Abstract
The Chk2 protein kinase protects genome integrity by promoting cell cycle arrest or apoptosis in response to DNA double-strand breaks, and Chk2 mutations are found in both familial and sporadic cancers. Exposure of cells to ionizing radiation (IR) or radiomimetic drugs induces Chk2 phosphorylation by ATM, followed by Chk2 oligomerization, auto-/transphosphorylation, and activation. Here we demonstrate that Chk2 is ubiquitinated upon activation and that this requires Chk2 kinase activity. Serine 379 (S379) was identified as a novel IR-inducible autophosphorylation site required for ubiquitination of Chk2 by a Cullin 1-containing E3 ligase complex. Importantly, S379 was required for Chk2 to induce apoptosis in cells with DNA double-strand breaks. Thus, auto-/transphosphorylation of S379 is required for Chk2 ubiquitination and effector function.
The integrity of chromosomal DNA is continuously monitored throughout the cell division cycle. DNA damage activates signaling pathways called checkpoints that induce cell cycle arrest or apoptosis. Defects in checkpoint control can result in genomic instability and cancer predisposition (15). The Chk2 protein kinase is a key mediator of the DNA damage checkpoint that responds to DNA double-strand breaks (DSBs) (7, 8, 31). DNA DSBs induce the activation of ATM (ataxia telangiectasia mutated), which subsequently phosphorylates Chk2 on T68 within the N-terminal SQ/TQ cluster domain (3, 32, 33). T68 phosphorylation is required for efficient oligomerization and subsequent auto-/transphosphorylation of Chk2 at other regulatory sites, including T383/387, S456, and S516 (2, 22, 25, 41, 53).
Heterozygous germ line mutations in CHK2 are associated with a p53-independent variant form of the Li-Fraumeni heritable cancer predisposition syndrome (6). Cells derived from Chk2-deficient mice exhibit defects in their ability to delay entry into S phase, sustain a G2 cell cycle arrest, and undergo apoptosis in response to DNA damage (16, 46). These effects have been attributed to a failure of p53 to be properly modified in Chk2-deficient cells. Chk2 phosphorylates p53 on S20 in vitro. S20 phosphorylation has been proposed to mediate p53 stabilization by preventing interaction of p53 with its negative regulator MDM2; however, the exact function for this phosphorylation event remains unclear (9, 10, 17, 20, 42, 52). In addition to p53, additional substrates of Chk2 include Brca1, MDMX2, PML, p73, and E2F1 (11, 26, 37, 45, 47, 54).
The covalent modification of cellular proteins by ubiquitin is best known for its role in mediating their proteasome-dependent proteolysis. However, recent studies have demonstrated that ubiquitination can also modify the enzymatic activity, trafficking, localization, and binding partners of the modified protein (21, 40). Here we show that Chk2 phosphorylates itself on S379 in response to DNA damage. This in turn, facilitates Chk2 ubiquitination by an E3 ligase complex containing Cullin 1 (Cul 1). Ubiquitination of Chk2 did not detectably alter its stability. Importantly, S379 was shown to play an important role in the ability of Chk2 to induce apoptosis in cells with DNA DSBs. Thus, ubiquitination of Chk2 may regulate Chk2 effector function.
MATERIALS AND METHODS
Antibodies and Western blotting.
Endogenous Chk2 was detected either with a mouse monoclonal antibody (Upstate, clone 7) or with a rabbit polyclonal antibody raised against bacterially produced glutathione S-transferase (GST)-Chk2 (14). Flag3-Chk2 fusion proteins were precipitated with anti-Flag M2 antibody-agarose affinity gel (Sigma Chemical Co.) and detected by Western blotting with anti-Flag M2 monoclonal antibody (Sigma Chemical Co.). Myc3-Chk2 fusion proteins were immunoprecipitated with anti-Myc mouse monoclonal 9E10-conjugated agarose (Santa Cruz Biotechnology) and detected by Western blotting with anti-c-Myc A14 rabbit polyclonal antibody (Santa Cruz Biotechnology). Bound primary antibodies were detected with horseradish peroxidase (HRP)-conjugated goat anti-mouse (ICN/CAPPEL), HRP-goat anti-mouse (Jackson Laboratories), HRP-donkey anti-goat (Santa Cruz Biotechnology), or HRP-goat anti-rabbit (Zymed) secondary antibodies, and proteins were visualized using ECL (GE Healthcare Life Sciences). Antibodies specific for Chk2 phosphorylated on S379 were generated by immunization of rabbits with the phosphopeptide CILGET-pS-LMR coupled to keyhole limpet hemocyanin. Antibodies specific for Chk2 phosphorylated on T68, T383/387, and S516 have been described previously (41). Ubiquitination assays carried out in vitro were probed with a ubiquitin-specific antibody (P4D1; Santa Cruz Biotechnology).
Cell culture and treatment of cells with DNA-damaging agents.
HEK-293 and U2OS cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% bovine growth serum, 100 units/ml (each) penicillin and streptomycin, and 1 mM l-glutamine (complete DMEM). Chk2 null mouse embryo fibroblasts (MEFs) (16) were maintained in DMEM supplemented with 10% fetal bovine serum, 100 units/ml penicillin and streptomycin, 2 mM l-glutamine, and 0.1 mM nonessential amino acids. In some cases, cells were exposed to 10 Gy from a 60Co source or were treated with complete DMEM containing 20 μM etoposide (VP16; Sigma Chemical Company).
Plasmids.
pET15b Chk2-WT (wild type) and -D368N; pcDNA3-Myc3Chk2-WT and -D368N; and pcDNA3-Flag3Chk2-WT, -D368N, -T68A, -T383/387A, and -S516A have been described previously (41). pET15b-Chk2-S379A was generated by site-directed mutagenesis of pET15b-Chk2-WT using the QuikChange site-directed mutagenesis kit (Stratagene) and the primers 5′-TCCAAGATTTTGGGAGAGACCGCTCTCATGAGAACCT-3′ (forward) and 5′-GGTTCCACATAAGGTTCTCATGAGAGCGGTCTCTCCC-3′ (reverse). The same primers were used to generate pcDNA3-Flag3-Chk2-S379A and pcDNA3-Myc3-Chk2-S379A by site-directed mutagenesis of pcDNA3-Flag3-Chk2-WT and pcDNA3-Myc3-Chk2-WT vectors, respectively. pcDNA5/FRT-Myc3-Chk2-WT was generated by inserting the KpnI/XhoI fragment of pcDNA3-Myc3-Chk2-WT encoding Myc3-Chk2 into the corresponding sites of pcDNA5/FRT (Invitrogen). pcDNA5/FRT-Myc3-Chk2-D368N and pcDNA5/FRT-Myc3-Chk2-S379A were generated in a similar manner. Sequences of all mutants were verified by DNA sequencing. Plasmids encoding Myc-tagged Cullin proteins and dominant-negative forms of Cul 1 and Cul 3 have been described previously (12, 36, 43). The first 393 amino acids of Cul 4B were cloned into the KpnI/XhoI-digested pcDNA3-Flag3 vector to generate a dominant-negative Cul 4B protein.
To generate pcDNA3-TAP-Chk2 constructs, pcDNA3-Flag3 was used to generate the backbone pcDNA3-TAP vector by sequential cloning of the protein A binding domain followed by two TEV protease cleavage sites upstream of the Flag3-coding sequence. Sequences encoding the protein A binding domain were amplified by PCR using the primers 5′-CCCAAGCTTATGGCGATTAAGGGTGAAGCTCA and 5′-GGGGTACCTTCTTTGCTCACCGAAGGATCG. The product was cloned into HindIII/KpnI-digested pcDNA3-Flag3. Oligonucleotides encoding the two TEV protease cleavage sites (5′-GGAATTCCGAAAATCTATATTTTCAAGGTACTGCTTCTCAGCACGAAAATCTATATTTTCAAGGTGAACTAGATATCCAG and its complement) were annealed and cloned into EcoRI/EcoRV-digested pcDNA3-protein A-Flag3, positioning the TEV cleavage sites between the protein A binding domain and the Flag3-coding sequence to generate pcDNA3-TAP-Chk2 (WT, D368N, and S379A), which was then cloned into BsiWI/EcoRV-digested pcDNA3-TAP.
Chk2 ubiquitination in vivo.
HEK-293 cells were cotransfected with plasmids encoding Flag3-tagged Chk2 (WT and mutants) and His6 Myc-ubiquitin using Superfect reagent (Qiagen). At 42 h posttransfection, cells were washed once with ice-cold phosphate-buffered saline and lysed in mammalian cell lysis buffer (MCLB) (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, 5 mM EDTA, 0.5% Nonidet P-40, and 2 mM dithiothreitol [DTT]) containing 1 mM sodium orthovanadate, 10 mM β-glycerophosphate, 1 μM microcystin, 2 mM phenylmethylsulfonyl fluoride (PMSF), 10 μg/ml aprotinin, and 5 μg/ml leupeptin. Ectopically expressed Chk2 was immunoprecipitated with anti-Flag M2 monoclonal antibody prebound to agarose (Sigma Chemical Co.). Immunoprecipitates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose, and immunoblotted with polyclonal antibodies specific for either the Myc epitope (Santa Cruz Biotechnology) or Chk2 (14).
Stability of endogenous and ectopic Chk2.
Asynchronously growing HEK-293 cells were treated with 20 μg/ml cycloheximide (Sigma Chemical Co.) in complete DMEM. Cells were then either mock irradiated or exposed to 10 Gy ionizing radiation (IR) and harvested at various times post-IR. Cell lysates were prepared in MCLB, resolved by SDS-PAGE, and transferred to nitrocellulose. For phosphatase treatment, cells were lysed in buffer containing 50 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.5% NP-40, 2 mM DTT, 1 mM PMSF, 5 μg/ml leupeptin, and 10 μg/ml aprotinin. Cleared lysates were incubated with λ phosphatase (1,200 units per mg of total cellular protein; New England Biolabs) at 30°C for 1 h prior to SDS-PAGE and Western blotting. Proteins were visualized using the ECL reagent (GE Healthcare Life Sciences), and densitometry analysis was performed using Image J software (1).
HEK-293 cells were transfected with plasmids encoding Flag3-tagged Chk2 (WT, D368N, and S379A) using Superfect transfection reagent (Qiagen). At 48 h posttransfection, cells were treated with 20 μg/ml cycloheximide and harvested at various time points. Cell lysates were prepared in MCLB and analyzed by Western blotting using anti-Flag antibody. Densitometry analysis was performed as described above.
Two-dimensional phosphopeptide mapping.
HEK-293 cells (1.5 × 106) were either mock transfected or transfected with plasmids encoding Flag3-tagged wild-type and mutant forms of Chk2 using Superfect transfection reagent (Qiagen). At 42 hours posttransfection, cells were incubated in phosphate-free DMEM containing 2.5 mCi of 32P-labeled inorganic phosphate per ml (Perkin-Elmer Life Sciences) and 25 μM VP16 for 2 h. Cells were washed once with ice-cold phosphate-buffered saline and lysed in MCLB containing 2 mM DTT, 1 μM microcystin, 1 mM PMSF, 5 μg/ml leupeptin, and 10 μg/ml aprotinin. Ectopically expressed Chk2 was immunoprecipitated with anti-Flag M2 monoclonal antibody prebound to agarose (Sigma Chemical Co.). Immunoprecipitates were subjected to SDS-PAGE on an 8% polyacrylamide gel, and proteins were transferred to nitrocellulose. Radiolabeled Chk2 was digested in a solution containing 0.2 mg/ml trypsin (Roche Applied Science) in 50 mM ammonium bicarbonate for 15 h. Tryptic phosphopeptides were separated in the first dimension by electrophoresis and in the second dimension by ascending chromatography in 200 ml of buffer consisting of 75 ml n-butanol, 50 ml pyridine,15 ml acetic acid, and 60 ml water (48).
Purification of bacterially produced Chk2 protein.
Escherichia coli strain BL21 was transformed with pET15b plasmids encoding His6-Chk2-WT, His6-Chk2-D368N, and His6-Chk2-S379A. Cultures were grown at 37°C to an optical density at 600 nm of 0.5, at which time isopropyl-1-thio-d-galactopyranoside (IPTG) was added to a final concentration of 0.5 mM and protein expression was induced at 37°C for 4 h. Bacterial pellets were resuspended in NP-40 lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 5 mM EGTA, 0.5% NP-40) supplemented with protease inhibitors (2 mM PMSF, 0.15 unit/ml of aprotinin, and 20 μM leupeptin), 1 μM microcystin, and 2 mg/ml lysozyme and then rocked at 4°C for 30 min. Cells were lysed by sonication (50% duty cycle for 20 bursts). Lysates were clarified by centrifugation (14,000 × g for 15 min). His6-tagged proteins were precipitated by incubation with Ni2+-nitrilotriacetic acid agarose (Qiagen) for 2 h at 4°C. Precipitated proteins were washed two times in NP-40 lysis buffer and two times in lysis buffer without NP-40. Bound proteins were eluted in elution buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1 mM DTT) containing 100 mM imidazole and dialyzed overnight at 4°C in dialysis buffer (50 mM Tris-HCl [pH 7.4], 2 mM DTT). Purified His-Chk2 proteins were concentrated using Vivaspin columns (Vivascience, Inc.).
Generation of U2OS/FRT stables lines.
The Flp-In system (Invitrogen) was used in order to create U2OS cell lines that stably express Myc3-tagged Chk2 proteins. U2OS cells were first stably transfected with pFRT/lacZeo using Lipofectamine 2000 (Invitrogen), and transfected colonies were selected in medium containing 50 μg/ml Zeocin (Invitrogen). Single colonies were grown up, their β-galactosidase activity was measured, and Southern analysis was performed to determine the number of FLP recombination target (FRT) sites inserted into the genome. The clone selected, U2OS/FRT16, had only one FRT site inserted into the genome. This clone was then cotransfected with pOG44, a vector for transient transfection of the Flp recombinase, and with pcDNA5/FRT-Myc3-Chk2-WT, -Myc3-Chk2-D368N, or -Myc3-Chk2-S379A using Lipofectamine 2000. The pcDNA5/FRT-Myc3-Chk2 vector contains the coding region for Chk2 (wild type or point mutants) and possesses an FRT site for homologous recombination. After insertion of the Chk2 gene into the FRT site, Chk2 transcription is driven by the human cytomegalovirus immediate-early enhancer/promoter. The pcDNA5/FRT vector also carries a hygromycin resistance gene to allow for selection of stable cells in 100 μg/ml hygromycin (Invitrogen). Homologous recombination of Myc3-Chk2 into the FRT site inactivates the lacZ-Zeocin fusion gene, and stable cells are no longer Zeocin resistant. Hygromycin-resistant colonies were grown as pools in DMEM containing 10% fetal bovine serum, 100 μg/ml hygromycin, 100 units/ml (each) penicillin and streptomycin, and 1 mM l-glutamine.
Phosphorylation experiments with Chk2 null MEFs.
Chk2 null MEFs were transfected with Flag3-Chk2 (WT, D368N, or S379A). At 44 h posttransfection, cells were harvested and lysed in MCLB. Lysates were immunoprecipitated with anti-Flag agarose for 2 h at 4°C. Immunoprecipitates were subsequently washed three times with MCLB. Bound proteins were released by boiling in SDS loading buffer, and then each immunoprecipitate was divided equally into three for resolution by SDS-PAGE. The resulting SDS gels were transferred to nitrocellulose and visualized with the indicated anti-phospho-Chk2 antibodies. The anti-phospho-Chk2 blots were then stripped and reprobed with an antibody specific for the Flag epitope.
Chk2 kinase assays.
U2OS/FRT16-Myc3-Chk2-WT, -D368N, and -S379A stable lines were either mock or gamma irradiated with 10 Gy IR. At 30 min post-IR, cells were harvested and lysed in MCLB containing protease and phosphatase inhibitors. Chk2 proteins were immunoprecipitated with anti-c-Myc agarose. The immunoprecipitates were washed three times with MCLB and once with incomplete kinase buffer (50 mM Tris HCl [pH 7.4], 1 mM DTT, 10 mM MgCl2). Kinase reactions with 50-microliter reaction mixtures were carried out in the presence of incomplete kinase buffer containing 10 μM ATP, 10 μCi of [γ32P]ATP (Perkin-Elmer Life Sciences), and 5 μg of soluble GST-Cdc25C(200-256) (34). Reaction mixtures were incubated at 30°C for 6 min and resolved by SDS-PAGE. Incorporated counts were quantified using a Molecular Dynamics Storm Imager (GE Healthcare Life Sciences). In the figures, error bars represent average counts of triplicate reactions performed for each mutant.
Etoposide-induced apoptosis.
Conditions for etoposide-induced apoptosis have been described previously (45). U2OS/FRT16-Myc3Chk2-WT, -D368N, and -S379A stable lines as well as parental U2OS/FRT16 cells were exposed to 20 μM etoposide for the indicated time periods. Cells were harvested at 24 and 48 h after treatment and fixed in 70% ethanol. Cells were then stained with 30 μg/ml of propidium iodide for 30 to 60 min and analyzed by flow cytometry using a Becton-Dickinson FACScan. The data were analyzed by using CELLQUEST software.
Purification of TAP-tagged Chk2 proteins.
Expression plasmids encoding TAP-tagged Chk2 proteins were transfected into HEK-293 cells using Superfect transfection reagent (Qiagen) for 48 h according to the manufacturer's instruction. Cells were lysed, and clarified lysates were incubated with rabbit immunoglobulin G (IgG) agarose (Sigma Chemical Co.) at 0.5 μg IgG agarose per mg of total cell protein for 3 h at 4°C. Bound Chk2 proteins were washed three times with MCLB followed by three times with TEV cleavage buffer (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.1% NP-40, 0.5 mM EDTA, 1 mM DTT). Washed beads were resuspended in 100 μl TEV cleavage buffer containing 10 units of TEV protease (Invitrogen) and incubated at 4°C overnight. Reaction mixtures were centrifuged at 250 × g for 3 min. Supernatants were removed and placed in a fresh tube, and pelleted IgG beads were suspended in 500 μl of MCLB, followed by centrifugation at 250 × g for 3 min. The resulting supernatants were combined with the first TEV elution, and then 50 μl of anti-Flag agarose was added and the mixture was incubated at 4°C for 2 h. Reaction mixtures were centrifuged at 250 × g for 3 min, and the precipitated Flag beads were washed five times with MCLB. Precipitates containing Flag3-Chk2 proteins were suspended in 100 μl of MCLB containing 0.35 mg/ml of Flag3 peptides (Peptide Synthesis Core Facility at Tufts University) for 1 h at 4°C to elute the Flag-tagged proteins.
Ubiquitination assays in vitro.
Ubiquitin ligase reactions were carried out in vitro as described previously (35). Purified ubiquitin, human recombinant E1 ubiquitin-activating enzyme, and E2 (UbcH5c) enzyme were purchased from Boston Biochem. TAP-tagged Chk2 proteins were purified as described above. Cullin ligase complexes were isolated from HEK-293 cells transfected with plasmids encoding Myc-Cul 1, Myc-Cul 3, or Myc-Cul 4B using Myc-agarose (Santa Cruz Biotechnology). Immunocomplexes were washed three times with MCLB and twice with ubiquitin reaction buffer (50 mM Tris-HCl [pH 7.4], 5 mM MgCl2, 2 mM NaF, 10 nM okadaic acid, 2 mM ATP, and 0.6 mM DTT). Washed immunocomplexes were added to a ubiquitin ligation reaction mixture (final volume, 30 μl) containing 15 μg purified TAP-tagged Chk2 proteins, 0.75 μg ubiquitin, 60 ng E1, and 300 ng E2 in ubiquitin reaction buffer. Reaction mixtures were incubated at 37°C for 60 min, reactions were terminated by addition of SDS loading buffer, mixtures were boiled for 5 min, and products were resolved by SDS-PAGE followed by Western blotting.
RESULTS
Autophosphorylation of Chk2 on S379 is required for Chk2 ubiquitination.
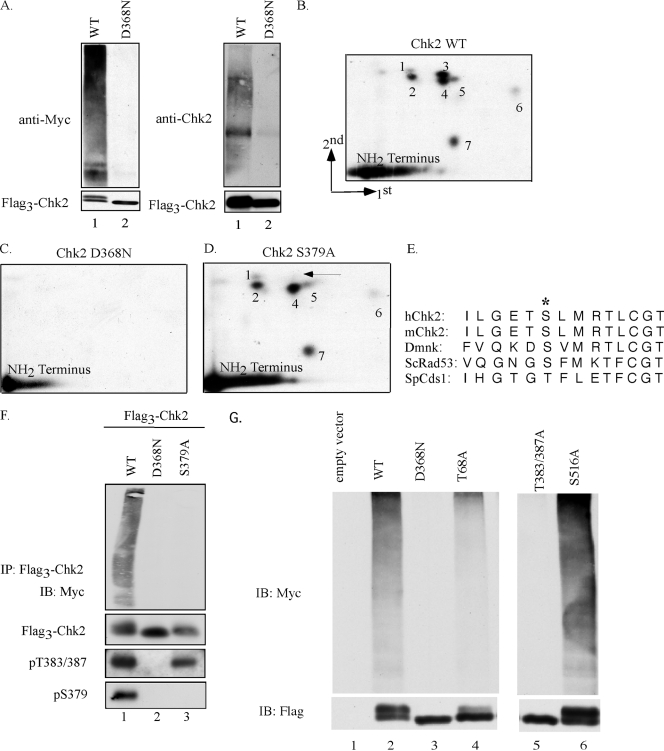
Following DNA damage, Chk2 oligomerizes in a phosphorylation-dependent manner, a process that ultimately results in its full activation (2, 41, 53). Previously we demonstrated that Chk2 oligomerizes and auto-/transphosphorylates in the absence of any exogenous DNA damage when expressed at high levels either in HEK-293 cells or in bacteria (41). As seen in Fig. 1A, a slower electrophoretic form of Chk2 was observed when wild-type (lane 1) but not kinase-inactive (lane 2) Chk2 was produced in HEK-293 cells, and this is due to Chk2 oligomerization followed by auto-/transphosphorylation (41). Strikingly, wild-type but not kinase-inactive Chk2 was ubiquitinated when coproduced with Myc-tagged ubiquitin in these cells. This was demonstrated using antibodies that recognize either the tagged form of ubiquitin (Fig. 1A, left) or Chk2 itself (Fig. 1A, right). These results demonstrate that Chk2 ubiquitination requires Chk2 kinase activity.
FIG. 1.
Chk2 autophosphorylation on S379 regulates ubiquitination. (A) HEK-293 cells were transfected with plasmids encoding Flag3-Chk2 (WT and D368N) and Myc-tagged ubiquitin. At 42 h posttransfection, cells were treated with MG132 for 2 h, and Chk2 proteins were immunoprecipitated using an antibody specific for the Flag epitope. Ubiquitinated proteins were visualized by immunoblotting with antibodies specific for either Myc (top left panel) or Chk2 (top right panel). Precipitated Flag3-Chk2 was detected with antibodies specific for Flag (bottom left panel) or Chk2 (bottom right panel). (B to D) Asynchronously growing HEK-293 cells expressing Flag3-Chk2-WT (B), Flag3-Chk2-D368N (C), or Flag3-Chk2-S379A (D) were incubated with 32P-labeled inorganic phosphate in the presence of 25 μM VP16. Radiolabeled Chk2 proteins were digested with trypsin, and peptides were subjected to two-dimensional phosphopeptide mapping. The arrow in panel D denotes the absence of a phosphopeptide that is present in the map of wild-type Chk2. (E) Alignment of sequences inclusive of and surrounding S379 in the indicated Chk2 orthologs. The asterisk denotes S379 in human Chk2 and conserved residues in other orthologs. (F) HEK-293 cells were transfected with plasmids encoding Flag3-Chk2 (WT, D368N, and S379A) and Myc-tagged ubiquitin. At 48 h posttransfection, cells were lysed and Chk2 proteins were immunoprecipitated (IP) using an antibody specific for the Flag epitope. Ubiquitinated proteins were visualized by immunoblotting (IB) with an antibody to the Myc tag. The Myc blots were then stripped and probed with an antibody specific for the Flag tag. Phosphorylation of Chk2 on T383/387 and S379 was examined on total lysates using antibodies specific for these sites. (G) HEK-293 cells were transfected with expression plasmids encoding Flag3-Chk2 (WT, D368N, T68A, T383/387A, or S516A) and Myc-tagged ubiquitin (Myc-Ub). Chk2 proteins were immunoprecipitated using an antibody specific for the Flag epitope. Ubiquitinated proteins were visualized by immunoblotting with an antibody to the Myc tag. The Myc blots were then stripped and probed with an antibody specific for the Flag tag.
To identify phosphorylation sites that might be required for Chk2 ubiquitination, HEK-293 cells were transfected with Flag3-Chk2 constructs, treated with etoposide (VP16), and radiolabeled with [32P]orthophosphate. The Flag3-tagged proteins were immunoprecipitated and used to generate tryptic phosphopeptide maps. Wild-type Chk2 gave rise to seven resolvable phosphopeptides (labeled 1 to 7 in Fig. 1B). Interestingly, all seven were absent when 32P-labeled kinase-inactive Chk2 (Chk2-D368N) was subjected to two-dimensional phosphopeptide mapping (Fig. 1C). These results demonstrate that phosphopeptides 1 to 7 require the kinase activity of Chk2. The identity of individual phosphopeptides was determined by phosphoamino acid analysis and site-directed mutagenesis. Mutation of serine 379 to alanine (S379A) resulted in loss of phosphopeptide 3 (Fig. 1D), whereas mutation of serine 516 to alanine (S516A) resulted in loss of phosphopeptide 7 (data not shown). Since the NH2 terminus of Chk2 lacks trypsin cleavage sites from amino acid 3 to 95, phosphopeptides from this region are not resolvable using this approach and are represented on the two-dimensional maps as unresolved phosphopeptides along the x axis (Fig. 1B to D). S379 was also identified as a Chk2 autophosphorylation site by mass spectroscopy (23). Interestingly, S379 is conserved in Chk2 orthologs from mice, flies, and budding yeast; a threonine residue is present in the equivalent site in the fission yeast Chk2 ortholog (Fig. 1E).
Ubiquitination assays were performed to determine if S379 was required for Chk2 ubiquitination. As seen in Fig. 1F, wild-type Chk2 was ubiquitinated in vivo (top panel, lane 1), and the kinase-inactive mutant (D368N) failed to be ubiquitinated (lane 2). Interestingly, the S379A Chk2 mutant also failed to be ubiquitinated in vivo (lane 3). In cells containing DNA DSBs, ATM initiates Chk2 activation by phosphorylating Chk2 on T68. T68 phosphorylation is required for efficient oligomerization and subsequent auto-/transphosphorylation of Chk2 at other regulatory sites, including T383/387. As seen in Fig. 1F, both wild-type Chk2 and S379A, but not kinase-inactive Chk2, were phosphorylated on T383/387. This suggests that the S379A mutant of Chk2 retains kinase activity. This conclusion is further supported by the fact that the electrophoretic mobility of S379A is slower than that of kinase-inactive Chk2 (Fig. 1F, Flag3-Chk2 panel).
Several Chk2 mutants were also expressed in HEK-293 cells to determine requirements for Chk2 ubiquitination (Fig. 1G). Two kinase-inactive mutants (D368N and T383/387A) failed to be ubiquitinated (lanes 3 and 5), demonstrating the importance of Chk2 kinase activity for ubiquitination. Wild-type Chk2, T68A, and S516A are all kinase active, although the T68A mutant fails to be activated to the same extent as wild-type Chk2 and S516A in response to DNA damage (3, 32, 33, 41, 57). As seen in Fig. 1G, wild-type Chk2, T68A, and S516A were all ubiquitinated in vivo (lanes 2, 4, and 6), and the ubiquitination of T68A was less that than that observed for wild-type Chk2 and S516A. These findings underscore the importance of both kinase activity and S379 phosphorylation for Chk2 ubiquitination.
Phosphorylation of S379 is IR inducible and requires Chk2 kinase activity.
To further characterize S379 phosphorylation, an antibody specific for Chk2 phosphorylated on this residue was generated (Fig. 1F and Fig. 2A). The antibody was then used to monitor endogenous Chk2 phosphorylation in vivo. As seen in Fig. 2B, endogenous Chk2 was inducibly phosphorylated on S379 within 30 minutes after IR treatment. Thus, S379 is a novel IR-inducible Chk2 phosphorylation site.
FIG. 2.
Phosphorylation and kinase activity of Chk2 S379A. (A) Lysates of HEK-293 cells expressing Flag3-Chk2-WT or Flag3-S379A were resolved by SDS-PAGE and blotted with an antibody specific for Chk2 phosphorylated on S379. The blot was then stripped and reprobed with an antibody specific for the Flag epitope. (B) Lysates from HEK-293 cells that had been mock irradiated (lane 1) or exposed to 10 Gy IR (lane 2) were resolved by SDS-PAGE and blotted with an antibody specific for Chk2 phosphorylated on S379 and an antibody specific for Chk2. (C) His6-Chk2-WT, -D368N, and -S379A were purified from bacteria. Matched amounts of protein were resolved by SDS-PAGE and blotted with the indicated antibodies. (D) Lysates from Chk2 null MEFs expressing Flag3Chk2-WT, -D368N, or -S379A were incubated with an antibody specific for the Flag epitope. Precipitates were resolved by SDS-PAGE and blotted with the indicated phospho-specific antibodies. The phospho-specific blots were then stripped and reprobed with an antibody specific for the Flag tag. (E) Myc3-tagged Chk2 proteins (WT and S379A) were precipitated from U2OS/FRT stable lines 30 min after mock irradiation (−) or exposure to 10 Gy IR (+). The precipitated Chk2 proteins were tested for their ability to phosphorylate GST-Cdc25C(200-256) in vitro. 32P-labeled GST-Cdc25C(200-256) was quantified using a Storm Imager. Error bars indicate the standard errors of the means for triplicate reactions performed for each Chk2 protein.
S379 is located within the activation loop of the Chk2 kinase domain. Thus, mutation of S379 could impair the kinase activity of Chk2. Therefore, S379A was tested for its ability to auto-/transphosphorylate in bacteria. As reported previously (41), overproduction of Chk2 in bacteria promotes Chk2 oligomerization and auto-/transphosphorylation (Fig. 2C, lane 1), whereas kinase-inactive Chk2 fails to be phosphorylated in bacteria (Fig. 2C, lane 2). Importantly, the S379A mutant displayed a shift in electrophoretic mobility and was phosphorylated on T68, T383/387, and S516 in bacteria (Fig. 2C, lane 3), demonstrating that mutation of S379 to alanine does not render Chk2 kinase inactive. The phosphorylation status of S379A was also monitored in CHK2 null MEFs (16) to eliminate any interactions between S379A and endogenous Chk2. As seen in Fig. 2D, wild-type Chk2 was phosphorylated on T68, T383/387, S379, and S516 (lane 1). Kinase-inactive Chk2 was phosphorylated on T68 but not on residues that require Chk2 kinase activity (T383/387 and S516). The finding that kinase-inactive Chk2 was not detectably phosphorylated on S379 demonstrates that phosphorylation of S379, like that of S516 and T383/387 but unlike that of T68, requires Chk2 kinase activity (Fig. 1F and 2D, lane 2). Notably, S379A was phosphorylated on T68, T383/387, and S516 in Chk2 null MEFs, demonstrating that S379A retains kinase activity in vivo (Fig. 2D, lane 3).
Next, kinase assays were performed to measure the activation of S379A following exposure of cells to IR (Fig. 2E). The kinase activity measured for S379A was similar to that of wild-type Chk2 in the absence of IR. IR induced an approximate 2.3-fold activation in the kinase activity of wild-type Chk2, whereas an approximate 1.5-fold activation in the kinase activity was measured for the S379A mutant. These results indicate that mutation of S379 does not inactivate the catalytic activity of Chk2 as does mutation of T383/387 that but mutation of S379 does partially impair Chk2 activation by IR. Taken together, our results demonstrate that IR induces phosphorylation of Chk2 on S379, that S379 phosphorylation is dependent on Chk2 kinase activity, and that phosphorylation of S379 is required for Chk2 ubiquitination.
Chk2 ubiquitination catalyzed by Cul 1-containing E3 ligase.
Next, experiments were performed to identify the E3 ubiquitin ligase responsible for Chk2 ubiquitination. Cullin-containing E3 ligases comprise the largest family of ubiquitin ligases in eukaryotic cells and consist of seven members (Cul 1, Cul 2, Cul 3, Cul 4A, Cul 4B, Cul 5, and Cul 7) (38). We first determined whether complexes between Chk2 and members of the Cullin family could be detected in vivo. As seen in Fig. 3A, cotransfection experiments revealed that Chk2 interacted with Cul 1, 3, and 4B (lanes 1, 3, and 5) but not Cul 2, 4A, or 5 (lanes 2, 4, and 6). Next, dominant-negative forms of Cul 1, 3, and 4B were tested for their ability to block Chk2 ubiquitination in vivo (12, 36, 43). As seen in Fig. 3B, ubiquitination of wild-type Chk2 was blocked by a dominant-negative Cul 1 mutant (lane 4) but not dominant-negative forms of Cul 3 or 4B (lanes 5 and 6). As expected, kinase-inactive Chk2 (lanes 7 to 10) and S379A (lanes 11 to 14) were not ubiquitinated (Fig. 3B) and did not associate with Cul 1 (Fig. 4A, lanes 2 and 3) in vivo. Finally, assays were performed in vitro to monitor the ability of Cul 1-, 3-, and 4B-containing complexes to catalyze Chk2 ubiquitination. As seen in Fig. 4B, only Cul 1-containing complexes ubiquitinated Chk2 in vitro (lane 4). Neither kinase-inactive Chk2 (lanes 7 to 9) nor S379A (lanes 10 to 12) was ubiquitinated in vitro. These findings indicate that a Cul 1-containing E3 ligase is responsible for Chk2 ubiquitination in vivo.
FIG. 3.
Chk2 ubiquitination by Cul 1 E3 ligase complex. (A) HEK-293 cells were transfected with plasmids encoding Flag3-Chk2-WT and the indicated Myc-tagged Cullin proteins for 48 h. Lysates were prepared and either resolved directly by SDS-PAGE (bottom panel) or first incubated with an antibody specific for the Flag epitope (top panel), and precipitates were resolved by SDS-PAGE. Western blotting was performed with Myc antibody to detect tagged Cullin proteins and with Flag antibody to detect precipitated Flag3-Chk2. (B) HEK-293 cells were transfected with plasmids encoding hemagglutinin-tagged ubiquitin (HA-Ub), Myc3-Chk2, and dominant-negative forms of Cul 1, 3, and 4B. Myc3-Chk2 was immunoprecipitated (IP), and ubiquitinated Chk2 was examined by Western blotting (IB) with HA-specific antibody. The blot was then stripped and blotted with Myc antibody. Total lysates were examined for Cullin levels by Western blotting with a Flag-specific antibody.
FIG. 4.
Chk2 is ubiquitinated in vitro by a Cul 1-containing E3 ligase. (A) HEK-293 cells were transfected with plasmids encoding Flag3-Chk2 (WT, D368N, or S379A) and Myc-Cul 1. Flag3-tagged proteins were isolated using Flag-agarose, and precipitates were examined for the presence of Myc-Cul 1 by Western blotting with antibodies specific for the Myc and Flag tags (top panel). Levels of Myc-Cul 1 were determined by Western blotting total cell lysates (bottom panel). (B) Chk2 proteins were purified as Flag-tagged proteins as described in Materials and Methods and incubated in vitro with purified ubiquitin, E1 (Ube1), and E2 (UbcH5c) in ubiquitin reaction buffer at 37°C for 1 h. Reactions were resolved by SDS-PAGE and subjected to Western blotting (IB) with antibodies specific for ubiquitin (top panel) or Flag (bottom panel).
Role of Chk2 S379 phosphorylation.
Endogenous Chk2 protein levels were monitored in both the presence and absence of DNA DSBs in HEK-293 cells (Fig. 5A). In the presence of cycloheximide, Chk2 levels remained stable for at least 2 h in mock-irradiated cells (Fig. 5A, lanes 1 to 4 and 9 to 12). IR induces Chk2 phosphorylation, which, in turn, causes Chk2 to migrate more slowly on SDS gels (Fig. 5A, lanes 5 to 8). Quantitation of Western blotting results suggested that IR also promoted Chk2 turnover. However, when cell lysates were treated with λ phosphatase prior to Western blotting (Fig. 5A, lanes 13 to 16), Chk2 was observed to be stable in IR-treated cells. These results demonstrate that the Chk2 antibody used for Western blotting inefficiently recognizes phosphorylated forms of Chk2 and that the half-life of endogenous Chk2 is not dramatically altered in cells exposed to IR. Next, the half-life of ectopically expressed Chk2 was measured in HEK-293 cells, as wild-type Chk2 is ubiquitinated when overproduced in these cells (Fig. 1A and F). HEK-293 cells expressing wild-type and mutant forms of Flag-tagged Chk2 were incubated with cycloheximide, and Chk2 levels were monitored by Western blotting using an antibody specific to the Flag tag. As seen in Fig. 5B, levels of wild-type and mutant Chk2 proteins remained stable over the 4-h incubation period.
FIG. 5.
Ubiquitination of Chk2 does not affect Chk2 stability. (A) HEK-293 cells were treated with cycloheximide and then either mock or gamma irradiated and collected at the indicated times post-IR. Lysates were incubated in the absence or presence of λ phosphatase. Chk2 protein levels were monitored by Western blotting using a Chk2 monoclonal antibody. An antibody specific for actin was used to verify similar loading in each lane. Densitometry analysis was performed using Image J software and normalized to the level of actin. Chk2 protein levels at each time point are shown relative to those in mock-irradiated samples (time zero is set at 1.0). (B) Flag3-Chk2 proteins (WT, D368N, and S379A) were expressed in HEK-293 cells. Cells were cultured in the presence of cycloheximide and collected at the indicated time points. Flag3-Chk2 protein levels were examined using an anti-Flag antibody. Densitometry analysis was performed as for panel A.
Protein ubiquitination regulates a plethora of biological activities in addition to proteosome-mediated degradation, including protein-protein interactions, enzymatic activity, and intracellular localization. Therefore, we tested whether S379 phosphorylation was required for Chk2 signaling. Chk2 is a critical regulator of DNA damage-induced apoptosis, particularly in response to agents that induce DNA DSBs (16, 19, 45, 46, 51, 54). To test whether S379 phosphorylation was required for Chk2-mediated apoptosis, U2OS cells stably expressing wild-type and mutant forms of Chk2 in the same genetic locus were generated. A FRT site was first introduced into U2OS cells to generate the U2OS/FRT parental line. Plasmids encoding Myc3-Chk2-WT, Myc3-Chk2-D368N, and Myc3-Chk2-S379A were then cotransfected with the Flp recombinase from S. cerevisiae to generate the U2OS/FRT stable cell lines expressing Myc3-Chk2 (WT and mutants) at the same locus. Densitometry analysis revealed that the Myc3-Chk2 proteins were expressed at approximately twofold the level of endogenous Chk2 in the U2OS/FRT lines (Fig. 6A).
FIG. 6.
Phosphorylation of S379 in activation of Chk2 and apoptosis induction. (A) U2OS/FRT parental cells were stably transfected with plasmids encoding Myc3-Chk2-WT, Myc3-Chk2-D368N, or Myc3-Chk2-S379A as described in Materials and Methods. Lysates were blotted with an antibody against Chk2, and the relative expression level of Myc3-Chk2 versus endogenous Chk2 for each cell line was quantified by chemiluminescence using a Storm Imager; the ratio of ectopic to endogenous Chk2 expression is indicated. (B) U2OS/FRT parental and Chk2-expressing cells were treated with etoposide for the indicated times. The percentage of cells containing sub-2N DNA content was assessed by propidium iodide staining and fluorescence-activated cell sorter analysis. Error bars denote standard errors of the means (n = 4). (C and D) U2OS/FRT parental and Chk2-expressing cells were exposed to IR. Lysates were prepared and Western blotting was performed with the indicated antibodies.
Apoptosis was assayed by treating the U2OS/FRT stable cell lines with etoposide and analyzing DNA content by flow cytometry (45). As seen in Fig. 6B, cells expressing D368N or S379A were less susceptible to etoposide-induced apoptosis than were parental cells or cells expressing wild-type Chk2. These results demonstrate that both kinase activity and S379 phosphorylation are required for the proapoptotic effector function of Chk2.
The phosphorylation status of endogenous Chk2 was monitored in each of the U2OS cell lines to determine whether D368N or S379A interfered with activation of endogenous Chk2. Chk2 activation can be determined indirectly by monitoring its mobility on SDS gels as well as by S516 phosphorylation. Part of the self-sustaining Chk2 activation cycle involves auto-/transphosphorylation of Chk2 on S516 as well as other residues, and this causes Chk2 to migrate more slowly on SDS gels. Phosphorylation of T68 is catalyzed by kinases in addition to Chk2 (3, 57), and as seen in Fig. 6C, IR induced the phosphorylation of endogenous Chk2 on T68 in each cell line (lanes 5 to 8). However, endogenous Chk2 failed to undergo mobility shifts (Fig. 6C, lane 7) and to be phosphorylated on S516 (Fig. 6D, lanes 8 and 9) in cells expressing D368N. This suggests that the kinase-inactive Chk2 mutant functions in a dominant-negative manner. In contrast, the activation and phosphorylation of endogenous Chk2 appeared normal in cells expressing S379A. Thus, expression of D368N blocked activation of endogenous Chk2, and this likely accounts for its ability to block DNA damage-induced apoptosis in U2OS cells. In contrast, results obtained for S379A indicate that this mutant interferes with pathways downstream of endogenous Chk2 activation.
DISCUSSION
In cells containing DNA DSBs, ATM initiates Chk2 activation by phosphorylating Chk2 on T68. This leads to Chk2 oligomerization, which facilitates Chk2 phosphorylation at additional sites through an auto-/transphosphorylation mechanism. Here we demonstrate that auto-/transphosphorylation of Chk2 on S379 also leads to its ubiquitination by a Cul 1-containing E3 ligase. To recruit specific substrates, Cul 1 binds the Skp1 adaptor protein (13, 44, 56). Skp1 then binds an F box protein, which in turn recruits substrates for ubiquitination. Additional studies will be required to identify the F box protein that targets Chk2 to the Cul 1 complex. In general, the recruitment of substrates to the Skp1-Cul 1-F box complex requires that the substrate first be phosphorylated. In the case of Chk2, S379 phosphorylation is required for its interactions with Cul 1 and its subsequent ubiquitination. Mutants of Chk2 that failed to be phosphorylated on S379 (kinase-inactive Chk2 and the S379A mutant) did not associate with Cul 1 and were not ubiquitinated in vivo. This suggests that S379 phosphorylation may regulate interactions between the Cul-containing E3 ligase complex and Chk2. We were unable to detect significant changes in the half-life of Chk2 after its ubiquitination, consistent with studies reporting that Chk2 remains stable during a DNA damage checkpoint response (4, 5, 28, 54). Kass et al. recently reported that dephosphorylation on S456 is necessary for Chk2 ubiquitination and that ubiquitination regulates Chk2 turnover (22). Differences in results between these studies may be due to differences in the cell lines used or the type or intensity of DNA damage.
Although proteasome-dependent degradation of ubiquitinated proteins is the best-characterized function for this modification, there have been several additional functions reported for protein ubiquitination over the past few years (21, 39). In particular, both Brca1 and RNF8 are E3 ubiquitin ligases that translocate to sites of DNA damage and ubiquitinate proteins to recruit additional proteins to sites of damage and to activate signaling pathways (18, 24, 30, 49). It has not been determined when Chk2 becomes ubiquitinated. The adaptor protein MDC1 recruits RNF8 to damaged chromatin in a phosphorylation-dependent manner, and the ubiquitin-interacting motif-containing protein RAP80 mediates the localization of Brca1 to DNA damage foci (18, 30, 49, 50). MDC1 associates with phosphorylated Chk2 at foci following DNA damage and contributes to Chk2-mediated apoptosis (27). It will be important to determine if Chk2 ubiquitination recruits Chk2 to foci by, for example, binding a ubiquitin-interacting motif-containing protein such as RAP80 at sites of DNA damage. This would enable Chk2 to interact with MDC1 and to phosphorylate Brca1 (26, 27). Alternatively, Chk2 may become ubiquitinated after its translocation to foci. Chk2 has also been shown to associate with USP28, a deubiquitinating enzyme, but the functional significance of this interaction requires further study (55).
The initial activation of Chk2 involves its translocation to sites of DNA damage. However, this relocalization is transient and Chk2 becomes rapidly redistributed throughout the nucleus in order to carry out its signaling function (29). The fact that the S379A mutant was not ubiquitinated and was impaired in its ability to induce apoptosis in cells with DNA DSBs suggests that Chk2 ubiquitination and signaling may be coupled. Expression of a kinase-inactive mutant of Chk2 blocked phosphorylation of endogenous Chk2 and impaired apoptosis in cells with DNA DSBs. Thus, kinase-inactive Chk2 may function as a dominant-negative mutant by oligomerizing with endogenous Chk2 and thereby preventing subsequent auto-/transphosphorylation events. In contrast, the S379A mutant did not detectably alter endogenous Chk2 phosphorylation. This suggests that proximal Chk2 activation events (Chk2 oligomerization and auto-/transphosphorylation) were not perturbed in S379A-expressing cells. However, the ability of DNA DSBs to induce apoptosis was impaired in cells expressing the S379A mutant. This may indicate that ubiquitination of Chk2 is an important step in licensing Chk2 for downstream signaling. Additional studies will be required to elucidate how ubiquitination couples Chk2 to its downstream substrates.
Acknowledgments
We thank Julie Schwarz for assistance in identifying Chk2 phosphorylation sites, Julia Cordero for assistance in preparing the U2OS/FRT stable cell lines, Janis Watkins for overproducing and purifying Chk2 from bacteria, Van Leung Pineda for help generating the TAP vector, and Jinwu Sun for generating plasmids encoding dominant-negative Cullin 4B and TAP-tagged Chk2 proteins. We thank Anurag Agarwal for developing purification conditions for the TAP-tagged proteins. In addition, we thank Ron Kopito (Stanford University) for providing the His6-Myc-ubiquitin expression construct and Tak Mak (University of Toronto) for providing the CHK2 null MEFs. Expression plasmids encoding Myc-tagged Cullin proteins were kindly provided by Yue Xiong (University of North Carolina, Chapel Hill), and plasmids encoding dominant-negative forms of Cullin 1 and Cullin 3 were kindly provided by Wade Harper (Harvard Medical School). We thank Emily Cheng for helpful suggestions throughout this study.
This work was supported by a grant from the National Institutes of Health. C.M.L. is a former member of the Medical Scientist Training Program at Washington University School of Medicine and was supported in part by a Pre- and Post-graduate Training in Molecular Hematology grant (NIH/NHLBI grant T32 HL07088). H.P.-W. is an Investigator and L.Y. is an Associate of the Howard Hughes Medical Institute.
Footnotes
Published ahead of print on 21 July 2008.
REFERENCES
- 1.Abramoff, M. D., P. J. Magelhaes, and S. J. Ram. 2004. Image processing with Image J. Biophotonics Int. 1136-42. [Google Scholar]
- 2.Ahn, J.-Y., X. Li, H. L. Davis, and C. E. Canman. 2002. Phosphorylation of threonine 68 promotes oligomerization and autophosphorylation of the Chk2 protein kinase via the Forkhead-associated (FHA) domain. J. Biol. Chem. 27719389-19395. [DOI] [PubMed] [Google Scholar]
- 3.Ahn, J.-Y., J. K. Schwarz, H. Piwnica-Worms, and C. E. Canman. 2000. Threonine 68 phosphorylation by ataxia telangiectasia mutated is required for efficient activation of Chk2 in response to ionizing radiation. Cancer Res. 605934-5936. [PubMed] [Google Scholar]
- 4.Bartek, J., and J. Lukas. 2001. Pathways governing G1/S transition and their response to DNA damage. FEBS Lett. 490117-122. [DOI] [PubMed] [Google Scholar]
- 5.Bartkova, J., J. Falck, E. Rajpert-De Meyts, N. E. Skakkebaek, J. Lukas, and J. Bartek. 2001. Chk2 tumour suppressor protein in human spermatogenesis and testicular germ-cell tumours. Oncogene 205897-5902. [DOI] [PubMed] [Google Scholar]
- 6.Bell, D. W., J. M. Variey, T. E. Szydlo, D. H. Kang, C. R. Wahrer-Doke, K. E. Shannon, M. Lubratovich, S. J. Versellls, K. J. Isselbacher, J. F. Fraumeni, J. M. Birch, F. P. Li, J. E. Garber, and D. A. Haber. 1999. Heterozygous germ line hCHK2 mutations in Li-Fraumeni syndrome. Science 2862528-2531. [DOI] [PubMed] [Google Scholar]
- 7.Brown, A., C.-H. Lee, J. K. Schwarz, N. Mitiku, D. Griffith, H. Piwnica-Worms, and J. H. Chung. 1999. A human Cds1-related kinase that functions downsteam of ATM in the cellular response to DNA damage. Proc. Natl. Acad. Sci. USA 963745-3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaturvedi, P., W. K. Eng, Y. Zhu, M. R. Mattern, R. Mishra, M. R. Hurle, X. Zhang, R. S. Annan, Q. Lu, L. F. Faucette, G. F. Scott, X. Li, S. A. Carr, R. K. Johnson, J. D. Winkler, and B.-B. S. Zhou. 1999. Mammalian Chk2 is a downstream effector of the ATM-dependent DNA damage checkpoint pathway. Oncogene 184047-4054. [DOI] [PubMed] [Google Scholar]
- 9.Chehab, N. H., A. Malikzay, M. Appel, and T. D. Halazonetis. 2000. Chk2/hCds1 functions as a DNA damage checkpoint in G1 by stabilizing p53. Genes Dev. 14278-288. [PMC free article] [PubMed] [Google Scholar]
- 10.Chehab, N. H., A. Malikzay, E. S. Stavridi, and T. D. Halazonetis. 1999. Phosphorylation of Ser-20 mediates stabilization of human p53 in response to DNA damage. Proc. Natl. Acad. Sci. USA 9613777-13782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, L., D. M. Gilkes, Y. Pan, W. S. Lane, and J. Chen. 2005. ATM and Chk2-dependent phosphorylation of MDMX contribute to p53 activation after DNA damage. EMBO J. 243411-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cullinan, S. B., J. D. Gordan, J. Jin, J. W. Harper, and J. A. Diehl. 2004. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol. Cell. Biol. 248477-8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feldman, R. M. R., C. C. Correll, K. B. Kaplan, and R. J. Deshaies. 1997. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 91221-230. [DOI] [PubMed] [Google Scholar]
- 14.Graves, P. R., L. Yu, J. K. Schwarz, J. Gales, E. A. Sausville, P. M. O'Connor, and H. Piwnica-Worms. 2000. The Chk1 protein kinase and the Cdc25C regulatory pathway are targets of the anticancer agent UCN-01. J. Biol. Chem. 2755600-5605. [DOI] [PubMed] [Google Scholar]
- 15.Hartwell, L. H., and M. B. Kastan. 1994. Cell cycle control and cancer. Science 2661821-1828. [DOI] [PubMed] [Google Scholar]
- 16.Hirao, A., A. Cheung, G. Duncan, P.-M. Girard, A. J. Elia, A. Wakeham, H. Okada, T. Sarkissian, J. A. Wong, T. Sakai, E.-d. Stanchina, R. G. Bristow, T. Suda, S. W. Lowe, P. A. Jeggo, S. J. Elledge, and T. W. Mak. 2002. Chk2 is a tumor suppressor that regulates apoptosis in both an ataxia telangiectasia mutated (ATM)-dependent and an ATM-independent manner. Mol. Cell. Biol. 226521-6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirao, A., Y. Y. Kong, S. Matsuoka, A. Wakeham, J. Ruland, H. Yoshida, D. Liu, S. J. Elledge, and T. W. Mak. 2000. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science 2871824-1827. [DOI] [PubMed] [Google Scholar]
- 18.Huen, M. S., R. Grant, I. Manke, K. Minn, X. Yu, M. B. Yaffe, and J. Chen. 2007. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131901-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jack, M. T., R. A. Woo, A. Hirao, A. Cheung, T. W. Mak, and P. W. Lee. 2002. Chk2 is dispensable for p53-mediated G1 arrest but is required for a latent p53-mediated apoptotic response. Proc. Natl. Acad. Sci. USA 999825-9829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson, M. W., M. K. Agarwal, M. L. Agarwal, A. Agarwal, P. Stanhope-Baker, B. R. Williams, and G. R. Stark. 2004. Limited role of N-terminal phosphoserine residues in the activation of transcription by p53. Oncogene 234477-4487. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, E. S. 2002. Ubiquitin branches out. Nat. Cell Biol. 4E295-E298. [DOI] [PubMed] [Google Scholar]
- 22.Kass, E. M., J. Ahn, T. Tanaka, W. A. Freed-Pastor, S. Keezer, and C. Prives. 2007. Stability of checkpoint kinase 2 is regulated via phosphorylation at serine 456. J. Biol. Chem. 28230311-30321. [DOI] [PubMed] [Google Scholar]
- 23.King, J. B., J. Gross, H. Rohrs, C. M. Lovly, H. Piwnica-Worms, and R. R. Townsend. 2006. Mass-driven analysis for the characterization of protein phosphorylation—a study of the human Chk2 protein kinase. Anal. Chem. 782171-2181. [DOI] [PubMed] [Google Scholar]
- 24.Kolas, N. K., J. R. Chapman, S. Nakada, J. Ylanko, R. Chahwan, F. D. Sweeney, S. Panier, M. Mendez, J. Wildenhain, T. M. Thomson, L. Pelletier, S. P. Jackson, and D. Durocher. 2007. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 3181637-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, C. H., and J. H. Chung. 2001. The hCds1 (Chk2)-FHA domain is essential for a chain of phosphorylation events on hCds1 that is induced by ionizing radiation. J. Biol. Chem. 27630537-30541. [DOI] [PubMed] [Google Scholar]
- 26.Lee, J. S., K. M. Collins, A. L. Brown, C. H. Lee, and J. H. Chung. 2000. hCds1-mediated phosphorylation of BRCA1 regulates the DNA damage response. Nature 404201-204. [DOI] [PubMed] [Google Scholar]
- 27.Lou, Z., K. Minter-Dykhouse, X. Wu, and J. Chen. 2003. MDC1 is coupled to activated Chk2 in mammalian DNA damage response pathways. Nature 421957-960. [DOI] [PubMed] [Google Scholar]
- 28.Lukas, C., J. Bartkova, L. Latella, J. Falck, N. Mailand, T. Schroeder, M. Sehested, J. Lukas, and J. Bartek. 2001. DNA damage-activated kinase Chk2 is independent of proliferation or differentiation yet correlates with tissue biology. Cancer Res. 614990-4993. [PubMed] [Google Scholar]
- 29.Lukas, C., J. Falck, J. Bartkova, J. Bartek, and J. Lukas. 2003. Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat. Cell Biol. 5255-260. [DOI] [PubMed] [Google Scholar]
- 30.Mailand, N., S. Bekker-Jensen, H. Faustrup, F. Melander, J. Bartek, C. Lukas, and J. Lukas. 2007. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131887-900. [DOI] [PubMed] [Google Scholar]
- 31.Matsuoka, S., Huang, M., and S. J. Elledge. 1998. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science 2821893-1897. [DOI] [PubMed] [Google Scholar]
- 32.Matsuoka, S., G. Rotman, A. Ogawa, Y. Shiloh, K. Tamai, and S. J. Elledge. 2000. Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc. Natl. Acad. Sci. USA 9710389-10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melchionna, R., X. B. Chen, A. Blasina, and C. H. McGowan. 2000. Threonine 68 is required for radiation-induced phosphorylation and activation of Cds1. Nat. Cell Biol. 2762-765. [DOI] [PubMed] [Google Scholar]
- 34.Ogg, S., B. Gabrielli, and H. Piwnica-Worms. 1994. Purification of a serine kinase that associates with and phosphorylates human Cdc25C on serine 216. J. Biol. Chem. 26930461-30469. [PubMed] [Google Scholar]
- 35.Ohta, T., J. J. Michel, A. J. Schottelius, and Y. Xiong. 1999. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol. Cell 3535-541. [DOI] [PubMed] [Google Scholar]
- 36.Ohta, T., and Y. Xiong. 2001. Phosphorylation- and Skp1-independent in vitro ubiquitination of E2F1 by multiple ROC-cullin ligases. Cancer Res. 611347-1353. [PubMed] [Google Scholar]
- 37.Pereg, Y., S. Lam, A. Teunisse, S. Biton, E. Meulmeester, L. Mittelman, G. Buscemi, K. Okamoto, Y. Taya, Y. Shiloh, and A. G. Jochemsen. 2006. Differential roles of ATM- and Chk2-mediated phosphorylations of Hdmx in response to DNA damage. Mol. Cell. Biol. 266819-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petroski, M. D., and R. J. Deshaies. 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 69-20. [DOI] [PubMed] [Google Scholar]
- 39.Pickart, C. M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70503-533. [DOI] [PubMed] [Google Scholar]
- 40.Pickart, C. M. 2001. Ubiquitin enters the new millennium. Mol. Cell 8499-504. [DOI] [PubMed] [Google Scholar]
- 41.Schwarz, J. K., C. M. Lovly, and H. Piwnica-Worms. 2003. Regulation of the Chk2 protein kinase by oligomerization-mediated cis- and trans-phosphorylation. Mol. Cancer Res. 1598-609. [PubMed] [Google Scholar]
- 42.Shieh, S. Y., J. Ahn, K. Tamai, Y. Taya, and C. Prives. 2000. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 14289-300. [PMC free article] [PubMed] [Google Scholar]
- 43.Shirogane, T., J. Jin, X. L. Ang, and J. W. Harper. 2005. SCFbeta-TRCP controls clock-dependent transcription via casein kinase 1-dependent degradation of the mammalian period-1 (Per1) protein. J. Biol. Chem. 28026863-26872. [DOI] [PubMed] [Google Scholar]
- 44.Skowyra, D., K. L. Craig, M. Tyers, S. J. Elledge, and J. W. Harper. 1997. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91209-219. [DOI] [PubMed] [Google Scholar]
- 45.Stevens, C., L. Smith, and N. B. La Thangue. 2003. Chk2 activates E2F-1 in response to DNA damage. Nat. Cell Biol. 5401-409. [DOI] [PubMed] [Google Scholar]
- 46.Takai, H., K. Naka, Y. Okada, M. Watanabe, N. Harada, S. Saito, C. W. Anderson, E. Appella, M. Nakanishi, H. Suzuki, K. Nagashima, H. Sawa, K. Ikeda, and N. Motoyama. 2002. Chk2-deficient mice exhibit radioresistance and defective p53-mediated transcription. EMBO J. 215195-5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Urist, M., T. Tanaka, M. V. Poyurovsky, and C. Prives. 2004. p73 induction after DNA damage is regulated by checkpoint kinases Chk1 and Chk2. Genes Dev. 183041-3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Geer, P., and T. Hunter. 1994. Phosphopeptide mapping and phosphoamino acid analysis by electrophoresis and chromatography on thin-layer cellulose plates. Electrophoresis 15544-554. [DOI] [PubMed] [Google Scholar]
- 49.Wang, B., and S. J. Elledge. 2007. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc. Natl. Acad. Sci. USA 10420759-20763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, B., S. Matsuoka, B. A. Ballif, D. Zhang, A. Smogorzewska, S. P. Gygi, and S. J. Elledge. 2007. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science 3161194-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu, X., and J. Chen. 2003. Autophosphorylation of checkpoint kinase 2 at serine 516 is required for radiation-induced apoptosis. J. Biol. Chem. 27836163-36168. [DOI] [PubMed] [Google Scholar]
- 52.Wu, Z., J. Earle, S. Saito, C. W. Anderson, E. Appella, and Y. Xu. 2002. Mutation of mouse p53 S23 and the response to DNA damage. Mol. Cell. Biol. 222441-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu, X., L. M. Tsvetkov, and D. F. Stern. 2002. Chk2 activation and phosphorylation-dependent oligomerization. Mol. Cell. Biol. 224419-4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang, S., C. Kuo, J. E. Bisi, and M. K. Kim. 2002. PML-dependent apoptosis after DNA damage is regulated by the checkpoint kinase hCds1/Chk2. Nat. Cell Biol. 4865-870. [DOI] [PubMed] [Google Scholar]
- 55.Zhang, D., K. Zaugg, T. W. Mak, and S. J. Elledge. 2006. A role for the deubiquitinating enzyme USP28 in control of the DNA-damage response. Cell 126529-542. [DOI] [PubMed] [Google Scholar]
- 56.Zheng, N., B. A. Schulman, L. Song, J. J. Miller, P. D. Jeffrey, P. Wang, C. Chu, D. M. Koepp, S. J. Elledge, M. Pagano, R. C. Conaway, J. W. Conaway, J. W. Harper, and N. P. Pavletich. 2002. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416703-709. [DOI] [PubMed] [Google Scholar]
- 57.Zhou, B. B., P. Chaturvedi, K. Spring, S. P. Scott, R. A. Johanson, R. Mishra, M. R. Mattern, J. D. Winkler, and K. K. Khanna. 2000. Caffeine abolishes the mammalian G(2)/M DNA damage checkpoint by inhibiting ataxia-telangiectasia-mutated kinase activity. J. Biol. Chem. 27510342-10348. [DOI] [PubMed] [Google Scholar]