Abstract
Raf-MEK-extracellular signal-regulated kinase (Erk) signaling initiated by growth factor-engaged receptor tyrosine kinases (RTKs) is modulated by an intricate network of positive and negative feedback loops which determine the specificity and spatiotemporal characteristics of the intracellular signal. Well-known antagonists of RTK signaling are the Sprouty proteins. The activity of Sprouty proteins is modulated by phosphorylation. However, little is known about the kinases responsible for these posttranslational modifications. We identify DYRK1A as one of the protein kinases of Sprouty2. We show that DYRK1A interacts with and regulates the phosphorylation status of Sprouty2. Moreover, we identify Thr75 on Sprouty2 as a DYRK1A phosphorylation site in vitro and in vivo. This site is functional, since its mutation enhanced the repressive function of Sprouty2 on fibroblast growth factor (FGF)-induced Erk signaling. Further supporting the idea of a functional interaction, DYRK1A and Sprouty2 are present in protein complexes in mouse brain, where their expression overlaps in several structures. Moreover, both proteins copurify with the synaptic plasma membrane fraction of a crude synaptosomal preparation and colocalize in growth cones, pointing to a role in nerve terminals. Our results suggest, therefore, that DYRK1A positively regulates FGF-mitogen-activated protein kinase signaling by phosphorylation-dependent impairment of the inhibitory activity of Sprouty2.
Signaling pathways initiated by receptor tyrosine kinases (RTKs) regulate fundamental cellular processes such as proliferation, differentiation, apoptosis, and cell migration (for a review, see reference 44). RTKs are activated by the binding of specific ligands, which induces their dimerization and autophosphorylation of tyrosine residues in their cytoplasmic tails. These events lead to the appearance of specific binding sites for a wide variety of proteins initiating intracellular signaling cascades (for a review, see reference 41). An essential pathway downstream of RTK signaling is the mitogen-activated protein kinase (MAPK) cascade, which is composed of members of the Raf family, MAPK kinases (MEK), and extracellular signal-regulated kinase (Erk) kinases. The signal output from the Erk cascade is finely modulated by positive and negative regulators that contribute to determining both the magnitude and the duration of Erk signaling (14). An important class of negative regulators of RTK-dependent pathways is the Sprouty (Spry) family of proteins. In Drosophila melanogaster, dSpry interferes with signaling by fibroblast growth factor (FGF) and epidermal growth factor (EGF) (8, 20). In mammals, four dSpry orthologs have been identified that function as negative regulators of RTK signaling in vivo (for a recent review, see reference 37). Several modes of action have been proposed for Spry proteins, but none of them satisfactorily explains the diversity of their inhibitory activities. These range from acting as an uncoupling factor at the level of FRS2/GRB2/Sos (22, 32) or Cbl/RTK complex formation (16, 42) to interfering in Ras or Raf activation (19, 43, 49). In addition, Spry activity is modulated at different levels by posttranslational modifications such as palmitoylation (28) and phosphorylation events in tyrosine, serine, and threonine residues (10, 16, 22, 32, 36).
Inappropriate spatial or temporal activation of MAPK cascades upon growth factor stimulation is often implicated in tumor progression and human developmental pathologies (for a recent review, see reference 45). One of the most common human developmental disorders is caused by trisomy of chromosome 21. Among the human chromosome 21 genes, DYRK1A has been proposed as a major contributor to Down syndrome (12, 15). The phenotypes associated with DYRK1A gene dosage imbalance in vivo are thought to be caused by alterations of specific gene expression patterns and/or changes in intracellular signaling outcomes (3, 6). DYRK1A encodes a protein kinase belonging to an evolutionarily conserved family of protein kinases known as DYRKs (dual-specificity tyrosine [Y] phosphorylation-regulated kinases). DYRK1A is highly expressed in the brain during development and also in adults, where it is detected at high levels in olfactory bulb, spinal cord, cerebellar cortex, and the majority of motor nuclei (35). Loss of Dyrk1A function is lethal at embryonic midstages, and these mice show severe developmental retardation (17). Heterozygotes are viable but present a reduced size at birth that is maintained at adult stages. Size reduction is most noticeable in organs such as liver and brain (17). Indeed, several brain structures are differently affected; for example, there is a reduction in the complexity of the neuronal dendritic tree in the cortex (5) and also a reduction in the number of neurons in the superior colliculus (17). These data, together with the expression pattern of the protein, suggest a role for DYRK1A in the development and homeostasis of the central nervous system. At the molecular level, several substrates and protein partners for DYRK1A have been reported (for a recent review, see reference 13). The list includes both cytosolic and nuclear proteins, suggesting specific roles for DYRK1A in different subcellular compartments. Regarding DYRK1A actions outside the nucleus, DYRK1A overexpression has been shown to potentiate the differentiation effects of nerve growth factor in the neuron-like cellular model PC12 (29). This effect, which is independent of kinase activity, correlates with increased levels of activated Erk1/2, a downstream effector of the neurogenic activity of nerve growth factor.
Despite all this, there is still a lack of understanding of the DYRK1A role in intracellular signaling pathways. Here, we show that the negative regulator of FGF signaling Spry2 is a novel DYRK1A partner and substrate. Based on our findings, we propose that DYRK1A promotes FGF signaling by acting as a negative regulator of RTK antagonistic Spry proteins.
MATERIALS AND METHODS
Plasmids.
Expression plasmids encoding the DYRK1A variants have been described previously: hemagglutinin (HA)-tagged variants (pHA derivatives), variants N-terminally fused to glutathione S-transferase (GST) (pGST derivatives), and the DYRK1A fragment 590-611 fused to the enhanced green fluorescent protein (GFP) (pGFP-Hrep) (1, 2). The expression plasmid pHA-DYRK1AΔHis was generated by site-directed mutagenesis. The expression plasmid for FGF receptor (FGFR; pMyc-FGFR1) (22) was kindly provided by E. Nishida (Department of Cell and Developmental Biology, Kyoto University, Japan). pSG.Gal-Elk (31) was a kind gift from P. E. Shaw (School of Biomedical Sciences, University of Nottingham, United Kingdom). pE1bGal-luc was as described previously (11), and pRL-Null was from Promega.
The Spry2 expression plasmid (pFlag-hSpry2) (33) was kindly provided by G. R. Guy (Institute of Molecular and Cell Biology, Singapore). To make plasmids pMyc-Spry2 and pGST-Spry2, the open reading frame (ORF) of human Spry2 was amplified by PCR and subcloned into the EcoRI and XhoI sites of pCDNA3-Myc or pGEX-5X-3 (Amersham Biosciences), respectively. For generation of pGST-Spry2-(1-164) and pGST-Spry2-(1-255), AflII/XhoI and NcoI/XhoI fragments, respectively, were removed from pGST-Spry2, and the ends were filled with T4 DNA polymerase and religated. To generate pFlag-Spry2Δ164-255, a DNA fragment encoding amino acids 255 to 315 of human Spry2 was amplified by PCR and then subcloned into the AflII/XhoI sites of pFlag-Spry2. Constructs for Y55F, T75A, T122A, and T126A Spry2 mutants were generated by site-directed mutagenesis. Plasmids pFlag-Spry1, pFlag-Spry3, and pFlag-Spry4 were generated by replacing a BamHI/XhoI fragment containing the Spry2 ORF in pFlag-Spry2 with DNA fragments containing the corresponding Spry ORFs. These fragments were obtained by PCR using IMAGE Consortium cDNA clones 6028920 (human Spry1), 2208245 (human Spry3), and 6821944 (mouse Spry4) as templates. The clones were purchased from the RZPD German Resource Center for Genome Research. Nucleotide changes and small deletions were made by oligonucleotide-directed mutagenesis using the QuikChange site-directed mutagenesis kit according to the manufacturer's instructions (Stratagene). All plasmids generated by PCR or site-directed mutagenesis, as well as all the in-frame fusions, were verified by DNA sequencing.
Yeast two-hybrid screening.
Full-length human DYRK1A cDNA (754-amino-acid isoform) was subcloned into pAS2 (Clontech) and used as bait to screen a fetal human brain cDNA library cloned into pACT2 (Clontech). Positive clones for HIS3 selection were rescreened for LacZ expression. Plasmids containing the prey sequences were rescued and checked by back-transformation with the bait. Library screening and β-galactosidase measurements were done according to Clontech protocols.
Cell culture and transfection.
HEK-293 and HEK-293T cell lines were maintained at 37°C in Dulbecco's modified Eagle's medium, supplemented with 10% fetal calf serum (FCS) and antibiotics (100 units/ml penicillin and 100 μg/ml streptomycin). Transient transfections were performed using the calcium phosphate method, and cells were processed 48 h after transfection. For preparation of dissociated cortical neuron cultures, mouse embryos at embryonic day 15 were obtained from anesthetized pregnant females. Cerebral cortices were isolated and mechanically dissociated in phosphate-buffered saline (PBS) with 5 mg/ml glucose. Cells were plated onto glass coverslips (80 cells/mm2) previously coated with 2 μg/ml poly-l-lysine (Sigma). Cells were maintained in neurobasal medium (Gibco) supplemented with B27 (Gibco) and 1% inactivated horse serum (Gibco).
Western blotting.
Samples were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred onto Hybond C membranes (Amersham), and blocked with 10% skim milk in 10 mM Tris-HCl (pH 7.5)-100 mM NaCl (TBS) plus 0.1% Tween 20 (TBS-T). Membranes were incubated with primary antibodies (in 5% skim milk in TBS-T) overnight at 4°C, except in the case of PY20 antibody, when bovine serum albumin replaced skim milk for both blocking and antibody incubation. After being washed with TBS-T, membranes were incubated for 45 min at room temperature with horseradish peroxidase-conjugated rabbit anti-mouse and goat anti-rabbit antibodies (Dako) (in 5% skim milk in TBS-T) and then washed again with TBS-T. Detection was by enhanced chemiluminescence with Supersignal West Pico (Pierce). Chemiluminescence was determined with a Fuji LAS-3000 image analyzer. Quantification of the data was performed using Multi Gauge software (v3; Fuji PhotoFilm). The following antibodies were used as primary antibodies: mouse monoclonal antibody (MAb) anti-HA (Covance), MAb anti-Flag M2 (Sigma), rabbit polyclonal antibody (rAb) anti-Myc (Santa Cruz), MAb anti-GFP JL-8 (BD Biosciences), rAb anti-lamin B1 (Santa Cruz), MAb anti-SNAP-25 (BD Biosciences), MAb antisynaptophysin (Sigma), rAb anticalretinin (Swant), rAb anti-Erk2 (Santa Cruz), MAb anti-phospho-Erk1/2 (Thr202/Tyr204) (Cell Signaling), rAb anti-B-Raf (Santa Cruz), rAb anti-c-Cbl (Santa Cruz), rAb anti-GRB2 (Santa Cruz), rAb anti-FGFR1 (Santa Cruz Biotech), rAb anti-DYRK1A (1), MAb anti-DYRK1A (Abnova), MAb anti-phospho-Tyr PY20 (BD Biosciences), and rAb anti-Spry2 (Upstate Biotechnology).
Mouse brain subfractionation.
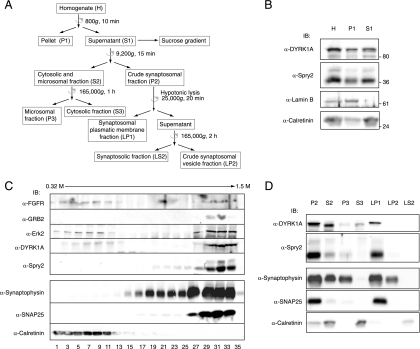
The fractionation procedures described here are summarized in Fig. 6A. All procedures were done at 4°C. Equal volumes of every fraction were resuspended in 6× Laemmli buffer (LB) and analyzed by Western blotting. For density gradient sucrose fractionation, two mouse brains were dissected and homogenized in 10 volumes of sucrose buffer (0.32 M sucrose, 10 mM HEPES [pH 7.4], 1 mM EGTA, 1.5 mM MgCl2, protease inhibitor cocktail [PIC; Roche], and phosphatase inhibitors [PI; 10 mM sodium orthovanadate, 30 mM sodium pyrophosphate]) with a glass-Teflon homogenizer using 10 up-and-down strokes at 700 rpm. The homogenate (H) was centrifuged for 10 min at 800 × g, yielding a pellet (P1) containing nuclei and any unbroken cells and a supernatant (S1) free of nuclei. The supernatant was placed at the top of a 0.4 to 2 M discontinuous sucrose gradient (nine 1-ml layers of 0.2 M increments) and fractionated by centrifugation for 3 h at 33,000 rpm in a SW41 rotor (Beckman). Aliquots of 0.2 ml were extracted from top to bottom of the gradient.
FIG. 6.
DYRK1A and Spry2 associate with the plasma membranes of synaptic terminals. (A) Schematic representation of the subcellular fractionation procedures used for the analyses in panels B to D. Centrifugation steps are indicated with a semicircular arrow. Full details are provided in Materials and Methods. (B) Homogenates from mouse brain were obtained as described in Materials and Methods. Equal volumes of the homogenate (H), the nuclear pellet (P1), and the postnuclear fraction (S1) were analyzed by Western blotting with the indicated antibodies. (C) The postnuclear fraction (S1) from mouse brain homogenates was fractionated by centrifugation in a discontinuous sucrose gradient. Equal volumes of the first 18 alternate fractions were analyzed by Western blotting with specific antibodies, as indicated. (D) The postnuclear fraction (S1) from mouse brain homogenates was fractionated by differential centrifugation. Equal volumes of the fractions obtained were analyzed by Western blotting with specific antibodies, as indicated. P2, crude synaptosomal fraction; S2, cytosolic and microsomal fraction; P3, microsomal fraction; S3, soluble cytosolic fraction; LP1, synaptosomal plasmatic membrane fraction; LP2, crude synaptosomal vesicle fraction; LS2, synaptosolic fraction.
Fractionation by differential centrifugation was performed essentially as described previously (26). The postnuclear S1 fraction was collected and centrifuged for 15 min at 9,200 × g. The supernatant (S2) was removed, and the pellet (P2) was washed by resuspension in 9 ml of sucrose buffer and centrifuged for 15 min at 10,200 × g, yielding a supernatant (S2′) and a pellet (P2′). The supernatants S2 and S2′ were pooled and centrifuged for 1 h at 165,000 × g, yielding a pellet (P3) and soluble fraction (S3). The pellet P2′ was resuspended in 1 ml of sucrose buffer, and 8 ml of cold water (supplemented with PIC and PI) was added. The whole suspension was subjected to seven up-and-down strokes at 1,500 rpm with a glass-Teflon homogenizer. The resulting P2′ lysate was rapidly transferred to a tube (to which was added 64 μl of 1 M HEPES, pH 7.4), kept on ice for 30 min, and then centrifuged for 20 min at 25,000 × g, yielding a lysate pellet (LP1) and a supernatant. The supernatant was collected and centrifuged for 2 h at 165,000 × g, yielding a lysate pellet (LP2) and lysate supernatant (LS2). All centrifugations were done with a TLA-100.3 rotor (Beckman).
Immunohistochemistry and immunofluorescence.
Animals were housed in the Animal Facility of the Parc de Recerca Biomedica de Barcelona and were sacrificed in accordance with the recommendations and protocols for the proper use of laboratory animals approved by the local ethics committee. Mice were anesthetized and transcardially perfused with cold PBS and 4% paraformaldehyde in 0.1 M phosphate buffer, pH 7.4. Brains were removed, postfixed in the same fixative for 24 h, and then cryoprotected in 30% sucrose in PBS. For immunohistochemistry, free-floating sections (40 μm) were blocked in blocking buffer (BB; 0.25% gelatin, 0.2% Triton X-100 in PBS) with 10% FCS and incubated overnight at 4°C with primary antibodies (rAb anti-DYRK1A or rAb anti-Spry2) diluted in BB with 1% FCS. For immunoperoxidase detection, sections were incubated with a goat anti-rabbit biotinylated secondary antibody (1:200; Vector Labs) followed by ABC complex (Vector Labs) and reacted with 0.03% diaminobenzidine (Sigma) and 0.003% hydrogen peroxide. Finally, sections were dehydrated and mounted on slides. For immunofluorescence detection, 20-μm sections on slides were blocked in BB with 10% FCS and then incubated overnight at 4°C with primary antibodies (MAb anti-DYRK1A and rAb anti-Spry2) diluted in BB with 1% FCS. Secondary antibodies used were goat anti-rabbit antibody-fluorescein isothiocyanate conjugate (Southern Biotech) and goat anti-mouse antibody-Texas Red conjugate (Jackson ImmunoResearch). Samples were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI) to visualize cell nuclei and mounted in Vectashield (Vector Labs). Samples were viewed under a Leica DMI 6000B microscope, and fluorescence images were acquired with a Leica DFC350 FX camera.
For immunolocalization studies in primary cortical neuron cultures, cells in coverslips were processed after 10 days in culture. Cells were fixed in 4% paraformaldehyde in PBS for 15 min at room temperature, and the immunofluorescence assay was performed as described above. Goat anti-mouse antibody-Alexa 555 and goat anti-rabbit antibody-Alexa 488 (Molecular Probes) were used as secondary antibodies. Samples were viewed under a Leica TSC SPE confocal microscope, and the images were captured in a consecutive confocal Z-sectioning of 0.37 μm (approximately six sections). The Z-stack projection image was generated using ImageJ processing software (http://rsb.info.nih.gov/ij).
GST fusion protein expression in bacteria and pull-down assays.
GST fusion expression constructs were transformed into Escherichia coli BL21(DE3)pLysS. Protein expression was induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside for 3 h at 37°C for unfused GST, 4 to 6 h at 22°C for GST-Spry2 proteins, and 8 h at 20°C for GST-DYRK1A. Induced recombinant proteins were bound to glutathione beads and, when required, were eluted with 10 mM reduced glutathione in 50 mM Tris-HCl, pH 8, and dialyzed against a buffer containing 50 mM HEPES (pH 7.4), 150 mM NaCl, and 2 mM EDTA. Protein concentration was determined with a colorimetric assay (bicinchoninic acid protein assay kit; Pierce) and/or by Coomassie blue staining of SDS-polyacrylamide gels and comparison with a standard.
For pull-down assays, soluble cell lysates were incubated overnight at 4°C with 5 μg of unfused GST or GST-Spry2 (full length or mutants) immobilized on glutathione-Sepharose beads that had been previously equilibrated in lysis buffer (50 mM HEPES [pH 7.4], 150 mM NaCl, 2 mM EDTA, 1% Nonidet P-40 [NP-40], PIC, and PI). After binding, beads were washed four times with lysis buffer, and the bound protein was eluted by boiling samples for 5 min in SDS buffer. Samples were resolved by SDS-PAGE, and proteins were detected by immunoblotting.
Immunoprecipitation.
For soluble cell lysates, cells were washed twice in PBS and lysed in buffer A (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 2 mM EDTA, 1% NP-40, 10% glycerol, PIC, and PI). Lysates were clarified by centrifugation and diluted with 2 volumes of buffer A without NP-40. Lysates were incubated for 6 h at 4°C with protein G-Sepharose beads (Amersham) prebound with 5 μg of anti-HA or anti-Flag antibodies, and then beads were washed with lysis buffer A.
For immunoprecipitation of endogenous DYRK1A protein from mouse brain, brains from 2- to 4-month-old mice were dissected and homogenized mechanically with a Dounce homogenizer in 5 ml of lysis buffer B (10 mM HEPES [pH 7.4], 150 mM NaCl, 1 mM EDTA, 0.1 mM MgCl2, 0.2% Triton X-100, 10% glycerol, PIC, and PI). The homogenates were sonicated with 5 pulses of 5 s with an intensity of 0.5 in a Sonifier 250 (Branson) and clarified by centrifugation for 10 min at 16,000 × g at 4°C. Ten milligrams of soluble lysates was precleared for 1 h at 4°C with equilibrated protein A-Sepharose and incubated overnight at 4°C with 10 μg of either rAb anti-DYRK1A or nonspecific rabbit immunoglobulin G (IgG; Sigma). Then, the immunocomplexes were incubated with protein A-Sepharose for 2 h at 4°C, and beads were washed with lysis buffer B. Finally, both lysates and the immunoprecipitates were resuspended in 6× LB and analyzed by Western blotting.
IVK assays.
The in vitro kinase (IVK) assays were performed with either immunocomplexes from transfected cells or bacterially expressed recombinant GST fusion proteins. For IVK assays from immunocomplexes, cell lysates were prepared in lysis buffer C (50 mM HEPES [pH 7.4], 75 mM NaCl, 1 mM EDTA, 1% NP-40, PIC, and PI). Cell lysates were incubated overnight at 4°C with anti-HA bound to protein G-Sepharose beads. Immunocomplexes were washed twice with buffer C with 0.1% NP-40 and twice with buffer C without NP-40. Anti-HA immunocomplexes were split into two aliquots, one for Western blotting and the other for IVK assay. Immunocomplexes were washed twice with kinase buffer (50 mM HEPES [pH 7.4], 0.5 mM dithiothreitol, 5 mM MgCl2, 5 mM MnCl2) and incubated for 20 min at 30°C in 30 μl of kinase buffer with a final concentration of 25 μM ATP and [γ-32P]ATP (1 × 10−2 μCi/pmol). For GST fusion proteins, 0.5 to 1 μg of eluted GST-Spry2 or its derivatives was incubated for 20 min at 30°C in 40 μl of kinase buffer with 50 μM ATP, [γ-32P]ATP (1 × 10−3 μCi/pmol), and 50 ng of purified GST-DYRK1A. Reactions were stopped by adding 6× LB, and samples were resolved by SDS-PAGE and then stained with Coomassie blue. 32P incorporation was detected by autoradiography of dried gels.
Luciferase reporter assay.
FGF-induced Erk-dependent transcriptional activation was measured by an Elk-1 reporter assay. HEK-293 cells were cotransfected with pSG.Gal-Elk1, pE1bGal-Luc, and the Renilla expression vector pRL-Null, together with effector plasmids. After 24 h, cells were serum starved (Dulbecco's modified Eagle's medium supplemented with 0.5% FCS) for 20 h, and then FGF2 (Peprotech) was added for 6 h. Each transfection point was done in triplicate. Luciferase and Renilla activities were determined in cell lysates by the dual-luciferase reporter assay system (Promega) and analyzed for bioluminescence with an AutoLumat LB 953 luminometer (Berthold Technologies).
Statistical analysis.
Significant differences between experiments were determined by the two-tailed Student t test. Data are presented as the means ± standard errors of the means (SEM) of independent experiments.
RESULTS
DYRK1A interacts with Spry2.
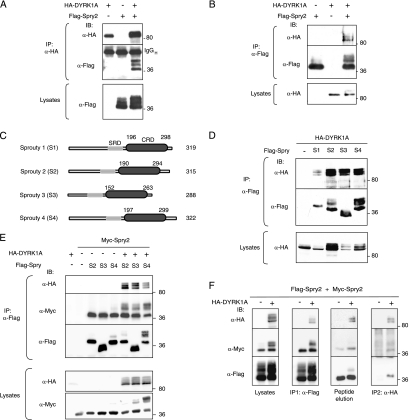
To identify partners involved in the possible effects of DYRK1A on signaling pathways, we performed a yeast two-hybrid screen of a human fetal brain cDNA library, using full-length DYRK1A as bait. One of the putative DYRK1A-interacting proteins identified was Spry2, a well-known antagonist of RTK signaling (37). To verify the interaction between DYRK1A and Spry2 in vivo, mammalian cells were transfected with plasmids encoding full-length HA-tagged DYRK1A and Flag-tagged Spry2. Cell extracts were subjected to immunoprecipitation with anti-HA or anti-Flag antibodies, and the presence of the proteins in the immunoprecipitates was analyzed by Western blotting. Flag-Spry2 was coimmunoprecipitated with HA-DYRK1A in anti-HA immunocomplexes but was not precipitated by anti-HA when expressed alone, thus indicating specific interaction between the two proteins (Fig. 1A). The complementary result was obtained when immunocomplexes were prepared with anti-Flag (Fig. 1B). These results thus demonstrate that DYRK1A and Spry2 can interact in vivo in mammalian cells.
FIG. 1.
DYRK1A interacts with the Spry family of RTK modulators. (A and B) HEK-293T cells were transfected with expression plasmids encoding HA-DYRK1A and Flag-Spry2, as indicated. At 48 h posttransfection, cell lysates were subjected to immunoprecipitation with anti-HA (A) or anti-Flag (B) antibodies. The presence of the large immunoglobulin (IgGH) is shown. (C) Schematic representation of the human Spry family members used in panels D and E. SRD, serine-rich domain. (D) Soluble extracts from cells expressing HA-DYRK1A and one of the four members of the mammalian Spry family, as indicated, were immunoprecipitated with anti-Flag, and DYRK1A and Spry proteins were detected in the complexes by Western blotting. (E) Cells were transfected as indicated with expression plasmids encoding HA-DYRK1A; Myc-Spry2; and either Flag-Spry2 (S2), Flag-Spry3 (S3), or Flag-Spry4 (S4), and cell extracts were subjected to immunoprecipitation with anti-Flag. (F) Cell lysates expressing HA-DYRK1A, Flag-Spry2, and Myc-Spry2 were subjected to immunoprecipitation with anti-Flag (IP1). The immunocomplexes were eluted from the beads with excess Flag peptide (peptide elution) and reimmunoprecipitated with anti-HA antibody (IP2). In all panels (A to F), the presence of the different proteins in both the immunoprecipitates (IP) and the total cell lysates was analyzed by Western blotting with the indicated antibodies. The positions of marker proteins (in kDa) are indicated in this and the other figures.
DYRK1A interacts with all mammalian Spry family members.
The human Spry family is composed of four members, Spry1, Spry2, Spry3, and Spry4, which have a high amino acid identity in the C-terminal region, in particular within the so-called cysteine-rich domain (CRD) (38) (Fig. 1C). We wondered therefore whether DYRK1A might interact with other Spry proteins. Coimmunoprecipitation assays confirmed that DYRK1A is able to interact with all known mammalian Spry proteins (Spry1 to Spry4) (Fig. 1D), indicating that DYRK1A is a common partner of the mammalian Spry family.
Spry proteins have been reported to form active homo- and heterodimers (22, 40). We therefore tested whether the interaction of DYRK1A with Spry family members affected their ability to form heterodimers with Spry2. In coimmunoprecipitation experiments, the presence of DYRK1A did not produce significant differences in the binding of Spry2 to other Spry family members (Fig. 1E). Thus, DYRK1A binding does not appear to interfere with heterodimer formation, and moreover, these results suggest that DYRK1A might interact with Spry dimers. To check this possibility, we performed a double round of immunoprecipitations, combining different epitope-tagged proteins and different antibodies, to isolate the putative trimeric complexes. First, we used anti-Flag to immunoprecipitate proteins bound to Flag-Spry2; both Myc-Spry2 and HA-DYRK1A were detected in the immunocomplexes (Fig. 1F, IP1 panel). The precipitated proteins were then eluted with an excess of competitor Flag peptide (Fig. 1F, peptide elution panel) and subsequently immunoprecipitated with anti-HA antibody; these immunocomplexes contained both Flag-Spry2 and Myc-Spry2 (Fig. 1F, IP2 panel). These results provide evidence of DYRK1A binding to at least the homodimer Spry2.
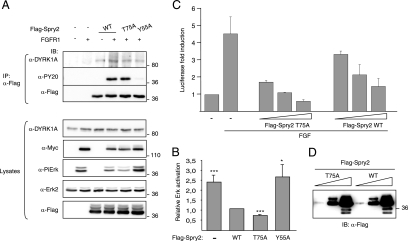
The interaction of DYRK1A with Spry2 is not affected by the activation status of the FGF pathway.
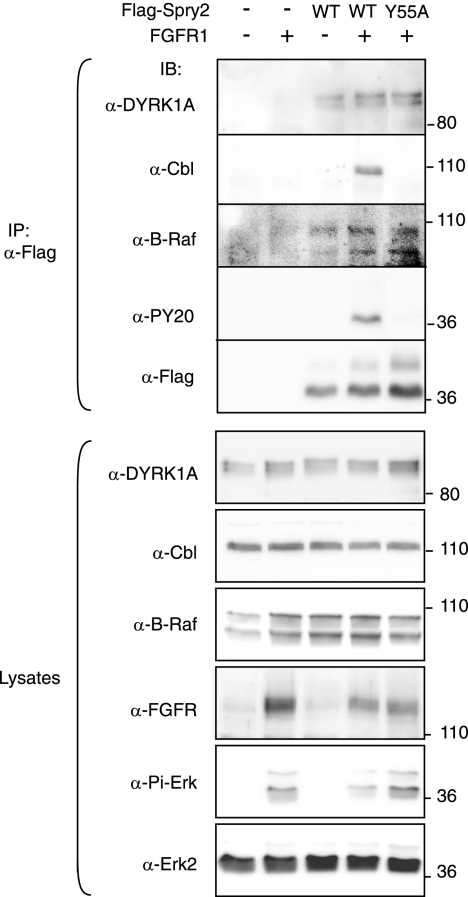
The interaction of some molecular partners with Spry2 or other Spry family members is modulated by RTK activation. This is the case with c-Cbl, which seems to require RTK-dependent phosphorylation of Spry2 at Tyr55 for binding (16, 21, 42). Conversely, other partners, such as Raf kinases, can interact directly with Spry proteins, independently of the activation state of RTK-dependent pathways (43, 47). In most cases, this differential behavior has been analyzed in the context of FGF signaling. Furthermore, we have recently demonstrated that the phosphorylation status of a regulatory serine residue in DYRK1A, involved in 14-3-3 binding, is modulated in vivo by stimulation with FGF but not EGF (1). Therefore, to investigate whether the DYRK1A/Spry2 interaction is modulated by RTK activation, we carried out coimmunoprecipitation experiments in cells overexpressing FGFR. This approach ensures constitutive and chronic receptor activation independently of ligand binding and mimics the continuous activation that occurs during some developmental processes and pathogenic situations. Thus, a clear increase in the levels of phospho-Erk was detected in cells overexpressing FGFR1, and this activation was inhibited by the expression of wild-type (wt) Flag-Spry2 but not by the dominant-negative Flag-Spry2-Y55A (Fig. 2, anti-Pi-Erk). The phosphorylation status on Tyr residues of Spry2-wt and Spry2-Y55A was checked by immunoblotting with anti-phospho-Tyr antibody, and the results confirmed that the major phospho-Tyr residue in Spry2 was Tyr55 (Fig. 2, anti-PY20).
FIG. 2.
DYRK1A interaction with Spry2 is not affected by FGF pathway activation. HEK-293 cells were transfected with expression plasmids encoding FGFR1 and either the wt or the Y55A mutant version of Flag-Spry2, as indicated. At 48 h posttransfection, soluble cell lysates were prepared and subjected to immunoprecipitation with anti-Flag antibody. Both the cell lysates and the immunoprecipitates (IP) were analyzed by Western blotting with the indicated antibodies (PY20, anti-phospo-Tyr antibody; Pi-Erk, anti-phospho-Erk1/2 Thr202/Tyr204).
Overexpression of FGFR1 did not alter the detectable levels of endogenous DYRK1A or other Spry2 partners such as c-Cbl and B-Raf (Fig. 2, lysates). Interaction of c-Cbl with Spry2-wt was detected only in FGFR1-overexpressing cells, and no interaction was detected with Spry2-Y55A (Fig. 2, anti-Cbl in IP). In contrast, interaction of Spry2 with B-Raf was detected with both Spry2-wt and the mutant Spry2-Y55A and was independent of the activation status of the pathway (Fig. 2, anti-B-Raf in IP). Similarly to B-Raf, endogenous DYRK1A protein was present in Spry2 immunocomplexes in all situations analyzed (Fig. 2, anti-DYRK1A in IP), thus indicating that DYRK1A interacts with Spry2 independently of phosphorylation on Tyr55 and that the activation status of the FGF pathway very likely does not affect the interaction.
DYRK1A is able to interact with Spry2 through their CRD.
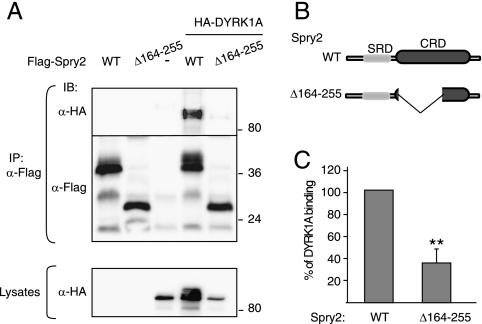
To define the region in Spry2 responsible for interaction with DYRK1A, we performed a yeast two-hybrid screen with DYRK1A as bait and several Spry2 deletion mutants as prey. This screen identified a region in Spry2 spanning amino acids 164 to 255 as necessary for the interaction with DYRK1A (see Fig. S1A in the supplemental material). These results were further tested by in vitro pull-down assays (see Fig. S1B in the supplemental material). To confirm the interaction data, we generated a deletion mutant of Flag-Spry2 lacking amino acids 164 to 255 (Flag-Spry2Δ164-255; scheme in Fig. 3B) and assayed its ability to bind DYRK1A in vivo. Compared with that of full-length Spry2, interaction of this mutant with DYRK1A was greatly compromised (Fig. 3A). Quantification of several experiments yielded a reduction in binding of about 70% (Fig. 3C). The deleted region overlaps with the CRD in Spry2, thus indicating that the integrity of the CRD in Spry2 is necessary for interaction with DYRK1A.
FIG. 3.
DYRK1A interacts with the CRD of Spry2. (A) HEK-293T cells were transfected with plasmids encoding HA-DYRK1A and either the wt or the Δ164-255 deletion mutant version of Flag-Spry2, as indicated. At 48 h posttransfection, cell extracts were prepared and immunoprecipitated with anti-Flag. Lysates and immunocomplexes (IP) were analyzed by Western blotting with the antibodies indicated. (B) Schematic representation of the Spry2 proteins used in panel A. SRD, serine-rich domain. (C) The histogram presents the proportion of DYRK1A protein bound to Spry2 (DYRK1A in immunocomplexes/DYRK1A in lysates) as a percentage of the value obtained with wt Spry2, which was set at 100%. Data are means ± SEM of three independent experiments (**, P ≤ 0.01).
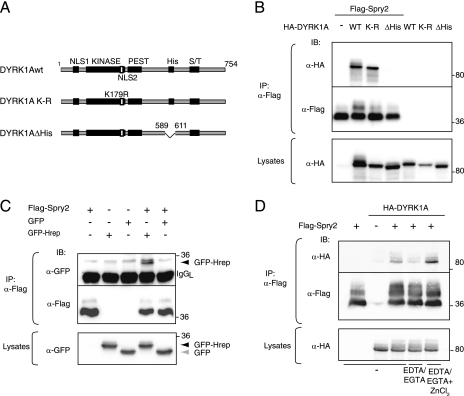
The polyhistidine segment in DYRK1A is necessary and sufficient for interaction with Spry2.
To identify the DYRK1A sequence responsible for binding to the Spry2 CRD, a yeast two-hybrid screen was performed with full-length and deletion mutant versions of DYRK1A as baits and Spry2 as prey (see Fig. S2A in the supplemental material). The screen identified a region in DYRK1A spanning amino acids 600 to 616 as being necessary for the interaction with Spry2. These results were confirmed by coimmunoprecipitation assays on cell extracts expressing different DYRK1A carboxy-terminal deletion mutants (see Fig. S2B in the supplemental material). The identified sequence corresponds closely to the distinctive polyhistidine segment in DYRK1A (590-HHHHGNSSHHHHHHHHHHHHH-611). Furthermore, a DYRK1A mutant in which only the polyhistidine stretch was deleted (DYRK1AΔHis) was unable to interact with Spry2 (Fig. 4B). Interestingly, an inactivating mutation in the DYRK1A kinase domain did not affect the interaction (Fig. 4B; see also Fig. S2 in the supplemental material). Thus, the interaction of DYRK1A with Spry2 requires the polyhistidine sequence but is independent of DYRK1A kinase activity.
FIG. 4.
The polyhistidine domain in DYRK1A is necessary and sufficient for interaction with Spry2. (A) Schematic representation of the DYRK1A proteins used in panel B. NLS, nuclear localization signal; KINASE, kinase catalytic domain; PEST, PEST domain; His, histidine repeat; S/T, serine/threonine-rich domain. (B) wt, kinase-dead (K-R), or histidine segment deletion (ΔHis) versions of DYRK1A were overexpressed with Flag-Spry2 in HEK-293T cells, and the presence of the different proteins was analyzed in anti-Flag immunoprecipitates. (C) HEK-293T cells were transfected with expression plasmids encoding Flag-Spry2 and either GFP or a fusion protein of GFP and the histidine domain of DYRK1A (GFP-Hrep), and cell lysates were immunoprecipitated with anti-Flag antibody. The positions of the GFPs are shown. (D) Cells were transfected with pHA-DYRK1A and pFlag-Spry2, as indicated. Soluble cell extracts were subjected to immunoprecipitation with anti-Flag antibody either under standard conditions (−) or in the presence of 5 mM each of EDTA and EGTA (EDTA/EGTA), with or without 20 mM ZnCl2 (EDTA/EGTA + ZnCl2). In panels B to D, both lysates and the immunoprecipitates (IP) were analyzed by Western blotting with the indicated antibodies.
To test whether the polyhistidine segment is sufficient for DYRK1A binding to Spry2, we fused it to a heterologous protein (GFP) and assayed it for interaction. Coimmunoprecipitation assays confirmed that the fusion protein (GFP-Hrep), but not GFP alone, interacted with Spry2 (Fig. 4C), indicating that the histidine segment itself forms the recognition motif for Spry2 binding. To further support this conclusion, we were able to show that interaction between DYRK1A and Spry2 is competed in a dosage-dependent manner by coexpressed GFP-Hrep fusion protein (see Fig. S2C in the supplemental material).
Histidine and cysteine residues commonly mediate the formation of structured protein domains around a coordinating metal, as occurs in zinc finger domains. It has been proposed that metal coordination can also mediate interaction between separate proteins, through the formation of an intermolecular zinc finger-like structure (24). We postulated that the interaction of DYRK1A and Spry2 might involve the coordination of metal ions by their respective histidine and cysteine residues. To test this hypothesis, we performed coimmunoprecipitation assays with standard cell extracts or extracts supplemented with cation chelators (EDTA/EGTA) with or without an excess of Zn ions. As Fig. 4D shows, chelation of divalent cations severely impaired the interaction of DYRK1A and Spry2; in addition, the presence of an excess of zinc not only overcame the effect of the chelators but enhanced the interaction between DYRK1A and Spry2. This result is consistent with the hypothesis that the DYRK1A polyhistidine segment and the Spry2 CRD interact by forming an intermolecular zinc finger-like structure coordinated by metal ions.
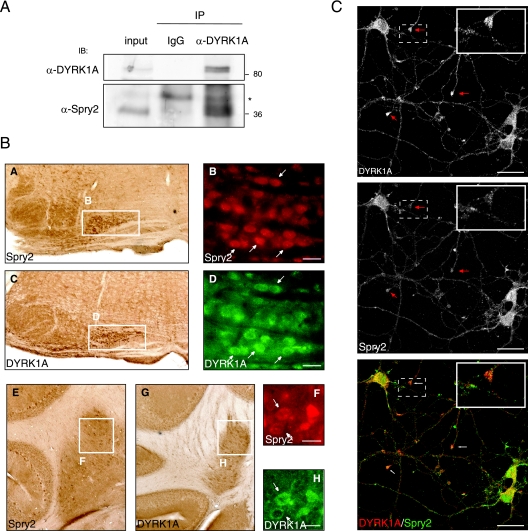
DYRK1A and Spry2 interact in the brain and codistribute in nerve endings.
Both DYRK1A and Spry2 mRNA transcripts are highly expressed in mouse brain in comparison with other organs or tissues (35, 46). To examine the physiological relevance of the DYRK1A-Spry2 interaction, we performed coimmunoprecipitation assays on mouse brain lysates. Spry2 was consistently detected in immunoprecipitates with a specific antibody against DYRK1A (Fig. 5A), supporting the idea of physiological interaction between Spry2 and DYRK1A.
FIG. 5.
DYRK1A and Spry2 are expressed in neurons and colocalized in the dendritic processes of cortical neurons. (A) Mouse brain extracts were subjected to immunoprecipitation with nonspecific rabbit IgGs (IgG) or with a specific rabbit anti-DYRK1A antibody. Both the mouse brain extract (input) and the immunoprecipitates (IP) were analyzed by Western blotting with the antibodies indicated. A cross-reacting band in the IP lanes is indicated with an asterisk. (B) Serial sections of adult mouse brain were processed for immunohistochemistry and immunofluorescence with antibodies against Spry2 (A, B, E, and F) and DYRK1A (C, D, G, and H). Coexpression of DYRK1A and Spry2 was detected in the processed tissue by optical microscopy (A, C, E, and G) or fluorescence microscopy (DYRK1A, green [D and H]; Spry2, red [B and F]). Subpanels A and C show coronal sections of ventral hindbrain; expression of DYRK1A and Spry2 is evident in the olive nuclei (boxed areas), and coexpression in cells of this region is shown in the corresponding immunofluorescence images in subpanels B and D (arrows). Subpanels E and G show coronal sections of cerebellum, and the boxed areas highlight coexpression of DYRK1A and Spry2 in the deep nuclei; cellular coexpression in this region is shown in the corresponding immunofluorescence images in subpanels F and H (arrows). Bars, 25 μm. (C) Double labeling of mouse primary cortical neurons for DYRK1A and Spry2, as indicated. A merged image is also shown (bottom panel). Arrows indicate neural growth cone-like structures where DYRK1A and Spry2 colocalize. Insets show a magnification of one of these structures. Bars, 25 μm.
Interaction between components of intracellular signaling pathways is clearly only of biological relevance if they are naturally expressed in the same cells. DYRK1A protein distribution has been studied in detail in the mouse adult brain (35), but the protein distribution of Spry2 in this tissue has not been reported previously. To identify sites of potential physiological DYRK1A/Spry2 interaction, we analyzed Spry2 expression by immunohistochemistry in brain structures where DYRK1A is known to be expressed. A summary of the results is presented in Fig. S3 in the supplemental material. In general, Spry2 immunostaining was detected throughout the brain, weakly in the neuropile and more strongly in cell somas and proximal projections. Although a complete overlap in DYRK1A and Spry2 expression patterns was not apparent, the main structures showing strong DYRK1A staining were also expressing Spry2. Figure 5B shows two illustrative examples. In hindbrain, Spry2 protein was detected in cells of the olive nuclei, where DYRK1A is also present (Fig. 5B, subpanels A and C). Coexpression was also seen in the deep nuclei of the cerebellum (Fig. 5B, subpanels E and G). A more detailed analysis by double immunofluorescence revealed that the two proteins are coexpressed in the majority of cells in these two structures (Fig. 5B, subpanels B, D, F, and H), confirming that DYRK1A and Spry2 are present in the same cells in specific structures of the adult brain.
To gain further details about the subcellular localization of both proteins in the central nervous system, we performed double colocalization studies in dissociated cortical neuron cultures. At 10 days in vitro, these cells are already mature, although numerous growth cone-like structures are still present (Fig. 5C). Using specific antibodies, we detected the presence of DYRK1A and Spry2 in the soma of cortical neurons in culture, not only in the cytosol but also in the nucleus (Fig. 5C). DYRK1A and Spry2 were also present across the neural processes, with a punctate staining that appeared enriched in the growth cone-like structures (Fig. 5C). At higher magnification, the overlay of red and green fluorescence revealed strong colocalization in the growth cones (Fig. 5C, lower panel). These results point to a high likelihood for a functional interaction of DYRK1A and Spry2 in neurons.
To get a better insight about DYRK1A and Spry2 codistribution in neural compartments, we subcellularly fractionated brain tissue by using two different approaches (Fig. 6A). In the first, subcellular compartments were separated according to their isopycnic equilibrium point on a discontinuous sucrose gradient; in the second, they were separated by mass and solubility. The following endogenous proteins were used as compartment markers to monitor the reliability of the procedures: lamin B1 for nuclei, calretinin for the soluble cytosolic fraction, synaptophysin for the vesicle fraction, and SNAP-25 for the plasma membrane fraction. The starting material for both procedures was a nucleus-free lysate (S1 fraction) obtained from detergent-free mouse brain homogenates (Fig. 6A). As shown in Fig. 6B, DYRK1A and Spry2 were both present in this postnuclear fraction. When the S1 fraction was fractionated by discontinuous sucrose gradient centrifugation (Fig. 6C), DYRK1A was separated into two populations, one in the soluble protein fractions (fractions 3 to 11) and the other in high-density fractions (fractions 27 to 33). Spry2 also separated into two protein pools, one in the light vesicle fractions (fractions 19 to 25) and the other codistributing with DYRK1A in the high-density fractions (fractions 29 to 33). Notably, other proteins involved in the FGF-MAPK signaling pathway, such as FGFR1, the adaptor protein GRB2, and the MAPK Erk2 were also detected in these fractions (Fig. 6C). In the second fractionation procedure, the postnuclear S1 fraction was separated by differential centrifugation into an S2 soluble/microsomal fraction and a P2 crude synaptosomal fraction, which were further fractionated (scheme in Fig. 6A). DYRK1A was detected in the soluble cytosolic fraction (S3), and both DYRK1A and Spry2 were detected in the microsomal (P3) fraction and the crude synaptosomal fraction (P2) (Fig. 6D). In addition, both proteins were enriched in the fraction corresponding to synaptosomal plasmatic membranes (LP1) but were barely detected in the synaptic vesicle fraction (LP2) (Fig. 6D).
Together, these results indicate that DYRK1A and Spry2 interact in brain and show that this interaction can take place close to membranes in nerve endings, corresponding to the active centers where cells initiate signaling cascades in response to growth factors.
DYRK1A is a kinase of Spry2.
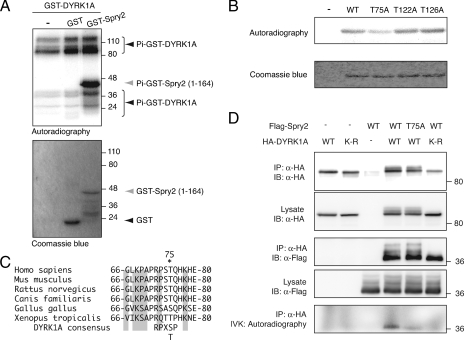
When analyzing the results of DYRK1A and Spry2 coexpression experiments, we observed changes in the electrophoretic mobility pattern of Spry2, as shown for instance in Fig. 1B and D. This mobility shift did not occur when Spry2 was coexpressed with the kinase-dead or DYRK1AΔHis DYRK1A mutants (Fig. 4B). The low-mobility bands are sensitive to alkaline phosphatase treatment (see Fig. S4A in the supplemental material), indicating that they are the result of phosphorylation events. All these data strongly suggest that DYRK1A increases the phosphorylation state of Spry2.
To verify the ability of DYRK1A to directly phosphorylate Spry2, we carried out IVK assays with bacterially expressed GST-DYRK1A as the source of enzyme and GST-Spry2 as substrate. Phosphate label incorporation was detected both in the DYRK1A fusion protein as a result of autophosphorylation and in different Spry2 fusion proteins (Fig. 7A; see also Fig. S4B in the supplemental material). These experiments confirmed that DYRK1A can directly phosphorylate Spry2 and showed that the putative phosphorylated residues are located within the 164-residue N-terminal portion. We could not find any consensus DYRK1A phosphorylation motifs (RxxS/TP [23]) within this region. Nevertheless, the Spry2 amino-terminal sequence does contain a TP pair and several serine/threonine residues preceded by arginine at position −3 (see Fig. S4C in the supplemental material). We generated GST-Spry2 mutants in which some of these residues were individually replaced by alanine (T75A, T122A, and T126A). Whereas phosphate label incorporation in vitro was unaffected by the T122A and T126A mutations, phosphorylation of the Spry2-T75A mutant was reduced compared with that of Spry2-wt (Fig. 7B). This phosphorylatable site is conserved among all vertebrate Spry2 proteins identified to date (Fig. 7C).
FIG. 7.
DYRK1A phosphorylates Spry2 at threonine residue 75. (A) IVK assay using purified bacterially expressed GST-DYRK1A as the enzyme source and GST-Spry2-(1-164) as substrate. Unfused GST was used as a control. The samples were resolved by SDS-PAGE, and the gel was stained with Coomassie blue and analyzed by autoradiography. The positions of the phosphorylated species are indicated. Note that the high-mobility bands that are present in all lanes correspond to phosphorylation of C-terminally truncated forms of GST-DYRK1A. (B) Equal amounts of wt, T75A, T122A, and T126A versions of bacterially expressed and purified GST-Spry2-(1-164) were used in IVK assays with purified GST-DYRK1A. The samples were analyzed as for panel A. (C) The amino acid sequences of Spry2 from human (NP_005833), mouse (NP_036027), rat (NP_001012046), dog (XP_542623), chicken (NP_990131), and frog (NP_001006932) are aligned in the conserved region surrounding T75 (marked with an asterisk). Numbers indicate first and last amino acids listed. Shading indicates residues conserved in all sequences listed. The DYRK1A phosphorylation consensus sequence is also included. (D) Cells were transfected with expression plasmids encoding HA-DYRK1A wt or the kinase-dead mutant HA-DYRK1A-K179R (K-R) and wt or T75A versions of Flag-Spry2. Cell extracts were subjected to immunoprecipitation with anti-HA antibody, followed by IVK assay. Samples were resolved by SDS-PAGE and analyzed by autoradiography and by Western blotting with the indicated antibodies.
The ability of DYRK1A to phosphorylate Spry2 was also checked in IVK assays with DYRK1A-specific immunocomplexes purified from transfected cells expressing DYRK1A and Spry2. In these experiments, phosphate was incorporated only into Spry2 coimmunoprecipitated with wt DYRK1A but not with the kinase-dead mutant (Fig. 7D, IVK). Likewise, Spry2-T75A showed a markedly reduced incorporation of radioactive phosphate in kinase assays from DYRK1A immunocomplexes (Fig. 7D, IVK), which cannot be due to differences in Spry2 interaction with DYRK1A (Fig. 7D, IP anti-HA). We noticed that the effect of mutating Spry2 T75 in the IVK assays was smaller when using bacterially purified proteins (Fig. 7B) than when using immunocomplexes from mammalian cells (Fig. 7D). This was due to the existence of Spry2 residues that are phosphorylated in the first type of assay but not in the second (data not shown). The different behavior could be explained by differences in the specificity of the DYRK1A proteins expressed in each system or by a restriction toward some phosphorylatable residues when the kinase and the substrate form a complex in the coimmunoprecipitation assay. We also observed that mutation T75A in Spry2 did not completely abrogate phosphate incorporation in the IVK assays (Fig. 7D), suggesting that other DYRK1A phosphorylated sites may exist. Altogether, these results indicate, therefore, that Spry2 is phosphorylated directly by DYRK1A on at least Thr75.
Spry2 activity is modulated by DYRK1A.
To study the functional role of this novel Spry2 phosphorylation site, we analyzed the ability of Spry2-T75A to inhibit FGF-MAPK signaling. No differences in the expression levels of Spry2-wt and Spry2-T75A were detected, either when expressed alone or in the presence of FGFR (Fig. 8A, lysates). Neither we did detect any significant difference in the level of tyrosine phosphorylation between the two proteins when coexpressed with FGFR (Fig. 8A, anti-PY20). However, compared with Spry2-wt, the nonphosphorylatable Spry2-T75A mutant induced a significantly increased inhibition of Erk1/2 activation in FGFR1-overexpressing cells (Fig. 8A, anti-PiErk, and quantification in Fig. 8B). These findings suggest an increased inhibitory efficiency of the mutant Spry2-T75A.
FIG. 8.
Phosphorylation of Spry2 modulates its activity as an RTK negative regulator. (A) HEK-293 cells were transfected with expression plasmids encoding FGFR1 and either the wt or the T75A mutant version of Flag-Spry2, as indicated. At 48 h posttransfection, soluble cell lysates were prepared and subjected to immunoprecipitation with anti-Flag antibody. Both the cell lysates and the immunoprecipitates (IP) were analyzed by Western blotting with the indicated antibodies (PY20, anti-phospo-Tyr antibody; PiErk, anti-phospho-Erk1/2 Thr202/Tyr204). (B) The histogram presents the relative Erk activation in the lysates (ratio of phospho-Erk to total Erk2 signals) as a proportion of the activation in cells transfected with wt Spry2 (set at 1). Data are the means ± SEM of four independent experiments (***, P ≤ 0.001). (C) Cells were cotransfected with pE1bGal4-luc, pGal4-Elk1, and pRL-Null plasmids, together with increasing amounts of expression plasmids encoding Spry2 wt (WT) and the nonphosphorylatable mutant T75A. Cells were stimulated with FGF2 (10 ng/ml) for 6 h, and luciferase activity was measured in triplicate plates. Values were corrected for transfection efficiency as measured by Renilla luciferase activity. Data are the means ± SEM of the induction of luciferase activity above the nonstimulated level from two independent experiments. (D) Total protein extracts from cells transfected with equivalent amounts of Spry2 expression plasmids as described for panel C were analyzed by Western blotting with anti-Flag.
To confirm this higher inhibitory capacity, we analyzed the ability of Spry2 to reduce the transcriptional activity of Elk-1, a nuclear target of Erk1/2, in FGF-stimulated cells. The high sensitivity of this approach allows detection of the inhibitory activity of overexpressed Spry2 at very low dosages. As Fig. 8C shows, the inhibitory activity of the Spry2-T75A mutant was more pronounced than that of the wt protein at all protein doses tested. Western blotting confirmed that there were no differences between the expression levels of wt and mutant Spry2 proteins in transfected cells (Fig. 8D). These results suggest that the phosphorylation of Spry2 at Thr75 negatively modulates the ability of Spry2 to antagonize FGF-MAPK signaling.
DISCUSSION
This study provides the first demonstration of a functional interaction between DYRK1A and Spry2. We have demonstrated that DYRK1A and Spry2 interact in mammalian cells when exogenously expressed and, more importantly, in the physiological context of the mouse brain. DYRK1A and Spry2 proteins are present in the same cells of different brain structures and codistribute in the same subcellular fractions, supporting the conclusion that DYRK1A and Spry2 are truly physiological partners. This finding has special relevance because, although the list of interacting proteins for DYRK1A has grown in recent years (for a recent review, see reference 13), most of these interactions have been detected only between exogenously overexpressed proteins.
The binding of some Spry interactors with Spry2, or with other Spry family members, is modulated by RTK activation. Our findings indicate that the interaction of DYRK1A with Spry2 is independent of the activation status of the FGF pathway, similarly to what has been reported for B-Raf (47). Interestingly, DYRK1A and B-Raf are both detected in Spry2 immunocomplexes, suggesting that binary interactions, such as Spry2/DYRK1A or Spry2/B-Raf, are mutually supportive. Given that B-Raf is a shared partner of both Spry family members (43, 47) and DYRK1A (29), it is possible that a trimeric complex involving all three proteins exists in cells.
Our current work identifies the interacting domains in DYRK1A and Spry2 as the polyhistidine domain and the CRD, respectively. Both regions are not only necessary but also sufficient for binding. We had previously shown that the DYRK1A polyhistidine sequence targets the kinase to the splicing factor compartment (2); however, a specific protein target for this activity has not yet been found. The present study therefore represents the first report showing that this amino acid sequence can act as a protein-protein interaction domain. There is growing evidence that kinase substrate specificity is achieved by many mechanisms in addition to direct recognition by the catalytic site (48). One such mechanism involves interactions outside the catalytic domain, known as docking interactions, which facilitate the discrimination of bona fide substrates for the kinase. Given that Spry2 is a DYRK1A substrate and that Spry2 does not get phosphorylated when coexpressed with a DYRK1A protein lacking the histidine stretch, it can be postulated that the polyhistidine segment is a docking site for Spry2; moreover, an appealing possibility is that the polyhistidine segment might act as a docking site for other DYRK1A substrates as well.
The CRD in Spry2 is highly conserved among all members of the Spry family. The conservation is not only restricted to its sequence but also functional, and several activities have been mapped to it. For example, the motif is responsible for Spry dimerization (22), harbors the region required for interaction with Raf kinases (43) and TESK (9), and is also required for plasma membrane localization (22, 33). CRD conservation would justify the ability of DYRK1A to interact with all four mammalian Spry proteins and not just Spry2. This finding places DYRK1A as a common partner of this family of signaling inhibitors, opening the possibility of intercepting RTK signaling at multiple levels. Furthermore, and given that DYRK1A interacts with at least Spry2 homodimers, our results suggest that the Spry CRD can recruit different partners simultaneously.
The special amino acid features of the DYRK1A and Spry2 binding regions led us to propose that the interaction occurs through the formation of an intermolecular zinc finger-like domain, in which metal ions are coordinated by histidine and cysteine residues at the interacting interface. Apart from the well-established function of Zn2+ as an organizer of intramolecular protein structures, for example in zinc fingers, ring fingers, or FYVE domains, there are also reports showing that this transition metal acts as an intermolecular linker between subunits in protein complexes. This kind of interaction features in the binding of the cytoplasmic tail of CD4 with the N-terminal region of Lck (25) and the formation of complexes between cyclin T1 and granulin (24). The finding that Zn2+ is required for the DYRK1A and Spry2 interaction is consistent with our hypothesis. It is thus easy to imagine how changes in intracellular Zn2+ concentration could provide a way of regulating the formation of DYRK1A-Spry2 complexes. Redox modifications of cysteines in Spry2 might also result in differential binding to DYRK1A.
Our present study is the first to show that Spry2 protein is expressed in several areas of the central nervous system in the mouse. In some of these areas, both Spry2 and DYRK1A are coexpressed. Moreover, proposals that DYRK1A and Spry2 interact physiologically in the brain are strengthened by the fact that the proteins codistribute in the same subcellular fractions and colocalize in the growth cone-like structures of cortical neurons in culture. The biochemical fractionation studies have revealed that both DYRK1A and Spry2 distribute in fractions enriched in molecules clearly involved in RTK signaling such as FGFR, the adaptor GRB2, or the MAPK Erk2. In addition, the analysis showed that both Spry2 and DYRK1A are present in the crude synaptosomal fraction, preferentially associated with the synaptic plasma membranes. Whereas Spry2 has been reported to localize to the plasma membrane through palmitoylation and/or caveolin binding (7, 28), nothing about DYRK1A membrane association has been reported or inferred from its primary structure. DYRK1A distribution in membrane fractions should depend therefore on its interaction with specific partners, one of them being Spry2. It is also worth mentioning that a small proportion of both proteins was detected in the free vesicle-containing fraction. This may indicate their presence in transport vesicles present in this fraction and is consistent with evidence that Spry proteins are associated with cytosolic vesicles (21, 28, 30) involved in endocytic events controlling receptor trafficking (30). In this context, DYRK1A has been shown to phosphorylate several components of the endocytic machinery, such as dynamin, synaptojanin, and amphiphysin I (39). Taken together, these findings would argue that DYRK1A/Spry2 interaction has the potential to contribute to the regulation of intracellular receptor trafficking. This suggestion is particularly interesting in the context of the regulation of intracellular signaling in neurons, in which the propagation of the signals received at axon terminals relies on the retrograde transport of signaling molecules to cell bodies (for a recent review, see reference 27).
Although Spry2 has been extensively reported as a cytosolic protein in cell lines (see, for instance, references 28 and 30), we have noticed that, apart from Spry2 being present in the neuritic processes, a fraction of Spry2 is detected in the neuronal cell nucleus. This has been observed by immunofluorescence analysis in brain sections as well as in dissociated cultures of cortical neurons. A similar staining pattern has been reported in primary cultures of cerebellar granule neurons (18), pointing to a not-yet-known nuclear role for Spry2.
An important finding of this work is that DYRK1A is able to directly phosphorylate Spry2. It is known that Spry2 activity is posttranslationally regulated by phosphorylation (22, 32), which modulates several aspects of its function; however, only two serine/threonine protein kinases have been described to functionally interact with Spry2. Phosphorylation by Mnk1 on Ser112 and Ser121 controls Spry2 degradation (10). In contrast, the alteration of the Spry2 phosphorylation status by TESK is due not to its kinase activity but to a displacement of Spry2 from binding to the phosphatase PP2A (9). Here, we have demonstrated that Spry2 is a substrate of DYRK1A and identified Thr75 as one of the residues phosphorylated by this kinase. Mutation of this threonine yields a Spry2 protein with an enhanced ability to inhibit Erk activation upon FGF signaling. Furthermore, we have also shown that phosphorylation of Thr75 impairs Spry2 inhibitory capacity without altering Tyr55 phosphorylation levels.
All together, our findings support a role for DYRK1A as a positive modulator of the FGF-dependent signaling pathway by antagonizing the activity of Spry2. It is worth mentioning that related phenotypes have been described in mouse models of increased expression of Spry2 or decreased expression of DYRK1A. Thus, in agreement with the proposal that the processes that shape the midbrain and cerebellum require distinct levels of FGF signaling (34), expression of a Spry2 gain-of-function allele in the embryonic midbrain results in the absence of the inferior colliculus (4). Interestingly, this phenotype is remarkably similar to that observed in Dyrk1A heterozygous mice that present a marked macroscopic reduction of the mesencephalic tectum (17). It is thus possible that the interplay between DYRK1A and the negative regulator Spry2 helps to ensure the appropriate strength and duration of the FGF-dependent signal.
Supplementary Material
Acknowledgments
We are especially grateful to M. L. Arbonés (CRG, Barcelona, Spain) for providing adult mice. We thank A. Raya and E. Ramírez for technical assistance, E. Martí for her advice on the immunohistochemical analysis, E. Balducci and M. J. Ballarobre for providing mouse primary cortical cultures, and S. Bartlett for English editorial work. We also thank G. R. Guy, E. Nishida, and P. E. Shaw for several of the plasmids used in the study.
This group is founded by the Centro de Investigación Biomédica en Red de Enfermedades Raras (CIBERER), ISCIII. S.A. and A.L. were FI predoctoral fellows (DURSI, Generalitat de Catalunya), and M.A. was a fellow of the Spanish Ministry of Health (BEFI Program). This work is supported by grants from the Spanish Ministry of Education and Science (BFU2004-01768 and BFU2007-61043/BMC) and the European Commission AnEUploidy grant.
Footnotes
Published ahead of print on 4 August 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Alvarez, M., X. Altafaj, S. Aranda, and S. de la Luna. 2007. DYRK1A autophosphorylation on serine residue 520 modulates its kinase activity via 14-3-3 binding. Mol. Biol. Cell 181167-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez, M., X. Estivill, and S. de la Luna. 2003. DYRK1A accumulates in splicing speckles through a novel targeting signal and induces speckle disassembly. J. Cell Sci. 1163099-3107. [DOI] [PubMed] [Google Scholar]
- 3.Arron, J. R., M. M. Winslow, A. Polleri, C. P. Chang, H. Wu, X. Gao, J. R. Neilson, L. Chen, J. J. Heit, S. K. Kim, N. Yamasaki, T. Miyakawa, U. Francke, I. A. Graef, and G. R. Crabtree. 2006. NFAT dysregulation by increased dosage of DSCR1 and DYRK1A on chromosome 21. Nature 441595-600. [DOI] [PubMed] [Google Scholar]
- 4.Basson, M. A., D. Echevarria, C. P. Ahn, A. Sudarov, A. L. Joyner, I. J. Mason, S. Martinez, and G. R. Martin. 2008. Specific regions within the embryonic midbrain and cerebellum require different levels of FGF signaling during development. Development 135889-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benavides-Piccione, R., M. Dierssen, I. Ballesteros-Yanez, M. Martinez de Lagran, M. L. Arbones, V. Fotaki, J. DeFelipe, and G. N. Elston. 2005. Alterations in the phenotype of neocortical pyramidal cells in the Dyrk1A+/− mouse. Neurobiol. Dis. 20115-122. [DOI] [PubMed] [Google Scholar]
- 6.Branchi, I., Z. Bichler, L. Minghetti, J. M. Delabar, F. Malchiodi-Albedi, M. C. Gonzalez, Z. Chettouh, A. Nicolini, C. Chabert, D. J. Smith, E. M. Rubin, D. Migliore-Samour, and E. Alleva. 2004. Transgenic mouse in vivo library of human Down syndrome critical region 1: association between DYRK1A overexpression, brain development abnormalities, and cell cycle protein alteration. J. Neuropathol. Exp. Neurol. 63429-440. [DOI] [PubMed] [Google Scholar]
- 7.Cabrita, M. A., F. Jaggi, S. P. Widjaja, and G. Christofori. 2006. A functional interaction between Sprouty proteins and caveolin-1. J. Biol. Chem. 28129201-29212. [DOI] [PubMed] [Google Scholar]
- 8.Casci, T., J. Vinos, and M. Freeman. 1999. Sprouty, an intracellular inhibitor of Ras signaling. Cell 96655-665. [DOI] [PubMed] [Google Scholar]
- 9.Chandramouli, S., C. Y. Yu, P. Yusoff, D. H. Lao, H. F. Leong, K. Mizuno, and G. R. Guy. 2008. Tesk1 interacts with Spry2 to abrogate its inhibition of ERK phosphorylation downstream of receptor tyrosine kinase signaling. J. Biol. Chem. 2831679-1691. [DOI] [PubMed] [Google Scholar]
- 10.DaSilva, J., L. Xu, H. J. Kim, W. T. Miller, and D. Bar-Sagi. 2006. Regulation of Sprouty stability by Mnk1-dependent phosphorylation. Mol. Cell. Biol. 261898-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de la Luna, S., K. E. Allen, S. L. Mason, and N. B. La Thangue. 1999. Integration of a growth-suppressing BTB/POZ domain protein with the DP component of the E2F transcription factor. EMBO J. 18212-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de la Luna, S., and X. Estivill. 2006. Cooperation to amplify gene-dosage-imbalance effects. Trends Mol. Med. 12451-454. [DOI] [PubMed] [Google Scholar]
- 13.Dierssen, M., and M. M. de Lagran. 2006. DYRK1A (dual-specificity tyrosine-phosphorylated and -regulated kinase 1A): a gene with dosage effect during development and neurogenesis. ScientificWorldJournal 61911-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ebisuya, M., K. Kondoh, and E. Nishida. 2005. The duration, magnitude and compartmentalization of ERK MAP kinase activity: mechanisms for providing signaling specificity. J. Cell Sci. 1182997-3002. [DOI] [PubMed] [Google Scholar]
- 15.Epstein, C. J. 2006. Down's syndrome: critical genes in a critical region. Nature 441582-583. [DOI] [PubMed] [Google Scholar]
- 16.Fong, C. W., H. F. Leong, E. S. Wong, J. Lim, P. Yusoff, and G. R. Guy. 2003. Tyrosine phosphorylation of Sprouty2 enhances its interaction with c-Cbl and is crucial for its function. J. Biol. Chem. 27833456-33464. [DOI] [PubMed] [Google Scholar]
- 17.Fotaki, V., M. Dierssen, S. Alcantara, S. Martinez, E. Marti, C. Casas, J. Visa, E. Soriano, X. Estivill, and M. L. Arbones. 2002. Dyrk1A haploinsufficiency affects viability and causes developmental delay and abnormal brain morphology in mice. Mol. Cell. Biol. 226636-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gross, I., O. Armant, S. Benosman, J. L. de Aguilar, J. N. Freund, M. Kedinger, J. D. Licht, C. Gaiddon, and J. P. Loeffler. 2007. Sprouty2 inhibits BDNF-induced signaling and modulates neuronal differentiation and survival. Cell Death Differ. 141802-1812. [DOI] [PubMed] [Google Scholar]
- 19.Gross, I., B. Bassit, M. Benezra, and J. D. Licht. 2001. Mammalian sprouty proteins inhibit cell growth and differentiation by preventing ras activation. J. Biol. Chem. 27646460-46468. [DOI] [PubMed] [Google Scholar]
- 20.Hacohen, N., S. Kramer, D. Sutherland, Y. Hiromi, and M. A. Krasnow. 1998. sprouty encodes a novel antagonist of FGF signaling that patterns apical branching of the Drosophila airways. Cell 92253-263. [DOI] [PubMed] [Google Scholar]
- 21.Hall, A. B., N. Jura, J. DaSilva, Y. J. Jang, D. Gong, and D. Bar-Sagi. 2003. hSpry2 is targeted to the ubiquitin-dependent proteasome pathway by c-Cbl. Curr. Biol. 13308-314. [DOI] [PubMed] [Google Scholar]
- 22.Hanafusa, H., S. Torii, T. Yasunaga, and E. Nishida. 2002. Sprouty1 and Sprouty2 provide a control mechanism for the Ras/MAPK signalling pathway. Nat. Cell Biol. 4850-858. [DOI] [PubMed] [Google Scholar]
- 23.Himpel, S., W. Tegge, R. Frank, S. Leder, H. G. Joost, and W. Becker. 2000. Specificity determinants of substrate recognition by the protein kinase DYRK1A. J. Biol. Chem. 2752431-2438. [DOI] [PubMed] [Google Scholar]
- 24.Hoque, M., T. M. Young, C. G. Lee, G. Serrero, M. B. Mathews, and T. Pe'ery. 2003. The growth factor granulin interacts with cyclin T1 and modulates P-TEFb-dependent transcription. Mol. Cell. Biol. 231688-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huse, M., M. J. Eck, and S. C. Harrison. 1998. A Zn2+ ion links the cytoplasmic tail of CD4 and the N-terminal region of Lck. J. Biol. Chem. 27318729-18733. [DOI] [PubMed] [Google Scholar]
- 26.Huttner, W. B., W. Schiebler, P. Greengard, and P. De Camilli. 1983. Synapsin I (protein I), a nerve terminal-specific phosphoprotein. III. Its association with synaptic vesicles studied in a highly purified synaptic vesicle preparation. J. Cell Biol. 961374-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ibanez, C. F. 2007. Message in a bottle: long-range retrograde signaling in the nervous system. Trends Cell Biol. 17519-528. [DOI] [PubMed] [Google Scholar]
- 28.Impagnatiello, M. A., S. Weitzer, G. Gannon, A. Compagni, M. Cotten, and G. Christofori. 2001. Mammalian sprouty-1 and -2 are membrane-anchored phosphoprotein inhibitors of growth factor signaling in endothelial cells. J. Cell Biol. 1521087-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelly, P. A., and Z. Rahmani. 2005. DYRK1A enhances the mitogen-activated protein kinase cascade in PC12 cells by forming a complex with Ras, B-Raf, and MEK1. Mol. Biol. Cell 163562-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim, H. J., L. J. Taylor, and D. Bar-Sagi. 2007. Spatial regulation of EGFR signaling by Sprouty2. Curr. Biol. 17455-461. [DOI] [PubMed] [Google Scholar]
- 31.Kortenjann, M., O. Thomae, and P. E. Shaw. 1994. Inhibition of v-raf-dependent c-fos expression and transformation by a kinase-defective mutant of the mitogen-activated protein kinase Erk2. Mol. Cell. Biol. 144815-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lao, D. H., P. Yusoff, S. Chandramouli, R. J. Philp, C. W. Fong, R. A. Jackson, T. Y. Saw, C. Y. Yu, and G. R. Guy. 2007. Direct binding of PP2A to Sprouty2 and phosphorylation changes are a prerequisite for ERK inhibition downstream of fibroblast growth factor receptor stimulation. J. Biol. Chem. 2829117-9126. [DOI] [PubMed] [Google Scholar]
- 33.Lim, J., E. S. Wong, S. H. Ong, P. Yusoff, B. C. Low, and G. R. Guy. 2000. Sprouty proteins are targeted to membrane ruffles upon growth factor receptor tyrosine kinase activation. Identification of a novel translocation domain. J. Biol. Chem. 27532837-32845. [DOI] [PubMed] [Google Scholar]
- 34.Liu, A., J. Y. H. Li, C. Bromleigh, Z. Lao, L. A. Niswander, and A. L. Joyner. 2003. FGF17b and FGF18 have different midbrain regulatory properties from FGF8b or activated FGF receptors. Development 1306175-6185. [DOI] [PubMed] [Google Scholar]
- 35.Marti, E., X. Altafaj, M. Dierssen, S. de la Luna, V. Fotaki, M. Alvarez, M. Perez-Riba, I. Ferrer, and X. Estivill. 2003. Dyrk1A expression pattern supports specific roles of this kinase in the adult central nervous system. Brain Res. 964250-263. [DOI] [PubMed] [Google Scholar]
- 36.Mason, J. M., D. J. Morrison, B. Bassit, M. Dimri, H. Band, J. D. Licht, and I. Gross. 2004. Tyrosine phosphorylation of Sprouty proteins regulates their ability to inhibit growth factor signaling: a dual feedback loop. Mol. Biol. Cell 152176-2188. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Mason, J. M., D. J. Morrison, M. A. Basson, and J. D. Licht. 2006. Sprouty proteins: multifaceted negative-feedback regulators of receptor tyrosine kinase signaling. Trends Cell Biol. 1645-54. [DOI] [PubMed] [Google Scholar]
- 38.Minowada, G., L. A. Jarvis, C. L. Chi, A. Neubuser, X. Sun, N. Hacohen, M. A. Krasnow, and G. R. Martin. 1999. Vertebrate Sprouty genes are induced by FGF signaling and can cause chondrodysplasia when overexpressed. Development 1264465-4475. [DOI] [PubMed] [Google Scholar]
- 39.Murakami, N., W. Xie, R. C. Lu, M. C. Chen-Hwang, A. Wieraszko, and Y. W. Hwang. 2006. Phosphorylation of amphiphysin I by minibrain kinase/dual-specificity tyrosine phosphorylation-regulated kinase, a kinase implicated in Down syndrome. J. Biol. Chem. 28123712-23724. [DOI] [PubMed] [Google Scholar]
- 40.Ozaki, K., S. Miyazaki, S. Tanimura, and M. Kohno. 2005. Efficient suppression of FGF-2-induced ERK activation by the cooperative interaction among mammalian Sprouty isoforms. J. Cell Sci. 1185861-5871. [DOI] [PubMed] [Google Scholar]
- 41.Pawson, T., and P. Nash. 2000. Protein-protein interactions define specificity in signal transduction. Genes Dev. 141027-1047. [PubMed] [Google Scholar]
- 42.Rubin, C., V. Litvak, H. Medvedovsky, Y. Zwang, S. Lev, and Y. Yarden. 2003. Sprouty fine-tunes EGF signaling through interlinked positive and negative feedback loops. Curr. Biol. 13297-307. [DOI] [PubMed] [Google Scholar]
- 43.Sasaki, A., T. Taketomi, R. Kato, K. Saeki, A. Nonami, M. Sasaki, M. Kuriyama, N. Saito, M. Shibuya, and A. Yoshimura. 2003. Mammalian Sprouty4 suppresses Ras-independent ERK activation by binding to Raf1. Nat. Cell Biol. 5427-432. [DOI] [PubMed] [Google Scholar]
- 44.Schlessinger, J. 2000. Cell signaling by receptor tyrosine kinases. Cell 103211-225. [DOI] [PubMed] [Google Scholar]
- 45.Schubbert, S., K. Shannon, and G. Bollag. 2007. Hyperactive Ras in developmental disorders and cancer. Nat. Rev. Cancer 7295-308. [DOI] [PubMed] [Google Scholar]
- 46.Tefft, J. D., M. Lee, S. Smith, M. Leinwand, J. Zhao, P. Bringas, Jr., D. L. Crowe, and D. Warburton. 1999. Conserved function of mSpry-2, a murine homolog of Drosophila sprouty, which negatively modulates respiratory organogenesis. Curr. Biol. 9219-222. [DOI] [PubMed] [Google Scholar]
- 47.Tsavachidou, D., M. L. Coleman, G. Athanasiadis, S. Li, J. D. Licht, M. F. Olson, and B. L. Weber. 2004. SPRY2 is an inhibitor of the ras/extracellular signal-regulated kinase pathway in melanocytes and melanoma cells with wild-type BRAF but not with the V599E mutant. Cancer Res. 645556-5559. [DOI] [PubMed] [Google Scholar]
- 48.Ubersax, J. A., and J. E. Ferrell, Jr. 2007. Mechanisms of specificity in protein phosphorylation. Nat. Rev. Mol. Cell Biol. 8530-541. [DOI] [PubMed] [Google Scholar]
- 49.Yusoff, P., D. H. Lao, S. H. Ong, E. S. Wong, J. Lim, T. L. Lo, H. F. Leong, C. W. Fong, and G. R. Guy. 2002. Sprouty2 inhibits the Ras/MAP kinase pathway by inhibiting the activation of Raf. J. Biol. Chem. 2773195-3201. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.