Abstract
Chicken DT40 cells deficient in the 9-1-1 checkpoint clamp exhibit hypersensitivity to a variety of DNA-damaging agents. Although recent work suggests that, in addition to its role in checkpoint activation, this complex may play a role in homologous recombination and translesion synthesis, the cause of this hypersensitivity has not been studied thoroughly. The immunoglobulin locus of DT40 cells allows monitoring of homologous recombination and translesion synthesis initiated by activation-induced deaminase (AID)-dependent abasic sites. We show that both the RAD9−/− and RAD17−/− mutants exhibit substantially reduced immunoglobulin gene conversion. However, the level of nontemplated immunoglobulin point mutation increased in these mutants, a finding that is reminiscent of the phenotype resulting from the loss of RAD51 paralogs or Brca2. This suggests that the 9-1-1 complex does not play a central role in translesion synthesis in this context. Despite reduced immunoglobulin gene conversion, the RAD9−/− and RAD17−/− cells do not exhibit a prominent defect in double-strand break-induced gene conversion or a sensitivity to camptothecin. This suggests that the roles of Rad9 and Rad17 may be confined to a subset of homologous recombination reactions initiated by replication-stalling lesions rather than those associated with double-strand break repair.
DNA replication is a complex and fragile reaction that is frequently stalled by damaged template strands. To alleviate replication blocks, cells have evolved two basic pathways: homologous recombination (HR) and translesion synthesis (TLS). HR facilitates a template switch to the intact sister chromatid. TLS involves the direct bypass of the damage by specialized DNA polymerases whose catalytic sites are flexible enough to accommodate damaged bases in the template DNA. This flexibility is accompanied by low fidelity. The recruitment of TLS polymerases is promoted by ubiquitylation of the polymerase processivity clamp (PCNA) at lysine 164 (19, 26, 48, 50). HR is also associated with DNA synthesis, which is facilitated by TLS polymerases such as Polη and Polζ (20, 41). These DNA polymerases thus perform a function in both TLS and HR. The role of posttranslational modification of PCNA in vertebrate HR has not been extensively studied.
The heterotrimeric checkpoint clamp, consisting of the Rad9, Hus1, and Rad1 subunits (corresponding to the Rad17, Mec3, and Ddc1 subunits in budding yeast), is structurally homologous to PCNA (39). It serves as a damage sensor in the S and G2/M phases, eventually facilitating cell cycle arrest in vertebrate cells (8, 39). In vitro, the 9-1-1 complex can be loaded onto DNA by a specific clamp loader, Rad17-RFC, which is analogous to the PCNA-RFC clamp-clamp loader system (5, 10).
Although these checkpoint factors do not appear to play a major role in mitotic HR in budding yeast, mammalian Rad9 and Rad17 seem to contribute to some HR reactions (e.g., repair of X-ray-induced double-strand breaks [DSBs] and gene targeting), though they are not required for spontaneous or damage-induced sister chromatid exchange (SCE) (7, 28). It was recently proposed that the yeast 9-1-1 complex also controls the access of translesion DNA polymerases to the 3′ termini (18, 32). The significance of these observations in terms of vertebrate TLS and HR has not been addressed.
The molecular mechanisms underlying the selective use of HR and TLS at replication blockages are difficult to investigate because of the paucity of phenotypic assays carried out to monitor the blockage and subsequent restart of individual replication forks. Analysis of immunoglobulin variable (Ig V) sequence diversification in the chicken DT40 B cell line provides a unique opportunity to study the decision-making cells that determine the use of HR or TLS in region-specific genomic DNA damage. Chicken B lymphocytes, including those of DT40, diversify their Ig V genes through both HR and single-nucleotide substitution (point mutation), both of which are triggered by activation-induced deaminase (AID)-mediated DNA damage (2, 16). The current model hypothesizes that AID catalyzes the deamination of deoxycytidine to form uracil, which is subsequently eliminated by a process involving hydrolysis by uracil DNA glycosylase, thus generating abasic sites (9, 29, 36). TLS across these sites depends on REV1 and (in DT40 cells at least) PCNA ubiquitination (3, 40). In birds, the abasic site can also stimulate Ig gene conversion, wherein tracts of genetic information from an array of upstream pseudogenes are nonreciprocally copied into the rearranged Vλ gene. The exact mechanism by which abasic sites stimulate Ig gene conversion remains unclear. Although it is known that apurinic/apyrimidinic endonuclease 1 (APE1/Ref-1) and bivalent glycosylases incise DNA strands at abasic sites during base excision repair (BER), it remains to be determined whether APE1 or other unidentified nucleases trigger the Ig gene conversion (reviewed in reference 45).
The balance between gene conversion and point mutation can be altered by deleting pseudo-V donor sequences or by disrupting HR or TLS factors. The loss of those proteins that play an early role in the establishment of an HR reaction (e.g., the RAD51 paralogs BRCA1 and BRCA2) results in an inhibition of gene conversion with a concomitant rise in point mutation (4, 17, 35), while HR factors (such as RAD54) that act later result in the loss of gene conversion without the stimulation of point mutation (6). Conversely, perturbing translesion synthesis triggered by the disruption of REV1 or by PCNA ubiquitylation results in diminished point mutation (3, 35). Both types of Ig V diversification are dependent on AID-mediated DNA damage and the subsequent formation of abasic sites (2, 16, 27). Collectively, Ig gene conversion and hypermutation are likely to reflect HR and TLS that are initiated by a common DNA lesion in a physiological context.
PCNAK164R (PCNAK164R/K164R), RAD9−/−, and RAD17−/− cells (3, 22) are hypersensitive to a wide variety of DNA-damaging agents, including γ rays, UV exposure, alkylating agents, and chemical cross-linkers. This hypersensitivity may be attributable, either singly or in combination, to defects in HR or TLS or to a DNA-damage checkpoint. To examine the contribution of the 9-1-1 complex to both HR and TLS, we modified the Ig diversification assay of DT40 cells to improve its sensitivity. By overexpressing AID from a recombinant retrovirus, we forced an increase in nontemplated point mutations at the G/C base pairs, as well as an increase in mutation at the A/T base pairs. The mutations at the A/T base pairs reflect short-tract gene conversion, allowing many more recombination events to be scored.
Despite previous reports of an interaction between the 9-1-1 complex and TLS (18, 32), we found that both the RAD9−/− and RAD17−/− cells were not compromised in their ability to undergo TLS, as illustrated by the high rates of Ig hypermutation. Remarkably, these mutants underwent virtually no Ig gene conversion events. In contrast, as previously reported (3), PCNAK164R cells showed a marked reduction in the rate of Ig hypermutation, as well as a generalized compromise of HR reactions, including Ig gene conversion, DSB repair in an artificial HR substrate, and cellular tolerance to camptothecin (1, 33). These observations indicate that the 9-1-1 complex and PCNA modification differentially contribute to HR and TLS and that the 9-1-1 complex may play a particularly prominent role in the subset of HR reactions initiated by replication-arresting DNA damage.
MATERIALS AND METHODS
Cell culture and DNA transfection.
Cells were cultured in RPMI 1640 medium supplemented with 10−5 M β-mercaptoethanol, 10% fetal calf serum, and 1% chicken serum (Sigma, St. Louis, MO) at 39.5°C. Methods for DNA transfection and genotoxic treatments were performed as described previously (42).
Real-time PCR.
Total RNA from DT40 cells expressing mouse AID and from bursae of Fabricius were extracted with Sepasol (Nakalai, Kyoto, Japan). The chicken or mouse AID and chicken β-actin (internal control) cDNAs were amplified using real-time PCR with the primers 5′-TTCCTACGCTACATCTCAG-3′ (forward) and 5′-CCCCTCAGGCTCAGCCTTG-3′ (reverse) for AID; and 5′-CATTGCTGACAGGATGCAGAAGG-3′ (forward) and 5′-TGCTTGCTGATCCACATCTGCTGG-3′ (reverse) for chicken β-actin. AID primers were designed from the conserved sequences of mouse and chicken AID.
AID overexpression by retrovirus infection.
For retrovirus infection, the pMSCV-IRES-GFP recombinant plasmid was constructed by ligating the 5.2-kb BamHI-Not1 fragment from pMSCVhyg (Clontech) with the 1.2-kb BamHI-Not1 fragment of pIRES2-EGFP (Clontech). Mouse AID cDNA (38) was inserted between the BglII and EcoR1 sites of pMSCV-IRES-GFP. Preparation and infection of the retrovirus were done as previously described (38). Expression of the green fluorescent protein (GFP) was confirmed by flow cytometry. The efficiency of infection was more than 90%, as assayed by GFP expression. Cells were subcloned into 96-well plates 1 day after infection, and clones displaying high levels of GFP were determined by a fluorescence-activated cell sorter.
Ig V mutation.
Genomic DNA was extracted at 14 days after subcloning (14 to 15 days postinfection). Using the primer 5′-CAGGAGCTCGCGGGGCCGTCACTGATTGCCG-3′ at the lead Vλ intron position and the primer 5′-GCGCAAGCTTCCCCAGCCTGCCGCCAAGTCCAAG-3′ at the back 3′ site of the JCλ intron, the rearranged Vλ segments were PCR amplified, cloned into the plasmid, and subjected to sequences analysis. To minimize PCR-introduced mutations, high-fidelity polymerase, Phusion (at 30 cycles of 94°C for 30 s, 60°C for 1 min, and 72°C for 1 min; Fynnzymes), was used for amplification. PCR products were cloned in a topo Zeroblunt vector (Invitrogen) and sequenced with M13 forward (−20) or M13 reverse primer. Sequence alignment with Genetyx-Mac (Software Development, Tokyo, Japan) allowed identification of the changes from the parental sequences in each clone.
As described previously (35), all sequence changes were assigned to one of three categories: gene conversion, point mutation (PM), or an ambiguous category (Amb). This discrimination rests on the published sequences of the Vλ pseudogenes that could act as donors for gene conversion. For each mutation, the database of the Vλ pseudogene was searched for a potential donor. If no pseudogene donor containing a string of more than 9 bp could be found, then the mutation was categorized as a nontemplated point mutation. If such a string was identified and there were further mutations that could be explained by the same donor, then all of these mutations were assigned to a single long-tract gene conversion event. If there were no further mutations, it was possible that the isolated mutation could have arisen through a conversion mechanism or could have been nontemplated and was therefore categorized as Amb.
Measurement of HR frequencies in artificial substrates.
The DR-GFP reporter construct was inserted into the OVALBUMIN locus of DT40 cells. An amount of 3 μg of I-SceI expression plasmid was transiently expressed by Amaxa (solution T; A-30 program), and the percentages of GFP-positive cells were counted by flow cytometry at 48 h after transfection.
Measurement of SCE.
The levels of SCE were measured as described previously (43).
RESULTS
Induction of single-nucleotide substitutions by AID overexpression.
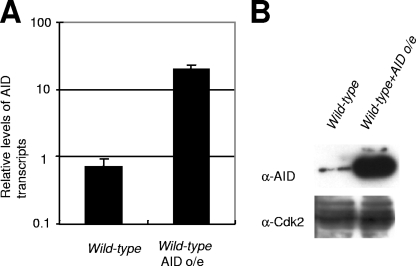
The rate of Ig gene conversion in DT40 cells (5 × 10−4/Vλ/division) is significantly lower than that of B precursor cells in the bursa of Fabricius (5 × 10−2 to 10 × 10−2/Vλ/division) (30). We wished to increase the number of events in the Ig loci of our cells to increase the sensitivity of our assay for Ig gene conversion and nontemplated mutation. To do this, we transduced DT40 cells with a retrovirus expressing AID as a bicistronic transcript with GFP. Expression of AID was assessed by measuring the GFP expression levels. To enrich AID-overexpressing cells, clones expressing the same high levels of GFP were isolated from infected populations of DT40 cells. RNA analysis of GFPhigh cells suggested that the infected cells expressed a level of AID that was about 20-fold higher than that of the endogenous AID protein (Fig. 1). The overexpression of AID had no effect on cellular proliferation (data not shown).
FIG. 1.
Expression level of AID in DT40 cells. (A) Reverse transcriptase PCR analysis of the AID transcripts in total RNA from the indicated cells. Primers for the AID transcripts were designed based on the conserved sequences of mouse and chicken cDNAs. Relative values were calculated by dividing the level of AID by that of β-actin (internal control). AID o/e, AID overexpression. (B) Quantification of the expression level of chicken AID by Western blotting. Cdk2 is shown as a loading control.
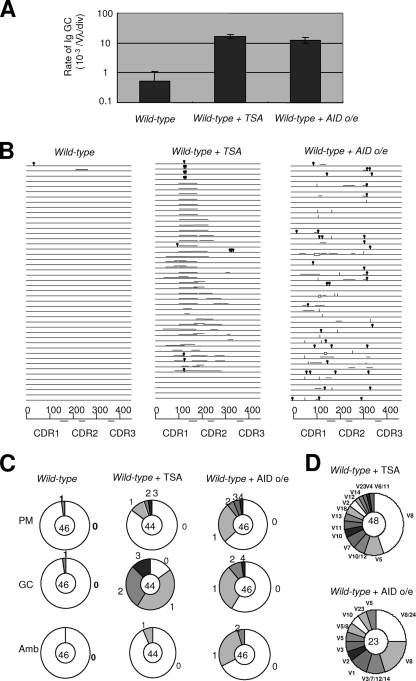
We examined Ig gene conversion and hypermutation by determining the nucleotide sequence of Ig Vλ in subclones at 14 days after infection (Fig. 2). The rate of Ig gene conversion was determined to be 0.54 × 10−3/Vλ/division in wild-type cells, given that DT40 cells divide three times per day at 39.5°C. Upon overexpression of a mouse AID transgene, the rate of Ig gene conversion rose to 12 × 10−3/Vλ/division, an increase of about 20-fold (Fig. 2). This rate is comparable to that of gene conversion induced by treatment with trichostatin A (TSA) (Fig. 2A), a histone-deacetylase inhibitor, which increases the rate of Ig gene conversion 50- to 100-fold (37). However, the use of the pseudogene induced by AID is significantly different from that induced by TSA (Fig. 2D). Remarkably, although point mutations were barely detectable in wild-type DT40 cells, AID overexpression induced point mutations in nearly all of the V segments analyzed, typically with multiple point mutations per sequence (Fig. 2B). The mutation rate was 6.0 × 10−5/Vλ/division, which is at least an order of magnitude greater than that of the parental DT40 cells (Fig. 3A) (35). Thus, the low AID expression of DT40 cells may, at least partially, account for the loss of Ig hypermutation.
FIG. 2.
Analysis of Vλ sequences following overexpression of mouse AID transgene. (A) Comparison of Ig gene conversion (Ig GC) rates in untreated, TSA-treated, and AID-overexpressing wild-type DT40 cells. (B) Ig gene conversion and single-base substitution events in wild-type parental DT40 cells, TSA-treated cells, and AID-overexpressing cells. Each horizontal line represents the rearranged Vλ gene (450 bp) with mutations classified, as described in the text, as nontemplated base substitution (lollipop shape), long-tract GC (horizontal bar above line), or single-nucleotide substitutions that could be the result of PMs or gene conversion (Amb, vertical bar). Deletions, duplications, and insertions are excluded. (C) Proportion of nontemplated single-base substitution (PM), long-tract GC, and Amb mutations. Segment sizes are proportional to the number of sequences, with the numbers of mutations indicated around the outsides of the pie charts. The total number of Vλ sequences analyzed is indicated in the center of each chart. (D) Proportion of ψV donor segments used in TSA-treated and AID-overexpressing wild-type cells. Data in panels B, C, and D are pooled from the analyses of more than four independent clones.
FIG. 3.
Point mutations at A/T base pairs are caused by short-tract gene conversion. (A) Rate of nontemplated single-base substitution (PM) compared to that of single-base substitution of Amb origin in wild-type and pseudo-V gene-deleted (ψV−; PCNA+/+/AID−/−/rAID/v-myb/ψV− in Table 1) DT40 cells. Cells were clonally expanded for 2 weeks. Two clones were analyzed from each data set. (B) Nucleotide substitution preferences (as a percentage of the total mutations) deduced from the Vλ sequence analysis of the clones shown in panel A. Preference is shown for mutations categorized as nontemplated base substitution (PM) and those categorized as Amb. Data are from reference 40.
Induction of mutations at A/T base pairs in AID-overexpressing DT40 cells.
Ig diversification in DT40 cells also differs from the in vivo diversification qualitatively in that the nontemplated point mutations associated with gene conversion are found predominantly at the G/C base pairs rather than at all four bases. Interestingly, a substantial fraction of the point mutations occurred at the A/T base pairs in AID-overexpressing wild-type cells (Fig. 3B). This observation is in marked contrast to the accumulation of point mutations found exclusively at the G/C pairs in the rad51 paralog mutants, as well as the brca2-deficient cells (17, 35). To investigate the molecular mechanism underlying the A/T mutations, we followed the previously described method (34, 35) for classifying Ig changes in the Ig gene sequence into one of three categories: a nontemplated PM; an Amb mutation, where an isolated mutation could have arisen through a short-tract gene conversion or could have been nontemplated; or a true gene conversion. As shown in Fig. 2C, the AID overexpression resulted in an increase in the rates of both the nontemplated PMs and those falling into the Amb category in wild-type cells.
In AID-overexpressing clones, nontemplated mutations accumulated predominantly at the G/C base pairs, whereas the Amb mutations accumulated at both the A/T and the G/C base pairs (Fig. 3B). This observation implies that A/T mutations are generated by a mechanism that differs from that of nontemplated mutations. To test whether point mutations at A/T are generated through DNA synthesis, using pseudo-V segments as a template, we overexpressed AID in DT40 cells in which the entire pseudo-V gene segments were artificially deleted (ψV−; PCNA+/+/AID−/−/rAID/v-myb/ψV− [Table 1]) (4). The rate of total point mutation in the ψV− cells was comparable to that in the wild-type cells (Fig. 3A). Strikingly, ψV− cells exhibited virtually no mutations at the A/T base pairs (Fig. 3B). We conclude that the point mutations at A/T depend on intact pseudo-V segments. This shows that the A/T mutations in the Amb category reflect Ig gene conversion, while nontemplated PMs at G/C likely reflects TLS past AP sites.
TABLE 1.
DT40 mutants used in this study
| Cell line | Selection marker(s)a | Reference(s); source |
|---|---|---|
| RAD17−/− | His | 22; this study |
| RAD9−/− | His/neo | 22 |
| POLη−/− | Bsr/puro | 20 |
| XRCC3−/− | Neo*/his/bsr | 35, 51 |
| RAD54−/− | Neo/his | 6 |
| PCNA+/+/AID−/−/rAID/v-myb | Neo*/his/puro | 4 |
| PCNAK164R/K164R/AID−/−/rAID/v-myb | Neo*/his/puro | 3 |
| PCNA+/+/AID−/−/rAID/v-myb/ψV− | Neo*/his/puro | 4 |
| PCNAK164R/K164R/AID−/−/rAID/v-myb/ψV− | Neo*/his/puro | 3 |
his, histidinol; bsr, blasticidin; puro, puromycin; hyg, hygromycin; neo, neomycin. Asterisk, ER-Cre transgene containing the neo cassette to remove the human XRCC3-IRES-GFP transgene or selection cassettes flanked by loxP.
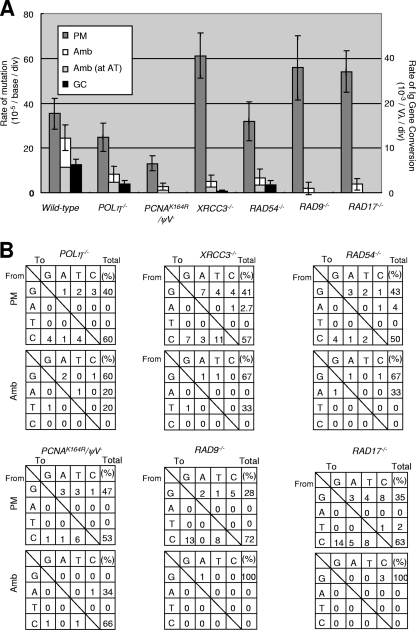
To verify that A/T point mutation was dependent on HR, we analyzed A/T mutations following AID overexpression in three mutants (POLη−/−, RAD54−/−, and XRCC3−/−) that are defective for Ig gene conversion (6, 20, 44). The rate of Ig gene conversion in POLη−/− cells was 3.8 × 10−3/Vλ/division (in agreement with results of a previous report [20]), which is threefold lower than that in wild-type cells (12 × 10−3/Vλ/division). Similarly, A/T mutations were substantially lower in the absence of Polη (Fig. 4B). The RAD54−/− and XRCC3−/− clones also showed significant decreases in the rates of mutation at the A/T base pairs (Fig. 4A). These observations confirm that the vast majority of the A/T mutations triggered by AID overexpression in DT40 cells are the products of HR, with pseudo-V segments serving as the donor template. Taking into account the ∼10% sequence divergence between the pseudo-V donor and the functional VJλ recipient (31), the length of the Ig gene conversion tract leading to these point mutations is likely to be relatively short. In summary, in AID-overexpressing DT40 cells, the Ig V PMs at A/T and nontemplated single-nucleotide substitutions reflect HR and TLS, respectively. Thus, AID overexpression in DT40 cells provides an opportunity for a more precise evaluation of TLS and HR.
FIG. 4.
Templated point mutations at A/T and gene conversion promoted by polymerase η, Rad9, and Rad17. (A) Rates are shown for nontemplated single-base substitution (PM), Amb point mutation, and Ig gene conversion (GC) in the indicated genotypes with AID overexpression. Clones were expanded for 2 weeks. More than two clones were analyzed from each data set. (B) Nucleotide substitution preference was deduced from Vλ sequence analysis of cells deficient in Polη, PCNA ubiquitylation, Xrcc3, Rad54, Rad9, and Rad17, as shown in Fig. 3B. Note that PCNAK164R/ψV− (PCNAK164R/K164R/AID−/−/rAID/v-myb/ψV− in Table 1) cells lack the whole pseudo-V genes on the rearranged allele.
Contribution of PCNA ubiquitylation to HR and TLS.
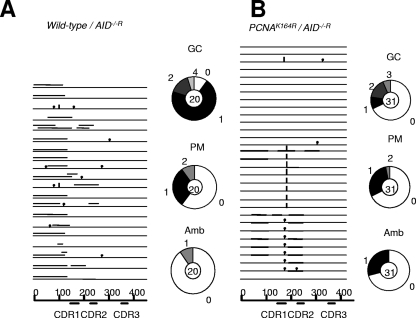
To test this phenotypic assay for TLS, we infected cells carrying both the PCNA K164R mutation and the deletion of pseudo-V segments (PCNAK164R/K164R/ψV−) with the AID-expressing retrovirus (Fig. 4). As shown in a previous study (3), the AID-overexpressing PCNAK164R cells exhibit a marked reduction in the frequency of nontemplated single-base substitutions (Fig. 4A) compared to that of wild-type cells expressing the same level of AID, which supports a role for monoubiquitylation of PCNA in TLS. Since the previous study (3) analyzed only the PCNAK164R/ψV− cells, the possible role of PCNA ubiquitylation in Ig gene conversion was not determined. To address this issue, we analyzed the PCNAK164R/AID−/−R (PCNAK164R/K164R/AID−/−/rAID/v-myb) and wild-type/AID−/−R (PCNA+/+/AID−/−/rAID/v-myb) clones (Table 1) (2), in which PCNA ubiquitination was disrupted but the pseudogene donors remained intact. The PCNA K164R mutation reduced the rate of Ig gene conversion to a level that is only one-third that found for the wild-type allele (Fig. 5). This suggests that the ubiquitylation of PCNA promotes Ig gene conversion as well as TLS-mediated Ig hypermutation.
FIG. 5.
Monoubiquitylation of PCNA promotes Ig gene conversion. Comparison of gene conversion rates and tracts in parental wild-type (A) and PCNAK164R (B) cells. Results are the same as those shown in Fig. 2. Cells were cultured for 6 weeks. Note that PCNAK164R/AID−/−R (PCNAK164R/K164R/AID−/−/rAID/v-myb) cells contain pseudo-V genes and were generated from the parental wild-type/AID−/−R (PCNA+/+/AID−/−/rAID/v-myb) cells shown in Table 1.
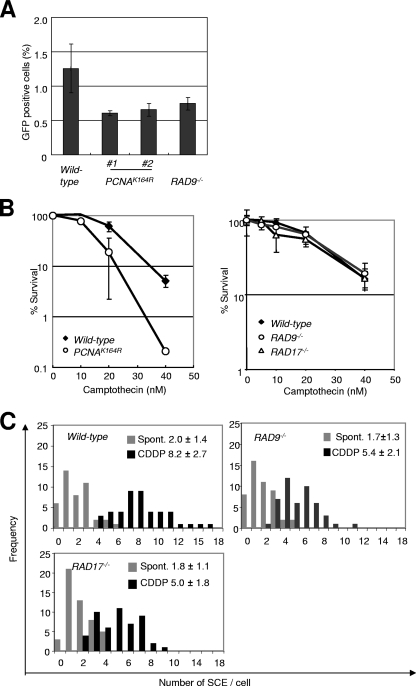
To explore whether or not the PCNA K164R mutation compromises general HR, we measured the following factors in the PCNAK164R cells: gene targeting frequency, HR-dependent repair of a site-specific DSB mediated by I-SceI acting on an artificial substrate inserted into the OVALBUMIN locus (12), and sensitivity to camptothecin, a topoisomerase I poison. We found that the frequency of gene targeting for the wild type was comparable to that for the PCNAK164R (PCNAK164R/K164R/AID−/−/rAID/v-myb/ψV− in Table 1) cells (Table 2). However, the efficiency of HR-dependent repair of an I-SceI-induced DSB was slightly lower in the PCNAK164R cells than in the wild-type cells (Fig. 6A). Camptothecin stabilizes a complex of topoisomerase I covalently linked to nicked DNA. Such complexes interrupt replication and cause DSBs in one of the sister chromatids. The resulting DSBs are repaired primarily by HR with the other (intact) sister chromatid (1, 33). In our study, the PCNAK164R cells exhibited a higher sensitivity to camptothecin than did their wild-type controls (Fig. 6B). We therefore conclude that the ubiquitylation of PCNA contributes to a subset of general HR reactions, as well as to Ig gene conversion.
TABLE 2.
Targeted integration frequency
| Genotype | No. of targeted clones/no. of clones analyzed (% of targeting frequency)a
|
||
|---|---|---|---|
| OVALBUMIN | Ig Vλ | CENP-H | |
| Wild type | 44/50 (88.0) | 10/24 (41.7)b | 56/72 (77.8)c |
| RAD9−/− | 16/44 (36.3) | 5/16 (31.2) | |
| RAD17−/− | 19/44 (43.2) | 10/36 (27.7) | 9/16 (56.2) |
| PCNAK164R | 30/48 (62.5) | 21/38 (55) | |
Cell lines were transfected with targeting constructs for the indicated loci. Data shown are the number of targeted clones divided by the number of drug-resistant clones analyzed. Note that the PCNAK164R genotype is PCNAK164R/K164R/AID−/−/rAID/v-myb/ψV−.
Data from reference 46.
Data from reference 20.
FIG. 6.
HR-mediated DSB repair compromised in RAD9−/−, RAD17−/− and PCNAK164R cells. (A) HR-mediated DSB repair was measured in cells containing the DR-GFP substrate inserted in the OVALBUMIN locus. GFP-positive fractions were determined by flow cytometry. (B) Colony survival in asynchronous populations of cells exposed to camptothecin, a topoisomerase I toxin. The left panel shows genotypes of the wild-type and PCNAK164R cells as PCNA+/+/AID−/−/rAID/v-myb/ψV and PCNAK164R/K164R/AID−/−/rAID/v-myb/ψV−, respectively. Error bars show the means ± standard deviations for at least three separate experiments. (C) Spontaneous (Spont.)- and cisplatin (CDDP)-induced SCE in RAD9−/− and RAD17−/− cells.
Dependence of Ig gene conversion on the 9-1-1 complex.
Phenotypic analysis of DT40 clones deficient in Rad9 and Rad17 has shown that these proteins are indeed required for a DNA damage checkpoint response, as are their yeast counterparts (22). To determine the functions of Rad9 and Rad17 in HR and TLS, we overexpressed AID in the RAD9−/− and RAD17−/− clones and examined Ig V diversification. Remarkably, although the level of nontemplated single-base substitutions was 50% higher than that of the wild-type cells in both mutants (Fig. 4), Ig gene conversion events and A/T mutations were virtually abolished. To confirm this result, we elevated the rate of Ig gene conversion by treating the cells with TSA. However, nucleotide sequence analysis of 30 Vλ segments, after an 8-week incubation with TSA, failed to detect any gene conversion events in RAD9−/− and RAD17−/− clones (data not shown). The analysis also revealed that very few A/T PMs had accumulated in these mutants. Given that the rate of Ig gene conversion events in the presence of TSA is more than 0.33 events/Vλ/week in wild-type cells, the loss of these damage checkpoint sensors reduced the rate of Ig gene conversion by more than 2 orders of magnitude. In summary, although Rad9 and Rad17 play an essential role in HR-dependent Ig gene conversion, they are dispensable for Ig hypermutation.
Due to the severe reduction of Ig gene conversion in the RAD9−/− and RAD17−/− clones, we analyzed their capabilities for more generalized HR by measuring gene targeting, HR-dependent repair of an artificial construct, sensitivity to camptothecin, and SCE (Fig. 6). Disruption of both RAD9 and RAD17 resulted in a modest reduction in gene targeting efficiency (Table 2). HR-dependent repair of an I-SceI-induced DSB was monitored in the RAD9−/− cells and revealed a slight reduction in repair efficiency (Fig. 6A). Thus, both HR-dependent repair of DSBs and gene targeting were compromised less significantly than Ig gene conversion in the 9-1-1 clamp mutants.
Since a previous study indicated that the level of SCE is not affected by the disruption of RAD17 in murine embryonic stem cells (7), we measured the frequency of SCE in DT40 clones. To induce SCE, we exposed cells to cisplatin, a chemical cross-linking agent. We found that the levels of spontaneous and induced SCE were only slightly diminished in RAD9−/− and RAD17−/ DT40 clones (Fig. 6C). To test HR-dependent DSB repair during replication, we measured sensitivity to killing by camptothecin. Surprisingly, and in contrast to the effect seen at the Ig locus, neither the RAD9−/− nor the RAD17−/− clones showed increased sensitivity to camptothecin (Fig. 6B). Consistent with the absence of a major effect on DSB-induced recombination, we found that the kinetics of γ-ray-induced Rad51 focus formation were within the normal range in the RAD9−/− and RAD17−/− clones (data not shown). We conclude that Rad9 and Rad17 are essential for Ig gene conversion, that they play a modest role in gene targeting and HR-dependent DSB repair, and that they are dispensable for cellular tolerance to camptothecin. This suggests that RAD9 and RAD17 play an important role in the initiation of HR from replication-stalling DNA damage but not from DSBs or collapsed replication forks.
DISCUSSION
Improved sensitivity of Ig diversification in DT40 cells as a method for monitoring HR and TLS.
The use of the constitutively diversifying Ig loci of DT40 cells as an assay to measure the induction of both HR and TLS by a defined physiologically created DNA lesion is well established. However, one problem with this approach is the relatively low level of gene conversion and the nontemplated mutation in wild-type cells. This means that long periods of culture and large sequence databases are needed for accurately determining the pattern of diversification.
We were able to achieve substantial improvements in the specificity and sensitivity of this assay by showing that the overexpression of AID can induce an increase in both TLS and short-tract gene conversion. Many of the latter group of mutations appear as single-base changes at A/T base pairs. However, their formation requires both an intact HR apparatus and intact V-pseudogene donors. Thus, although it seems unlikely that they are formed by the same mechanism that is used in mammalian Ig somatic hypermutation at A/T base pairs (23, 24, 29), they provide an additional and useful marker for HR events, thereby increasing the dynamic range of the Ig diversification assay in DT40 cells.
Role of PCNA ubiquitination in TLS and HR.
Using the improved assay described above, we analyzed PCNAK164R cells and clones deficient in Rad9 or Rad17 to understand the role these clamp and clamp-loader proteins play in TLS and HR. In PCNAK164R cells, we found a moderate decrease in the rate of Ig gene conversion, indicating that the ubiquitylation of PCNA contributes to HR as well as to TLS in Ig V diversification. This is consistent with the previously demonstrated role of Polη (20), which is known to be recruited by PCNA ubiquitination (19, 50). We confirmed the finding that PCNA modification contributes slightly to cellular tolerance to camptothecin and HR-dependent DSB repair in an artificial construct. In contrast with the significant decrease in the rate of Ig hypermutation in PCNAK164R cells, we conclude that neither Rad9 nor Rad17 is required for Ig hypermutation, despite recent reports of an interaction between the 9-1-1 complex and some components of the TLS apparatus (18, 32).
A key role for the 9-1-1 complex in Ig gene conversion.
A key finding of this study is the role that the vertebrate 9-1-1 complex plays in gene conversion initiated by abasic sites in the Ig loci. Although general HR factors such as Rad51 and Brca1/2 contribute to every type of HR reaction (i.e., Ig gene conversion, I-SceI-induced DSB repair, camptothecin-induced DSB repair, gene targeting and SCE), RAD9−/− and RAD17−/− cells displayed a severe defect only in Ig gene conversion and not in other types of HR reactions. Thus, these early checkpoint factors may facilitate Ig gene conversion by mechanisms other than general HR. There are a number of possible models for these mechanisms.
Biochemical studies suggest that the 9-1-1 complex may promote recruitment of molecules involved in BER, including APE1, Polβ, and FEN1 (11, 13-15, 47, 49). Indeed, the RAD9−/− and RAD17−/− cells exhibit a marked hypersensitivity to methyl methanesulfonate (22), an agent that induces DNA lesions that are repaired by BER. The idea that a defective 9-1-1 clamp might reduce the activity of APE1 is consistent with the 50% increase observed in the level of Ig nontemplated point mutations in RAD9−/− and RAD17−/− cells (Fig. 4A). Moreover, if an APE1-mediated incision at an abasic site stimulates Ig gene conversion, the reduced activity of APE1 might be expected to diminish the rate of Ig gene conversion. Nonetheless, because of the essential requirement of this endonuclease for cellular proliferation, we believe that the absence of detectable Ig gene conversion events in RAD9−/− and RAD17−/− cells is unlikely to be caused by the loss of APE1 activity (25).
Our data suggest that the 9-1-1 complex plays a specific role in the initiation of recombination from a replication stall that is created, for example, by a replication fork encountering an abasic site in the Ig locus or by a methyl methanesulfonate adduct in the genome, but that it does not play a role in recombination induced by a fork collapse, such as that induced by camptothecin. This model is consistent with the apparently early roles played by Rad9 and Rad17 in the Ig gene conversion reaction. This is suggested by the swing toward a nontemplated PM in these mutants, similar to that seen in mutants of RAD51 paralogs or BRCA2 (17, 35). This notion is consistent with a number of observations made in this study. However, it does not fully explain the decreased HR-dependent repair by gene conversion of an I-SceI-induced DSB, although this defect is modest in comparison with that seen in a major HR mutant such as one of the RAD51 paralogs or BRCA2 (51). In addition, the defect does not mimic exactly the phenotypes seen following the loss of the Rad51 paralogs or Brca2. For example, in terms of the RAD51-focus formation induced by infrared exposure, unlike mutants of the Rad51 paralogs and Brca2, RAD9−/− and RAD17−/− cells display normal Rad51-focus formation following exposure to γ irradiation.
A related possibility assumes that Rad9 and Rad17 are absolutely required for single-strand gap-induced HR but are largely dispensable for DSB-induced HR such as camptothecin-induced fork collapse and gene conversion of an I-SceI-induced DSB. Since the abortive HR in a Rad51 paralog or a Brca2-deficient background causes a shift of Ig V diversification from HR- to TLS-mediated hypermutation, Ig gene conversion may be triggered by single-stranded lesions (17, 35). This requires that the template strand, including the abasic site, remains intact, although the extent to which Ig gene conversion is triggered directly by replication stalling at an abasic site or by a postreplicative single-strand gap remains to be determined.
Last, the 9-1-1 complex is required if two homologous duplex DNAs contain heterologous sequences, such as the selection markers in gene targeting constructs or the I-SceI recognition sequences, neither of which is shared by its gene conversion donor (21). This hypothesis would explain why Rad9 and Rad17 are required for Ig gene conversion, where a sequence divergence of ∼10% is present between the Ig gene conversion donor and the recipient V segments (31). Although our demonstration of a role for the 9-1-1 complex in HR opens up a range of possible roles for this complex in vertebrates, elucidation of its precise function in this context is clearly going to require further work.
Acknowledgments
We thank Y. Sato for technical assistance.
Financial support was provided in part by a Core Research for Evolutional Science and Technology (CREST) grant from the Japan Science and Technology Corporation; by a Center of Excellence (COE) grant for Scientific Research; and by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology of the Japanese Government. J. E. Sale is supported by the Medical Research Council and the Association for International Cancer Research.
REFERENCES
- 1.Adachi, N., S. So, and H. Koyama. 2004. Loss of nonhomologous end joining confers camptothecin resistance in DT40 cells. Implications for the repair of topoisomerase I-mediated DNA damage. J. Biol. Chem. 27937343-37348. [DOI] [PubMed] [Google Scholar]
- 2.Arakawa, H., J. Hauschild, and J. M. Buerstedde. 2002. Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science 2951301-1306. [DOI] [PubMed] [Google Scholar]
- 3.Arakawa, H., G. L. Moldovan, H. Saribasak, N. N. Saribasak, S. Jentsch, and J. M. Buerstedde. 2006. A role for PCNA ubiquitination in immunoglobulin hypermutation. PLoS Biol. 4e366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arakawa, H., H. Saribasak, and J. M. Buerstedde. 2004. Activation-induced cytidine deaminase initiates immunoglobulin gene conversion and hypermutation by a common intermediate. PLoS Biol. 2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bermudez, V. P., L. A. Lindsey-Boltz, A. J. Cesare, Y. Maniwa, J. D. Griffith, J. Hurwitz, and A. Sancar. 2003. Loading of the human 9-1-1 checkpoint complex onto DNA by the checkpoint clamp loader hRad17-replication factor C complex in vitro. Proc. Natl. Acad. Sci. USA 1001633-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bezzubova, O., A. Silbergleit, Y. Yamaguchi-Iwai, S. Takeda, and J. M. Buerstedde. 1997. Reduced X-ray resistance and homologous recombination frequencies in a RAD54-/- mutant of the chicken DT40 cell line. Cell 89185-193. [DOI] [PubMed] [Google Scholar]
- 7.Budzowska, M., I. Jaspers, J. Essers, H. de Waard, E. van Drunen, K. Hanada, B. Beverloo, R. W. Hendriks, A. de Klein, R. Kanaar, J. H. Hoeijmakers, and A. Maas. 2004. Mutation of the mouse Rad17 gene leads to embryonic lethality and reveals a role in DNA damage-dependent recombination. EMBO J. 233548-3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delacroix, S., J. M. Wagner, M. Kobayashi, K. Yamamoto, and L. M. Karnitz. 2007. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 211472-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di Noia, J. M., and M. S. Neuberger. 2007. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 761-22. [DOI] [PubMed] [Google Scholar]
- 10.Ellison, V., and B. Stillman. 2003. Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5′ recessed DNA. PLoS Biol. 1E33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedrich-Heineken, E., M. Toueille, B. Tannler, C. Burki, E. Ferrari, M. O. Hottiger, and U. Hubscher. 2005. The two DNA clamps Rad9/Rad1/Hus1 complex and proliferating cell nuclear antigen differentially regulate flap endonuclease 1 activity. J. Mol. Biol. 353980-989. [DOI] [PubMed] [Google Scholar]
- 12.Fukushima, T., M. Takata, C. Morrison, R. Araki, A. Fujimori, M. Abe, K. Tatsumi, M. Jasin, P. K. Dhar, E. Sonoda, T. Chiba, and S. Takeda. 2001. Genetic analysis of the DNA-dependent protein kinase reveals an inhibitory role of Ku in late S-G2 phase DNA double-strand break repair. J. Biol. Chem. 27644413-44418. [DOI] [PubMed] [Google Scholar]
- 13.Gembka, A., M. Toueille, E. Smirnova, R. Poltz, E. Ferrari, G. Villani, and U. Hubscher. 2007. The checkpoint clamp, Rad9-Rad1-Hus1 complex, preferentially stimulates the activity of apurinic/apyrimidinic endonuclease 1 and DNA polymerase beta in long patch base excision repair. Nucleic Acids Res. 352596-2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guan, X., H. Bai, G. Shi, C. A. Theriot, T. K. Hazra, S. Mitra, and A. L. Lu. 2007. The human checkpoint sensor Rad9-Rad1-Hus1 interacts with and stimulates NEIL1 glycosylase. Nucleic Acids Res. 352463-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guan, X., A. Madabushi, D. Y. Chang, M. E. Fitzgerald, G. Shi, A. C. Drohat, and A. L. Lu. 2007. The human checkpoint sensor Rad9-Rad1-Hus1 interacts with and stimulates DNA repair enzyme TDG glycosylase. Nucleic Acids Res. 356207-6218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris, R. S., J. E. Sale, S. K. Petersen-Mahrt, and M. S. Neuberger. 2002. AID is essential for immunoglobulin V gene conversion in a cultured B cell line. Curr. Biol. 12435-438. [DOI] [PubMed] [Google Scholar]
- 17.Hatanaka, A., M. Yamazoe, J. E. Sale, M. Takata, K. Yamamoto, H. Kitao, E. Sonoda, K. Kikuchi, Y. Yonetani, and S. Takeda. 2005. Similar effects of Brca2 truncation and Rad51 paralog deficiency on immunoglobulin V gene diversification in DT40 cells support an early role for Rad51 paralogs in homologous recombination. Mol. Cell. Biol. 251124-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kai, M., and T. S. Wang. 2003. Checkpoint activation regulates mutagenic translesion synthesis. Genes Dev. 1764-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kannouche, P. L., J. Wing, and A. R. Lehmann. 2004. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell 14491-500. [DOI] [PubMed] [Google Scholar]
- 20.Kawamoto, T., K. Araki, E. Sonoda, Y. M. Yamashita, K. Harada, K. Kikuchi, C. Masutani, F. Hanaoka, K. Nozaki, N. Hashimoto, and S. Takeda. 2005. Dual roles for DNA polymerase eta in homologous DNA recombination and translesion DNA synthesis. Mol. Cell 20793-799. [DOI] [PubMed] [Google Scholar]
- 21.Kikuchi, K., Y. Taniguchi, A. Hatanaka, E. Sonoda, H. Hochegger, N. Adachi, Y. Matsuzaki, H. Koyama, D. C. van Gent, M. Jasin, and S. Takeda. 2005. Fen-1 facilitates homologous recombination by removing divergent sequences at DNA break ends. Mol. Cell. Biol. 256948-6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kobayashi, M., A. Hirano, T. Kumano, S. L. Xiang, K. Mihara, Y. Haseda, O. Matsui, H. Shimizu, and K. Yamamoto. 2004. Critical role for chicken Rad17 and Rad9 in the cellular response to DNA damage and stalled DNA replication. Genes Cells 9291-303. [DOI] [PubMed] [Google Scholar]
- 23.Langerak, P., A. O. Nygren, P. H. Krijger, P. C. van den Berk, and H. Jacobs. 2007. A/T mutagenesis in hypermutated immunoglobulin genes strongly depends on PCNAK164 modification. J. Exp. Med. 2041989-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martomo, S. A., W. W. Yang, R. P. Wersto, T. Ohkumo, Y. Kondo, M. Yokoi, C. Masutani, F. Hanaoka, and P. J. Gearhart. 2005. Different mutation signatures in DNA polymerase eta- and MSH6-deficient mice suggest separate roles in antibody diversification. Proc. Natl. Acad. Sci. USA 1028656-8661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitra, S., T. Izumi, I. Boldogh, K. K. Bhakat, R. Chattopadhyay, and B. Szczesny. 2007. Intracellular trafficking and regulation of mammalian AP-endonuclease 1 (APE1), an essential DNA repair protein. DNA Repair (Amsterdam) 6461-469. [DOI] [PubMed] [Google Scholar]
- 26.Moldovan, G. L., B. Pfander, and S. Jentsch. 2007. PCNA, the maestro of the replication fork. Cell 129665-679. [DOI] [PubMed] [Google Scholar]
- 27.Muramatsu, M., K. Kinoshita, S. Fagarasan, S. Yamada, Y. Shinkai, and T. Honjo. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102553-563. [DOI] [PubMed] [Google Scholar]
- 28.Pandita, R. K., G. G. Sharma, A. Laszlo, K. M. Hopkins, S. Davey, M. Chakhparonian, A. Gupta, R. J. Wellinger, J. Zhang, S. N. Powell, J. L. Roti Roti, H. B. Lieberman, and T. K. Pandita. 2006. Mammalian Rad9 plays a role in telomere stability, S- and G2-phase-specific cell survival, and homologous recombinational repair. Mol. Cell. Biol. 261850-1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rada, C., J. M. Di Noia, and M. S. Neuberger. 2004. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol. Cell 16163-171. [DOI] [PubMed] [Google Scholar]
- 30.Reynaud, C. A., V. Anquez, H. Grimal, and J. C. Weill. 1987. A hyperconversion mechanism generates the chicken light chain preimmune repertoire. Cell 48379-388. [DOI] [PubMed] [Google Scholar]
- 31.Reynaud, C. A., B. Bertocci, A. Dahan, and J. C. Weill. 1994. Formation of the chicken B-cell repertoire: ontogenesis, regulation of Ig gene rearrangement, and diversification by gene conversion. Adv. Immunol. 57353-378. [DOI] [PubMed] [Google Scholar]
- 32.Sabbioneda, S., B. K. Minesinger, M. Giannattasio, P. Plevani, M. Muzi-Falconi, and S. Jinks-Robertson. 2005. The 9-1-1 checkpoint clamp physically interacts with polzeta and is partially required for spontaneous polzeta-dependent mutagenesis in Saccharomyces cerevisiae. J. Biol. Chem. 28038657-38665. [DOI] [PubMed] [Google Scholar]
- 33.Saberi, A., H. Hochegger, D. Szuts, L. Lan, A. Yasui, J. E. Sale, Y. Taniguchi, Y. Murakawa, W. Zeng, K. Yokomori, T. Helleday, H. Teraoka, H. Arakawa, J. M. Buerstedde, and S. Takeda. 2007. RAD18 and poly(ADP-ribose) polymerase independently suppress the access of nonhomologous end joining to double-strand breaks and facilitate homologous recombination-mediated repair. Mol. Cell. Biol. 272562-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sale, J. E. 2004. Immunoglobulin diversification in DT40: a model for vertebrate DNA damage tolerance. DNA Repair (Amsterdam) 3693-702. [DOI] [PubMed] [Google Scholar]
- 35.Sale, J. E., D. M. Calandrini, M. Takata, S. Takeda, and M. S. Neuberger. 2001. Ablation of XRCC2/3 transforms immunoglobulin V gene conversion into somatic hypermutation. Nature 412921-926. [DOI] [PubMed] [Google Scholar]
- 36.Saribasak, H., N. N. Saribasak, F. M. Ipek, J. W. Ellwart, H. Arakawa, and J. M. Buerstedde. 2006. Uracil DNA glycosylase disruption blocks Ig gene conversion and induces transition mutations. J. Immunol. 176365-371. [DOI] [PubMed] [Google Scholar]
- 37.Seo, H., M. Masuoka, H. Murofushi, S. Takeda, T. Shibata, and K. Ohta. 2005. Rapid generation of specific antibodies by enhanced homologous recombination. Nat. Biotechnol. 23731-735. [DOI] [PubMed] [Google Scholar]
- 38.Shinkura, R., S. Ito, N. A. Begum, H. Nagaoka, M. Muramatsu, K. Kinoshita, Y. Sakakibara, H. Hijikata, and T. Honjo. 2004. Separate domains of AID are required for somatic hypermutation and class-switch recombination. Nat. Immunol. 5707-712. [DOI] [PubMed] [Google Scholar]
- 39.Shiomi, Y., A. Shinozaki, D. Nakada, K. Sugimoto, J. Usukura, C. Obuse, and T. Tsurimoto. 2002. Clamp and clamp loader structures of the human checkpoint protein complexes, Rad9-1-1 and Rad17-RFC. Genes Cells 7861-868. [DOI] [PubMed] [Google Scholar]
- 40.Simpson, L. J., and J. E. Sale. 2003. Rev1 is essential for DNA damage tolerance and non-templated immunoglobulin gene mutation in a vertebrate cell line. EMBO J. 221654-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sonoda, E., T. Okada, G. Y. Zhao, S. Tateishi, K. Araki, M. Yamaizumi, T. Yagi, N. S. Verkaik, D. C. van Gent, M. Takata, and S. Takeda. 2003. Multiple roles of Rev3, the catalytic subunit of polzeta in maintaining genome stability in vertebrates. EMBO J. 223188-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sonoda, E., M. S. Sasaki, J. M. Buerstedde, O. Bezzubova, A. Shinohara, H. Ogawa, M. Takata, Y. Yamaguchi-Iwai, and S. Takeda. 1998. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 17598-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sonoda, E., M. S. Sasaki, C. Morrison, Y. Yamaguchi-Iwai, M. Takata, and S. Takeda. 1999. Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol. Cell. Biol. 195166-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takata, M., M. S. Sasaki, S. Tachiiri, T. Fukushima, E. Sonoda, D. Schild, L. H. Thompson, and S. Takeda. 2001. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol. Cell. Biol. 212858-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang, E. S., and A. Martin. 2007. Immunoglobulin gene conversion: synthesizing antibody diversification and DNA repair. DNA Repair (Amsterdam) 61557-1571. [DOI] [PubMed] [Google Scholar]
- 46.Tauchi, H., J. Kobayashi, K. Morishima, D. C. van Gent, T. Shiraishi, N. S. Verkaik, D. vanHeems, E. Ito, A. Nakamura, E. Sonoda, M. Takata, S. Takeda, S. Matsuura, and K. Komatsu. 2002. Nbs1 is essential for DNA repair by homologous recombination in higher vertebrate cells. Nature 42093-98. [DOI] [PubMed] [Google Scholar]
- 47.Toueille, M., N. El-Andaloussi, I. Frouin, R. Freire, D. Funk, I. Shevelev, E. Friedrich-Heineken, G. Villani, M. O. Hottiger, and U. Hubscher. 2004. The human Rad9/Rad1/Hus1 damage sensor clamp interacts with DNA polymerase beta and increases its DNA substrate utilisation efficiency: implications for DNA repair. Nucleic Acids Res. 323316-3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ulrich, H. D. 2006. Deubiquitinating PCNA: a downside to DNA damage tolerance. Nat. Cell Biol. 8303-305. [DOI] [PubMed] [Google Scholar]
- 49.Wang, W., P. Brandt, M. L. Rossi, L. Lindsey-Boltz, V. Podust, E. Fanning, A. Sancar, and R. A. Bambara. 2004. The human Rad9-Rad1-Hus1 checkpoint complex stimulates flap endonuclease 1. Proc. Natl. Acad. Sci. USA 10116762-16767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watanabe, K., S. Tateishi, M. Kawasuji, T. Tsurimoto, H. Inoue, and M. Yamaizumi. 2004. Rad18 guides polzeta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 233886-3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yonetani, Y., H. Hochegger, E. Sonoda, S. Shinya, H. Yoshikawa, S. Takeda, and M. Yamazoe. 2005. Differential and collaborative actions of Rad51 paralog proteins in cellular response to DNA damage. Nucleic Acids Res. 334544-4552. [DOI] [PMC free article] [PubMed] [Google Scholar]