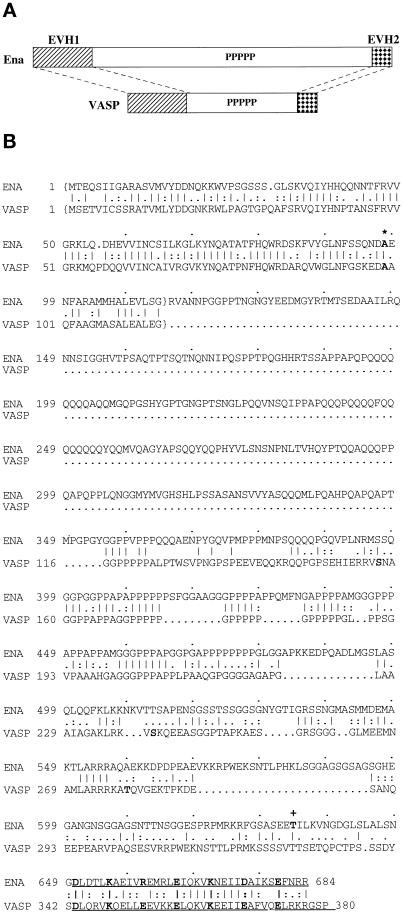
Figure 1.
Ena and VASP have structural domain and sequence similarity, and the ena 210 and ena 23 mutations map to these regions of similarity. (A) Comparison of the overall structure and structural domain organization of Ena and VASP. The conserved EVH1 domain (hatched box) and EVH2 domain (checked box) are separated by a central domain of variable length, which contains a region rich in prolines. (B) Comparison of the deduced amino acid sequences and sequence motifs of Drosophila Ena and human VASP. The central regions of both Ena and VASP have multiple polyproline motifs. The C-terminal 35 amino acids contain a mixed charge cluster containing a charged residue at every fifth position (marked in bold). The conserved alanine that is altered in ena 210 mutants is in bold and marked with an asterisk. The EVH1 domains are enclosed in brackets, and the EVH2 domain is underlined. The location of the stop codon in ena23 is marked by a plus sign. Sequence alignment was generated using the Genetics Computer Group (Madison, WI) BestFit program. Amino acid identity is indicated by vertical lines, and similarity is indicated by dots.
Table 1.
Rescue of ena mutant flies with ena and VASP transgenes
| Transgene | Experiment | Expected no. of ena mutant fliesa | Observed no. of rescued ena mutant flies | Rescue (%)b | Average rescue per line (%) |
|---|---|---|---|---|---|
| wt1A | 1 | 140 | 107 | 76 | |
| 2 | 82 | 63 | 77 | 79 | |
| 3 | 135 | 112 | 83 | ||
| wt1B | 1 | 118 | 116 | 98 | |
| 2 | 96 | 102 | 106 | 100 | |
| 3 | 131 | 128 | 98 | ||
| wt10A | 1 | 115 | 93 | 81 | |
| 2 | 87 | 76 | 87 | 87 | |
| 3 | 78 | 72 | 92 | ||
| VASP.A | 1 | 116 | 94 | 81 | |
| 2 | 90 | 82 | 91 | 85 | |
| 3 | 123 | 101 | 82 | ||
| VASP.3 | 1 | 65 | 32 | 49 | |
| 2 | 61 | 18 | 30 | 40 | |
| VASP.K | 1 | 65 | 16 | 25 | |
| 2 | 104 | 25 | 24 | 24 | |
| 3 | 87 | 20 | 23 | ||
| VASP.C | 1 | 116 | 33 | 28 | |
| 2 | 117 | 23 | 20 | 24 | |
| 3 | 102 | 24 | 24 | ||
| VASP.F | 1 | 118 | 39 | 33 | |
| 2 | 115 | 32 | 27 | 26 | |
| 3 | 92 | 16 | 17 |
Expected number of ena mutants was calculated as one-half of the observed ena heterozygotes.
Percent rescue was determined by dividing the observed number of Cy+ ena homozygotes by the expected number of ena mutants.
Ena and VASP Are Colocalized to Focal Adhesions and Actin Stress Fibers
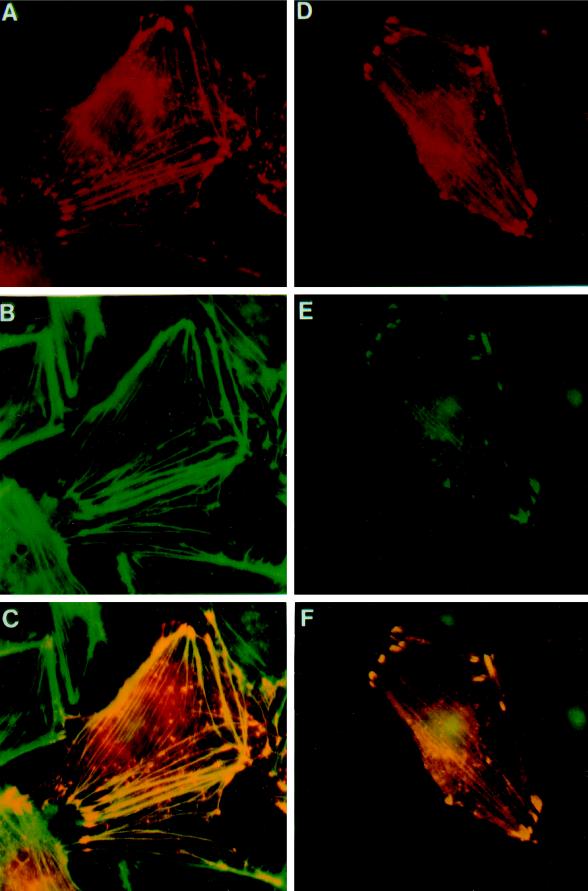
VASP is localized to focal adhesions, stress fibers, and cell–cell contacts (Reinhard et al., 1992). The Ena protein is localized to the axonal tracts of the developing Drosophila embryonic CNS (Gertler et al., 1995), but the small size of these cells makes higher-resolution localization difficult. Because Ena and VASP have some conserved function in vivo, we compared their subcellular distribution when expressed in mammalian cells. Both Ena and VASP were detected at actin filaments and focal adhesion contacts. Coexpression of Ena and VASP resulted in an essentially identical pattern of expression (Figure 2). These results indicate that both proteins have domains sufficient to permit binding in vivo to proteins in the actin cytoskeleton and focal adhesions.
Figure 2.
Colocalization of Ena and VASP proteins to stress fibers and focal contacts in transfected PtK2 cells. PtK2 cells were transiently transfected with pCMV/ena (A–C) or pCMV/ena and pVSV-VASP (D–F) and processed for double-label immunofluorescence microscopy. Ena was detected with rabbit polyclonal antibodies (A and D), VASP with a monoclonal mouse anti-VSV antibody (E), and F-actin by fluorescein-labeled phalloidin (B). A merge of A and B is shown in C, and a merge of D and E is shown in F. Fluorescent staining was by TRITC for Ena and DTAF for VASP. Ena expressed alone is localized either in focal contacts and stress fibers or is also variably found in spot-like structures, which also stain for F-actin (A–C). Coexpression of Ena and human VASP results in an identical localization of the two proteins in focal contacts and stress fibers (D–F). Controls show no cross-reactivity of the anti-Ena antibodies with VASP. Nuclear fluorescence is nonspecific background because of the secondary antibody.
Identification of Lethal Mutations in the EVH1 and EVH2 Domains of Drosophila Enabled
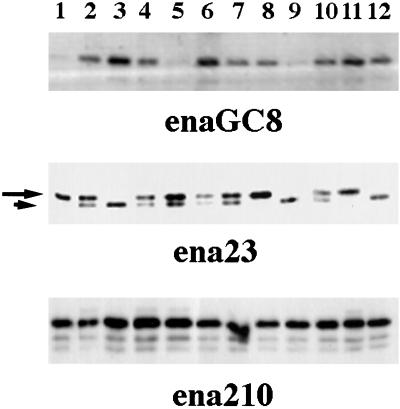
Mutant ena alleles that were generated by mutagenesis with ethyl-nitroso-urea during the genetic screen that identified ena as a dominant genetic suppressor of abl mutations result in 100% lethality when expressed over an ena deficiency (Table 2). Because ethyl-nitroso-urea normally introduces single-base changes, we tested for the expression of Ena protein from these mutant alleles by Western blot analysis of single embryos from stocks of flies heterozygous for the ena mutation of interest. A chromosomal inversion (enaGC8) known to be null for Ena protein was used as a control. Because the parents were heterozygous for the ena mutant allele, 25% of the embryos picked were expected to be homozygous for the ena mutations, 25% for the wild-type ena allele, and 50% heterozygous for both the mutant and wild-type alleles. As expected, 25% of the embryos from the enaGC8/wild-type parents did not express the Ena protein (Figure 3, top panel). Half of the embryos from the ena23/wild-type parents expressed two sizes of Ena protein, and 25% had only the slower-migrating species, identical to the wild-type protein. The other 25% of the embryos were presumably homozygous ena23 mutant embryos that expressed a faster-migrating form consistent with the presence of a smaller protein (Figure 3, middle panel). Embryos from the ena210/wild-type parents all expressed a full-length protein (Figure 3, bottom panel).
Table 2.
Viability of ena210 and ena23 mutant flies
| Genotypes | Experiment | Expected no. of Ena mutant fliesa | Observed no. of Ena mutant flies |
|---|---|---|---|
| enaGC1/enaGC5 | 1 | 120 | 0 |
| 2 | 115 | 0 | |
| ena23/enaGC1 | 1 | 120 | 0 |
| 2 | 117 | 0 | |
| ena210/enaGC1 | 1 | 126 | 0 |
| 2 | 101 | 0 |
The entire progeny was scored for each cross. The expected number of Ena mutant flies was calculated as one-half the number of the balanced, heterozygous siblings.
Figure 3.
Analysis of ena mutant alleles. Single-embryo Western blot analysis of heterozygous ena mutant stocks. Embryos were collected for 2 h and then aged 5 h. Single embryos were picked and lysed in sample buffer and Western blotted. For the enaGC8 chromosomal inversion, 25% of the embryos were null for Ena protein (top panel, lanes 1, 5, and 9). This blot was stripped and reprobed with an anti-fax antibody to demonstrate the presence of embryonic proteins in all 12 lanes. The ena 23 allele appears to encode an Ena protein with a small truncation, because 25% of the embryos encode only a smaller protein (small arrow) of ∼80 kDa (middle panel, lanes 3, 9, and 12), and 50% of the embryos encode both full-length Ena (large arrow) and the truncated protein. The ena 210 allele appears to encode a full-length protein, because all embryos picked express full-length Ena.
To explain the embryonic lethality associated with ena210 and ena23, we speculated that these mutant Ena proteins harbored lesions in critical functional domains. Identification of these mutations could be informative about the function of other Ena/VASP family members, because they have a high degree of functional conservation with Ena. To analyze the nature of these mutations, the coding regions of ena210 and ena23 were sequenced after reverse transcription PCR of total RNA from Drosophila pupae that were heterozygous for the ena mutation being examined. A single C→T mutation was identified in the EVH1 domain of ena210 that results in an A97V change (Figure 1B). Examination of the EVH1 domains from previously cloned family members demonstrated complete conservation of this alanine in all reported Ena/VASP family members (Gertler et al., 1996). Two changes were found in ena23. First was an A→G change in the proline rich-domain of Ena changing N379F. The second was an A→T mutation that introduced a stop codon 52 amino acids from the C terminus of Ena that deletes the EVH2 domain (Figure 1B) and encodes a protein consistent with the smaller protein seen in the single-embryo Western blots.
The N379F mutation identified in ena23 involves an amino acid that is not conserved in any other Ena/VASP family member (Gertler et al., 1996). However, the introduction of the stop codon (K636Stop) deletes the conserved EVH2 domain. Consequently, we focused on this mutation and tested whether the K636Stop described above contributed to the lethality of ena23. The K636Stop mutation was generated by site-directed mutagenesis in an Ena P element transposon, and transgenic flies were generated. The UAS/GAL4 binary expression system was used to ubiquitously express wild-type or mutant Ena K636Stop in Ena null Drosophila embryos. Two Ena K636Stop transgenic lines were tested and rescued from 0 to 8% of the expected homozygous ena mutant flies. Both lines expressed truncated Ena K636Stop protein as assessed by Western blots of lysates from transgenic flies induced by GAL4 to express the UAS-ena23 transgene (our unpublished observations). This indicates that the removal of the EVH2 domain is in part responsible for the lethality seen in the ena23 mutant flies. The identification of lethal mutations in the EVH1 and EVH2 domains of Ena is strong evidence that these domains are critical for in vivo Ena function and by analogy for in vivo functions of the Ena/VASP family of proteins.
Drosophila Enabled Binding to Zyxin Is Disrupted In ena210
Previous studies with VASP and Mena have demonstrated that the EVH1 domain is involved in binding the focal adhesion–associated protein zyxin (Reinhard et al., 1995a; Gertler et al., 1996). Based on these observations, we speculated that the EVH1 domain from Drosophila Ena would also bind zyxin. We examined the zyxin binding properties of Ena with a filter binding assay and found that Ena, like VASP and Mena, bound to the zyxin protein (Figure 4A). The zyxin protein consists of three highly conserved LIM domains and a proline-rich domain. We expressed full-length zyxin and the LIM and proline-rich domains of zyxin individually as GST fusion proteins and tested in solution-binding assays for binding to Ena or to human VASP. As expected, both proteins bound to the full-length zyxin protein. Both proteins also bound to the proline-rich region of zyxin but not to the LIM domains (Figure 4B), consistent with results obtained for VASP and Mena (Niebuhr et al., 1997) and indicating that the ability to bind zyxin is conserved in Drosophila Ena and mediated through the same region of zyxin. To investigate whether Ena and zyxin can associate in vivo, we compared their subcellular distribution in mammalian cells transfected with Ena. Both Ena and zyxin are detected at focal adhesion contacts and on microfilaments (see Figure 9, 1A and 1B).
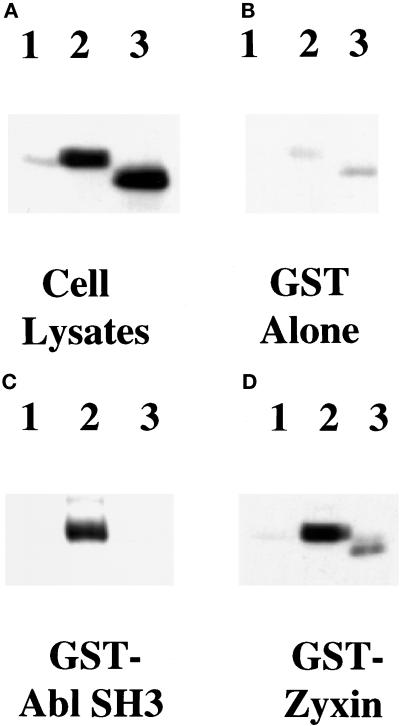
Figure 4.
Binding of Ena and VASP proteins to zyxin and mapping of the binding domain in zyxin. (A) Ena and VASP overlays were carried out essentially as previously described (Reinhard et al., 1995a). Fifty micrograms of total protein from human platelets (lane 1) and 100 ng of purified porcine platelet zyxin (lane 2) were separated by SDS-PAGE and transferred to nitrocellulose. The blot was overlaid with Ena purified from baculovirus-infected cells, and bound Ena was detected by the anti-Ena antibody followed by 125I-protein A (left panel). The control (right panel) was treated identically, except that the overlay solution contained no Ena protein. Ena protein bound to an 83-kDa protein present in platelet lysates (lane 1), which comigrates with full-length zyxin (lane 2), whereas the control panel gave no detectable signal (right panel). (B) S2 cells were transfected with Ena or VASP, and proteins were bound to GST fusion proteins as follows: full-length chicken zyxin (lane 1), zyxin proline domain (lane 2), zyxin LIM domains (lane 3) (Schmeichel and Beckerle, 1994), and GST alone (lane 4). Retained proteins were detected with anti-Ena N-terminal (left panel) or anti-VASP (right panel) polyclonal antibodies. Both proteins bound to full-length zyxin and to the zyxin proline domain (lanes 1 and 2). Neither protein bound to the zyxin LIM domain (lane 3). The faint signal seen in lanes 3 and 4 in the anti-VASP blot is due to background binding between VASP and the GST fusion proteins. Aliquots of transfected cell lysates were analyzed for expression of Ena or VASP protein (lanes 5). These results are representative of three independent experiments.
Figure 9.
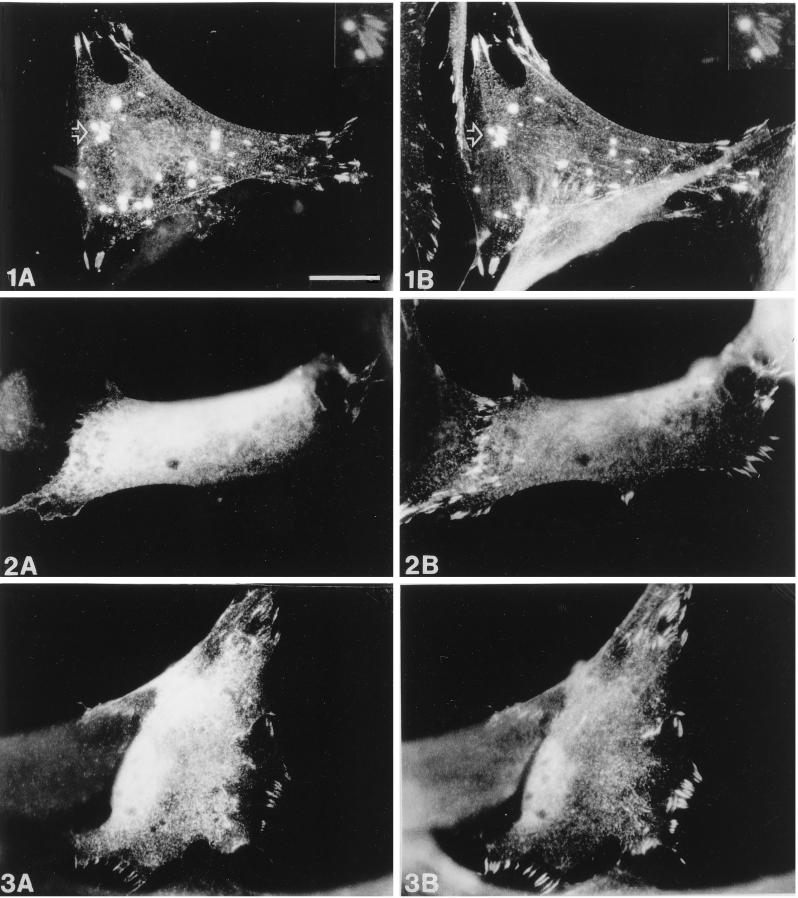
Comparison of the subcellular distribution of zyxin and wild-type or mutant Ena. Human fibroblasts were transiently transfected with pCMV/Ena (1), pCMV/EnaA97V (2), or pCMV/EnaK636Stop (3) and processed for double-label immunofluorescence microscopy. Ena staining is shown in A, staining of endogenous zyxin in B. After transfection of wild-type Ena (1A and 1B), colocalization of both Ena and zyxin is found in focal contacts, on microfilaments and in spot-like structures. A magnification of the area indicated by the arrow showing two “spots” and several focal contacts is seen at the top right (insets in 1A and 1B). In contrast, EnaA97V (2A and 2B) is diffusely distributed throughout the cytoplasm and totally absent from focal contacts, which are labeled by the zyxin antibody. EnaK636Stop (3A and 3B) is also diffusely distributed in the cytoplasm but shows some residual focal contact staining, although less pronounced than that of the wild-type Ena protein. Bar in 1A, 20 μm (valid for all panels).
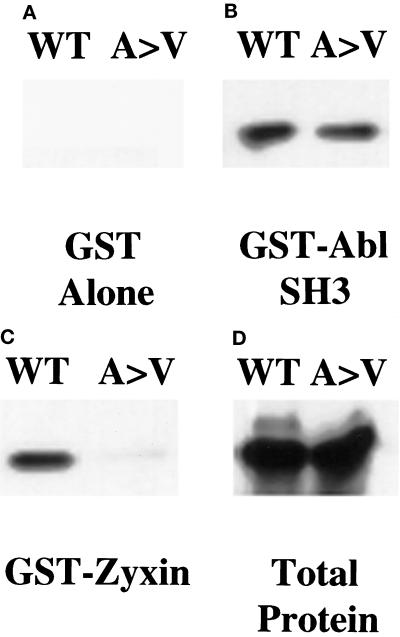
To determine whether the A97V change affected zyxin binding to the Ena EVH1 domain, we examined the zyxin binding properties of wild-type and A97V mutant Ena by generating this mutation in the Ena cDNA and cloning these cDNAs into the pCite expression vector. Wild-type or Ena A97V proteins were produced using a combined in vitro transcription–translation reaction and tested in solution-binding assays for their ability to bind GST-zyxin. Although equal amounts of the wild-type and mutant proteins were expressed, less mutant Ena A97V protein was pulled down by the GST-zyxin fusion protein when compared with the wild-type Ena protein (Figure 5C). In contrast, both proteins bound equally well to the GST Abl SH3 domain, demonstrating that the mutation is specific for an EVH1 domain function and does not disrupt the proline-rich central region of Ena (Figure 5B). This suggests that the mutation does not cause an overall disruption of the Ena protein structure. To further confirm that the A97V change in Ena affects zyxin binding, we examined the ability of this mutant protein to colocalize with zyxin in transfected mammalian cells. In contrast to wild-type Ena and zyxin, which are clearly present in the focal contacts, the A97V mutant Ena exhibits diffuse staining and is absent from the focal contacts (see Figure 9, 2A and 2B). We conclude that EVH1 binding domain function is required for Ena’s function in Drosophila.
Figure 5.
ena 210 mutation disrupts zyxin binding. Full-length wild-type Ena (WT) or ena210 (A97V) proteins were prepared using an in vitro transcription–translation reaction and bound to GST alone (A), GST-AblSH3 (B), or GST-zyxin (C). Retained proteins were detected with anti-Ena N-terminal polyclonal antibody. Both proteins were expressed equally well (D). As expected, the GST negative control did not bind detectably to either protein (A). Both proteins bound equally well to the GST-AblSH3 domain (B). The wild-type Ena protein bound to the GST-zyxin protein, whereas binding to the A97V mutant protein was reduced. (C) These results are representative of three independent experiments.
The EVH2 Domain of Drosophila Enabled Mediates Multimerization
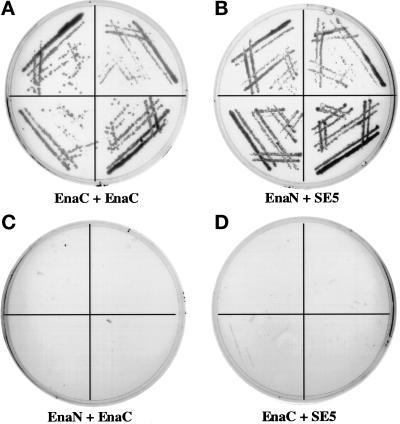
Truncation of the EVH2 domain by the mutation identified in Ena K636Stop results in embryonic lethality, suggesting that this region of Ena may mediate a critical function of the Ena protein. Initial clues to this function came by the isolation of the EVH2 domain of Ena in a yeast two-hybrid screen for Drosophila proteins that interact with the C-terminal 243 amino acids of Ena. The C-terminal 243 amino acids of Ena were fused to the DNA binding domain of the yeast transcription factor Gal4 and used to screen a third instar larval library whose inserts were fused to the activation domain of Gal4. The separately expressed domains are unable to activate transcription of the reporter genes HIS3 and LacZ unless a protein–protein interaction takes place (Chien et al., 1991). Clones (20.5 million) were screened, 9 of which interacted with Ena as assessed by expression of both the HIS and LacZ reporter genes. Two of these clones were partial Ena cDNAs encoding Ena amino acids 628–684 and 634–684, respectively. The interaction was specific, because a construct with the Ena N-terminal domain fused to the DNA binding domain of Gal4 did not interact with these same isolated clones (Figure 6). Seven additional unique sequences were also identified that will be described in a later report. We thus speculated that the EVH2 domain may be involved in mediating multimerization of Ena/VASP proteins.
Figure 6.
Interaction of the Ena EVH2 domain with the C-terminal 243 amino acids of Ena in the yeast two-hybrid system. The first 235 amino acids of Ena (GAL4DB-Ena1–235) and last 243 amino acids of Ena (GAL4DB-Ena441–684) were fused to the sequence encoding the GAL4 DNA-binding domain in the pAS1-CYH2 vector. These two constructs were cotransformed with GAL4AD-Ena628–684, a clone in which amino acids 628–684 of Ena are fused to the sequence encoding the GAL4 activation domain and that was isolated in a yeast two-hybrid screen using the C terminal 243 amino acids of Ena as bait. Four independent colonies from each of the cotransformations were spread on medium lacking histidine and analyzed for growth. GAL4DB-Ena441–684 containing the C-terminal region of the Ena protein can interact with GAL4AD-Ena628–684 (A, EnaC + EnaC), whereas GAL4DB-Ena1–235 cannot (C, EnaN + EnaC). As a control, GAL4DB-Ena1–235 and GAL4DB-Ena401–684 were also tested for their ability to interact with GAL4AD-SE5, a clone identified from the same larval library as GAL4AD-Ena628–684 by its ability to interact with GAL4DB-Ena1–235. As expected, GAL4DB-Ena1–235 can interact with GAL4AD-SE5 (B, EnaN + SE5), whereas GAL4DB-Ena401–684 cannot (D, EnaC + SE5).
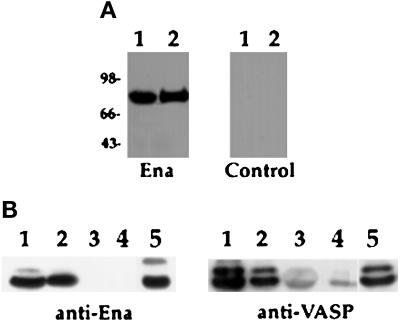
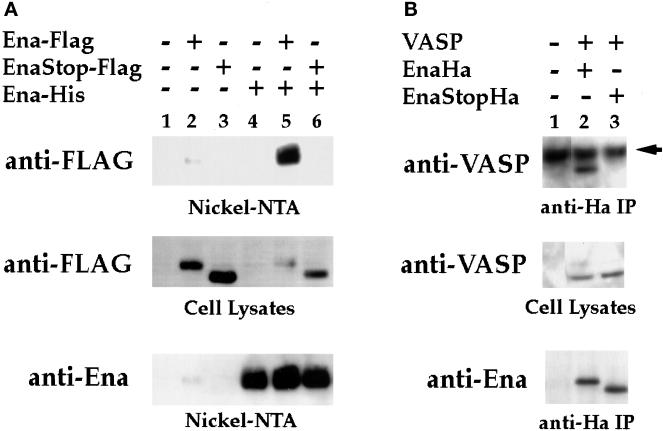
To assess the ability of Ena to multimerize, Ena expression vectors were generated in which full-length Ena was tagged at the C terminus with a 6-histidine tag for purification on Ni-NTA resin. Hemaglutinin or FLAG epitopes were then added to the C terminus of either full-length Ena or Ena K636Stop. The tagged Ena and Ena K636Stop proteins were recognized by commercially available mAbs directed against FLAG (M2) or Ha (12CA5) epitopes. S2 cells were transfected with the FLAG-tagged Ena or Ena K636Stop expression vectors in the presence or absence of the His-tagged Ena expression vector. Transfected cell lysates were purified on Ni-NTA resin, and isolated complexes were analyzed by Western blot using the anti-FLAG mAb. When the His-tagged Ena was expressed in the presence of the full-length FLAG-tagged Ena, a strong FLAG signal was seen, suggesting that the two differentially tagged proteins formed a complex in cultured cells (Figure 7A, lane 5). However, there was no evidence of complex formation between the His-tagged Ena and the FLAG-tagged Ena K636Stop protein, indicating that this interaction required the Ena EVH2 domain (Figure 7A, lane 6).
Figure 7.
The EVH2 domain mediates multimerization. (A) S2 cells were transfected with the following expression constructs: no DNA (lane 1), Ena-FLAG (lane 2), Ena K636Stop-FLAG (EnaStopFlag; lane 3), Ena-His (lane 4), Ena-FLAG + Ena-His (lane 5), and Ena K636Stop-FLAG (EnaStopFlag) + Ena-His (lane 6). Cells were lysed, and extracted proteins were purified on Ni-NTA agarose and blotted with anti-FLAG (top panel) or anti-Ena (bottom panel) antibodies. Aliquots of cell lysates representing <2% of the total protein used in the purifications were analyzed with the anti-FLAG antibody for expression of the FLAG-tagged proteins, and both the Ena-FLAG and EnaK636Stop-FLAG proteins were expressed in the presence of Ena-His (middle panel). However, only full-length Ena-FLAG copurified with the Ena-His on the nickel NTA resin (top panel, compare lanes 5 and 6). A small amount of contaminating Ena-FLAG was purified with the Nickel-NTA resin (top panel, lane 2). (B) S2 cells were transfected with the following expression constructs: no DNA (lane 1), VASP + Ena-Ha (lane 2), and VASP + Ena K636Stop-Ha (EnaStopHa; lane 3). Transfected cell lysates were immunoprecipitated with 5 μg of anti-Ha antibody. Complexes were subsequently analyzed by Western Blot with an anti-VASP antibody (top panel) or anti-Ena antibody (bottom panel). Aliquots of cell lysates representing <2% of the total protein used for the immunoprecipitations were analyzed for expression of the VASP protein with an anti-VASP antibody (middle panel). VASP was expressed in the presence of Ena-Ha and Ena K636Stop-Ha (middle panel, lanes 2 and 3), but VASP only copurified with full-length Ena-Ha in the anti-Ha IPs (compare lanes 2 and 3, top panel). The dark band present in all lanes in the top panel is the heavy-chain antibody from the anti-Ha IPs cross-reacting with the secondary antibody of the Western blot (arrow).
Ena and the Ena/VASP family of proteins share similarity in the EVH2 domain. To determine whether this sequence conservation translated to functional conservation, we looked for complex formation between VASP and Ena or Ena K636Stop. S2 cells were transfected with VASP and Ha-tagged Ena or Ena K636Stop expression vectors. Transfected cell lysates were immunoprecipitated with 5 μg of anti-Ha antibody, and complexes were analyzed by Western blot using an anti-VASP antibody. As seen with Ena, VASP associated with full-length Ena (Figure 7B, lane 2), whereas no evidence of complex formation was detected with Ena K636Stop, which lacks the EVH2 domain (Figure 7B, lane 3). Taken together, the coprecipitation and yeast two-hybrid data suggest that the EVH2 domain is necessary and sufficient to mediate Ena multimerization with both Ena and VASP. Therefore, this EVH2 function appears to be conserved within the Ena/VASP family and may be essential for Ena’s in vivo effects.
The EVH2 Domain Is Important for Ena’s Ability to Bind the Abl-SH3 Domain and Zyxin
Because the ability to multimerize may be an important property of the Ena protein, we speculated that it may be required for other known functions of Ena. Studies in other laboratories have indicated that multimerization of proteins can alter their ligand-binding capacity (Oh et al., 1997; Seiffert, 1997). We thus decided to test the effect of removing the Ena EVH2 domain on its affinity for the Abl-SH3 domains and zyxin. S2 cells were transfected with Ena or Ena K636Stop expression vectors. Full-length zyxin and the Abl SH3 domain were expressed as GST fusion proteins and bound in solution to the transfected cell lysates, and bound complexes were analyzed by Western blot analysis using the anti-Ena antibody. Although the mutant and wild-type Ena proteins were expressed equally well, less of the Ena K636Stop protein was pulled down by both the Abl SH3 domain (Figure 8C, lane 3) and zyxin (Figure 8D, lane 3) when compared with the full-length Ena protein (Figure 8, C and D, lanes 2). To further confirm that the Ena K636Stop change in Ena affects its ability to bind zyxin, we examined the ability of this mutant protein to colocalize with zyxin in transfected mammalian cells. In contrast to wild-type Ena and zyxin, which are clearly present in the focal contacts, K636Stop mutant Ena exhibits more diffuse staining with reduced staining in the focal contacts (Figure 9, 3A and 3B).
Figure 8.
Ena K636Stop is impaired in its ability to bind zyxin and the Abl-SH3 domain. S2 cells were transfected with no DNA (lanes 1), full-length Ena (lanes 2), or Ena K636Stop (lanes 3) expression vectors. Transfected cell lysates were incubated with GST-Zyxin (D) or Abl SH3 (C) domain fusion proteins or with GST alone as a negative control (B). Cells were lysed, and aliquots of the cell lysates were compared to confirm that equal amounts of the full-length and truncated Ena proteins were expressed (A). Lysates were incubated with the GST fusion proteins, and retained proteins were detected with anti-Ena N-terminal polyclonal antibody. Little protein bound to the GST negative control (B). A decrease in binding of the truncated Ena K636Stop protein was seen with both zyxin and Abl SH3 when compared with the full-length Ena protein (C and D, compare lanes 2 and 3). These results are representative of three independent experiments.
DISCUSSION
In this report, we have taken advantage of the Drosophila system to demonstrate in vivo that there is a striking degree of functional conservation between Drosophila Ena and human VASP, both members of the recently described Ena/VASP family of proteins. Specifically, we have demonstrated that human VASP can partially substitute for a loss of Ena in the developing Drosophila embryo. This is particularly striking in light of the limited sequence identity that these two proteins share, primarily in the EVH1 and EVH2 domains. In addition, two lethal Ena mutations that map to the EVH1 and EVH2 domains of Drosophila Ena have been described and characterized. Identification of functional mutations in the EVH1 and EVH2 domains of Ena are of particular interest because of the high degree of similarity in both sequence and function between different Ena/VASP family members, which, by analogy, implicates these protein domains as critical functional domains in other Ena/VASP family members.
One of the shared properties of Ena and VASP that may account for the observed rescue of Ena mutant flies by expression of human VASP is the ability to bind in vitro to the focal adhesion protein zyxin. Previous studies demonstrated that the focal adhesion–associated protein zyxin binds VASP and Mena through the EVH1 domain (Reinhard et al., 1995b; Gertler et al., 1996). Here we demonstrated that this binding is conserved in Ena and is mediated by the zyxin proline-rich domain, consistent with recent peptide-mapping and competition studies of the Ena/VASP binding domain in zyxin (Niebuhr et al., 1997). This would leave the zyxin LIM domains, which have been shown to mediate protein–protein interactions, available to link the zyxin-Ena/VASP complexes to other cellular proteins (Sadler et al., 1992; Schmeichel and Beckerle, 1994; Arber and Caroni, 1996). Binding to zyxin may provide a specific mechanism for localization of Ena and VASP to focal adhesions that may permit Ena/VASP proteins to direct actin assembly to sites of contact between the cell and the extracellular environment. Consistent with this is our report that Ena, like VASP, is localized to focal adhesions and our observation that Ena colocalizes with endogenous zyxin when expressed in cultured mammalian cells.
We have also shown that zyxin binding is disrupted by the A97V mutation identified in the EVH1 domain of Ena in ena210. This alanine is conserved between Drosophila Ena and VASP, as well as in murine Ena, EVL, and Drosophila AE33 (Haffner et al., 1995; DeMille et al., 1996; Gertler et al., 1996; Symons et al., 1996). Taken together, these data suggest that this alanine is important for the overall structure of the EVH1 domain and that this domain is critical for Ena function. A Drosophila protein related to zyxin has been identified (our unpublished results). It will be interesting to determine whether this protein is a ligand for the Ena EVH1 domain, and further studies will be necessary to determine this. However, because this Drosophila protein has not yet been purified and antibodies directed against it are not available, it is not presently possible to test this possibility. It is also possible that Ena may bind a different Drosophila protein through this same highly conserved, critical binding domain. In fact, in addition to zyxin, VASP and Mena have also been shown to bind to both vinculin and bacterial Act A through the EVH1 domain, bolstering the argument that multiple proteins may interact with this domain to influence cytoskeletal assembly (Gertler et al., 1996; Reinhard et al., 1996). Efforts are currently under way to definitively identify all the potential binding partners for this critical domain of Drosophila Ena. Thus, although it is not yet clear that Ena/VASP proteins share the same binding specificity, our in vivo rescue data suggest that the EVH1 domains from these proteins recognize an overlapping set of proteins in vivo.
A second Ena mutation was identified that results in truncation of the Ena EVH2 domain. Although several potential functions have been proposed for this conserved protein domain, we provide compelling in vitro evidence that it may be involved in multimer formation. This function of the EVH2 domain is conserved among family members, because Ena also forms multimers with VASP, and this multimerization requires the Ena EVH2 domain. Additionally, loss of the EVH2 domain reduces Ena’s ability to bind to zyxin and to the Abl SH3 domain in vitro. Because the zyxin binding domain has already been mapped to the EVH1 domain (Gertler et al., 1996; Niebuhr et al., 1997) and the Abl-SH3 binding domain has been mapped to the Ena proline-rich sequences (Gertler et al., 1995), we propose that this decrease in zyxin and Abl-SH3 binding may be due to a loss of ability to multimerize.
It is possible that removal of the Ena EVH2 domain disrupts the overall structure of the Ena protein, thus resulting in the reduction of zyxin and SH3 binding. This seems unlikely, however, in light of several observations. First, truncated forms of Ena lacking the EVH2 domain are expressed stably in both S2 cells and in Drosophila and are efficiently phosphorylated by the Abl tyrosine kinase (Gertler et al., 1995). Second, Gertler et al. (1996) have shown that an isolated EVH1 domain is able to localize to focal adhesions in cultured cells when injected as a GST fusion protein, suggesting that removal of the Mena C terminus does not grossly alter the structure of the EVH1 domain (Gertler et al., 1996). Because GST domains are reported to form multimers (Reinemer et al., 1991), it is possible that GST-mediated multimerization may have compensated for the lack of EVH2 domain in the GST-EVH1 fusion protein in these experiments (Gertler et al., 1996). Thus, we propose that loss of the EVH2 domain may reduce binding to Ena ligands through a loss of ability to multimerize. This would be similar to the effect of multimerization on the ligand-binding properties of both vitronectin and syndecan-4 (Oh et al., 1997; Seiffert, 1997).
Zyxin binding is thought to be a mechanism for localizing Ena/VASP proteins to focal adhesions. Thus, multimerization of Ena/VASP proteins may be necessary for proper subcellular localization of these proteins to focal adhesions and the actin cytoskeleton. This would be consistent with results demonstrating that removal of the C-terminal 100 amino acids of VASP, which includes the EVH2 domain, results in an absence of VASP in the focal adhesions (Haffner, et al. 1995). Interestingly, our results indicate that both the EnaA97V and the EnaK636Stop mutants show a rather diffuse intracellular localization and were clearly absent from focal contacts when expressed in mammalian cells.
Localization of the Ena/VASP family of proteins at focal adhesions places them at cellular structures where bidirectional signal transduction takes place (Machesky, 1997). Thus, it is of particular interest that in addition to being cytoskeletal proteins, this family of proteins represents docking sites and substrates for signal transduction molecules. Several kinases phosphorylate Ena, Mena, and VASP, indicating that multiple signaling pathways act on these proteins to regulate cytoskeleton assembly (Halbrugge, et al., 1990; Gertler et al., 1996; Comer et al., 1997). A biological function has been demonstrated for the Abl-mediated phosphorylation of Drosophila Ena, because mutation of the Ena tyrosine phosphorylation sites to phenylalanine partially impairs the ability of Ena to restore viability to Ena mutant animals (Comer et al., 1997). It is interesting to note that this unphosphorylatable form of Ena is only partially impaired in function and does retain some ability to rescue Ena mutants in light of the fact that VASP, a protein that is not a substrate of Abl, can also partially rescue a loss of ena function in Drosophila. Perhaps phosphorylation by Abl is only most important for fine tuning of Ena function. Thus, VASP, like the unphosphorylatable form of Ena, would substitute for Ena’s basic functions, such as profilin and zyxin binding, ability to multimerize, and proper subcellular localization. However, it would be unresponsive to signals from Abl that more subtly regulate its activity and would thus not able to rescue as well as a wild-type Ena protein.
Previous studies have demonstrated that two members of the growing Ena/VASP family of proteins, VASP and Mena, are linked to cytoskeletal assembly (Machesky, 1997). We now demonstrate the functional conservation of these proteins to Drosophila Ena by the ability of human VASP to partially compensate for a loss of Drosophila Ena in vivo and the identification of lethal Ena mutations in the most conserved domains of the Ena/VASP family. Despite their striking similarities, it is clear that Ena/VASP proteins regulate multiple distinct cellular processes and respond to different signal transduction pathways because VASP is involved in regulation of platelet adhesion and aggregation, and Ena and Mena are associated with neural development and function. We propose that these proteins could provide a bridge between signal transduction and the cytoskeleton. Further study of this new family of proteins promises to provide new insights into the regulation of cytoskeleton dynamics during development.
ACKNOWLEDGMENTS
We thank F. Fogerty, G. Panganiban, and M. Reinhard for critical reading of the manuscript and Ping Hua for excellent technical assistance. This work was supported by National Institutes of Health grant CA49582 (to F.M.H.), NIH grant GM50877 and a Faculty Research Award from the American Cancer Society (to M.C.B.), and a grant from the Deutsche Forchungsgemeinschaft (to U.W.). Postdoctoral fellowship support was provided by NIH postdoctoral training grant CA09681 (to S.M.A.-D. and A.R.C.). S.M.A. was supported by a fellowship from the National Cancer Institute, and A.R.C. is a Leukemia Society postdoctoral fellow (5156-94).
REFERENCES
- Arber S, Caroni P. Specificity of single LIM motifs in targeting and LIM/LIM interactions in situ. Genes Dev. 1996;10:289–300. doi: 10.1101/gad.10.3.289. [DOI] [PubMed] [Google Scholar]
- Bennett RL, Hoffmann FM. Increased levels of the Drosophila Abelson tyrosine kinase in nerves and muscles: subcellular localization and mutant phenotypes imply a role in cell-cell interactions. Development. 1992;116:953–966. doi: 10.1242/dev.116.4.953. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Chakraborty T, et al. A focal adhesion factor directly linking intracellularly motile Listeria monocytogenes and Listeria ivanovii to the actin-based cytoskeleton of mammalian cells. EMBO J. 1995;14:1314–1321. doi: 10.1002/j.1460-2075.1995.tb07117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien C-T, Bartel PL, Sternglanz R, Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comer AR, Ahern-Djamali SM, Juang J-L, Jackson PD, Hoffmann FM. Phosphorylation of Enabled by the Drosophila Abelson tyrosine kinase regulates the in vivo function and protein:protein interactions of Enabled. Mol Cell Biol. 1997;18:152–160. doi: 10.1128/mcb.18.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damsky C, Sutherland A, Fisher S. Extracellular matrix 5: adhesive interactions in early mammalian embryogenesis, implantation and placentation. FASEB J. 1993;7:1320–1329. doi: 10.1096/fasebj.7.14.8224605. [DOI] [PubMed] [Google Scholar]
- DeMille MM, Kimmel BE, Rubin GM. A Drosophila gene regulated by rough and glass shows similarity to Ena and VASP. Gene. 1996;183:103–108. doi: 10.1016/s0378-1119(96)00506-9. [DOI] [PubMed] [Google Scholar]
- Deng WP, Nickoloff JA. Site-directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal Biochem. 1992;200:81–88. doi: 10.1016/0003-2697(92)90280-k. [DOI] [PubMed] [Google Scholar]
- Derry JMJ, Kerns JA, Weinberg KI, Ochs HD, Volpini V, Estivill X, Walker AP, Francke U. WASP gene mutations in Wiskott-Aldrich syndrome and X-linked thrombocytopenia. Hum Mol Genet. 1995;4:1127–1135. doi: 10.1093/hmg/4.7.1127. [DOI] [PubMed] [Google Scholar]
- Gertler FB, Bennett RL, Clark MJ, Hoffmann FM. Drosophila abl tyrosine kinase in embryonic CNS axons: a role in axonogenesis is revealed through dosage-sensitive interactions with disabled. Cell. 1989;58:103–113. doi: 10.1016/0092-8674(89)90407-8. [DOI] [PubMed] [Google Scholar]
- Gertler FB, Comer AR, Juang J-L, Ahern SM, Clark MJ, Liebl EC, Hoffmann FM. Enabled, a dosage-sensitive suppressor of mutations in the Drosophila Abl tyrosine kinase, encodes an Abl substrate with SH3-binding properties. Genes Dev. 1995;9:521–533. doi: 10.1101/gad.9.5.521. [DOI] [PubMed] [Google Scholar]
- Gertler FB, Doctor JS, Hoffmann FM. Genetic suppression of mutations in the Drosophila abl proto-oncogene homolog. Science. 1990;248:857–860. doi: 10.1126/science.2188361. [DOI] [PubMed] [Google Scholar]
- Gertler FB, Hill KK, Clark MJ, Hoffmann FM. Dosage-sensitive modifiers of Drosophila abl tyrosine kinase function: prospero, a regulator of axonal outgrowth, and disabled, a novel tyrosine kinase substrate. Genes Dev. 1993;7:441–453. doi: 10.1101/gad.7.3.441. [DOI] [PubMed] [Google Scholar]
- Gertler F, Niebuhr K, Reinhard M, Wehland J, Soriano P. Mena, a relative of VASP and Drosophila Enabled, is implicated in the controlof microfilament dynamics. Cell. 1996;87:227–239. doi: 10.1016/s0092-8674(00)81341-0. [DOI] [PubMed] [Google Scholar]
- Golsteyn RM, Beckerle MC, Koay T, Friederich E. Structural and functional similarities between the human cytoskeletal protein, zyxin, and the ActA protein of Listeria monocytogenes. J Cell Sci. 1997;110:1893–1906. doi: 10.1242/jcs.110.16.1893. [DOI] [PubMed] [Google Scholar]
- Haffner C, Jarchau T, Reinhard M, Hoppe J, Lohmann SM, Walter U. Molecular cloning, structural analysis and functional expression of the proline-rich focal adhesion and microfilament-associated protein VASP. EMBO J. 1995;14:19–27. doi: 10.1002/j.1460-2075.1995.tb06971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbrugge M, Friedrich C, Eigenthaler M, Schanzenbacher P, Walter U. Stoichiometric and reversible phosphorylation of a 46-kDa protein in human platelets in response to cGMP and cAMP-elevating vasodilators. J Biol Chem. 1990;265:3088–3093. [PubMed] [Google Scholar]
- Hill KK, Bedian V, Juang J-J, Hoffmann FM. Genetic interactions between the Drosophila Abelson (Abl) tyrosine kinase and Failed Axon Connevtions (Fax), a novel protein in axon bundles. Genetics. 1995;141:595–606. doi: 10.1093/genetics/141.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolluri R, Shehabeldin A, Peacocke M, Lamhonwah AM, Teichert-Kuliszewska K, Weissman SM, Siminovitch KA. Identification of WASP mutations in patients with Wiskott-Aldrich syndrome and isolated thrombocytopenia reveals allelic heterogeneity at the WAS locus. Hum Mol Genet. 1995;4:1119–1126. doi: 10.1093/hmg/4.7.1119. [DOI] [PubMed] [Google Scholar]
- Kwan S-P, Hagemann TL, Radtke BE, Blaese RM, Rosen FS. Identification of mutations in the Wiskott-Aldrich syndrome gene and characterization of a polymorphic dinucleotide repeat at DXS6940, adjacent to the disease gene. Proc Natl Acad Sci USA. 1995;92:4706–4710. doi: 10.1073/pnas.92.10.4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM. Cell motility: complex dynamics at the leading edge. Curr Biol. 1997;7:R164–R167. doi: 10.1016/s0960-9822(97)70079-4. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- Montell DJ. Moving right along: regulation of cell migration during Drosophila development. Trends Genet. 1994;10:59–62. doi: 10.1016/0168-9525(94)90150-3. [DOI] [PubMed] [Google Scholar]
- Niebuhr K, Ebel F, Frank R, Reinhard M, Domann E, Carl UD, Walter U, Gertler FB, Wehland J, Chakraborty T. A novel proline-rich motif present in ActA of Listeria monocytogenes and cytoskeletal proteins is the ligand for the EVH1 domain, a protein module present in the Ena/VASP family. EMBO J. 1997;16:5433–5444. doi: 10.1093/emboj/16.17.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh ES, Woods A, Couchman JR. Multimerization of the cytoplasmic domain of syndecan-4 is required for its ability to activate protein kinase C. J Biol Chem. 1997;272:11805–11811. doi: 10.1074/jbc.272.18.11805. [DOI] [PubMed] [Google Scholar]
- Pistor S, Chakraborty T, Walter U, Wehland J. The bacterial actin nucleator protein ActA of Listeria monosytogenes contains multiple binding sites for host microfilament proteins. Curr Biol. 1995;5:1–10. doi: 10.1016/s0960-9822(95)00104-7. [DOI] [PubMed] [Google Scholar]
- Pollard T. Actin cytoskeleton. Missing link for intracellular bacterial motility? Curr Biol. 1995;5:837–840. doi: 10.1016/s0960-9822(95)00167-9. [DOI] [PubMed] [Google Scholar]
- Reinemer P, Dirr HW, Ladenstein R, Schaffer J, Gallay O, Huber R. The three-dimensional structure of class p glutathione S-transferase in complex with glutathione sulfonate at 2.3 A resolution. EMBO J. 1991;10:1997–2005. doi: 10.1002/j.1460-2075.1991.tb07729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard M, Giehl K, Abel K, Haffner C, Jarchau T, Hoppe V, Jockusch BM, Walter U. The proline-rich focal adhesion and microfilament protein VASP is a ligand for profilins. EMBO J. 1995a;14:1583–1585. doi: 10.1002/j.1460-2075.1995.tb07146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard M, Halbrugge M, Scheer U, Wiegand C, Jockusch BM, Walter U. The 46/50 kDa phosphoprotein VASP purified from human platelets is a novel protein associated with actin filaments and focal contacts. EMBO J. 1992;11:2063–2070. doi: 10.1002/j.1460-2075.1992.tb05264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard M, Jouvenal K, Tripier D, Walter U. Identification, purification, and characterization of a zyxin-related protein which binds the focal adhesion and microfilament protein VASP. Proc Natl Acad Sci USA. 1995b;92:7956–7960. doi: 10.1073/pnas.92.17.7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard M, Rudiger M, Jockusch BM, Walter U. VASP interaction with vinculin: a recurring theme of interactions with proline-rich motifs. FEBS Lett. 1996;399:103–107. doi: 10.1016/s0014-5793(96)01295-1. [DOI] [PubMed] [Google Scholar]
- Rubinfeld B, Souza B, Albert I, Muller O, Chamberlain SH, Masiarz FR, Munemitsu S, Polakis P. Association of the APC gene product with beta-catenin. Science. 1993;262:1731–1734. doi: 10.1126/science.8259518. [DOI] [PubMed] [Google Scholar]
- Sadler I, Crawford AW, Michelson JW, Beckerle MC. Zyxin and cCRP: two interactive LIM domain proteins associated with the cytoskeleton. J Cell Biol. 1992;119:1573–1587. doi: 10.1083/jcb.119.6.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazuka T, Tomooka Y, Kathju S, Ikawa Y, Noda M, Kumar S. Identification of a developmentally regulated gene in the mouse central nervous system which encodes a novel proline rich protein. Biochim Biophys Acta. 1992;1132:240–247. doi: 10.1016/0167-4781(92)90156-t. [DOI] [PubMed] [Google Scholar]
- Schmeichel KL, Beckerle MC. The LIM domain is a modular protein binding interface. Cell. 1994;79:211–219. doi: 10.1016/0092-8674(94)90191-0. [DOI] [PubMed] [Google Scholar]
- Seiffert D. The glycosaminoglycan binding site governs ligand binding to the somatomedin B domain of vitronectin. J Biol Chem. 1997;272:9971–9978. doi: 10.1074/jbc.272.15.9971. [DOI] [PubMed] [Google Scholar]
- Su L-K, Vogelstein B, Kinzler KW. Association of the APC tumor suppressor protein with catenins. Science. 1993;262:1734–1737. doi: 10.1126/science.8259519. [DOI] [PubMed] [Google Scholar]
- Symons M, Derry JDJ, Karlak B, Jiang S, Lemahieu V, McCormick FFU, Abo A. Wiskott-Aldrich syndrome protein, a novel effector for the GTPase CDC42Hs, is implicated in actin polymerization. Cell. 1996;84:723–734. doi: 10.1016/s0092-8674(00)81050-8. [DOI] [PubMed] [Google Scholar]
- Tanaka E, Sabry J. Making the connection: cytoskeletal rearrangements during growth cone guidance. Cell. 1995;83:171–176. doi: 10.1016/0092-8674(95)90158-2. [DOI] [PubMed] [Google Scholar]
- Villa A, et al. X-linked thrombocytopenia and Wiskott-Aldrich syndrome are allelic diseases with mutations in the WASP gene. Nat Genet. 1995;9:414–417. doi: 10.1038/ng0495-414. [DOI] [PubMed] [Google Scholar]
- Wengler GS, Notarangelo LD, Berardelli S, Pollonni G, Mella P, Fasth A, Ugazio AG, Parolini O. High prevalence of nonsense, frame-shift and splice-site mutations in 16 patients with full-blown Wiskott-Aldrich syndrome. Blood. 1995;86:3684–3654. [PubMed] [Google Scholar]
- Zhu Q, Zhang M, Blaese RM, Derry JMJ, Junker A, Francke U, Chen SH, Ochs HD. The Wiskott-Aldrich syndrome and X-linked congenital thrombocytopenia are caused by mutations of the same gene. Blood. 1995;86:3797–3804. [PubMed] [Google Scholar]