Abstract
DNA methylation is involved in gene silencing and genomic stability in mammals, plants, and fungi. Genetics studies of Neurospora crassa have revealed that a DNA methyltransferase (DIM-2), a histone H3K9 methyltransferase (DIM-5), and heterochromatin protein 1 (HP1) are required for DNA methylation. We explored the interrelationships of these components of the methylation machinery. A yeast two-hybrid screen revealed that HP1 interacts with DIM-2. We confirmed the interaction in vivo and demonstrated that it involves a pair of PXVXL-related motifs in the N-terminal region of DIM-2 and the chromo shadow domain of HP1. Both regions are essential for proper DNA methylation. We also determined that DIM-2 and HP1 form a stable complex independently of the trimethylation of histone H3K9, although the association of DIM-2 with its substrate sequences depends on trimethyl-H3K9. The DIM-2/HP1 complex does not include DIM-5. We conclude that DNA methylation in Neurospora is largely or exclusively the result of a unidirectional pathway in which DIM-5 methylates histone H3K9 and then the DIM-2/HP1 complex recognizes the resulting trimethyl-H3K9 mark via the chromo domain of HP1.
In most eukaryotic organisms, genomic DNA is modified by the enzymatic conversion of certain cytosines to 5-methylcytosines. DNA methylation directly and indirectly influences gene expression and plays crucial roles in fundamental biological processes, such as X chromosome inactivation, genomic imprinting, and silencing of selfish DNA (5, 17, 21, 25, 29, 44). DNA methylation is also thought to play key roles in tumorigenesis; inappropriate methylation can silence tumor-suppressor genes and/or activate oncogenes (14). Aberrant DNA methylation is associated with other diseases, most notably immunodeficiency, centromere instability, and facial anomalies syndrome in humans (2). Although information on the relationship between DNA methylation and human diseases is accumulating, basic questions, such as how DNA methylation is controlled and how it functions, remain unanswered.
The results of research on DNA methylation using the filamentous fungus Neurospora crassa revealed that DNA methylation is controlled by the state of underlying chromatin proteins (17). In eukaryotic cells, DNA is wrapped around a protein octamer composed of histones (H2A, H2B, H3, and H4), which are subject to a variety of posttranslational modifications, including methylation, acetylation, phosphorylation, and ubiquitylation (3). The results of genetic studies in Neurospora demonstrated that condensed chromatin (“heterochromatin”) associated with methylated lysine 9 of histone H3 (H3K9) is involved in DNA methylation. Mutations in either dim-5 or hpo, which, respectively, encode the DIM-5/KMT1 histone methyltransferase (HMTase) and heterochromatin protein 1 (HP1), eliminate all detectable DNA methylation, just like null mutations in the DNA methyltransferase (DMTase) gene dim-2 (16, 30, 51). The DIM-5 HMTase specifically trimethylates H3K9, and trimethyl-H3K9 (H3K9me3) is preferentially associated with methylated DNA (53). Moreover, HP1 is localized to heterochromatic foci in a DIM-5-dependent manner (16). These data suggest that HP1 serves as an adaptor between methylated H3K9 and the DNA methylation machinery. Interestingly, dim-5 and hpo mutants show growth defects, unlike dim-2 mutants, suggesting that dim-5 and hpo are required for additional important cellular processes, perhaps including chromosomal segregation (16, 30, 51).
A role of H3K9 methylation in the control of DNA methylation has been conserved evolutionally (17). In Arabidopsis thaliana, dimethylation of H3K9 resulting from the action of the HMTase KRYPTONITE/SUVH4/KMT1 and other HMTases (11) is critical for DNA methylation by the DMTase CHROMOMETHYLASE 3 (CMT3) (27). Curiously, although the only known Arabidopsis HP1 homolog (LHP1) interacts with CMT3 in vitro, it is not required for DNA methylation (33, 35). The CMT3 chromo domain can directly interact with methylated H3K9 (33). This situation, however, appears to be unique to plants, because mammalian and Neurospora DMTases do not contain chromo domains (21). In mice, pericentric heterochromatin is enriched in H3K9me3 produced by the HMTases Suv39h1/KMT1A and Suv39h2/KMT1B, and Suv39h double-null mouse embryonic stem cells display loss of DNA methylation within these heterochromatic regions (32). In addition, mono- and dimethylated H3K9 resulting from the activity of the HMTase G9a/KMT1C is localized specifically to silent sites within uncondensed chromatin (“euchromatin”) (40, 41), and G9a null mouse embryonic stem cells show loss of DNA methylation at the Prader-Willi syndrome imprinting center and at Oct-3/4 (15, 56).
The three HP1 paralogues of mammals (HP1α, HP1β, and HP1γ) are found largely, but not exclusively, in heterochromatin. HP1α localizes specifically to pericentric heterochromatin sites, but HP1β and HP1γ are found in both heterochromatic and euchromatic sites (24, 34). Evidence for direct or indirect interactions among mammalian HP1, DMTases, and HMTases has been reported (1, 12, 13, 18, 19, 49), but further studies are needed to determine whether any mammalian HP1 plays a direct role in DNA methylation.
We used a combination of biochemical and genetic methods to elucidate the connections of the Neurospora DMTase DIM-2 with HP1 and histone methylation. We identified Neurospora HP1 as an interacting partner for DIM-2 by using a yeast two-hybrid screen and confirmed that DIM-2 interacts directly with HP1 in vivo. The results of mutational analyses revealed that amino acid substitutions within DIM-2 or HP1 that perturb their interaction cause loss of DNA methylation, implying that the direct interaction is essential for DNA methylation.
MATERIALS AND METHODS
Plasmid constructs for the yeast two-hybrid system.
The primers used in this study are listed in Table SI in the supplemental material. For two-hybrid bait vectors, DNA fragments corresponding to residues 1 to 366, 290 to 566, and 431 to 847 of the N-terminal domain of DIM-2 were amplified by PCR and inserted into pGBDU-C1 (28) to yield pGBDU-dim-21-366, pGBDU-dim-2290-566, and pGBDU-dim-2431-847, respectively. To construct pGBDU-dim-22-74, a DNA fragment containing the corresponding residues was amplified by PCR and inserted into pGBDU-C1. All the other deletion constructs of DIM-2 diagramed in Fig. 1 were generated in a similar manner. The open reading frame of the hpo cDNA was amplified by reverse transcription-PCR and inserted into pGAD-C1 to yield pGAD-HP12-266. All the other deletion constructs of HP1 diagramed in Fig. 1 were generated in a similar manner. The open reading frame of the dim-5 cDNA was amplified by reverse transcription-PCR and inserted into pGBDU-C1 to yield pGAD-Dim-52-338. Substitutions of AXAXA for IXVXL (hereafter abbreviated as IVL/AAA) or PXSXL (hereafter abbreviated as PSL/AAA) in DIM-2 and replacements of Tyr-244 with Glu or Ile-250 with Glu in HP1 were made with a QuikChange site-directed mutagenesis kit (Stratagene).
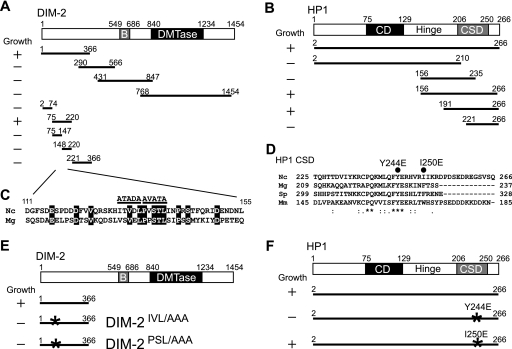
FIG. 1.
Mapping of the domains required for the direct interaction between DIM-2 and HP1 with yeast two-hybrid assays. Amino acid coordinates are indicated, numbered from the N termini. Abbreviations are as follows: B, bromo adjacent homology domain; DMTase, catalytic domain of known DMTases; CD, chromo domain; Hinge, hinge region. (A) Schematic diagram of tested truncated derivatives of DIM-2 fused to the GAL4 DNA binding domain. All constructs were cotransformed with the pGAD-HP1156-266 prey vector into PJ69-4A yeast cells. Transformants were tested on SD agar plates without Ade, His, Leu, and Ura; growth results are shown at left. (B) Schematic diagram of truncated HP1 derivatives fused to the GAL4 activation domain and tested for interaction with DIM-21-366 fused to the GAL4 DNA binding domain as the “bait.” Growth results are shown at left. (C) Sequence comparison of the HP1-interacting fragment of DIM-2 from N. crassa (Nc) and the corresponding region of its Magnaporthe grisea (Mg) counterpart (accession number XP_368355). Identical residues are indicated in white on a black background. Two regions including sequences similar to the PXVXL consensus sequence implicated in HP1 binding are indicated by the horizontal lines. Residues in the two putative PXVXL motifs were replaced by alanines (PSL/AAA and IVL/AAA) as shown. (D) Comparison of the CSDs of HP1 proteins among N. crassa (Nc), Magnaporthe grisea (Mg), Schizosaccharomyces pombe (Sp) and Mus musculus (Mm). Residues that are identical in all HP1 proteins are indicated by asterisks. Colons and periods indicate strong and weak conservation, respectively. The two black circles indicate point-mutated sites (Y244E and I250E). (E) Summary of results from yeast two-hybrid tests of interactions between the binding domain fusions of the DIM-2 mutant protein and the activation domain fusion of HP1156-266. Growth results are shown at left. The asterisks show mutated sites. (F) Summary of results from yeast two-hybrid tests of interactions between the activation domain fusions of an HP1 point mutant and the binding domain fusion of DIM-21-366. Growth results are shown at left. The asterisks show mutated sites.
Yeast two-hybrid screen.
A HybriZAP two-hybrid system (Stratagene) was used according to the manufacturer's instructions using Saccharomyces cerevisiae strain PJ69-4A as the host strain as described by James et al. (28). An N. crassa conidia cDNA library constructed in the HybriZap vector was obtained from the Fungal Genetic Stock Center (FGSC). In brief, the HybriZAP lambda library was converted into a pAD-GAL4 plasmid library by in vivo mass excision. The yeast reporter strain PJ69-4A transformed with the bait vectors were grown on SD agar plates without uracil (Ura). All yeast transformations described here were performed by heat shock with target DNAs in one-step buffer (40% polyethylene glycol 4000, 0.2 LiAc, 0.1 M dithiothreitol, 20 μg/ml single-stranded DNA) at 42°C for 1 h. The yeast cells containing the bait plasmid were transformed with 0.5 mg of the Neurospora cDNA library and selected for growth on SD agar plates lacking histidine (His), leucine (Leu), and Ura and containing 2.5 mM 3-aminotrizole. The surviving colonies were then streaked on SD agar plates lacking adenine (Ade), His, Leu, and Ura and were further tested for expression of the Gal-4-dependent reporter lacZ by a filter lift assay. Library plasmids from positive colonies were recovered in Escherichia coli cells and sequenced. To verify their interaction and to identify the domain required for their interaction, the bait and pray vectors described above were introduced into yeast strain PJ69-4A and selected on SD agar plates lacking Leu and Ura. The cotransformants were then streaked onto SD agar plates lacking Ade, His, Leu, and Ura and assayed for growth to test the interactions.
Neurospora strains and isolation of genomic DNA.
The N. crassa strains used in this study are listed in Table SII in the supplemental material and were grown, maintained, and crossed according to standard published procedures (10). For isolation of genomic DNA, Neurospora strains were grown with shaking in Vogel's minimal medium N with required supplements at 32°C for 3 days; genomic DNA was isolated and used for PCR and Southern hybridizations as described previously (16).
Gene disruption of dap1.
We followed the Neurospora gene knockout procedure described by Colot et al. (8), using a construct in which we placed an hph cassette contained between dap-1 flanking sequences. We took advantage of the mutation of mus52, the homologue of human Ku80, which abolishes nonhomologous end-joining and thereby facilitates gene knockouts (38). Briefly, the 5′ and 3′ flanking fragments and the hph gene were amplified by LA Taq (Takara). The three PCR products and XhoI- plus XbaI-digested yeast vector pRS416 were transformed into the yeast strain PJ64-4A and assembled in yeast by using the endogenous homologous recombination system (39). The plasmid containing the linear knockout cassette was recovered in E. coli cells. The cassette was cut out by digestion with AscI and KpnI and transformed into a Δmus-52 strain (strain N2930) by electroporation (36).
Genetic engineering to add epitope tags to proteins expressed from genes at their native chromosomal location.
We modified the gene knock-out procedure described by Colot et al. (8) to serve to “knock-in” Neurospora genes (S. Honda and E. U. Selker, unpublished data). The constructions involved cassettes with 10-glycine “tails” followed by one of three different epitope tags (three FLAGs, three hemagglutinins [HA], or green fluorescent protein [GFP]) and the hph gene flanked by loxP sequences. For the DIM-2-FLAG knock-in construct, we PCR amplified a 1-kb segment, including the end of the dim-2 coding region but without the stop codon and a 500-bp segment of the 3′ flanking region. The cassettes containing the three-FLAG segment and the hph gene were isolated from p3xFLAG::hph::loxP by digestion with KpnI and XhoI. pRS416 was linearized by digestion with BamHI and EcoRI. The four fragments were introduced into the yeast strain PJ64-4A for assembly in yeast using the endogenous homologous recombination system (39). The cassette was cut out by digestion with XhoI and XbaI and transformed into the Δmus-52 Neurospora strain (strain N2930) (38) by electroporation. The HP1-GFP and DIM-5-HA knock-in constructs were generated similarly.
Complementation of dim-2 and hpo mutations.
For the DIM-2 construct, a 500-bp segment including the promoter region of dim-2 was amplified by PCR. The PCR products were digested with NotI and XbaI. The pCCG::C-Gly::3xFLAG vector, which contains a 10-glycine tail followed by the three-FLAG tag and is driven by the ccg-1 promoter, was digested with NotI and XbaI and ligated with the inserts to replace the ccg-1 promoter with the dim-2 promoter. The vector was digested with XbaI and PacI, ligated with a fragment containing the coding region of the dim-2 gene, and digested with XbaI and PacI. For the HP1 construct, a 500-bp promoter region of hpo was amplified by PCR. The coding region of hpo was removed from pMF308 by digestion with BamHI and XbaI. The fragments were inserted into pCCG::C-Gly::GFP, which contains 10 glycines followed by the GFP tag and is driven by the ccg-1 promoter. The HP1-GFP and DIM-2-FLAG constructs were linearized and inserted into the his-3 locus of the hpo strain N3395 or the dim-2 null mutant N3396, respectively, by the gene replacement method previously described (36).
Coimmunoprecipitation (Co-IP) assay.
N. crassa strains were grown at 32°C with shaking in 20- by 150-mm glass test tubes containing 7 ml Vogel's minimal medium N with 1.5% sucrose and supplements required based on their genotypes. After 16 h, tissue was harvested by filtration, washed in phosphate-buffered saline (PBS), and suspended in 1 ml of ice-cold lysis buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 10% glycerol, 0.02% NP-40, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride [PMSF; Sigma], 1 μg/ml leupeptin [Roche], 1 μg/ml pepstatin [Roche], and 1 μg/ml E-64 [Roche]). Extracts were sonicated by using a sonicator (Branson Sonifier-450) three times, at 10-min intervals, for 20 s with a duty cycle of 80 and output set to 2. After centrifugation at 12,000 rpm for 10 min, aliquots of the supernatants were incubated with 2 μl of mouse monoclonal anti-FLAG M2 antibody (F3165; Sigma), 25 μl mouse monoclonal anti-HA antibody (University of Oregon Monoclonal Antibody Facility), or 2 μl of rabbit polyclonal anti-GFP antibody (ab290; Abcam) for 16 h, followed by protein A-agarose (for rabbit antibodies) or protein G-agarose (for mouse antibodies) for 2 h. Immune complexes were washed twice in lysis buffer and suspended in sodium dodecyl sulfate (SDS) sample buffer. Samples were separated by SDS-polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes in 48 mM Tris, 39 mM glycine (pH 9.2) containing 20% methanol at 150 mA for 2 h. Membranes were blocked in 10 mM Tris, pH 7.5, 150 mM NaCl, 0.05% Tween 20 (TBS-Tween) containing 3% skim milk powder for 30 min and incubated for 2 h at room temperature with anti-FLAG M2 antibody in TBS-Tween containing 3% skim milk; anti-GFP antibody in TBS-Tween containing 3% bovine serum albumin and rat monoclonal anti-HA antibody (Roche); and purified anti-DIM-5 antibody (7) in TBS-Tween. Immunoprecipitates were detected by using horseradish peroxidase-conjugated secondary antibodies and SuperSignal West Femto chemiluminescent substrate (Pierce) as described in the manufacturer's instructions.
Visualization of HP1-GFP by fluorescence microscopy.
A suspension of conidia was spotted on a slide glass and immediately covered by a glass coverslip. Pictures were taken using an Axioplan2 fluorescence microscope and AxioCam HRm digital camera (Carl Zeiss, Thornwood, NY).
Histone extraction and analysis.
We modified the procedures for the extraction of histone from Neurospora spheroplasts described by Goff (20) and from plants described by Green et al. (22). Strains were grown in 125-ml flasks containing 25 ml of Vogel's medium with 1.5% sucrose and 1.5% agar at 32°C for 7 days. The conidia were resuspended and grown with shaking in 250-ml flasks containing 60 ml of Vogel's medium with 1.5% sucrose at 32°C overnight. Cells were harvested by filtration and frozen in liquid nitrogen. The frozen tissue was ground with a mortar and pestle to a fine powder and resuspended in 50 ml of ice-cold lysis buffer (0.3 M sucrose, 0.04 M NaHSO3, 25 mM Tris-HCl [pH 7.4], 0.01 M MgSO4, 0.5 mM EDTA, 0.5% NP-40, 1 mM PMSF, 1 μg/ml leupeptin, 1 μg/ml pepstatin). The nuclear fraction was pelleted by centrifugation (8000 × g), washed in the same buffer, and again recovered by centrifugation. It was then suspended in 2.0 ml of ice-cold CW buffer (0.15 M NaCl, 10 mM Tris-HCl [pH 8.0], 1 mM β-mercaptoethanol, 1 mM PMSF, 1 μg/ml leupeptin, 1 μg/ml pepstatin), mixed with 2 ml 0.4 M H2SO4, and incubated with rotation overnight at 4°C with to elute the histones. Debris was removed by centrifugation at 12,000 rpm for 10 min in a microcentrifuge, and the supernatant, containing the histones, was mixed with 1/4 volume 100% trichloroacetic acid and incubated on ice for 1 h. After centrifugation again at 12,000 rpm for 10 min, precipitated histones were washed in ice-cold acetone twice and dried at room temperature. The purified histones were suspended in sample buffer with SDS and subjected to SDS-polyacrylamide gel electrophoresis and Western blotting using antibodies against H3K9me3 (catalog no. 39161; Active Motif) and H3K4me2 (catalog no. 39141; Active Motif).
Chromatin immunoprecipitation (ChIP) assay.
Neurospora strains were grown at 32°C with shaking in 20- by 150-mm glass tubes containing 7 ml Vogel's minimal medium N with 1.5% sucrose and any required supplements. After 16 h, tissue was harvested by filtration and washed in PBS. For chromatin fixation, tissues were suspended in 10 ml of PBS containing 1% formaldehyde and incubated at 32°C for 30 min with shaking. The subsequent steps were performed as described previously (53). Anti-H3K9me3 antibody (9), anti-FLAG M2 antibody, and anti-GFP antibody were used for immunoprecipitation of H3K9me3, DIM-2-FLAG, and HP-GFP, respectively.
RESULTS
Identification of HP1 and DAP1 as DIM-2-interacting proteins by yeast two-hybrid screening.
The DMTase DIM-2 encodes a 1,454-amino-acid protein that includes a C-terminal 600-amino-acid domain that corresponds to the catalytic domain of known DMTases (Fig. 1). Although the large N-terminal region of DIM-2 shows a few sequence features, including a bromo adjacent homology domain and a putative nuclear localization signal, except for its fungal homologues, it shows no significant similarity to any proteins in current databases. Studies with mammalian DMTases revealed evidence of interactions of their N-terminal regions with other proteins, including HP1, SUV39H1 HMTase (18, 19), G9a HMTase (13), the ATP-dependent chromatin remodeling enzyme SNF2H (19), the DNA replication factor PCNA (6), the tumor suppressor protein Rb (42), histone deacetylase 2 (HDAC2), and the DNMT1-associated protein DMAP1 (43). As a step to elucidate the control of DNA methylation in Neurospora, we used the yeast two-hybrid system to search for proteins that interact with the N-terminal region of DIM-2. In preliminary work, we found that the N-terminal domain of DIM-2 fused with the GAL4 DNA binding domain strongly autoactivated reporter genes (data not shown). Fortunately, when the domain was broken into three overlapping fragments, autonomous activation of the reporter genes was not observed (Fig. 1A). We therefore used these for full screens using an N. crassa cDNA library in a two-hybrid vector. Positive colonies were only obtained using the most-N-terminal fragment (residues 1 to 366), and not with fragments including residues 290 to 566 or 431 to 847. Sixty-nine positive colonies were identified by the activation of HIS3, ADE2, and LacZ reporter genes from 2 × 106 cotransformants. Analysis of the corresponding plasmids showed that 62 encoded the C terminus of the ATP-synthase β chain mitochondrial precursor (accession number XP_965645); five encoded the C terminus of a 1,752-amino-acid hypothetical protein (GenBank accession number XP_961249), which we named dim-2-associated protein 1 (dap-1); and the remaining two colonies encoded the C terminus of HP1, including the hinge region and the chromo shadow domain (CSD). The interaction of DIM-2 with ATP-synthase α chain protein was deemed artifactual based on its localization in mitochondria. Although DAP-1 shows no significant similarity to known proteins, a computer program for the predication of protein localization sites in cells, PSORT (37), predicted that it is nuclear. The possibility that DAP-1 interacts with DIM-2 in Neurospora prompted us to test if the protein plays a role in DNA methylation. The dap-1 gene was replaced by homologous recombination with the bacterial hph gene, which confers hygromycin resistance in Neurospora (data not shown). Strains lacking dap-1 grew poorly and showed a slight loss of DNA methylation at the Ψ63 region but normal DNA methylation in other methylated regions, unlike dim-2 strains, which show essentially normal growth but no DNA methylation (data not shown). These results indicate that DAP-1 is unlikely to contribute to DNA methylation through an interaction with DIM-2.
Domains involved in the interaction between DIM-2 and HP1.
To define the domains involved in the DIM-2-HP1 interaction, we prepared a series of fragments of DIM-2 and HP1 and tested them in the yeast two-hybrid assay. A construct with amino acids 75 to 220 of DIM-2 allowed for growth on the nutritional selection medium when combined with an HP1 construct containing amino acids 191 to 266, which includes the CSD (Fig. 1A and B). These data suggest that residues within these domains mediate the DIM-2-HP1 interaction. Some proteins that have been found to interact with the CSD in other systems contain a PXVXL motif, and structural studies on HP1 have identified residues in the CSD that interact with this motif (4, 50, 54). DIM-2 does not have a perfect PXVXL motif but does contain a tandem pair of similar sequences (ITVDL at amino acids 130 to 134 and PVSTL at amino acids 135 to 139) in the fragment that showed interaction with the CSD in the two-hybrid assay. Notably, these PXVXL-like sequences fall in a region of DIM-2 that is highly conserved in the fungus Magnaporthe grisea (Fig. 1C). To test the possibility that DIM-2 and HP1 interact with each other through one of the tandem PXVXL-related motifs and the CSD, we made substitutions of AXAXA for IXVXL (IVL/AAA) or PXSXL (PSL/AAA) in DIM-2 (Fig. 1D) and tested the mutant proteins in various ways. We also made and tested two mutations in the region of the HP1 CSD that has been implicated in binding to PXVXL motifs in other systems (4, 54). In particular, we made Tyr-244-to-Glu (Y244E) and Ile-250-to-Glu (I250E) mutations (Fig. 1D). The corresponding Y244E substitution in mice has been reported to both abolish the dimerization of HP1 and influence its interaction with PXVXL motif-containing peptides, whereas the mutation in mouse HP1 corresponding to the I250E substitution (W170E in mice) has been reported to perturb the PXVXL-HP1 interaction without affecting HP1 dimerization (4). We found that both the DIM-2IVL/AAA and the DIM-2PSL/AAA mutations abolished growth on the nutritional selection medium in the yeast two-hybrid assay (Fig. 1E). These data suggest that both PXVXL-related motifs in the N-terminal region of DIM-2 are involved in interaction with the CSD of HP1. We also found that the HP1Y244E mutation, but not the HP1I250E mutation, interfered with interaction with the DIM-2 construct (Fig. 1F), supporting our expectation that the highly conserved Y at position 244 is critical for the binding of the CSD to DIM-2.
Interaction between DIM-2 and HP1 in vivo.
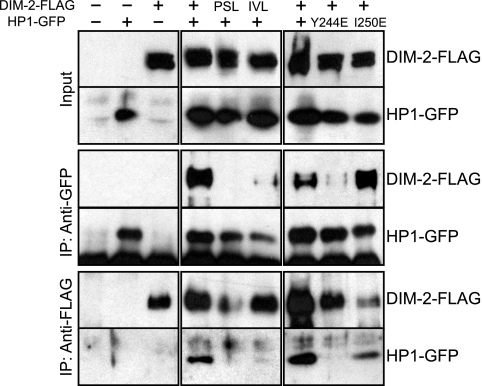
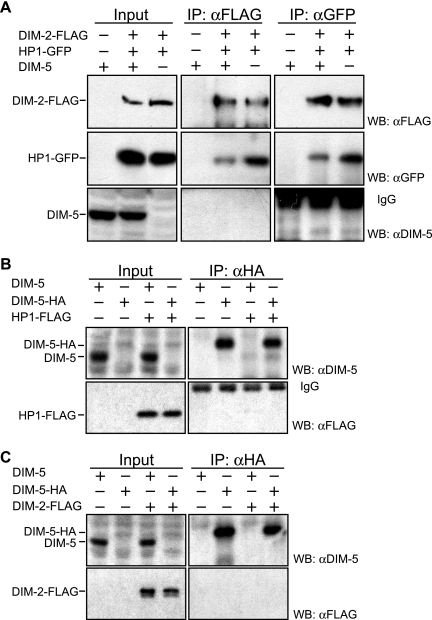
In the yeast two-hybrid assay, the N-terminal region of DIM-2 served as a binding site for HP1. To confirm the interaction between DIM-2 and HP1 in Neurospora, we built and coexpressed constructs with epitope-tagged wild-type and mutant dim-2 and hpo alleles and performed Co-IP assays. The dim-2 alleles, which sported C-terminal three-FLAG tags (dim-2-FLAG, dim-2PSL/AAA-FLAG, or dim-2IVL/AAA-FLAG) and were driven by their endogenous promoters, were targeted to the his-3 locus of a dim-2 null mutant containing a C-terminal GFP-tagged hpo gene at its native locus (strain N3396). In addition, we built strains with wild-type or mutant GFP-tagged hpo alleles (hpo-gfp, hpoY244E-gfp, or hpoI250E-gfp) driven by the endogenous promoter at the his-3 locus of strain N3395, which is an hpo null mutant (hpoRIP2) containing three-FLAG-tagged dim-2 (dim-2-FLAG) at its native locus. We found that the strains expressing DIM-2-FLAG or HP1-GFP without the mutations showed nearly normal levels of DNA methylation (see Fig. 3) and normal growth rates (see Fig. 4), indicating that both DIM-2-FLAG and HP1-GFP are functional. The results of a Co-IP assay revealed that DIM-2-FLAG was pulled down with HP1-GFP using anti-GFP antibodies and vice versa (Fig. 2), revealing stable interaction of DIM-2 and HP1 in vivo. Interestingly, the interaction was completely abolished by DIM-2PSL/AAA and significantly reduced by DIM-2IVL/AAA or HP1Y244E, but not HP1I250E (Fig. 2). Mutant proteins were expressed at levels equivalent to those of the wild-type proteins (Fig. 2, input). These data strongly support the results of the yeast two-hybrid assays, indicating that DIM-2 interacts directly with HP1 through the PXVXL-related motifs in DIM-2 and the CSD of HP1.
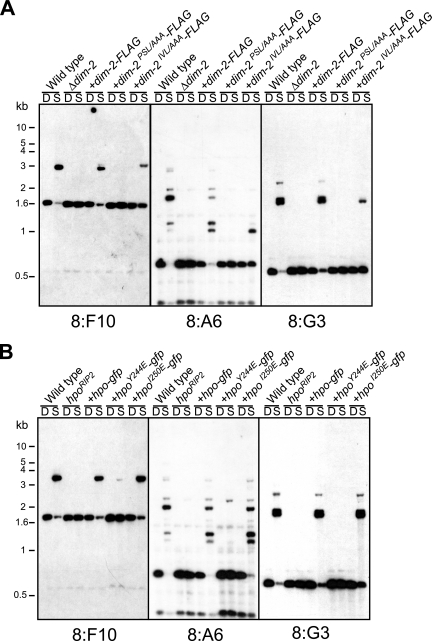
FIG. 3.
The tandem PXVXL-related motifs of DIM-2 and the CSD of HP1 are required for DNA methylation. Both wild-type and modified genes were driven by their own promoters and targeted to the his-3 locus. (A) To test dim-2 mutants, genomic DNA samples were tested from a dim-2+ strain (N623; wild type), a Δdim-2 strain used as the transformation host (N3396), and transformants with dim-2-FLAG (N3415), dim-2PSL/AAA-FLAG (N3417), or dim-2IVL/AAA-FLAG (N3416). DNA samples were digested with 5′-methyl-cytosine-sensitive Sau3AI (S) or its 5′-methyl-cytosine-insensitive isoschizomer, DpnII (D), and used for Southern hybridizations, with the indicated probes, for methylated regions (8:F10, 8:A6, and 8:G3; see Selker et al. [48]). (B) To test hpo mutants, an hpoRIP2 host strain and transformants with hpo-gfp (N3418), hpoY244E-gfp (N3419), or hpoI250E-gfp (N3420) were analyzed in the same way. Positions of size standards are shown at left.
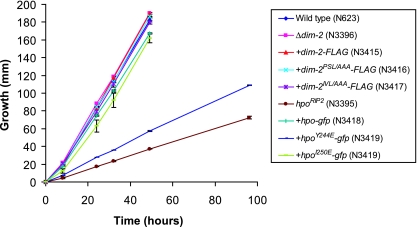
FIG. 4.
Slow growth of hpo null mutant is partially alleviated by introduction of the hpoY244E-gfp gene. Growth rates are shown for a dim-2+ hpo+ strain (N623), a dim-2− mutant (N3396), transformants of a dim-2− strain (N3396) with dim-2-FLAG (N3416), dim-2PSL/AAA-FLAG (N3417), or dim-2IVL/AAA-FLAG (N3416), an hpo− mutant (N3395), and transformants of an hpo− strain (N3395) with hpo-gfp (N3418), hpoY244E-gfp (N3419), or hpoI250E-gfp (N3420). Linear growth rates were determined at 32°C by using race tubes containing Vogel's minimal medium agar with histidine. The data represent the averages of two independent experiments. The modified forms of dim-2 and hpo were introduced into dim-2 and hpo null mutant strains, respectively (indicated by +). The error bars represent ranges.
FIG. 2.
The results of Co-IP assays reveal in vivo interaction between DIM-2 and HP1 mediated by the tandem PXVXL-related motifs of DIM-2 and the CSD of HP1. The following strains were tested: N623, N3322, N3323, N3415, N3416, N3417, N3418, N3419, and N3420. Extracts from strains with (+) or without (−) the FLAG-tagged dim-2 gene or dim-2 mutant genes (PSL/AAA and IVL/AAA) and/or the GFP-tagged hpo gene or hpo point-mutated genes (Y244E and I250E) were immunoprecipitated with anti-FLAG antibodies or anti-GFP antibodies. Input and IP samples were fractionated, transferred to polyvinylidene difluoride membranes, and immunoblotted with anti-FLAG antibodies or anti-GFP antibodies as indicated.
HP1-DIM-2 interaction is required for DNA methylation in vivo.
To assess the functional consequences of the HP1-DIM-2 interaction detected in the yeast two-hybrid and Co-IP experiments, we tested the effects of the DIM-2 and HP1 mutations on DNA methylation in vivo. We digested genomic DNA samples of various strains with methyl-cytosine-sensitive and methyl-cytosine-insensitive endonucleases (Sau3AI and DpnII, respectively) and analyzed them by probing Southern blots for three genomic regions that are normally methylated in N. crassa, namely, the 8:F10, 8:A6, and 8:G3 relics of repeat-induced point mutation (RIP) (48). DNA methylation was restored to nearly the wild-type level by the introduction of the FLAG-tagged dim-2 gene into the dim-2 mutant (Fig. 3A). In contrast, the DIM-2PSL/AAA-FLAG construct failed to complement the methylation defect, demonstrating the importance of this PXVXL-related region of DIM-2 in vivo (Fig. 3A). Interestingly, the DIM-2IVL/AAA-FLAG construct showed partial restoration of DNA methylation (Fig. 3A), which is consistent with the partial interaction between this mutant and HP1 (Fig. 2), suggesting that the second PXVXL-related motif is less important. HP1-GFP and HP1I250E-GFP almost fully restored the DNA methylation of the hpo mutant, but the HP1Y244E-GFP protein did not (Fig. 3B). The results of Western blotting demonstrated that the expression levels of the altered proteins were comparable to those of the wild-type proteins (Fig. 2, input). These data imply that the direct interaction between DIM-2 and HP1 detected in the two-hybrid and Co-IP assays is required for DNA methylation.
Nuclear localization of HP1-GFP.
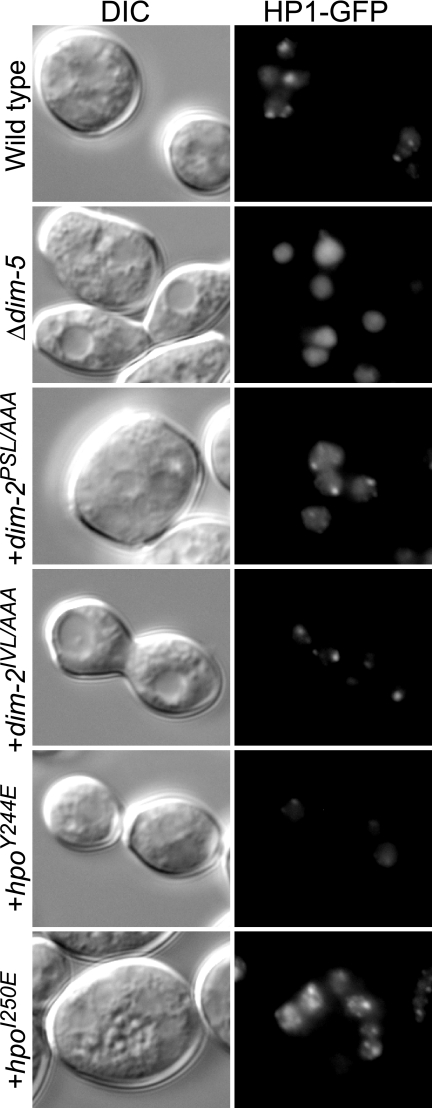
HP1 is required for processes besides DNA methylation, as illustrated by the fact that an hpo null strain, but not a dim-2 null strain, grows abnormally slowly (compare the growth of strains N3395 and N3396 in Fig. 4) (16). We found that a strain expressing HP1Y244E showed a severe growth defect, although the growth defect was not as great as that of the untransformed hpo strain (Fig. 4). In contrast, strains bearing the other altered HP1 or either of the altered DIM-2 proteins grew normally. Given that strains with dim-2 grow well and that DIM-2IVL/AAA showed disruption of the HP1-DIM-2 interaction equivalent to that shown by HP1Y244E, it seemed likely that the alteration in HP1Y244E causes at least one deficiency in addition to its defect in interacting with DIM-2. It has been demonstrated that a single-amino acid change in the CSD of mouse HP1β (I161E) that disrupts binding to PXVXL motif-containing proteins also influences the recognition of methylated H3K9 (54). We therefore used fluorescence microscopy to assess the possible effect of HP1Y244E on binding to methylated H3K9 in Neurospora. HP1-GFP is localized in heterochromatic foci in a wild-type background, whereas it is dispersed throughout the nucleus in the dim-5 mutant, which lacks H3K9 methylation (16). Interestingly, HP1Y244E-GFP localization appeared partially dispersed in the dim-5+ background (Fig. 5), suggesting that the HP1Y244E alteration interferes with normal recognition of methylated H3K9. This may contribute to the severe DNA methylation defect of this mutant. As expected, DIM-2PSL/AAA, DIM-2IVL/AAA, and HP1I250E did not affect HP1-GFP localization (Fig. 5).
FIG. 5.
Localization of HP1-GFP is not affected by dim-2 mutations. Fresh conidia of the following strains were examined: a dim-2+ hpo+ dim-5+ strain (N3418; wild type), a dim-5 mutant (N3436), transformants of a dim-2− strain (N3396) with a dim-2PSL/AAA-FLAG construct (N3417) or a dim-2IVL/AAA-FLAG construct (N3416), and/or transformants of an hpo− strain (N3395) with an hpoY244E-gfp construct (N3419) or an hpoI250E-gfp construct (N3420). Differential interference contrast (DIC) and fluorescence (HP1-GFP) images are shown. The modified forms of dim-2 and hpo were introduced into dim-2 and hpo null mutant strains, respectively (indicated by +).
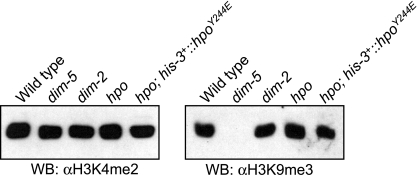
Global level of H3K9me3 in hpo mutants.
The results of our previous study showed that mutation of dim-2 does not change the global level of H3K9me3 (53). In contrast to the situation in a dim-2 null strain or in the strains with the DIM-2PSL/AAA and DIM-2IVL/AAA mutations, the hpo null mutant and the strain with HP1Y244E exhibited severe growth defects, similar to that of a dim-5 mutant. To examine the possibility that the growth defects of HP1 mutants are due to loss of H3K9me3, we isolated histones from the mutants and analyzed the global levels of H3K9me3 by Western blotting. As expected from our previous observations (53), H3K9me3 was significantly decreased in a dim-5 strain, but not in a dim-2 strain (Fig. 6). Interestingly, the strain with HP1Y244E and an hpo null strain also exhibited normal levels of H3K9me3 (Fig. 6). This suggests that lack of HP1 or interaction between HP1 and other unidentified PXVXL motif-containing proteins causes the growth defect, rather than loss of H3K9me3.
FIG. 6.
Global level of H3K9me3 does not change in dim-2 and hpo mutants. Histones were extracted from a dim-2+ hpo+ dim-5+ strain (N623; Wild type), a dim-5 mutant (N3436), a dim-2 mutant (N3396), an hpo mutant (N3395), and a transformant of an hpo− strain with an hpoY244E-gfp construct at the his-3 locus (N3419) and subjected to Western blotting (WB) using antibodies against H3K9me3 and H3K4me2. α, anti.
The localization of DIM-2 depends on DIM-5 and HP1.
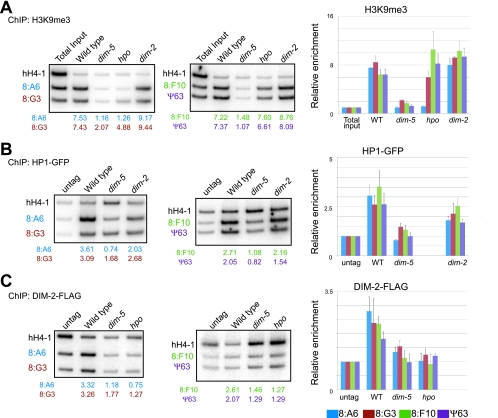
To further explore the relationship between H3K9 methylation and DNA methylation, we used ChIP assays to examine the distribution of H3K9me3, HP1-GFP, and DIM-2-FLAG at four relics of RIP that are normally methylated (8:A6, 8:G3, 8:F10, and Ψ63) (48), in various genetic backgrounds. As expected from the results of prior work from our laboratory (53), we found that H3K9me3 was enriched at all four RIPed regions in a wild-type strain but absent in a dim-5 strain (Fig. 7A). The dim-2 mutant showed essentially normal distribution of H3K9me3, implying that this histone mark does not depend on feedback from DNA methylation. Similar results were obtained with the hpo mutant but, interestingly, one of the four regions examined (8:A6) did show reduced H3K9me3 (Fig. 7A), suggesting limited feedback from HP1 on H3K9me3 localization. The levels of HP1-GFP were reduced substantially in the dim-5 mutant but only slightly or not at all in the dim-2 mutant (Fig. 7B), which is consistent with the localization of HP1-GFP observed by fluorescent microscopy (16). We successfully detected an enrichment of DIM-2-FLAG at all four RIPed regions and found that its localization was dependent on both DIM-5 and HP1 (Fig. 7C).
FIG. 7.
Methylation of H3K9 depends on DIM-5, but neither HP1 nor DIM-2, and binding of HP1 depends on DIM-5, but not DIM-2, while binding of DIM-2 depends on both DIM-5 and HP1. Possible interdependency was investigated by ChIP assays using antibodies recognizing H3K9me3 (A), HP1-GFP (GFP) (B), and DIM-2-FLAG (FLAG) (C). The following strains were tested by ChIP assays: N3395, N3396, N3415, and N3436. Total inputs or DNA isolated from strains without tagged protein (untag) were used for controls. Samples were subjected to PCR to amplify a gene lacking DNA methylation (hH4-1; see Tamaru et al. [53]) and four methylated DNA regions (8:A6, blue; 8:G3, brown; 8:F10, green; and Ψ63, purple), two at a time. The ratios of intensities of the methylated regions to hH4-1 between the ChIP samples from the tagged strains and the samples from the untagged strains or total inputs were used to calculate the relative enrichment, shown below the lanes. Results from duplicate ChIP experiments (one set shown) were averaged and are presented graphically (right). Error bars indicate ranges. −, absent; WT, wild type.
The DIM-2-HP1 complex does not depend on H3K9me3 in Neurospora and does not contain detectable DIM-5.
The ChIP results revealed that the association of DIM-2 with the regions that it methylates depends on the binding of HP1 to chromatin containing H3K9me3. In principle, binding of DIM-2 to HP1 might only occur on chromatin, i.e., after HP1 binds to H3K9 methylation. Although the yeast two-hybrid experiments provided evidence that HP1 and DIM-2 can bind in the absence of chromatin, we wished to test this possibility in Neurospora. To do so, we carried out Co-IP experiments to determine whether DIM-2 and HP1 bind equivalently in the presence or absence of the methylated H3K9 mark created by DIM-5. We confirmed that the expression levels of DIM-2-FLAG and HP1-GFP were not affected by mutation of dim-5 (Fig. 8A, input) and went on to find that DIM-2 and HP1 can form a stable complex independent of H3K9me3 (Fig. 8A, middle and right panels). Interestingly, we could not detect the DIM-5 protein in the samples pulled down with the DIM-2-HP1 complex (Fig. 8), suggesting that the HMTase is not part of the complex.
FIG. 8.
DIM-2 and HP1 form a stable complex independent of H3K9me3 and are not associated with DIM-5. (A) The following strains were used for Co-IP assays: N623, N3415, and N3436. Extracts from dim-5+ or dim-5− strains with (+) or without (−) a FLAG-tagged dim-2 gene and a GFP-tagged hpo gene were immunoprecipitated with antibodies against FLAG or GFP as indicated at the top, fractionated for Western blotting, and probed with antibodies indicated on the right. (B) The following strains were used for Co-IP assays: N623, N3315, N3320, and N3452. Extracts from strains with (+) or without (−) an HA-tagged dim-5 gene and/or a FLAG-tagged hpo gene were immunoprecipitated with anti-HA antibodies, processed for Western blotting, and probed with antibodies against DIM-5 or FLAG, as indicated. (C) The following strains were used for Co-IP assays: N623, N3315, N3323, and N3451. Extracts from strains with (+) or without (−) an HA-tagged dim-5 gene and/or a FLAG-tagged dim-2 gene were immunoprecipitated with anti-HA antibodies and analyzed in the same way. Bands in the bottom right quadrants of panels A and B are immunoglobulins (IgG) from the primary antibodies. WB, Western blot; α, anti.
This observation contrasts with evidence of associations between mammalian HMTases and both HP1 and DMTases (1, 13, 18, 19, 45). To investigate the possibility that a low level of DIM-5 was in the Neurospora HP1-DIM-2 complex, we reversed the Co-IP experiments, i.e., we tested whether the DIM-2-HP1 complex could be pulled down with DIM-5. To guard against the possibility that anti-DIM-5 antibodies might interfere with the association of DIM-5 and potential interacting proteins, we replaced the wild-type dim-5 gene with a modified form encoding DIM-5-HA and used anti-HA antibodies for the pull downs. Although the level of DIM-5-HA expression was somewhat low compared with that of untagged DIM-5 (Fig. 8B and C, input), strains expressing DIM-5-HA proteins showed normal DNA methylation and normal growth (data not shown). Thus, the HA tag did not interfere with DIM-5 function and its expression was sufficient for DNA methylation. Importantly, we found that neither HP1-FLAG nor DIM-2-FLAG was pulled down with DIM-5-HA (Fig. 8B and C). We also failed to find any evidence of interaction(s) of DIM-5 with HP1 and/or DIM-2 in yeast two-hybrid assays (data not shown). Finally, in separate work we purified an HP1 complex, subjected it to analysis by mass spectrometry, and found evidence of DIM-2, but not DIM-5 (our unpublished data). Altogether, our findings suggest that the DIM-2-HP1 complex does not feed back on histone methylation by DIM-5 in Neurospora.
DISCUSSION
Direct interaction between DIM-2 and HP1 is essential for DNA methylation.
DNA methylation, histone H3K9 methylation, and HP1 binding have all been implicated in heterochromatin formation and gene silencing (21, 31). Although associations between HP1s and DMTases have been reported in a variety of organisms (1, 13, 18, 19, 45), interesting differences are becoming evident. In Arabidopsis, one of several DMTases, CMT3, interacts in vitro with the only known HP1 homolog, LHP1 (27), but LHP1 does not appear to be involved in DNA methylation (33, 35) and is apparently directed by H3K27me3 (55, 57). In mammals, the DMTases DNMT1 and DNMT3a/b have been reported to associate with HP1s in vitro (18, 19, 49) but there is limited evidence that the DMTases are directed by H3K9me3 and HP1. Accumulation of HP1 in Oct3/4, a gene that is subject to de novo DNA methylation, is observed after the accumulation of G9a-mediated H3K9 methylation (15). Overexpression of mammalian HP1 fused with the GAL4 DNA binding domain triggers DNA methylation mediated by DNMT1 and silences a reporter gene in vivo (49). It is not yet clear, however, whether any of the mammalian HP1 proteins are normally involved in DNA methylation.
In our study, we used Neurospora to explore potential interactions of the essential components of the DNA methylation machinery. First, to identify proteins that interact with DIM-2, we screened a yeast two-hybrid library and identified HP1, which is predicted to be an adaptor protein between DNA methylation and H3K9 methylation (16). The results of a deletion analysis showed that regions containing the tandem PXVXL-related motifs of DIM-2 and the CSD of HP1 are required for the proteins' interaction. We confirmed that altered PXVXL-related motifs of DIM-2 (DIM-2PSL/AAA or DIM-2IVL/AAA) and the single-amino acid substitution of HP1Y244E perturbed both DIM-2-HP1 interaction and DNA methylation in vivo, showing that direct interaction between HP1 and DIM-2 is required for DNA methylation. In addition, we found that this interaction is independent of DIM-5 and confirmed that the localization of HP1 and DIM-2 to methylated DNA regions, as well as DNA methylation, depends on the H3K9me3 mark left by DIM-5. Thus, the HP1-DIM-2 complex serves to directly link H3K9me3 with DNA methylation.
Interaction between the HP1 CSD and the tandem PXVXL-like motif of DIM-2.
The results of our deletion and mutagenesis studies revealed that the CSD of HP1 binds directly to the PXVXL-like motifs in the N-terminal region of DIM-2. The CSD is less conserved than the chromo domain (16), but based on a structural analysis of the CSD in mouse HP1 (54), we predict that Neurospora HP1 forms a dimer and this creates a binding pocket for the PXVXL-like sequences. The Y244E change in HP1 was expected to disrupt both dimerization and binding to DIM-2. Indeed, we observed both reduced binding to DIM-2 and reduced localization at heterochromatin even though its methylated H3K9 binding domain, the chromo domain, was intact. These findings are similar to observations for mice except that the corresponding mutation in mouse cells showed complete disruption of HP1 binding and localization (54). This distinction may arise from sequence differences surrounding the critical Tyr of the Neurospora and mouse CSDs (Fig. 1D). Similarly, our observation that another HP1 CSD mutation (I250E) did not cause noticeable defects in heterochromatin formation or DNA methylation presumably reflects the lack of conservation of Ile-250 and its neighboring residues in Neurospora and other species (Fig. 1D). The results of a structural study of a cocrystal of the human HP1β CSD and the EMSY protein revealed that the CSD binds to a PXVXL-related motif plus residues neighboring EMSY (26). This is one indication that the interaction of the CSDs with the PXVXL-like region(s) may be more complex than that predicted based on the results of the structural study of mouse HP1 (54). Similarly, the binding of the Drosophila HP1 CSD to Hip (HP1-interacting protein) was reported to differ from that of the mouse consensus PXVXL motif (47). Structural studies of HP1s in other organisms might reveal interesting differences in the binding mechanics of HP1s from various organisms.
Other HP1 interactions.
It is noteworthy that a Neurospora DIM-2 mutant that abolishes the interaction of DIM-2 with HP1 does not influence HP1-GFP localization or show a growth defect comparable to that observed in Neurospora strains with hpo mutations. This suggests that other, yet-unidentified proteins interact with Neurospora HP1, presumably through a domain with a PXVXL-like sequence(s). It is proposed that HP1 serves as an adaptor to connect between methylated H3K9 and a variety of effecter proteins for transcriptional silencing, chromosomal segregation, and high-order chromatin architecture, etc. (23). Using a biochemical approach, we have purified a Neurospora HP1 complex and identified several putative effector proteins that contain PXVXL-like sequences (our unpublished data). It will be interesting to learn whether fundamental chromosome processes that involve HP1 are also involved in DNA methylation.
Predominantly unidirectional pathway of DNA methylation in Neurospora.
The requirement of H3K9me3 for DNA methylation is well established in Neurospora, but the various effects of DNA methylation, DIM-2, and HP1 on H3K9 methylation are not well understood. In mammals, DNMT1 directly binds the HMTase G9a and the interaction stimulates both enzymatic activities (13). The methyl-CpG binding protein MBD1 directly recruits the H3K9 HMTase SETDB1 to facilitate the methylation of H3K9 during DNA replication (46). In Drosophila, HP1 interacts with the H3K9 HMTase SU(VAR)3-9 in vitro and the interaction facilitates HP1 binding to methylated H3K9 in a reconstituted chromatin system (12). In contrast, we could not detect associations between DIM-5 and HP1 or DIM-2 in vivo or in the yeast two-hybrid assay, suggesting that the limited effect of HP1 or DIM-2 on DIM-5 action is indirect.
We found that a dim-2 mutant showed a wild-type distribution of H3K9me3 at the four regions tested that are normally subject to DNA methylation. Tamaru and colleagues noted loss of this histone mark at the methylated amRIP8 allele in a dim-2 mutant, although the total level of H3K9me3 in the genome appeared normal (53). It may be relevant that this allele, generated by RIP, is transcribed in dim-2, but not dim+, strains (44). This limited feedback of DNA methylation on H3K9 methylation may involve an unidentified methyl-DNA binding protein. We found that loss of HP1 led to a reduced level of H3K9me3 at one of the four regions (the RIP relic 8:A6). It is noteworthy that Western blots show no significant global change in the level of H3K9me3 in an hpo mutant. We confirmed that the localization of HP1 depends entirely on DIM-5. We conclude that Neurospora HP1 feeds back on H3K9 methylation in a minority of genomic regions and that the histone/DNA methylation pathway is predominantly unidirectional in Neurospora.
Recruitment of DIM-5 to methylated regions.
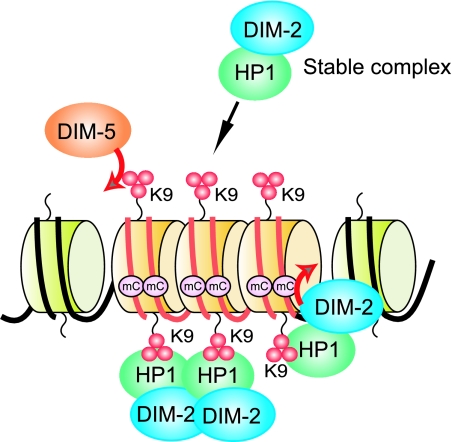
Most methylated regions in Neurospora are relics of transposons with numerous G:C to A:T mutations generated by the genomic defense system, RIP (48). Our observations that the global level of H3K9me3 is not grossly affected by mutation of either hpo or dim-2 indicate that the recruitment of DIM-5 is largely independent of the DIM-2-HP1 complex. The observations that many A:T-rich sequences trigger DNA methylation and that the AT hook analogue dystamycin inhibits DNA methylation (52) suggest that an unidentified A:T-rich binding protein (or proteins), perhaps containing one or more AT-hook motifs, can read the DNA level signals for de novo DNA methylation and, directly or indirectly, recruits DIM-5 (Fig. 9). The results of the work described here indicate that a stable DIM-2-HP1 complex then binds to the H3K9me3 mark made by DIM-5. Finally, DIM-2 methylates DNA on the nucleosomes bearing methylated H3K9 (Fig. 9). A full understanding of the control of DNA methylation in Neurospora will require the identification of proteins operating upstream of DIM-5.
FIG. 9.
Basic model for control of DNA methylation in Neurospora. The HMTase DIM-5, recruited to chromatin regions, such as those including AT-rich DNA (red lines) resulting from the genome defense system RIP, trimethylates H3K9 (red spheres). HP1 recruits DIM-2 and binds H3K9me3, resulting in DNA methylation at sites associated with heterochromatin.
Supplementary Material
Acknowledgments
We thank Gregory Kothe for the construction of yeast two-hybrid vectors pGBDU-dim-21-366, pGBDU-dim-2290-566, and pGBDU-dim-2431-847 and Glen Cronan and Tamir Khlafallah for advice on fluorescence microscopy. We also thank Zachary Lewis, Keyur Adhvaryu, and T.K. for comments on the manuscript.
This work was supported by grant GM025690-22 to E.U.S. from the National Institutes of Health.
Footnotes
Published ahead of print on 4 August 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Aagaard, L., G. Laible, P. Selenko, M. Schmid, R. Dorn, G. Schotta, S. Kuhfittig, A. Wolf, A. Lebersorger, P. B. Singh, G. Reuter, and T. Jenuwein. 1999. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 181923-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bestor, T. H. 2000. The DNA methyltransferases of mammals. Hum. Mol. Genet. 92395-2402. [DOI] [PubMed] [Google Scholar]
- 3.Bhaumik, S. R., E. Smith, and A. Shilatifard. 2007. Covalent modifications of histones during development and disease pathogenesis. Nat. Struct. Mol. Biol. 141008-1016. [DOI] [PubMed] [Google Scholar]
- 4.Brasher, S. V., B. O. Smith, R. H. Fogh, D. Nietlispach, A. Thiru, P. R. Nielsen, R. W. Broadhurst, L. J. Ball, N. V. Murzina, and E. D. Laue. 2000. The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J. 191587-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan, S. W., I. R. Henderson, and S. E. Jacobsen. 2005. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat. Rev. Genet. 6351-360. [DOI] [PubMed] [Google Scholar]
- 6.Chuang, L. S., H. I. Ian, T. W. Koh, H. H. Ng, G. Xu, and B. F. Li. 1997. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science 2771996-2000. [DOI] [PubMed] [Google Scholar]
- 7.Collins, R. E., M. Tachibana, H. Tamaru, K. M. Smith, D. Jia, X. Zhang, E. U. Selker, Y. Shinkai, and X. Cheng. 2005. In vitro and in vivo analyses of a Phe/Tyr switch controlling product specificity of histone lysine methyltransferases. J. Biol. Chem. 2805563-5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colot, H. V., G. Park, G. E. Turner, C. Ringelberg, C. M. Crew, L. Litvinkova, R. L. Weiss, K. A. Borkovich, and J. C. Dunlap. 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 10310352-10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cowell, I. G., R. Aucott, S. K. Mahadevaiah, P. S. Burgoyne, N. Huskisson, S. Bongiorni, G. Prantera, L. Fanti, S. Pimpinelli, R. Wu, D. M. Gilbert, W. Shi, R. Fundele, H. Morrison, P. Jeppesen, and P. B. Singh. 2002. Heterochromatin, HP1 and methylation at lysine 9 of histone H3 in animals. Chromosoma 11122-36. [DOI] [PubMed] [Google Scholar]
- 10.Davis, R. H. 2000. Neurospora: contributions of a model organism. Oxford University Press, New York, NY.
- 11.Ebbs, M. L., and J. Bender. 2006. Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. Plant Cell 181166-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eskeland, R., A. Eberharter, and A. Imhof. 2007. HP1 binding to chromatin methylated at H3K9 is enhanced by auxiliary factors. Mol. Cell. Biol. 27453-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esteve, P. O., H. G. Chin, A. Smallwood, G. R. Feehery, O. Gangisetty, A. R. Karpf, M. F. Carey, and S. Pradhan. 2006. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev. 203089-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feinberg, A. P. 2007. Phenotypic plasticity and the epigenetics of human disease. Nature 447433-440. [DOI] [PubMed] [Google Scholar]
- 15.Feldman, N., A. Gerson, J. Fang, E. Li, Y. Zhang, Y. Shinkai, H. Cedar, and Y. Bergman. 2006. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat. Cell Biol. 8188-194. [DOI] [PubMed] [Google Scholar]
- 16.Freitag, M., P. C. Hickey, T. K. Khlafallah, N. D. Read, and E. U. Selker. 2004. HP1 is essential for DNA methylation in Neurospora. Mol. Cell 13427-434. [DOI] [PubMed] [Google Scholar]
- 17.Freitag, M., and E. U. Selker. 2005. Controlling DNA methylation: many roads to one modification. Curr. Opin. Genet. Dev. 15191-199. [DOI] [PubMed] [Google Scholar]
- 18.Fuks, F., P. J. Hurd, R. Deplus, and T. Kouzarides. 2003. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 312305-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geiman, T. M., U. T. Sankpal, A. K. Robertson, Y. Zhao, and K. D. Robertson. 2004. DNMT3B interacts with hSNF2H chromatin remodeling enzyme, HDACs 1 and 2, and components of the histone methylation system. Biochem. Biophys. Res. Commun. 318544-555. [DOI] [PubMed] [Google Scholar]
- 20.Goff, C. G. 1976. Histones of Neurospora crassa. J. Biol. Chem. 2514131-4138. [PubMed] [Google Scholar]
- 21.Goll, M. G., and T. H. Bestor. 2005. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 74481-514. [DOI] [PubMed] [Google Scholar]
- 22.Green, G. R., L. C. Gustavsen, and D. L. Poccia. 1990. Phosphorylation of plant H2A histones. Plant Physiol. 931241-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grewal, S. I., and S. Jia. 2007. Heterochromatin revisited. Nat. Rev. Genet. 835-46. [DOI] [PubMed] [Google Scholar]
- 24.Hediger, F., and S. M. Gasser. 2006. Heterochromatin protein 1: don't judge the book by its cover! Curr. Opin. Genet. Dev. 16143-150. [DOI] [PubMed] [Google Scholar]
- 25.Hermann, A., H. Gowher, and A. Jeltsch. 2004. Biochemistry and biology of mammalian DNA methyltransferases. Cell. Mol. Life Sci. 612571-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang, Y., M. P. Myers, and R. M. Xu. 2006. Crystal structure of the HP1-EMSY complex reveals an unusual mode of HP1 binding. Structure 14703-712. [DOI] [PubMed] [Google Scholar]
- 27.Jackson, J. P., A. M. Lindroth, X. Cao, and S. E. Jacobsen. 2002. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416556-560. [DOI] [PubMed] [Google Scholar]
- 28.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 1441425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klose, R. J., and A. P. Bird. 2006. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 3189-97. [DOI] [PubMed] [Google Scholar]
- 30.Kouzminova, E., and E. U. Selker. 2001. dim-2 encodes a DNA methyltransferase responsible for all known cytosine methylation in Neurospora. EMBO J. 204309-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krauss, V. 2008. Glimpses of evolution: heterochromatic histone H3K9 methyltransferases left its marks behind. Genetica 13393-106. [DOI] [PubMed] [Google Scholar]
- 32.Lehnertz, B., Y. Ueda, A. A. Derijck, U. Braunschweig, L. Perez-Burgos, S. Kubicek, T. Chen, E. Li, T. Jenuwein, and A. H. Peters. 2003. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 131192-1200. [DOI] [PubMed] [Google Scholar]
- 33.Lindroth, A. M., D. Shultis, Z. Jasencakova, J. Fuchs, L. Johnson, D. Schubert, D. Patnaik, S. Pradhan, J. Goodrich, I. Schubert, T. Jenuwein, S. Khorasanizadeh, and S. E. Jacobsen. 2004. Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. EMBO J. 234286-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maison, C., and G. Almouzni. 2004. HP1 and the dynamics of heterochromatin maintenance. Nat. Rev. Mol. Cell Biol. 5296-304. [DOI] [PubMed] [Google Scholar]
- 35.Malagnac, F., L. Bartee, and J. Bender. 2002. An Arabidopsis SET domain protein required for maintenance but not establishment of DNA methylation. EMBO J. 216842-6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Margolin, B. S., M. Freitag, and E. U. Selker. 1997. Improved plasmids for gene targeting at the his-3 locus of Neurospora crassa by electroporation. Fungal Genet. Newsl. 4434-36. [Google Scholar]
- 37.Nakai, K., and P. Horton. 1999. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 2434-36. [DOI] [PubMed] [Google Scholar]
- 38.Ninomiya, Y., K. Suzuki, C. Ishii, and H. Inoue. 2004. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc. Natl. Acad. Sci. USA 10112248-12253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oldenburg, K. R., K. T. Vo, S. Michaelis, and C. Paddon. 1997. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 25451-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peters, A. H., S. Kubicek, K. Mechtler, R. J. O'Sullivan, A. A. Derijck, L. Perez-Burgos, A. Kohlmaier, S. Opravil, M. Tachibana, Y. Shinkai, J. H. Martens, and T. Jenuwein. 2003. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell 121577-1589. [DOI] [PubMed] [Google Scholar]
- 41.Rice, J. C., S. D. Briggs, B. Ueberheide, C. M. Barber, J. Shabanowitz, D. F. Hunt, Y. Shinkai, and C. D. Allis. 2003. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol. Cell 121591-1598. [DOI] [PubMed] [Google Scholar]
- 42.Robertson, K. D., S. Ait-Si-Ali, T. Yokochi, P. A. Wade, P. L. Jones, and A. P. Wolffe. 2000. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 25338-342. [DOI] [PubMed] [Google Scholar]
- 43.Rountree, M. R., K. E. Bachman, and S. B. Baylin. 2000. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet. 25269-277. [DOI] [PubMed] [Google Scholar]
- 44.Rountree, M. R., and E. U. Selker. 1997. DNA methylation inhibits elongation but not initiation of transcription in Neurospora crassa. Genes Dev. 112383-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sampath, S. C., I. Marazzi, K. L. Yap, A. N. Krutchinsky, I. Mecklenbrauker, A. Viale, E. Rudensky, M. M. Zhou, B. T. Chait, and A. Tarakhovsky. 2007. Methylation of a histone mimic within the histone methyltransferase G9a regulates protein complex assembly. Mol. Cell 27596-608. [DOI] [PubMed] [Google Scholar]
- 46.Sarraf, S. A., and I. Stancheva. 2004. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol. Cell 15595-605. [DOI] [PubMed] [Google Scholar]
- 47.Schwendemann, A., T. Matkovic, C. Linke, A. Klebes, A. Hofmann, and G. Korge. 2008. Hip, an HP1-interacting protein, is a haplo- and triplo-suppressor of position effect variegation. Proc. Natl. Acad. Sci. USA 105204-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Selker, E. U., N. A. Tountas, S. H. Cross, B. S. Margolin, J. G. Murphy, A. P. Bird, and M. Freitag. 2003. The methylated component of the Neurospora crassa genome. Nature 422893-897. [DOI] [PubMed] [Google Scholar]
- 49.Smallwood, A., P. O. Esteve, S. Pradhan, and M. Carey. 2007. Functional cooperation between HP1 and DNMT1 mediates gene silencing. Genes Dev. 211169-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smothers, J. F., and S. Henikoff. 2000. The HP1 chromo shadow domain binds a consensus peptide pentamer. Curr. Biol. 1027-30. [DOI] [PubMed] [Google Scholar]
- 51.Tamaru, H., and E. U. Selker. 2001. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414277-283. [DOI] [PubMed] [Google Scholar]
- 52.Tamaru, H., and E. U. Selker. 2003. Synthesis of signals for de novo DNA methylation in Neurospora crassa. Mol. Cell. Biol. 232379-2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tamaru, H., X. Zhang, D. McMillen, P. B. Singh, J. Nakayama, S. I. Grewal, C. D. Allis, X. Cheng, and E. U. Selker. 2003. Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat. Genet. 3475-79. [DOI] [PubMed] [Google Scholar]
- 54.Thiru, A., D. Nietlispach, H. R. Mott, M. Okuwaki, D. Lyon, P. R. Nielsen, M. Hirshberg, A. Verreault, N. V. Murzina, and E. D. Laue. 2004. Structural basis of HP1/PXVXL motif peptide interactions and HP1 localisation to heterochromatin. EMBO J. 23489-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Turck, F., F. Roudier, S. Farrona, M. L. Martin-Magniette, E. Guillaume, N. Buisine, S. Gagnot, R. A. Martienssen, G. Coupland, and V. Colot. 2007. Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 3e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xin, Z., M. Tachibana, M. Guggiari, E. Heard, Y. Shinkai, and J. Wagstaff. 2003. Role of histone methyltransferase G9a in CpG methylation of the Prader-Willi syndrome imprinting center. J. Biol. Chem. 27814996-15000. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, X., S. Germann, B. J. Blus, S. Khorasanizadeh, V. Gaudin, and S. E. Jacobsen. 2007. The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat. Struct. Mol. Biol. 14869-871. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.