Abstract
The E2 ubiquitin-conjugating enzyme UBC13 plays pivotal roles in diverse biological processes. Recent studies have elucidated that UBC13, in concert with the E3 ubiquitin ligase RNF8, propagates the DNA damage signal via a ubiquitylation-dependent signaling pathway. However, mechanistically how UBC13 mediates its role in promoting checkpoint protein assembly and its genetic requirement for E2 variants remain elusive. Here we provide evidence to support the idea that the E3 ubiquitin ligase complex RNF8-UBC13 functions independently of E2 variants and is sufficient in facilitating ubiquitin conjugations and accumulation of DNA damage mediator 53BP1 at DNA breaks. The RNF8 RING domain serves as the molecular platform to anchor UBC13 at the damaged chromatin, where localized ubiquitylation events allow sustained accumulation of checkpoint proteins. Intriguingly, we found that only a group of RING domains derived from E3 ubiquitin ligases, which have been shown to interact with UBC13, enabled UBC13-mediated FK2 and 53BP1 focus formation at DNA breaks. We propose that the RNF8 RING domain selects and loads a subset of UBC13 molecules, distinct from those that exist as heterodimers, onto sites of double-strand breaks, which facilitates the amplification of DNA damage signals.
Genomic stability and its maintenance entail proper DNA damage recognition, signal propagation, and effector activation (7). Cells have evolved elaborate networks to coordinate cellular processes that, in response to DNA damage, combine to ensure faithful transmission of genetic materials (3). The ability to temporally and spatially control these components is endowed by a growing list of posttranslational modifications, which when covalently attached to their substrates, govern processes including protein-protein interactions, protein localization and activities, and protein degradation (10).
Phosphorylation of the histone variant H2AX by the phosphoinositide-3-kinase-related protein kinase ATM/ATR constitutes one of the initial signals upon damage detection and is required for the subsequent accumulation of DNA damage sensors and mediators in the vicinity of DNA lesions (17). Formation of discernible nuclear focus structures has been suggested to enable the amplification of the initial damage signal to elicit the proper cellular response to DNA damage. Indeed, mouse models with deficiencies in H2AX and MDC1, proteins of which are instrumental in regulating the assembly of numerous DNA damage-responsive factors at DNA breaks, show growth defects, chromosomal instability, DNA repair defects, and radiation sensitivity (4, 5, 14, 19). Mechanistically how these downstream DNA damage-responsive proteins, including the tumor suppressors BRCA1 and 53BP1, are retained at DNA breaks is beginning to emerge with the recent identification of the ubiquitin ligase RNF8 (11, 13, 15, 26). RNF8 relocalizes to DNA damage sites via a phospho-dependent interaction with MDC1. Importantly, the chromatin-associated RNF8 mediates substrate ubiquitylation at sites of DNA breaks, which is essential for the ionizing radiation (IR)-induced focus (IRIF) formation of BRCA1 and 53BP1. These observations revealed an unprecedented role of ubiquitylation in damage signaling and indicated that ubiquitylation plays an intimate part in cell proliferation and survival in response to genotoxic stress. Similar to RNF8-associated deficits, depletion of the E2 UBC13 resulted in abolished BRCA1 and 53BP1 IRIF, suggesting that RNF8 acts in concert with UBC13 in the propagation of the DNA damage response (11, 13, 26, 31).
UBC13 catalyzes noncanonical Lys63-based ubiquitin chains and forms heterodimers with its E2 variants MMS2 and UEV1A in vivo (9, 25). Interestingly, the pairing of UBC13 with either of the E2 variants has been suggested to dictate UBC13 function in a diverse number of processes, including translesion bypass, misfolded protein degradation, NF-κB activation, and endolysosomal degradation (2, 6, 8, 21, 27, 29, 30). Given the possibility that RNF8 functions as a complex with UBC13 in the DNA damage response, we decided to examine how RNF8 facilitates the UBC13-catalyzed ubiquitylation events at sites of DNA breaks. Interestingly, despite the current view of a requirement for E2 variants, we found that MMS2 and UEV1A are dispensable for UBC13 function in licensing the assembly of checkpoint proteins at double-strand breaks (DSBs). Our findings indicate that, via selection of UBC13 molecules distinct from those that exist as heterodimers, the RNF8 RING domain signals UBC13 to sites of damage, which is sufficient for DNA damage signal transduction.
MATERIALS AND METHODS
Antibodies.
Antibodies recognizing γH2AX and 53BP1 were described previously (11). The anti-FK2 antibody was obtained from Upstate Cell Signaling. Antihemagglutinin (anti-HA) and anti-Myc antibodies were purchased from Covance. Antiactin and anti-Flag (M2) were obtained from Sigma. Anti-MMS2/UEV1A and anti-UBC13 antibodies were from Zymed Laboratories and Anaspec, respectively. Anti-phospho-H3 was described previously (11).
Cell culture, transfection, and siRNAs.
HeLa and 293T cells were cultured in RPMI 1640 supplemented with 5% fetal calf serum, 5% bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin and maintained in 5% CO2 at 37°C. Cell transfection was performed using Lipofectamine 2000 or Oligofectamine (Invitrogen) by following the manufacturer's protocol. The small interfering RNAs (siRNAs) against MMS2 and UEV1A were described previously (2).
Culture of MEFs and retroviral infection.
Mouse embryonic fibroblasts (MEFs) derived from knockout strains were cultured in Dulbecco's modified Eagle's medium supplemented with 15% fetal calf serum, 100 U/ml penicillin, and 100 μg/ml streptomycin and maintained in 5% CO2 at 37°C. For viral particle packaging, BOSC23 cells were transiently transfected with the pCL-Ampho vector and expression constructs. Viral supernatant was collected 48 h posttransfection and was used for infection. Stable pools of infected MEFs were selected in the presence of 2 μg/ml puromycin.
Expression constructs.
All cDNAs were subcloned into pDON201 as entry clones and were subsequently transferred to gateway-compatible destination vectors for N-terminal Flag- or myc-tagged fusion protein expression. Site-directed mutagenesis was performed according to standard procedures, and clones were sequenced to verify desirable mutations. Chimeric proteins were generated by overlap PCR according to standard procedures. For tandem-affinity purification of UBC13, UBC13 was N-terminally tagged with a streptavidin binding peptide and an S-protein sequence (SBP-S-UBC13) (12).
Immunostaining procedure.
To visualize IRIF, cells were cultured on coverslips and treated with 10 Gy IR followed by recovery for 6 h. Cells were then washed in phosphate-buffered saline, incubated in 3% paraformaldehyde for 12 min, and permeabilized in 0.5% Triton solution for 5 min at room temperature. Samples were blocked with 5% goat serum and then incubated with primary antibody for 60 min. Samples were washed three times and incubated with secondary antibody for 60 min. Cells were then counterstained with DAPI (4′,6-diamidino-2-phenylindole) to visualize nuclear DNA. The coverslips were mounted onto glass slides with antifade solution and visualized using a Nikon ECLIPSE E800 fluorescence microscope.
Yeast two-hybrid assay and G2/M checkpoint assay.
Entry clones were transferred to destination vectors, and interaction studies were performed according to the ProQuest two-hybrid system (Gateway Technology) instruction manual. The procedures for the G2/M checkpoint assay were described previously (11).
RESULTS
Interaction of RNF8 and UBC13 is essential for 53BP1 and FK2 IRIF formation.
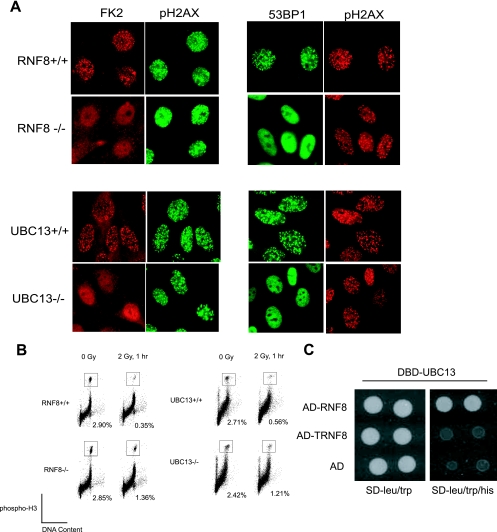
The E3 ubiquitin ligase RNF8, in concert with the E2 enzyme UBC13, was implicated in mediating the formation of ubiquitin conjugates (immunodetected by the anti-ubiquitin FK2 antibody and referred to hereafter as FK2) at DSBs and IRIF of tumor suppressor proteins BRCA1 and 53BP1. Indeed, examination of spontaneously immortalized MEFs deficient in RNF8 or UBC13 revealed a clear requirement for these proteins in facilitating local accumulation of ubiquitin conjugates at sites of DNA breaks (Fig. 1A). Moreover, similar to previous studies using siRNAs, 53BP1 foci were abolished in the absence of RNF8 or UBC13, suggesting that RNF8 and UBC13 might function as a complex in promoting checkpoint protein assembly. Accumulation of checkpoint proteins at DSBs is required for optimal checkpoint activation. Accordingly, deficiencies in RNF8 and UBC13, which result in abrogated IRIF of mediator/checkpoint proteins, including 53BP1 and BRCA1 (11, 13, 15, 26, 31), correlated with a failure for proper G2/M arrest in response to DNA damage (Fig. 1B). Consistent with the requirement for the RNF8 RING domain in FK2 and 53BP1 IRIF, our yeast two-hybrid assay showed that RNF8 interacted with UBC13 in a RING-dependent manner (Fig. 1C).
FIG. 1.
RNF8 and UBC13 are required for damage-induced accumulation of ubiquitin conjugates and 53BP1. (A) Wild-type, RNF8−/−, or UBC13−/− MEFs were irradiated and processed as described in Materials and Methods. Localizations of ubiquitin conjugates and 53BP1 were visualized by immunostaining with FK2 and polyclonal anti-53BP1 antibody, respectively. (B) MEFs deficient in RNF8 or UBC13, and their respective wild-type counterparts, were irradiated, allowed to recover, and fixed with 70% ethanol. Cells were stained with polyclonal anti-H3 S10P and were subjected to fluorescence-activated cell sorter analysis for the determination of the percentage of mitotic cells. Experiments were repeated twice, and representative results are shown. (C) The interaction between UBC13 and RNF8 or its RING domain deletion mutant was tested using a yeast two-hybrid assay as described in Materials and Methods. AD, activation domain; DBD, DNA binding domain.
RNF8 functions in concert with UBC13 in promoting checkpoint protein assembly.
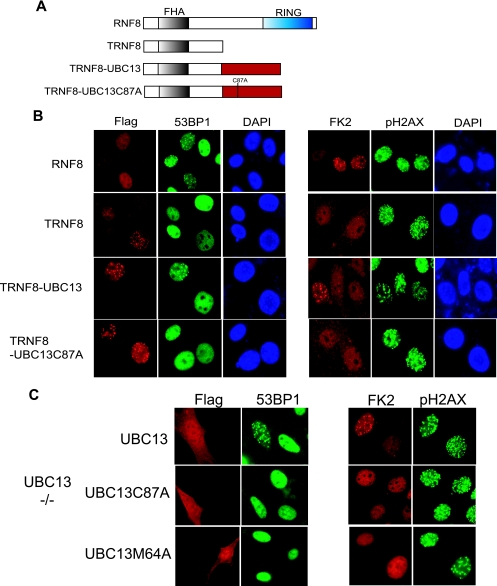
RNF8 localizes at DSBs via a Forkhead-associated (FHA) domain-dependent interaction with phosphorylated MDC1 (11, 13, 15). Since RNF8 interacts with UBC13 in a RING-dependent manner, to directly test whether the recruitment of UBC13 to sites of DNA breaks is sufficient for its role in checkpoint protein assembly, we generated chimeric fusions containing genes that encode the RNF8 FHA domain with UBC13 (Fig. 2A), The RNF8 FHA domain allowed IR-dependent DSB localization of these ectopically expressed proteins (see Fig. S1A in the supplemental material). In RNF8-deficient MEFs, reconstitution of wild-type RNF8 restored IRIF of 53BP1 and FK2, and the focal accumulation of these entities manifested a requirement for the RNF8 RING domain (Fig. 2B). More importantly, focus formation of 53BP1 and FK2 was restored in TRNF8-UBC13-expressing cells but not those expressing the catalytically inactive UBC13 C87A mutant. Similar to the requirement for FK2 IRIF, IR-induced H2AX ubiquitylation was restored in cells reconstituted with either wild-type RNF8 (11) or TRNF8-UBC13, but not in TRNF8 or TRNF8-UBC13 C87A-expressing cells (see Fig. S1B in the supplemental material). Together, these results suggest that RNF8 serves to anchor UBC13 specifically at DSBs to facilitate localized ubiquitylation events required for checkpoint protein assembly.
FIG. 2.
RNF8 recruits UBC13 via its RING domain to promote checkpoint protein assembly. (A) Schematic illustration of full-length RNF8, the truncated RNF8 mutant, and the chimera fusion protein genes that encode the RNF8 FHA domain and UBC13. (B) RNF8-deficient fibroblasts were reconstituted with expression constructs as described in the legend to panel A, and focus formation of FK2 and 53BP1 was examined by immunostaining. Representative results are shown. (C) UBC13-deficient cells were reconstituted with wild-type UBC13, the catalytically inactive UBC13 C87A mutant, or the UBC13 M64A mutant. IRIF of 53BP1 and FK2 were examined after IR treatment.
Structural and functional studies have shown that the M64 residue on UBC13 is important for its ability to interact with E3 ligases (22, 23, 28). To further verify that UBC13 is recruited to DSBs in an RNF8-dependent manner to promote FK2 and 53BP1 IRIF, we reconstituted UBC13-deficient fibroblasts with wild-type UBC13 or its mutants (i.e., C87A and M64A). In agreement with the proposed model in which RNF8-UBC13 functions as a complex in DNA damage-signaling pathway, only cells that express wild-type UBC13 restored FK2 and 53BP1 IRIF, but not those carrying either the C87A or M64A mutant of UBC13 (Fig. 2C). Together, these data further substantiate the notion that RNF8 acts with UBC13 to propagate the DNA damage signals.
Localized recruitment of UBC13 to DSBs is required for 53BP1 and FK2 focus accumulation.
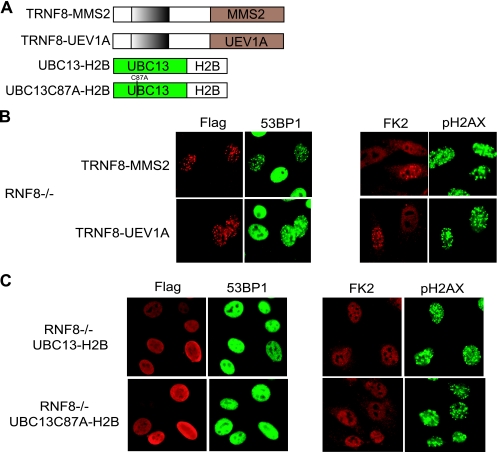
UBC13 forms heterodimers with E2 variants MMS2 and UEV1A in vivo (25, 27). It was proposed that heterodimer formation enables differential regulation of cellular processes, including DNA repair and immune receptor signaling. However, which of the E2 variants is required for UBC13 function in the DNA damage-signaling pathway remains elusive. Given that the RNF8 FHA domain is sufficient to localize UBC13 at DSBs, we wondered whether expression of chimeric fusion proteins encoding the RNF8 FHA domain with either MMS2 or UEV1A in RNF8-deficient cells might shed light on the requirement for a specific E2 variant in DNA damage signaling. Surprisingly, both MMS2 and UEV1A, when fused with the DSB-specific variant conferring the RNF8 FHA domain, localized at DSBs and restored FK2 and 53BP1 IRIF (Fig. 3B and see Fig. S2A in the supplemental material), suggesting that the mere recruitment of UBC13 to sites of DNA breaks is sufficient for its role in checkpoint protein assembly. We did not observe endogenous UBC13 focus formation following IR (see Fig. S2B in the supplemental material). Similarly, UBC13 IRIF was not detected in the RNF8-MMS2- or RNF8-UEV1A-expressing cells (see Fig. S2B in the supplemental material), implying that the recruitment of UBC13 to DNA breaks may be transient.
FIG. 3.
Recruitment of UBC13 to DSBs is sufficient for 53BP1 and FK2 IRIF formation. (A) Schematic illustration of chimeras of the RNF8 FHA domain fused with MMS2 or UEV1A or the UBC13-histone H2B fusion protein. (B) The chimeras of the RNF8 FHA domain fused with MMS2 or UEV1A were ectopically expressed in RNF8-deficient cells, and localization of FK2 and 53BP1 was determined by immunostaining. Representative results are shown. (C) The UBC13-H2B fusion was ectopically expressed in RNF8-deficient MEFs, and IRIF of FK2 and 53BP1 were examined by immunostaining as indicated. (D) RNF8-deficient cells were infected with retroviral constructs expressing chimeras of the RNF8 FHA domain fused with the respective RING domains derived from a series of E3 ubiquitin ligases, as indicated.
Next, we decided to investigate whether the fusion of the histone H2B with UBC13 might be sufficient in promoting checkpoint protein assembly. Functionality of the fusion construct was confirmed in UBC13-deficient cells, where expression of UBC13-H2B, but not the catalytically inactive UBC13 C87A-H2B, partially restored 53BP1 and FK2 focus formation in response to IR (∼50%; see Fig. S2C in the supplemental material). To circumvent the possibility that the residual nucleoplasmic fraction of the UBC13-H2B fusion protein might be targeted to DSBs by RNF8, we used RNF8-deficient cells and found that 53BP1 and ubiquitin conjugates failed to accumulate at γH2AX-marked sites in cells expressing UBC13-H2B fusion proteins (Fig. 3C), indicating that simple chromatin association of UBC13 is not sufficient for its function.
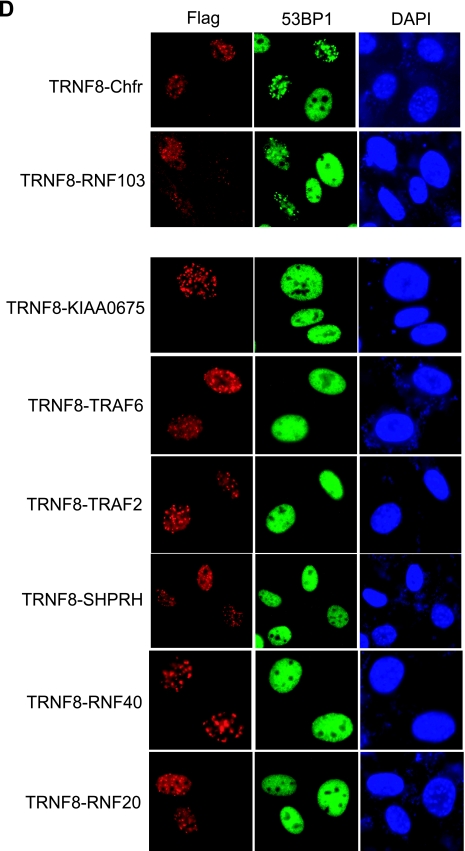
To extend this observation, we generated RNF8 FHA domain fusion proteins that harbor the RING domain of a number of E3 ubiquitin ligases that were previously reported to interact with UBC13. These included Chfr, RNF103, KIAA0675, TRAF2, TRAF6, and SHPRH. We also included RING domains derived from E3 ligases RNF20 and RNF40 as controls. These fusion proteins localized to DSBs in response to IR (see Fig. S3 in the supplemental material). Interestingly, while the Chfr and RNF103 RING domain fusion proteins were sufficient in mediating 53BP1 IRIF formation, fusion constructs containing genes that encode their respective RING domain cysteine point mutants (TRNF8-Chfr C292S and TRNF8-RNF103 C621S) or fusion proteins that harbor the RING domains derived from other E3 ubiquitin ligases did not restore RNF8 function in the DNA damage-signaling pathway (Fig. 3D and see Fig. S4 in the supplemental material). We speculated that the RNF8 RING domain, like those derived from Chfr and RNF103, might specifically recruit a subset of UBC13 for its function in vivo.
UBC13 acts independently of E2 variants MMS2 and UEV1A in the DNA damage-signaling pathway.
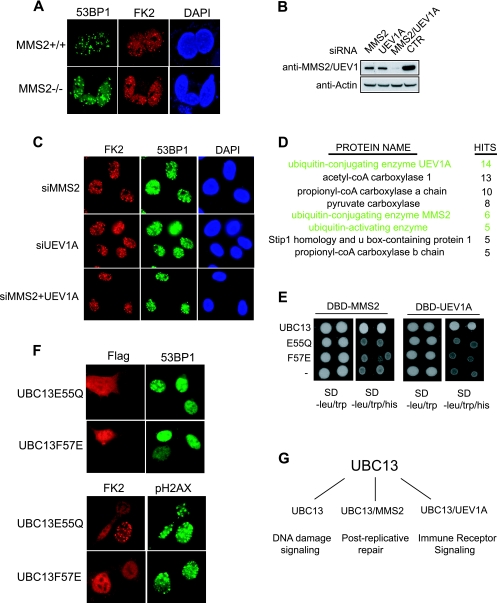
It is not clear which of the two known E2 variants might work with UBC13 in the DNA damage-signaling pathway, since TRAF2 and TRAF6 have been reported to function with UEV1A (18, 20, 27) and SHPRH is known to work with MMS2 (24). From the above experiments, none of these E3 RING domains can substitute for RNF8 RING domain function. Because MMS2 has been implicated in DNA damage and repair, we decided first to determine whether MMS2 would be required for this RNF8/UBC13-dependent early DNA damage response. We generated MMS2-null embryonic stem (ES) cells (data not shown) and found that 53BP1 and FK2 IRIF were intact in MMS2−/− cells (Fig. 4A), suggesting that MMS2 is not essential for UBC13 function in DNA damage signaling.
FIG. 4.
UBC13 function in the DNA damage-signaling pathway is independent of E2 variants MMS2 and UEV1A. (A and C) FK2 and 53BP1 IRIF were not affected in MMS2-deficient cells (A) or by the depletion of MMS2 and UEV1A in HeLa cells (C). (B) Western blot analysis verified siRNA-mediated depletion of MMS2, UEV1A, or both in HeLa cells. CTR, control. (D) Identification of UBC13-associated proteins. A list of proteins that copurified with UBC13 using 293T cells expressing tagged UBC13 is presented. coA, coenzyme A. (E and F) Interaction between UBC13 and its E2 variants is not essential for its function in promoting FK2 and 53BP1 IRIF. The yeast two-hybrid assay verified the interaction or absence of interaction between wild-type or mutant UBC13 with MMS2 or UEV1A (E). The formation of 53BP1 and FK2 IRIF was evaluated in UBC13-deficient cells reconstituted with UBC13 E55Q or F57E mutants (F). (G) Model of UBC13 function in diverse biological processes.
To further probe whether the E2 variants might be functionally redundant for focal accumulation of checkpoint proteins at DSBs, we depleted HeLa cells of MMS2, UEV1A, or both (Fig. 4B). Interestingly, neither MMS2 nor UEV1A is required for 53BP1 and FK2 IRIF (Fig. 4C). Depletion of both MMS2 and UEV1A together also failed to show any defects in FK2 or 53BP1 focus formation (Fig. 4C).
Knowing that UBC13 exhibits high-affinity binding with its E2 variants in vivo, we adopted a tandem-affinity purification approach in an attempt to identify additional E2 variants that might function with UBC13 in the DNA damage-signaling pathway. Mass spectrometry analysis of UBC13 complex revealed peptides that corresponded to UEV1A and MMS2 but did not identify additional potential E2 variants that might form stable heterodimers with UBC13 (Fig. 4D).
Since MMS2 and UEV1A were the only E2 variants that formed stable heterodimers with UBC13 and were not required for UBC13 function in the DNA damage-signaling pathway, to rule out the requirement for E2 variants, we employed the UBC13 E55Q and F57E mutations, which disrupt the binding of UBC13 to MMS2 or UEV1A (16) (Fig. 4E and see Fig. S5 in the supplemental material). We reasoned that if UBC13 functions independently of E2 variants, these mutants would mimic wild-type UBC13 in mediating checkpoint protein localization at DNA breaks. To test this idea, we reconstituted UBC13-deficient cells with the UBC13 mutants. Intriguingly, despite a clear deficit for both mutants (i.e., E55Q and F57E) in binding with its E2 variants, while the UBC13 E55Q mutation restored the ability of UBC13 in promoting 53BP1 and FK2 IRIF, the F57E mutation did not (Fig. 4F). We do not yet know why the F57E mutant displayed a defect in DNA damage signaling: however, the fact that both MMS2 and UEV1A, when fused with the RNF8 FHA domain, mediated damage-induced accumulation of 53BP1 and FK2 in an RNF8-null background further supports the notion that docking of UBC13 alone at DSBs is sufficient for DNA damage signal transduction (Fig. 3B). Furthermore, based on our analysis of UBC13-associated proteins in vivo (Fig. 4D) and the fact that depletion of both E2 variants had no effect on the ability of UBC13 to promote FK2 and 53BP1 focus formation, we favor the idea that UBC13 functions independently of E2 variants in the DNA damage-signaling pathway (Fig. 4G).
DISCUSSION
Recent studies have uncovered an important role of protein ubiquitylation in the cellular responses to DNA damage. Specifically, RNF8-dependent amplification of damage signals at the proximity of DNA lesions is crucial for proper activation of the cell cycle checkpoint and maintenance of genome stability. Using 53BP1 and FK2 IRIF as markers, we provide strong evidence to support that UBC13 harbors the enzymatic activity that facilitates the RNF8-mediated checkpoint protein assembly. This is accomplished in part by the RNF8 FHA domain, which enables the specific loading of UBC13 onto DNA lesions. Moreover, we showed that the RNF8 RING domain is essential for bringing UBC13 to the proximity of sites of DNA breaks. Given the exquisite coordination of the RNF8-UBC13 complex and its instrumental role in damage signal amplification, it would be interesting to examine whether dysregulation of such complex formation might contribute to tumorigenesis.
UBC13 plays diverse roles in biological processes through its ability to interact with multiple substrate specificity-conferring E3 ubiquitin ligases. Intriguingly, we found that only a subset of RING domains derived from these E3 ubiquitin ligases was competent in anchoring and facilitating UBC13 function at DSBs. Given that UBC13 forms stable heterodimers with E2 variants MMS2 and UEV1A, one might speculate that these RING domains might structurally conform to target a specific heterodimer for function. Although it is possible that an unidentified E2 variant might exist and only transiently associate with UBC13 in response to DNA damage, the fact that both MMS2 and UEV1A are dispensable for the UBC13-mediated checkpoint protein assembly, together with the fact that the binding of UBC13 to E2 variants is not absolutely required for its function, suggests that UBC13 might function independently of E2 variants.
Our observation that UBC13 at DSBs is required for checkpoint protein assembly (see Fig. S6A and B in the supplemental material), which in turn is required for optimal cellular response to DNA damage, is also illustrated by the prolonged accumulation of pH2AX in UBC13−/− cells (see Fig. S6C in the supplemental material). Although MMS2 and UEV1A are dispensable for UBC13 function in DNA damage signaling, depletion of MMS2 also led to sustained basal DNA damage (see Fig. S7A, B, and C in the supplemental material [as indicated by the elevated percentages of 53BP1 and FK2 IRIF and pH2AX-positive cells in untreated cells]) and manifested prolonged recovery upon IR. Since MMS2 is pivotal for postreplication repair, these MMS2-associated deficits might reflect a failure of proper DNA repair in these cells.
Recent evidence suggests that UBC13 acts in concert with RNF8 in promoting checkpoint protein assembly. Our studies using a chimeric fusion protein gene that encodes the RNF8 FHA and UBC13 and was sufficient to restore RNF8 function in the DNA damage response are entirely consistent with this notion. It remains possible, however, that an unidentified E3 ubiquitin ligase may participate in substrate ubiquitylation at DSBs. Similar to RNF8, the E3 ubiquitin ligase RAD18 has been shown to promote DSB repair (1). However, unlike RNF8, RAD18 is not required for FK2 and 53BP1 IRIF (unpublished observations and see Fig. S8A and B in the supplemental material).
In summary, we have provided strong evidence that the UBC13 gene encodes the enzymatic module which is essential for the RNF8-dependent propagation of DNA damage signals. We propose that the RNF8 RING domain selects a subset of UBC13 in vivo that enables substrate ubiquitylation that differs from those catalyzed by UBC13-MMS2 and UBC13-UEV1A heterodimers. Ubiquitylation events at DSBs subsequently allow sustained accumulation of checkpoint proteins, which in turn is essential for proper cellular response to genotoxic stress.
Supplementary Material
Acknowledgments
This work was supported in part by grants from the National Institutes of Health (CA089239, CA092312, and CA100109 to J.C.). J.C. is a recipient of an Era of Hope Scholar award from the Department of Defense and a member of the Mayo Clinic Breast SPORE program (P50 CA116201). M.S.Y.H. is supported by the Anna Fuller Fund Fellowship.
Footnotes
Published ahead of print on 4 August 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Andersen, P. L., F. Xu, and W. Xiao. 2008. Eukaryotic DNA damage tolerance and translesion synthesis through covalent modifications of PCNA. Cell Res. 18162-173. [DOI] [PubMed] [Google Scholar]
- 2.Andersen, P. L., H. Zhou, L. Pastushok, T. Moraes, S. McKenna, B. Ziola, M. J. Ellison, V. M. Dixit, and W. Xiao. 2005. Distinct regulation of Ubc13 functions by the two ubiquitin-conjugating enzyme variants Mms2 and Uev1A. J. Cell Biol. 170745-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakkenist, C. J., and M. B. Kastan. 2004. Initiating cellular stress responses. Cell 1189-17. [DOI] [PubMed] [Google Scholar]
- 4.Bassing, C. H., H. Suh, D. O. Ferguson, K. F. Chua, J. Manis, M. Eckersdorff, M. Gleason, R. Bronson, C. Lee, and F. W. Alt. 2003. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell 114359-370. [DOI] [PubMed] [Google Scholar]
- 5.Celeste, A., S. Difilippantonio, M. J. Difilippantonio, O. Fernandez-Capetillo, D. R. Pilch, O. A. Sedelnikova, M. Eckhaus, T. Ried, W. M. Bonner, and A. Nussenzweig. 2003. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell 114371-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duncan, L. M., S. Piper, R. B. Dodd, M. K. Saville, C. M. Sanderson, J. P. Luzio, and P. J. Lehner. 2006. Lysine-63-linked ubiquitination is required for endolysosomal degradation of class I molecules. EMBO J. 251635-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harper, J. W., and S. J. Elledge. 2007. The DNA damage response: ten years after. Mol. Cell 28739-745. [DOI] [PubMed] [Google Scholar]
- 8.Hoege, C., B. Pfander, G. L. Moldovan, G. Pyrowolakis, and S. Jentsch. 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419135-141. [DOI] [PubMed] [Google Scholar]
- 9.Hofmann, R. M., and C. M. Pickart. 1999. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96645-653. [DOI] [PubMed] [Google Scholar]
- 10.Huen, M. S., and J. Chen. 2008. The DNA damage response pathways: at the crossroad of protein modifications. Cell Res. 188-16. [DOI] [PubMed] [Google Scholar]
- 11.Huen, M. S., R. Grant, I. Manke, K. Minn, X. Yu, M. B. Yaffe, and J. Chen. 2007. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131901-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim, H., J. Chen, and X. Yu. 2007. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science 3161202-1205. [DOI] [PubMed] [Google Scholar]
- 13.Kolas, N. K., J. R. Chapman, S. Nakada, J. Ylanko, R. Chahwan, F. D. Sweeney, S. Panier, M. Mendez, J. Wildenhain, T. M. Thomson, L. Pelletier, S. P. Jackson, and D. Durocher. 2007. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 3181637-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lou, Z., K. Minter-Dykhouse, S. Franco, M. Gostissa, M. A. Rivera, A. Celeste, J. P. Manis, J. van Deursen, A. Nussenzweig, T. T. Paull, F. W. Alt, and J. Chen. 2006. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol. Cell 21187-200. [DOI] [PubMed] [Google Scholar]
- 15.Mailand, N., S. Bekker-Jensen, H. Faustrup, F. Melander, J. Bartek, C. Lukas, and J. Lukas. 2007. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131887-900. [DOI] [PubMed] [Google Scholar]
- 16.Pastushok, L., T. F. Moraes, M. J. Ellison, and W. Xiao. 2005. A single Mms2 “key” residue insertion into a Ubc13 pocket determines the interface specificity of a human Lys63 ubiquitin conjugation complex. J. Biol. Chem. 28017891-17900. [DOI] [PubMed] [Google Scholar]
- 17.Paull, T. T., E. P. Rogakou, V. Yamazaki, C. U. Kirchgessner, M. Gellert, and W. M. Bonner. 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10886-895. [DOI] [PubMed] [Google Scholar]
- 18.Shi, C. S., and J. H. Kehrl. 2003. Tumor necrosis factor (TNF)-induced germinal center kinase-related (GCKR) and stress-activated protein kinase (SAPK) activation depends upon the E2/E3 complex Ubc13-Uev1A/TNF receptor-associated factor 2 (TRAF2). J. Biol. Chem. 27815429-15434. [DOI] [PubMed] [Google Scholar]
- 19.Stucki, M., J. A. Clapperton, D. Mohammad, M. B. Yaffe, S. J. Smerdon, and S. P. Jackson. 2005. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 1231213-1226. [DOI] [PubMed] [Google Scholar]
- 20.Sun, L., L. Deng, C. K. Ea, Z. P. Xia, and Z. J. Chen. 2004. The TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MALT1 in T lymphocytes. Mol. Cell 14289-301. [DOI] [PubMed] [Google Scholar]
- 21.Torres-Ramos, C. A., S. Prakash, and L. Prakash. 2002. Requirement of RAD5 and MMS2 for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 222419-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ulrich, H. D. 2003. Protein-protein interactions within an E2-RING finger complex. Implications for ubiquitin-dependent DNA damage repair. J. Biol. Chem. 2787051-7058. [DOI] [PubMed] [Google Scholar]
- 23.Ulrich, H. D., and S. Jentsch. 2000. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 193388-3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Unk, I., I. Hajdu, K. Fatyol, B. Szakal, A. Blastyak, V. Bermudez, J. Hurwitz, L. Prakash, S. Prakash, and L. Haracska. 2006. Human SHPRH is a ubiquitin ligase for Mms2-Ubc13-dependent polyubiquitylation of proliferating cell nuclear antigen. Proc. Natl. Acad. Sci. USA 10318107-18112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.VanDemark, A. P., R. M. Hofmann, C. Tsui, C. M. Pickart, and C. Wolberger. 2001. Molecular insights into polyubiquitin chain assembly: crystal structure of the Mms2/Ubc13 heterodimer. Cell 105711-720. [DOI] [PubMed] [Google Scholar]
- 26.Wang, B., and S. J. Elledge. 2007. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc. Natl. Acad. Sci. USA 10420759-20763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang, C., L. Deng, M. Hong, G. R. Akkaraju, J. Inoue, and Z. J. Chen. 2001. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412346-351. [DOI] [PubMed] [Google Scholar]
- 28.Wooff, J., L. Pastushok, M. Hanna, Y. Fu, and W. Xiao. 2004. The TRAF6 RING finger domain mediates physical interaction with Ubc13. FEBS Lett. 566229-233. [DOI] [PubMed] [Google Scholar]
- 29.Yamamoto, M., T. Okamoto, K. Takeda, S. Sato, H. Sanjo, S. Uematsu, T. Saitoh, N. Yamamoto, H. Sakurai, K. J. Ishii, S. Yamaoka, T. Kawai, Y. Matsuura, O. Takeuchi, and S. Akira. 2006. Key function for the Ubc13 E2 ubiquitin-conjugating enzyme in immune receptor signaling. Nat. Immunol. 7962-970. [DOI] [PubMed] [Google Scholar]
- 30.Zhang, M., M. Windheim, S. M. Roe, M. Peggie, P. Cohen, C. Prodromou, and L. H. Pearl. 2005. Chaperoned ubiquitylation—crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol. Cell 20525-538. [DOI] [PubMed] [Google Scholar]
- 31.Zhao, G. Y., E. Sonoda, L. J. Barber, H. Oka, Y. Murakawa, K. Yamada, T. Ikura, X. Wang, M. Kobayashi, K. Yamamoto, S. J. Boulton, and S. Takeda. 2007. A critical role for the ubiquitin-conjugating enzyme Ubc13 in initiating homologous recombination. Mol. Cell 25663-675. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.