Abstract
Edc3 is an enhancer of decapping and serves as a scaffold that aggregates mRNA ribonucleoproteins together for P-body formation. Edc3 forms a network of interactions with the components of the mRNA decapping machinery and has a modular domain architecture consisting of an N-terminal Lsm domain, a central FDF domain, and a C-terminal YjeF-N domain. We have determined the crystal structure of the N-terminally truncated human Edc3 at a resolution of 2.2 Å. The structure reveals that the YjeF-N domain of Edc3 possesses a divergent Rossmann fold topology that forms a dimer, which is supported by sedimentation velocity and sedimentation equilibrium analysis in solution. The dimerization interface of Edc3 is highly conserved in eukaryotes despite the overall low sequence homology across species. Structure-based site-directed mutagenesis revealed dimerization is required for efficient RNA binding, P-body formation, and likely for regulating the yeast Rps28B mRNA as well, suggesting that the dimeric form of Edc3 is a structural and functional unit in mRNA degradation.
The controlled turnover of eukaryotic mRNA is crucial for regulating gene expression (11, 25, 29, 42). Two general pathways exist to degrade eukaryotic mRNAs, both of which are initiated by deadenylation (23, 35, 39). Following deadenylation, mRNAs can be degraded 3′ to 5′ by the cytoplasmic exosome or, more commonly, are decapped by the Dcp1/Dcp2 decapping enzyme and degraded by the 5′-to-3′ exonuclease, Xrn1p. The formation of an mRNA ribonucleoprotein (mRNP) capable of decapping correlates with the mRNA ceasing translation and forming a translationally repressed mRNA (13, 38). These translationally repressed mRNPs can aggregate in the cytoplasm to form processing bodies (P-bodies), which are dynamic cytoplasmic RNA granules. P-bodies are of interest, as the mRNPs within them have been implicated in translation repression (13, 26), general mRNA decay (14, 44), nonsense-mediated mRNA decay (45, 49), microRNA-mediated translational repression (32, 41), and mRNA storage (5, 6).
An important aspect of understanding the process of mRNA decapping and P-body formation is to understand the interactions and functions of the proteins that form the translationally repressed mRNP capable of decapping and P-body localization. A variety of genetic, biochemical, cell biology, and genomic analyses have indicated that the mRNP capable of decapping and P-body localization contains a variety of conserved proteins that interact with each other and RNA, including the decapping enzyme Dcp1p/Dcp2p, the decapping activators Dhh1p, Lsm1-7p, Pat1p, and Edc3p, the 5′-3′ exoribonuclease Xrn1p, and the Ccr4p-Pop2-Not1p deadenylase complex (20, 38). It has also been shown that P-body formation can be affected by the pool of nontranslating mRNAs (20, 38) and that proteins such as RCK/p54/Dhh1 and Pat1p contribute to P-body formation by increasing the pool of nontranslating mRNAs (13, 40). Recently, Dcp2p was found to contribute to P-body formation, presumably through its multiple interactions with Dcp1p, Dhh1p, Edc3p, and Pat1p, which could act to cross-link and stabilize individual mRNPs, thereby contributing to their aggregation into a larger P-body (46).
One conserved and interesting component of the decapping/P-body mRNP is the Edc3 protein. Edc3p was initially identified as an enhancer of decapping in Saccharomyces cerevisiae (31). Further genomic studies indicated that Edc3p plays a role as a regulator of the degradation of specific mRNAs. In yeast, Edc3p was found to play an important role in the autoregulatory systems of Rps28b, a ribosomal protein (4), and Yra1p, an mRNA export factor (17). In humans, Edc3 interacts with tristetraprolin, an activator of the AU-rich-element-containing mRNA decay pathway, enhancing the decapping and subsequent turnover of AU-rich-element-containing mRNA (21). In humans and plants, Edc3 associates with Hedls and its homolog Varicose in the multiprotein decapping complex (21, 50). A recent study on the mechanism of P-body formation in yeast revealed a novel function of Edc3p as a scaffold for decapping proteins during P-body assembly (15). Given these roles, an understanding of Edc3 structure and function will be important in elucidating the control of mRNA degradation and P-body formation.
Edc3 belongs to the Lsm16 family of proteins, which possess three functional domains: an N-terminal divergent Lsm (Sm-like) domain, a central FDF domain (which contains a conserved FDF amino acid motif), and a C-terminal YjeF-N domain as found in the N-terminal domain of the protein YjeF (Fig. 1A) (1, 3). The Lsm domain is required for Edc3 to promote P-body assembly (15, 48). Analyses with recombinant proteins have demonstrated that the Lsm domain of Edc3p interacts with the Nudix domain of Dcp2p, whereas the FDF domain interacts with the Dhh1 protein as well as Dcp2p (15). Recently, the structures of the Lsm domains of Drosophila melanogaster and human Edc3 have been determined (48). These studies revealed that the Lsm domain of Edc3 adopts a divergent Sm fold that lacks the characteristics of a classical (L)Sm domain for RNA binding and that a conserved patch of surface residues in this domain is required for coimmunoprecipitation with Dcp1 in cell extracts, but not for P-body localization (48).
FIG. 1.
hEdc3 proteins form dimers in solution. (A) Domain architecture of the hEdc3 protein, showing the N-terminal Lsm domain, a central FDF domain, the C-terminal YjeF-N domain, and a putative low-complexity linker region between the Lsm and FDF domains. Numbers below the schematic protein outline represent the amino acid positions for the domain boundaries. (B and C) hEdc3-250C wild type (B) and hEdc3-250C E306A V310A (C) were analyzed by sedimentation equilibrium and fitted to an ideal single-species model. Representative fits for each protein are shown. (D) The graph shows the molar mass distribution, c(M), of the hEdc3-250C wild-type and hEdc3-250C E306A V310A mutant proteins. (E) The graph shows the molar mass distribution, c(M), of hEdc3FL wild-type and hEdc3FL E306A V310A mutant proteins.
All Lsm16 members possess an additional C-terminal YjeF-N domain whose function is still unclear. However, the YjeF-N domain was recently found to be required for Edc3p to promote P-body formation (15). Moreover, based on two-hybrid and phage display analyses, the YjeF-N domain has been suggested to function as a self-interaction domain (15, 22, 33). Several crystal structures of the YjeF-N-containing proteins have been determined. These include an uncharacterized protein, YNL200C, in yeast, Mus musculus apolipoprotein A-I binding protein (AI-BP) (28), a hypothetical protein (TM0922) from Thermotoga maritima, and methylene-tetrahydromethanopterin dehydrogenase (MtdA) from Methylobacterium extorquens AM1 (19). All these structures revealed that YjeF-N domains adopt a Rossmann-like fold, which is a very common protein fold present in many enzyme families, suggesting that YjeF-N-containing proteins may possess enzymatic activity and may be involved in metabolic reactions and pathways.
In this paper, we determined the crystal structure of an N-terminally truncated human Edc3 at a resolution of 2.2 Å. The structure revealed that the YjeF-N domain adopts a divergent Rossmann fold topology. Structural data combined with mutagenesis results indicated that the self-association of hEdc3 is mediated by its YjeF-N domain. hEdc3 is a dimer in solution, and dimerization of hEdc3 is crucial for its functions in RNA binding and P-body aggregation.
MATERIALS AND METHODS
Construction and mutagenesis of hEdc3 and scEdc3 plasmids.
The DNA sequences encoding hEdc3FL, hEdc3-203C (residues 203 to 508), hEdc3-250C (residues 250 to 508), hEdc3-N236 (residues 1 to 236), and hEdc3-N250 (residues 1 to 250) proteins were PCR amplified from a human cDNA clone with primers containing suitable restriction sites and cloned into a vector, pGEX-6P-1, resulting in their fusion to a glutathione S-transferase (GST) sequence. Point mutations in hEdc3FL, hEdc3-250C, and pRP1433 (Flag-tagged Edc3 in yeast centromere plasmid) (15) were created using the QuikChange mutagenesis method (Stratagene). The nucleotide sequences of all the clones were verified by automated DNA sequencing. Plasmids with deletions of scEdc3 domains have been described by Decker et al. (15).
Protein expression and purification.
GST-tagged wild-type and mutant hEdc3 proteins were expressed in Escherichia coli strain BL21(DE3) star (Novagen) and purified using glutathione-Sepharose 4B, MonoQ, and Superdex-200 columns (Amersham). Selenomethionine (SeMet)-substituted hEdc3-250C was purified in the same way except that 20 mM dithiothreitol was used.
Crystallization, data collection, and structure determination.
hEdc3-250C crystals were grown at 20°C by hanging drop vapor diffusion. An equal volume of protein was mixed with the precipitant solution (8% [wt/vol] polyethylene glycol 6000 [PEG 6000] and 0.05 M sodium citrate, pH 5.0). SeMet-substituted crystals were grown under the same conditions. Before data collection, crystals were transferred stepwise to a cryobuffer (25% glycerol, 8% [wt/vol] PEG 6000, and 0.05 M sodium citrate, pH 5.0) and flash-frozen in liquid nitrogen. The crystals belonged to space group C2221 with four molecules per asymmetric unit. The cell parameters were a = 94.4 Å, b = 169.2 Å, c = 163.6 Å, α = β = γ = 90°. SeMet single-wavelength anomalous dispersion (SAD) data (λ = 0.9798Å) were collected at beamline BM14 (European Synchrotron Radiation Facility, Grenoble, France) and were processed with CCP4 programs (10). Twelve selenium sites were located by using the program SnB (36). Refinement of the heavy atom sites and phasing were done with SHARP (16). After density modification and solvent flattening, a partial model built with RESOLVE (47) was used for manual model building with the program COOT (18). The model was refined with CNS (7) and REFMAC5 (37). The final model has a good stereochemistry with a free R-factor of 25.6% and an R-factor of 22.8%.
Native hEdc3-203C crystals were grown at 4°C by hanging drop vapor diffusion. An equal volume of protein was mixed with the precipitant solution (8% [wt/vol] PEG 6000, 0.05 M sodium cacodylate, pH 6.5, and 0.4 M magnesium acetate). Before data collection, crystals were transferred stepwise to a cryobuffer (25% glycerol, 8% [wt/vol] PEG 6000, 0.05 M sodium cacodylate, pH 6.5, and 0.4 M magnesium acetate) and flash-frozen in liquid nitrogen. The crystals belonged to space group I422 with one molecule per asymmetric unit. The cell parameters are a = 119.74 Å, b = 119.74 Å, c = 93.92 Å, α = β = γ = 90°. Native SAD data were collected at beamline ID14-4 (European Synchrotron Radiation Facility, Grenoble, France) and were processed with CCP4 programs (10). The structure was solved at a resolution of 2.8 Å by molecular replacement with Phaser (34) using the coordinates of hEdc3-250C as the search model. The data collection and refinement statistics of hEdc3-250C and hEdc3-203C are shown in Table 1.
TABLE 1.
Data collection, phase determination, and refinement statistics
| Parameter | hEdc3-250C, SeMet SAD data | hEdc3-203C, native data |
|---|---|---|
| Data collection | ||
| Space group | C2221 | I422 |
| Cell dimensions | ||
| a/b c (Å) | 94.40/169.20/163.60 | 119.74/119.74/93.92 |
| α/β/γ (°) | 90/90/90 | 90/90/90 |
| Resolution (Å) | 2.2 | 2.8 |
| Rmerge (%)a | 8.4 (47.5) | 7.2 (28.0) |
| I/σ | 6.5 (1.5) | 7.2 (2.0) |
| Unique reflections (N) | 66,636 | 9,673 |
| Completeness (%) | 100.0 (100.0) | 100.0 (100.0) |
| Redundancy | 8.2 (8.2) | 7.7 (7.9) |
| No. of sites | 12 | |
| Figure of merit after density modification | 0.8 | |
| Refinement | ||
| Resolution range | 20.0-2.2 | 20.0-2.8 |
| No. of reflections used | 63,120 | 8,273 |
| Rworkb/Rfreec | 22.8, 25.6 | 22.9, 29.0 |
| Nonhydrogen atoms | ||
| Proteins (N) | 7,212 | 1,792 |
| Water (N) | 349 | 30 |
| RMS deviations | ||
| Bond length (Å) | 0.008 | 0.006 |
| Bond angle (°) | 1.102 | 0.978 |
Rmerge = ∑h∑j lIj(h) − <I(h)>l/∑h∑j Ij(h), where Ij(h) is the jth measurement of reflection of indices h and <I(h)> is the mean intensity.
Rwork = ∑h‖Fo(h)|−|Fc(h)‖/∑h|F0(h)|, where Fo(h) and Fc(h) are the observed and calculated structure factor amplitudes, respectively, for reflection h calculated against all data included in the refinement.
Rfree was calculated similarly to Rwork for 5.0% of randomly chosen reflections not included at any stage in the refinement.
Analytical ultracentrifugation.
The experiments were performed using a Beckman ProteomeLab XL-A analytical ultracentrifuge equipped with a UV-VIS optics detection system, using an An-60 Ti rotor at 20°C. The viscosity (η = 1.0259E-2) and density (ρ = 1.00704) of the buffer used and the partial specific volume (ν) of the hEdc3-250C wild-type and mutant proteins were determined using the Sednterp program (version 1.09; http://www.rasmb.bbri.org). The ν values of hEdc3FL, hEdc3-250C wild-type, and hEdc3-250C E306A V310A mutant proteins were calculated from the amino acid content to be 0.7310 ml/g and 0.7455 ml/g. The molecular masses of hEdc3FL, hEdc3-250C wild-type, and hEdc3-250C E306A V310A mutant proteins were calculated to be 56,077 Da, 28,554 Da, and 28,468 Da, respectively. Prior to centrifugation, the protein samples were dialyzed exhaustively against the buffer blank, which consisted of 20 mM Tris-HCl, pH 8.5, 200 mM NaCl, and 0.5 mM Tris (2-carboxyethyl) phosphine hydrochloride.
Sedimentation velocity.
Sedimentation velocity experiments were run at 42,000 rpm in quartz cells fitted with double-sector centerpieces. Absorption measurements were made at 280 nm, collected every 300 s until the boundaries were at the cell bottom. The data were analyzed by two methods, the c(S) and c(M) methods of the SEDFIT program (43).
Sedimentation equilibrium.
Sedimentation equilibrium experiments were performed using quartz cells fitted with six-channel centerpieces. The sedimentation equilibrium runs were carried out at multiple speeds (12,000, 15,000, and 18,000 rpm) and wavelengths (230, 250, and 280 nm). The samples were run for 22 h at each speed plus an additional 2 h for the collection of the scans. After the equilibrium scans, a high-speed centrifugation run (42,000 rpm) was done to determine any residual absorbance to set initial baseline offset values. The extinction coefficient at 280 nm of 24,430 M−1 cm−1 was calculated from the amino acid composition, using the ProtParam tool of the ExPASy website (http://au.expasy.org/cgi-bin/protparam). Weight-average buoyant molecular weights of hEdc3-250C and its mutant were determined by fitting data to a Monomer-Nmer model and Ideal species model, respectively, using the HeteroAnalysis program (http://www.biotech.uconn.edu/auf/) (12).
In vitro RNA binding assay.
Surface plasmon resonance (SPR) was carried out on a Biacore 2000 instrument. The 5′-end biotin-labeled single-stranded RNA (TGTCATTCGAGTACAGTCTGTTCAGCTAGTCTCC), purchased from CureVAc, was attached to a streptavidin-coated sensor chip (Biacore). A buffer of 10 mM Tris, pH 8.5, 150 mM NaCl, 3 mM EDTA, and 0.005% (vol/vol) Tween 20 was flowed across the chip until the trace leveled off. The biotin-labeled RNA was then attached to flow cell 2 by injecting 20 μl of 100 nM RNA in 0.3 M NaCl at a flow rate 5 μl/min. After immobilization, flow cell 2 and reference flow cell 1 were blocked with 100 μl of 1 mg/ml biotin at 5 μl/min. A binding buffer of 20 mM Tris, pH 8.5, 100 mM NaCl, 2 mM MgCl2, 2 mM dithiothreitol, and 0.002% (vol/vol) Tween 20 was flowed across flow cells 1 and 2. Typically, 100 μl of a 400 nM protein sample was injected into the same buffer across the chip at 50 μl/min. The data were analyzed and fitted to the different models, the 1:1 binding mode mass transfer, bianalyte model, and two-state conformation change, using the software program BIAevaluation 3.1.
Yeast two-hybrid assay.
Two-hybrid fusion plasmids of scEdc3 mutant alleles were constructed by homologous recombination of PCR products containing the mutant alleles into pOAD or pOBD-2 in yeast strains PJ69-4A and PJ69-4α (27) as described in reference 8. Two-hybrid plasmids and strains were obtained from the Yeast Resource Center (provided by S. Fields, University of Washington, Seattle). Construction of Dcp2 (residues 102 to 300) and wild-type full-length scEdc3 two-hybrid plasmids was described previously (15). Interactions were measured by β-galactosidase plate assays in diploids containing pOAD and pOBD-2 derivatives. Growth of diploids containing the pOAD and pOBD-2 derivatives was assayed on minimal medium lacking leucine, tryptophan, and histidine and containing 100 mM 3-aminotriazole.
Microscopy.
Strains yRP2340 (edc3Δ lsm4ΔC Dcp2GFP), yRP2232 (edc3Δ Lsm1GFP), yRP2240 (edc3Δ Pat1GFP), and yRP2248 (edc3Δ Xrn1GFP) (15) containing pRS200 (empty plasmid), pRP1433 (Flag-tagged Edc3 plasmid), or pRP1601 (Flag-tagged R360A H361A Edc3 plasmid) were grown at 30°C in synthetic complete medium supplemented with appropriate amino acids and 2% glucose to an optical density at 600 nm (OD600) of 0.3 to 0.4, collected by centrifugation, washed, and resuspended in synthetic complete medium without glucose. Cells were incubated in a flask in a shaking water bath for 10 min and then analyzed by microscopy using a Deltavision RT (Applied Precision, Inc., Issaquah, WA), with an Olympus 100×, 1.4 numerical aperture objective. Images were collected as 512 by 512 pixel files with a CoolSnap camera (Photometrics, Tucson, AZ) using 1 by 1 binning. Images are a Z-series compilation of 15 images made using Image J, and all images from the same strain were adjusted to the same contrast range. Quantitation of the number of foci per cell was performed using Image J.
Western blot analysis.
The level of Flag-tagged Edc3 protein in extracts from yRP2340 containing pRS200, pRP1433, or pRP1601 was detected using anti-Flag M2 monoclonal antibody (Sigma).
Rps28B mRNA analysis.
yRP2340 containing the scEdc3 plasmids was grown to an optical density at 600 nm of 0.3 to 0.4 and collected by centrifugation, total RNA was extracted (9), and the amount of Rps28B mRNA was quantified by Northern analysis using an oligonucleotide complementary to Rps28B (5′-CATCATTGACTATTTCGACGCATTTG-3′). Loading corrections were performed as described previously using SCR1 RNA, the RNA component of the signal recognition particle, as a control (9).
Protein structure accession numbers.
The coordinates and structure factors for hEdc3-203C and hEdc3-250C have been deposited in the Protein Data Bank (PDB; accession codes 3D3K and 3D3J).
RESULTS
hEdc3 proteins are dimeric in solution.
We expressed and purified full-length human Edc3 (hEdc3FL) and two N-terminal-truncated proteins denoted as hEdc3-203C and hEdc3-250C. hEdc3-203C (residues 203 to 508) contains the central FDF and the C-terminal YjeF-N domains, while hEdc3-250C contains just the C-terminal YjeF-N domain (Fig. 1A). Since the conserved YjeF-N domain of yeast Edc3p has been implicated in mediating Edc3 self-interaction (15, 22, 33), we set out to examine the oligomeric state of hEdc3-250C and hEdc3FL in solution. The calculated molecular mass of monomeric hEdc3-250C is 28,554 Da, based on its amino acid composition. However, gel filtration showed that hEdc3-250C is dimeric in solution (data not shown). To confirm the gel filtration result, this protein was further characterized using analytical ultracentrifugation. hEdc3-250C was analyzed by sedimentation equilibrium over a wide concentration range. The data for this protein fit well to a Monomer-Nmer model and gave very similar weight-averaged molecular weights to the formula mass of a dimer, showing that hEdc3-250C dimerizes via its YjeF-N domain (Fig. 1B). No concentration dependence was seen over the concentration range used for this protein, indicating that the dimer interface has high affinity. Sedimentation equilibrium analysis on hEdc3FL showed the protein was somewhat aggregated in solution (data not shown).
Both hEdc3-250C and hEdc3FL were also analyzed by sedimentation velocity to confirm that both the full-length and truncated proteins are dimeric. The sedimentation velocity data were analyzed using direct fitting of the boundaries using numeric solutions of the Lamm equation to derive the sedimentation coefficient distribution, c(S), which can then be transformed to a molar mass distribution, c(M), that allows the estimation of the molecular weight of the protein (43). The sedimentation velocity data of the hEdc3-250C protein fit well and gave rise to a single peak at 3.6S, which corresponds to a molecular mass of 51,000 Da (Fig. 1D). This value is very close to a dimeric hEdc3-250C, which is calculated to be 57,108 Da. A single species observed in the distribution profiles indicates the monodispersity of the protein sample. The frictional ratio is calculated to be 1.26, which estimates that the shape of the hEdc3-250C dimer is ellipsoidal. The major peak in the c(S) of the hEdc3FL protein yields a sedimentation coefficient of 5.3S, which corresponds to a molecular mass of 94,500 Da (Fig. 1E), which is close to a dimeric hEdc3FL protein (the calculated molecular mass of the hEdc3FL dimer is 112,154 Da). These results demonstrate that the hEdc3 protein exists as a dimer in solution and the YjeF-N domain is sufficient for dimer formation.
Structure determination.
Both full-length human and N-terminal-truncated proteins were screened for crystallization conditions, and the constructs hEdc3-250C and hEdc3-203C yielded diffraction quality crystals. The hEdc3-250C protein was crystallized in space group C2221, with four molecules (A, B, C, and D) in the crystallographic asymmetric unit. The structure was solved by SAD, using the data obtained from SeMet-substituted crystals. These SAD data yielded an easily interpretable electron density map at a resolution of 2.2 Å, allowing 95% of the model to be built automatically. Some regions of the polypeptide chain are not visible in the electron density map and are assumed to be disordered, namely, residues 250 to 257, 265 to 267, 324 to 332, and 448 to 452 in each molecule in AU. As no substantial differences were observed between the structures of molecules A, B, C, and D (root mean square [RMS] deviation of 0.60 Å for all the equivalent Cα atoms), all subsequent analyses reported here will be based on molecule A. The final refined model has an R-factor value of 22.8% and an Rfree value of 25.6% with very good stereochemistry. The statistics of structural determination of hEdc3-250C are shown in Table 1.
The hEdc3-203C protein was crystallized in space group I422 with one molecule in the crystallographic asymmetric unit. The structure was solved at a resolution of 2.8 Å by molecular replacement with Phaser (34) using the coordinates of hEdc3-250C as the search model. The N-terminal region encompassing the FDF motif (amino acids 203 to 258) is disordered. As the rest of the structure is identical to the structure of hEdc3-250C, only the structure of hEdc3-250C will be used for the subsequent analysis and discussion.
Overall structure description.
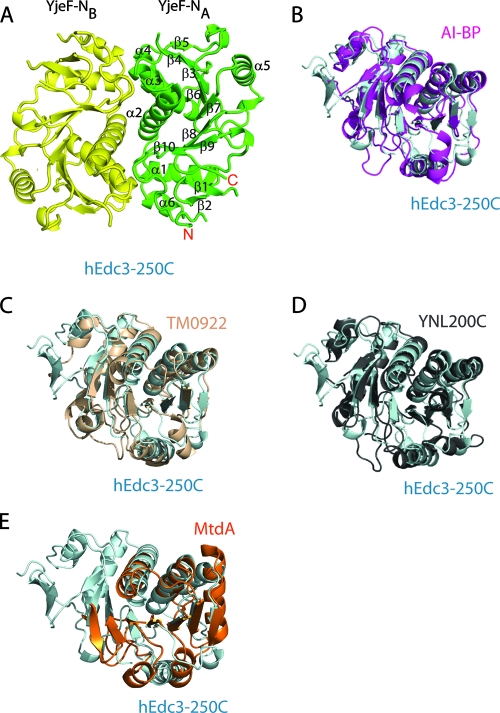
hEdc3-250C forms a tight dimer with each subunit related by a twofold noncrystallographic symmetry. The overall structure of hEdc3-250C resembles a Rossmann-like fold with each subunit consisting of six α-helices, three 310-helices, and 10 β-strands. A notable feature of the tertiary structure is the formation of a three-layer α-β-α sandwich with a central eight-stranded β-sheet surrounded by six α-helices and three 310-helices (Fig. 2A). The core β-sheet is curved around helices α2, α3, and α4 to form a half-barrel structure. The β-strands 3, 4, 5, 6, 7, 8, and 9 are arranged in a parallel fashion with strand β10 antiparallel to all others. In addition to the core β-sheet, strands β1 and β2 form a small β-hairpin which packs against helix α6. This region precedes the YjeF-N domain and may belong to part of the FDF domain, which is disordered in the structure.
FIG. 2.
Structure of hEdc3-250C and comparison of it with other YjeF-N proteins. (A) Ribbon diagram of the hEdc3-250C homodimer formed by molecules A (green) and B (yellow). Secondary structure elements are labeled. (B) Diagram of superimposed structures of hEdc3-250C (cyan) and AI-BP (PDB 2o8n; magenta). (C) Diagram of superimposed structures of hEdc3-250C (cyan) and a hypothetical protein (TM0922) from Thermotoga maritima (PDB 2ax3) (wheat colored). (D) Diagram of superimposed structures of hEdc3-250C (cyan) and a yeast hypothetical protein, YNL200C (PDB 1jzt) (gray). (E) Diagram of superimposed structures of hEdc3-250C (cyan) and methylene tetrahydromethanopterin dehydrogenase (MtdA) (PDB 1lua) (orange). The view of hEdc3-250C is the same in panels B, C, D, and E.
Structural comparison with other YjeF-N-containing proteins.
A search in the Protein Data Bank using the DALI server revealed two structural homologs, AI-BP from Mus musculus (PDB entry 2o8n; Z-score, 23.8) and a hypothetical protein (TM0922) from Thermotoga maritima (PDB 2ax3; Z-score, 20.6). Further search by removal of the N-terminal β-hairpin region identified two more structural homologs, a hypothetical protein, YNL200C, from S. cerevisiae (PDB 1jzt; Z-score, 22.8) and methylene-tetrahydromethanopterin dehydrogenase (MtdA) from Methylobacterium extorquens (PDB 1lua; Z-score, 8.4). Although the sequence of human Edc3 shows only ∼12% identity with AI-BP, TM0922, YNL200C, and MtdA, superposition of the equivalent Cα atoms of hEdc3-250C with these YjeF-N-containing proteins gives rise to RMS deviation values of 1.4 Å, 1.5 Å, 1.4 Å, and 1.6 Å for AI-BP (Fig. 2B), TM0922 (Fig. 2C), YNL200C (Fig. 2D), and MtdA (Fig. 2E), respectively, suggesting the core regions of the YjeF-N domains in these structures are very similar. The best match is the central β-sheet, and the largest deviation is in the loop regions. The unique feature of hEdc3-250C is the presence of the β-hairpin in the N-terminal region, which is not present in other YjeF-N-containing proteins.
MtdA is an NADP-dependent methylene-H4MPT dehydrogenase, an enzyme that catalyzes the dehydrogenation of methylene-tetrahydromethanopterin and methylene-tetrahydrofolate, using NADP as the cosubstrate (19). The structural similarity of Edc3 with MtdA raises the possibility that Edc3 might bind a nucleotide or nucleotide-containing cofactor. However, structural comparison showed that binding of NADP or a similar cofactor to Edc3p (data not shown) would require substantial structural changes, suggesting that Edc3 may bind, if anything, a different cofactor.
Comparison of the related structures suggests the YjeF-N domain of Edc3 may bind a small molecule that affects its function. Specifically, in the structure of AI-BP, a sulfate ion has been found to be situated in a deep pocket (Fig. 3B). Similarly positioned pockets with a bound chloride ion and a glycerol molecule have been observed in YNL200C (Fig. 3C) and TM0922 (Fig. 3D), respectively. Structural superposition showed that hEdc3-250C has a similar pocket (Fig. 3E) which is located adjacent to the dimer interface and overlaps with the NADP binding site of MtdA (Fig. 3A). The importance of this pocket in Edc3 is underscored by the fact that it contains three invariant residues, E295, E415, and D443. Similarly, the pocket in AI-BP also contains a negatively charged invariant residue, D188. The existence of a common pocket in these YjeF-N domain-containing proteins suggests that they may commonly bind small molecules in their function.
FIG. 3.
Surface view of hEdc3 and its structural homologs, showing the putative ligand binding pockets. (A) MtdA (orange) with bound NADP (in stick model). (B) AI-BP (magenta) with a bound sulfate ion (in stick model) in the putative binding pocket. (C) YNL200C (gray) with a bound chloride ion (in yellow dot model). (D) The YjeF-N domain of TM0922 (in wheat) with a bound glycerol molecule (in stick model). (E) hEdc3-250C (cyan) with a close-up view showing the residues located in the pocket.
The YjeF-N domain mediates dimerization of Edc3.
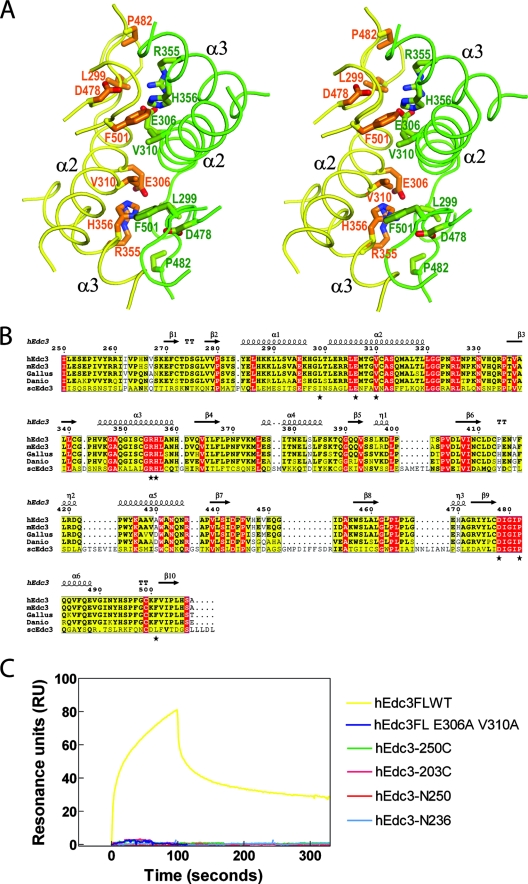
The structure of the YjeF-N domains suggested a possible basis for dimer formation. hEdc3-250C forms a tight homodimer in a crystallographic asymmetric unit, which is consistent with our gel filtration results (data not shown), sedimentation velocity, and equilibrium analyses (Fig. 1B and D), showing this domain of Edc3 can form a dimer. In the dimer interface observed in the structure, the YjeF-N domains interact extensively with a buried accessible surface area of 3,737.44 Å2. Specifically, helices α2 and α3 in each subunit interact with three loop regions connecting α1 and α2, β9 and α6, and α6 and β10 in the other subunit through hydrophobic, van der Waals, and hydrogen-bonding interactions (Fig. 4A). The extensive dimer interface and the involvement of some highly conserved residues (e.g., L299, E306, V310, R355, H356, D478, P482, and F501) (Fig. 4B) in the dimer interface suggest that this interface is the basis of Edc3 dimerization.
FIG. 4.
Dimer interface of the YjeF-N domain of hEdc3-250C. (A) Stereo diagram with the YjeF-N domain dimer interface of hEdc3-250C is shown in green for chain A and yellow for chain B. Residues involved in dimerization are shown in stick models and labeled in dark green and brown for chains A and B. (B) Alignment of amino acid sequences of human, mouse, chicken, zebrafish, and S. cerevisiae Edc3 proteins. The secondary structures shown above the sequences are for hEdc3. Mutated residues involved in YjeF-N domain dimerization are marked with an asterisk. Invariant residues are represented in white letters on a red background, similar residues are in black with bold and unbold letters on a yellow background, and others are in black and unbold letters. Residues with similarities between 0.7 and 1.0 as defined in EScript (24) are boxed in blue. (C) Sensorgram showing single-stranded RNA binding activity of 400 nM hEdc3FL, hEdc3FL E306A V310A, hEdc3-N250, hEdc3-N236, hEdc3-203C, and hEdc3-250C.
To directly test whether residues in the interface are important for dimerization, we mutated residues R355, H356, D478, P482, F501, L299, E306, and V310 to Ala in the context of the full-length human Edc3 protein. Double or triple mutants were generated and expressed in E. coli. However, only the double mutant E306A V310A was soluble, with the rest of the mutants present in the insoluble fraction. Importantly, analytical ultracentrifugation analysis showed that the E306A V310A double mutant is present in monomeric form in solution, in contrast to the dimeric form of the wild-type protein (Fig. 1E). Similarly, mutation of E306 and V310 to Ala in hEdc3-250C also converted this truncated protein from a dimeric to a monomeric form (Fig. 1C and D). Moreover, CD spectroscopy showed that the wild type and double mutant for both full-length and hEdc3-250C proteins had very similar spectra, suggesting that the double mutation had no significant effect on the general folding or stability of the Edc3 protein (see Fig. S1 in the supplemental material). These results indicated that residues Glu306 and Val310 play an important role in dimerization and argue that the dimer interface seen in the structure is the basis of Edc3 dimerization in solution.
To examine further if the residues in the dimer interface are important for self-interaction of the YjeF-N domain, we took advantage of the fact that Edc3 is a conserved protein with a similar role in mRNA decay from yeast to mammals. Given this, we asked if mutations in the Edc3 interface disrupted the ability of S. cerevisiae Edc3p to interact with itself in a two-hybrid assay as previously reported (15). Two triple mutants, I303A E310A V314A and D518A P522A F541A, and one double mutant, R360A H361A, in S. cerevisiae Edc3p were generated, which corresponded to the human mutants L299A E306A V310A, D478A P482A F501A, and R355A H356A, respectively. Two-hybrid plasmids were constructed to express activation domain and binding domain fusions of the full-length Edc3 protein with the mutant alleles.
Given the difficulty of expressing Edc3 variants with mutations in the interface, we first determined if the Edc3 variants would form functional proteins by testing their ability to interact with Dcp2 in a two-hybrid assay, which requires the Lsm domain (15). Any mutant that fails to interact with Dcp2 due to a lesion in the YjeF-N domain is likely to be misfolded or unstable, and therefore its ability to interact with Edc3p cannot be reliably interpreted. We observed that only the double mutant R360A H361A retained the ability to interact with Dcp2 (residues 102 to 300) (Fig. 5A and data not shown). Interestingly, the R360A H361A mutation reduced the ability of Edc3 to interact with itself, similar to what is observed when the entire Yjef-N domain is deleted (Fig. 5A), providing additional evidence that the interface seen in the structure provides the basis for Edc3 dimerization. Collectively, the results from the yeast two-hybrid and the analytical ultracentrifugation experiments demonstrated that the YjeF-N domain mediates the dimerization of Edc3 protein.
FIG. 5.
Effect of point mutations in the dimer interface of the YjeF-N domain on the two-hybrid interaction of Edc3 with itself and on P-body aggregation in S. cerevisiae. (A) Two-hybrid interaction of Gal4 binding domain (BD) fusions of wild-type full-length Edc3p, Edc3p lacking the C-terminal Yjef-N domain (16), and the R360A H361A mutant of Edc3p with Gal4 activation domain (AD) fusions of full-length wild-type Edc3p, the catalytic domain of Dcp2p, and the activation domain alone, assayed by growth on plates containing 100 mM 3-aminotriazole. (B) Western analysis showing the levels of the Flag-tagged R360A H361A version of Edc3p relative to wild-type Flag-tagged Edc3p in edc3Δ lsm4ΔC cells. (C) Localization of GFP-tagged proteins to P-bodies after 10 min of glucose depletion in strains lacking Edc3p and the C-terminal domain of Lsm4p (edc3Δ lsm4ΔC) or Edc3p alone (edc3Δ) expressing wild-type or mutant Flag-tagged versions of Edc3p. The average number of GFP foci per cell was 3.0 ± 0.0 in the wild type and 1.1 ± 0.9 in the R360A H361A mutant for Dcp2GFP, 0.7 ± 0.5 in the wild type and 0.4 ± 0.3 in the R360A H361A mutant for Pat1GFP, 1.9 ± 0.7 in the wild type and 0.2 ± 0.3 in the R360A H361A mutant for Lsm1GFP, and 2.4 ± 0.2 in the wild type and 0.7 ± 0.2 in the R360A H361A mutant for Xrn1GFP.
Dimerization of Edc3 is required for RNA binding.
An unresolved issue is whether the Edc3 protein can bind RNA. Based on their features and relationship to other proteins, the Lsm and FDF domains have been postulated to be involved in RNA binding (1, 2, 30, 51). However, analysis of the Lsm domain suggests that it is not capable of binding RNA, although the ability of full-length Edc3 to bind RNA was not examined (48). To examine whether Edc3 can bind RNA we examined the interaction of full-length hEdc3 with a single-stranded 34-nucleotide RNA oligomer by SPR assay. The results (Fig. 4C) showed that hEdc3FL exhibits RNA binding with an estimated Kd of 60 μM, demonstrating that Edc3 is an RNA binding protein.
To determine what domains of Edc3 were involved in RNA binding, we examined the ability of different Edc3 fragments or point mutants to bind RNA (Fig. 4C). We observed that the N-terminal 250 residues which encompass the Lsm and FDF domains, hEdc3-N250, had no RNA binding at all at a protein concentration of 400 nM (Fig. 4C) and showed very weak RNA binding when the protein concentration was increased to 3,000 nM (see Fig. S2 in the supplemental material). This observation suggests that this region of Edc3 has some intrinsic RNA binding capabilities that are enhanced by the presence of the YjeF-N domain. Consistent with the primary region that interacts with RNA being the Lsm and FDF domains, hEdc3-250C, which contains only the YjeF-N domain, failed to bind RNA. Moreover, a construct with the Lsm domain and only a part of the FDF domain (hEdc3-N236) and a construct with just the FDF domain and the YjeF-N domain (hEdc3-203C) failed to bind to RNA. These latter results suggest that both the Lsm and FDF domains are required for RNA binding. Bioinformatics analysis showed that the FDF domain is enriched in polar and charged residues with few hydrophobic residues and is predicted to adopt an entirely α-helical structure with multiple exposed hydrophilic loops (3). These features of the FDF domain are consistent with it having a role in RNA binding. Taken together, these results indicate that the Edc3 protein can bind RNA in a manner dependent on both the Lsm and FDF domains and enhanced by the presence of the YjeF-N domain.
One possible role of the YjeF-N domain in enhancing RNA binding would be to dimerize the protein and thereby possibly create a larger or more stable RNA binding surface. To test this possibility, we examined how the E306A V310A mutations, which prevent dimerization, affected RNA binding of the full-length Edc3. Using the SPR assay, we observed that the double mutant E306A V310A, which is monomeric in solution, is unable to bind RNA (Fig. 4C). We interpret this observation to indicate that dimerization of Edc3 enhances RNA binding.
Dimerization of Edc3p is required for efficient P-body aggregation in yeast.
To examine the role of Edc3 dimerization in vivo, the R360A H361A interface mutant was made in a Flag-tagged version of scEdc3 on a centromere-based plasmid, and its effects on the protein levels and P-body formation in Saccharomyces cerevisiae were examined. The effect of the R360A H361A mutant on P-body aggregation was first assessed in a strain that is severely defective for P-body aggregation because it carries a deletion of Edc3 and the C-terminal prion-like domain of Lsm4p (Dcp2-GFP edc3Δ lsm4ΔC) (15). The accumulation of Dcp2GFP into P-bodies in response to glucose deprivation was reduced in cells expressing the R360A H361A mutant compared to those expressing the wild-type version of Edc3 (Fig. 5C, top panel). The R360A H361A mutant protein was expressed at twofold-lower levels than the wild-type protein (Fig. 5B); however, it is unlikely that this modest effect on protein levels is responsible for the defect in P-body accumulation, given that deletion of the FDF domain was previously found to result in a similar decrease in protein levels but had no effect on P-body formation (15). The reduced accumulation of Dcp2p in P-bodies in the edc3Δ lsm4ΔC strain in the presence of the R360A H361A mutant suggested that dimerization of Edc3 is required for efficient P-body assembly.
To confirm that dimerization of Edc3p is important for P-body assembly, we examined whether the localization of additional proteins to P-bodies was defective in the presence of the R360A H361A mutant. The effect of the R360A H361A mutant on the accumulation of the decapping activators Pat1p and Lsm1p and the 5′-to-3′ exonuclease Xrn1p in P-bodies in response to glucose depletion was examined in edc3Δ strains containing green fluorescent protein (GFP)-tagged versions of these proteins integrated in the genome. The accumulation levels of Pat1GFP, Lsm1GFP, and Xrn1GFP were all reduced in cells expressing the mutant protein compared to cells expressing the wild-type version of Edc3 (Fig. 5C). The partial defect in the P-body localization of all four proteins examined is consistent with the hypothesis that dimerization of Edc3p is required for efficient P-body assembly in yeast.
The YjeF-N Edc3p domain is required for regulation of Rps28B mRNA in yeast.
The above results indicate that the YjeF-N domain of Edc3 plays an important role in the dimerization of Edc3p which can then affect its ability to assemble P-bodies. Previous work showed that the YjeF-N domain of Edc3p is not required for Edc3p to be able to enhance the activity of Dcp2p in vivo, which is consistent with Dcp2 interacting with the Lsm and FDF domains (15). Edc3p also functions as an mRNA-specific regulator of decapping and enhances the decapping rate of the Rps28B mRNA (4). Given this specific role of Edc3p in controlling the Rps28B mRNA, we examined what domains of Edc3p were required for this mRNA-specific function.
We observed that the level of Rps28B mRNA was increased about twofold in an edc3Δ lsm4ΔC strain carrying an empty vector plasmid compared to the strain with Edc3 expressed from a centromere plasmid (Fig. 6). This result is similar to what was reported by Badis et al. (4) when they compared Rps28B mRNA levels in wild-type and edc3Δ strains, although they observed a larger increase in Rps28B mRNA levels (four- to fivefold). Moreover, we observed that Edc3 variants lacking the Lsm or YjeF-N domains led to a similar increase in the Rps28B mRNA levels, while a variant lacking the FDF domain showed normal levels of Rps28B mRNA (Fig. 6). These results indicate that the YjeF-N domain and the Lsm domain of Edc3 are required for its role in the regulation of the Rps28B mRNA. Moreover, the R360A and H361A mutant in the YjeF-N domain increased Rps28B mRNA levels about twofold (Fig. 6), which implies that Edc3p dimerization may be required for Rps28B mRNA regulation.
FIG. 6.

Effect of deletion of Edc3p domains and point mutations in the dimer interface of the YjeF-N domain on Rps28B mRNA levels in S. cerevisiae. Northern analysis results show Rps28B mRNA levels in the edc3Δ lsm4ΔC strain with no Edc3p, wild-type Flag-tagged Edc3p, or mutant versions of Edc3p expressed from a centromere plasmid. Scr1RNA, the RNA component of the signal recognition particle, was used as a loading control.
DISCUSSION
The Edc3 protein is a modular protein involved in mRNA degradation and assembly of P-bodies. Computational analysis identified three distinct domains within Edc3 (1, 3). At the N terminus is an Lsm domain which might be anticipated to bind RNA based on its relationship with other Sm and Lsm domains. However, the structure of the isolated Lsm domain of Edc3 protein showed that it lacks the features that canonical Lsm domains require for RNA binding (48), implying that it is unlikely to bind to RNA on its own. Indeed, analysis of the Drosophila melanogaster and human Edc3 Lsm domains (48) indicates that the Lsm region is insufficient for binding RNA. Instead, the Lsm domain of Edc3 has been shown to directly interact with Dcp2p (15) and to coimmunoprecipitate from cells with Dcp1p (48), suggesting the possibility of a direct interaction with Dcp1p as well. The Lsm domain, most likely through its interaction with Dcp2p, is required for Edc3p's function in P-body assembly (15, 48) and for targeting Edc3p to P-bodies (15, 48). The central FDF domain binds to Dhh1 and Dcp2 (15), whereas the C-terminal domain is predicted to self-interact (15, 22, 33).
Several lines of evidence now demonstrate that Edc3p forms a dimer through the YjeF-N domain. First, the structure of the YjeF-N domain reveals an extensive and conserved interface between two molecules (Fig. 2). Second, gel filtration, sedimentation equilibrium, and sedimentation velocity all demonstrate that both the full-length Edc3 protein and the isolated YjeF-N domain are dimers in solution (Fig. 1). Third, mutations in the YjeF-N interface disrupt dimer formation in hEdc3 in solution and reduce two-hybrid interactions between S. cerevisiae Edc3 molecules (Fig. 1 and 5A).
Our analysis indicates that Edc3p can bind RNA and that dimer formation is important for RNA binding. The key observation is that full-length hEdc3 binds to RNA while mutations that disrupt dimer formation also abolish RNA binding (Fig. 4). Interestingly, the full-length Edc3 binds RNA while the YjeF-N domain alone cannot bind RNA (Fig. 4), suggesting a role for the central FDF domain and/or the Lsm domain in RNA binding. Consistent with this view, a construct containing both the Lsm and FDF domains showed very weak RNA binding at a high protein concentration (Fig. 4; see also Fig. S2 in the supplemental material). One possible explanation for these observations is that the dimerization of the YjeF-N domain brings the Lsm and/or FDF domain from both subunits into close proximity to form a large channel or platform for RNA binding.
The Edc3p protein has been shown to function as a scaffolding protein that provides a site for bringing the components of the decapping complex together to form mRNPs for P-body assembly (15, 48). The YjeF-N domain is critical to P-body assembly and has been speculated to function to bring together different complexes to aggregate mRNPs (15). This hypothesis is now supported by the demonstration that the YjeF-N domain of Edc3p can interact with itself and form dimers in solution (Fig. 1). Moreover, this dimer formation appears to be important for P-body assembly, since mutations that disrupt this interface also reduce P-body assembly in S. cerevisiae (Fig. 5). The simplest interpretation of this observation is that dimerization of Edc3 molecules links together individual mRNPs and contributes to assembly of higher-order aggregates. However, it is also possible that the role of the YjeF-N domain in P-body assembly is to enhance RNA binding by Edc3, which might lead to cross-bridging interactions between mRNPs. Future work should be able to resolve these possibilities.
We have also found that the Yjef-N domain of scEdc3p is required for the regulation of Rps28B mRNA levels. In contrast, the Yjef-N domain is not required for stimulating decapping of the MFA2 or PGK1 mRNAs (15). There are two possible explanations for why the Yjef-N domain is specifically required for the regulation of Rps28B mRNA and not for Edc3p to stimulate decapping in general. First, the ability of the Edc3 protein to dimerize and/or form P-bodies may be required for the regulation of this particular mRNA. Alternatively, since the level of Rps28B mRNA is regulated by the binding of the Rps28b protein to the 3′ untranslated region of its mRNA, which then recruits Edc3p (4), it could be that the Yjef-N domain is required for Edc3p to interact with the Rps28b protein. The observation that the R360A H361A mutation disrupts Rps28b mRNA regulation is more consistent with the hypothesis that dimerization and/or P-body assembly is important for the regulation of the decay of this specific mRNA.
In summary, the results presented here show that the YjeF-N domain mediates the dimerization of Edc3p, and this dimerization greatly enhances Edc3p's ability to bind RNA. The self-interaction of the YjeF-N domain may also contribute to P-body assembly by linking mRNPs together through the direct interaction of decapping proteins with the Lsm and FDF domains. Our current structural and mutational work provides a framework for further crystallographic and biochemical studies of the full-length Edc3p to elucidate its role in mRNA decay, and it opens a way for structural studies on the Dcp1-Dcp2-Edc3 complex and/or the Dhh1-Edc3 complex.
Supplementary Material
Acknowledgments
This work was financially supported by the Agency for Science, Technology and Research in Singapore (H.S.) and by the Howard Hughes Medical Institute (R.P.).
Footnotes
Published ahead of print on 4 August 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Albrecht, M., and T. Lengauer. 2004. Novel Sm-like proteins with long C-terminal tails and associated methyltransferases. FEBS Lett. 56918-26. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 3.Anantharaman, V., and L. Aravind. 2004. Novel conserved domains in proteins with predicted roles in eukaryotic cell-cycle regulation, decapping and RNA stability. BMC Genomics 545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Badis, G., C. Saveanu, M. Fromont-Racine, and A. Jacquier. 2004. Targeted mRNA degradation by deadenylation-independent decapping. Mol. Cell 155-15. [DOI] [PubMed] [Google Scholar]
- 5.Bhattacharyya, S. N., R. Habermacher, U. Martine, E. I. Closs, and W. Filipowicz. 2006. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 1251111-1124. [DOI] [PubMed] [Google Scholar]
- 6.Brengues, M., D. Teixeira, and R. Parker. 2005. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310486-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54905-921. [DOI] [PubMed] [Google Scholar]
- 8.Cagney, G., P. Uetz, and S. Fields. 2000. High-throughput screening for protein-protein interactions using two-hybrid assay. Methods Enzymol. 3283-14. [DOI] [PubMed] [Google Scholar]
- 9.Caponigro, G., D. Muhlrad, and R. Parker. 1993. A small segment of the MATα1 transcript promotes mRNA decay in Saccharomyces cerevisiae: a stimulatory role for rare codons. Mol. Cell. Biol. 135141-5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.CCP4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50760-763. [DOI] [PubMed] [Google Scholar]
- 11.Chabas, A., and R. Happle. 2006. Understanding the biology of X-linked diseases. Acta Paediatr. Suppl. 959-10. [DOI] [PubMed] [Google Scholar]
- 12.Cole, J. L. 2004. Analysis of heterogeneous interactions. Methods Enzymol. 384212-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coller, J., and R. Parker. 2005. General translational repression by activators of mRNA decapping. Cell 122875-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cougot, N., S. Babajko, and B. Seraphin. 2004. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 16531-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Decker, C. J., D. Teixeira, and R. Parker. 2007. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 179437-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De la Fortelle, E., and G. Bricogne. 1997. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 276472-494. [DOI] [PubMed] [Google Scholar]
- 17.Dong, S., C. Li, D. Zenklusen, R. H. Singer, A. Jacobson, and F. He. 2007. YRA1 autoregulation requires nuclear export and cytoplasmic Edc3p-mediated degradation of its pre-mRNA. Mol. Cell 25559-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 602126-2132. [DOI] [PubMed] [Google Scholar]
- 19.Ermler, U., C. H. Hagemeier, A. Roth, U. Demmer, W. Grabarse, E. Warkentin, and J. A. Vorholt. 2002. Structure of methylene-tetrahydromethanopterin dehydrogenase from methylobacterium extorquens AM1. Structure 101127-1137. [DOI] [PubMed] [Google Scholar]
- 20.Eulalio, A., I. Behm-Ansmant, and E. Izaurralde. 2007. P bodies: at the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 89-22. [DOI] [PubMed] [Google Scholar]
- 21.Fenger-Gron, M., C. Fillman, B. Norrild, and J. Lykke-Andersen. 2005. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol. Cell 20905-915. [DOI] [PubMed] [Google Scholar]
- 22.Fromont-Racine, M., A. E. Mayes, A. Brunet-Simon, J. C. Rain, A. Colley, I. Dix, L. Decourty, N. Joly, F. Ricard, J. D. Beggs, and P. Legrain. 2000. Genome-wide protein interaction screens reveal functional networks involving Sm-like proteins. Yeast 1795-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garneau, N. L., J. Wilusz, and C. J. Wilusz. 2007. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 8113-126. [DOI] [PubMed] [Google Scholar]
- 24.Gouet, P., and E. Courcelle. 2002. ENDscript: a workflow to display sequence and structure information. Bioinformatics 18767-768. [DOI] [PubMed] [Google Scholar]
- 25.Hollams, E. M., K. M. Giles, A. M. Thomson, and P. J. Leedman. 2002. MRNA stability and the control of gene expression: implications for human disease. Neurochem. Res. 27957-980. [DOI] [PubMed] [Google Scholar]
- 26.Holmes, L. E., S. G. Campbell, S. K. De Long, A. B. Sachs, and M. P. Ashe. 2004. Loss of translational control in yeast compromised for the major mRNA decay pathway. Mol. Cell. Biol. 242998-3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 1441425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jha, K. N., I. A. Shumilin, L. C. Digilio, O. Chertihin, H. Zheng, G. Schmitz, P. E. Visconti, C. J. Flickinger, W. Minor, and J. C. Herr. 2008. Biochemical and structural characterization of apolipoprotein A-I binding protein, a novel phosphoprotein with a potential role in sperm capacitation. Endocrinology 1492108-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khajavi, M., K. Inoue, and J. R. Lupski. 2006. Nonsense-mediated mRNA decay modulates clinical outcome of genetic disease. Eur. J. Hum. Genet. 141074-1081. [DOI] [PubMed] [Google Scholar]
- 30.Khusial, P., R. Plaag, and G. W. Zieve. 2005. LSm proteins form heptameric rings that bind to RNA via repeating motifs. Trends Biochem. Sci. 30522-528. [DOI] [PubMed] [Google Scholar]
- 31.Kshirsagar, M., and R. Parker. 2004. Identification of Edc3p as an enhancer of mRNA decapping in Saccharomyces cerevisiae. Genetics 166729-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, J., M. A. Valencia-Sanchez, G. J. Hannon, and R. Parker. 2005. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 7719-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marino-Ramirez, L., and J. C. Hu. 2002. Isolation and mapping of self-assembling protein domains encoded by the Saccharomyces cerevisiae genome using lambda repressor fusions. Yeast 19641-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCoy, A. J., R. W. Grosse-Kunstleve, L. C. Storoni, and R. J. Read. 2005. Likelihood-enhanced fast translation functions. Acta Crystallogr. D 61458-464. [DOI] [PubMed] [Google Scholar]
- 35.Meyer, S., C. Temme, and E. Wahle. 2004. Messenger RNA turnover in eukaryotes: pathways and enzymes. Crit. Rev. Biochem. Mol. Biol. 39197-216. [DOI] [PubMed] [Google Scholar]
- 36.Miller, R., S. M. Gallo, H. G. Khalak, and C. M. Weeks. 1994. SnB: crystal structure determination via shake-and-bake. J. Appl. Crystallogr. 27613-621. [Google Scholar]
- 37.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53240-255. [DOI] [PubMed] [Google Scholar]
- 38.Parker, R., and U. Sheth. 2007. P bodies and the control of mRNA translation and degradation. Mol. Cell 25635-646. [DOI] [PubMed] [Google Scholar]
- 39.Parker, R., and H. Song. 2004. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 11121-127. [DOI] [PubMed] [Google Scholar]
- 40.Pilkington, G. R., and R. Parker. 2008. Pat1 contains distinct functional domains that promote P-body assembly and activation of decapping. Mol. Cell. Biol. 281298-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pillai, R. S., S. N. Bhattacharyya, C. G. Artus, T. Zoller, N. Cougot, E. Basyuk, E. Bertrand, and W. Filipowicz. 2005. Inhibition of translational initiation by Let-7 microRNA in human cells. Science 3091573-1576. [DOI] [PubMed] [Google Scholar]
- 42.Scholzova, E., R. Malik, J. Sevcik, and Z. Kleibl. 2007. RNA regulation and cancer development. Cancer Lett. 24612-23. [DOI] [PubMed] [Google Scholar]
- 43.Schuck, P. 2000. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 781606-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sheth, U., and R. Parker. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300805-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sheth, U., and R. Parker. 2006. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell 1251095-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teixeira, D., and R. Parker. 2007. Analysis of P-body assembly in Saccharomyces cerevisiae. Mol. Biol. Cell 182274-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Terwilliger, T. C. 2002. Automated structure solution, density modification and model building. Acta Crystallogr. D 581937-1940. [DOI] [PubMed] [Google Scholar]
- 48.Tritschler, F., A. Eulalio, V. Truffault, M. D. Hartmann, S. Helms, S. Schmidt, M. Coles, E. Izaurralde, and O. Weichenrieder. 2007. A divergent Sm fold in EDC3 proteins mediates DCP1 binding and P-body targeting. Mol. Cell. Biol. 278600-8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson, J. R. Knight, D. Lockshon, V. Narayan, M. Srinivasan, P. Pochart, A. Qureshi-Emili, Y. Li, B. Godwin, D. Conover, T. Kalbfleisch, G. Vijayadamodar, M. Yang, M. Johnston, S. Fields, and J. M. Rothberg. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403623-627. [DOI] [PubMed] [Google Scholar]
- 50.Xu, J., J. Y. Yang, Q. W. Niu, and N. H. Chua. 2006. Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell 183386-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang, W. H., J. H. Yu, T. Gulick, K. D. Bloch, and D. B. Bloch. 2006. RNA-associated protein 55 (RAP55) localizes to mRNA processing bodies and stress granules. RNA 12547-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.