Abstract
In situ gene expression studies are promising approaches for improving our understanding of the cheese microbial flora. This requires efficient RNA extraction methods, but studies of cheeses are scarce. The objective of the present study was to determine whether RNA samples compatible with quantitative mRNA transcript analyses can be obtained without separating the cells from the cheese matrix. In the method that we describe, the cellular processes are stopped at the very beginning of the procedure. When cheeses were produced with Lactococcus lactis LD61 as the only starter microorganism, the integrity of the purified RNA was good, even for 2-week-old cheeses that had been incubated at 30°C. In addition, the RNA samples did not contain any traces of RNases, and the amount of genomic DNA was negligible. A good level of reproducibility could also be achieved. When real-time reverse transcription-PCR analyses were normalized to the total RNA concentration, the amounts of 16S and 23S rRNA transcripts were constant during the 2-week incubation period, whereas the amount of tuf mRNA transcripts decreased substantially. RNA samples obtained using the method described in this study were compared to samples obtained using the method described by Ulvé et al. (J. Appl. Microbiol., in press), which is based on separation of the cells from the cheese matrix. For most of the 29 genes investigated, the transcript abundance was the same for both types of samples. Differences were observed mainly for genes whose expression has previously been shown to be modified by heat, acid, or osmotic stresses, such as busAA and glnQ.
Many species of bacteria, yeasts, and molds are involved in the production of cheese. The presence of these organisms is required to generate the typical sensory properties of the final product, such as texture, taste, aroma, and color. The composition of the cheese microbial flora varies as a function of the type of cheese and the manufacturing stage. In order to improve our understanding of the activity of the cheese microbial flora, one promising approach is in situ analysis of the mRNA transcripts. Indeed, more and more genomic sequences of cheese microorganisms are becoming available, and it is therefore possible to design primers or probes that specifically target mRNA transcripts from a given species. Quantitative analyses can be performed by using real-time reverse transcription-PCR, and semiquantitative analyses can be performed for a large number of genes using DNA microarrays.
However, even though procedures for extraction of DNA from cheese are being used increasingly in research laboratories (1, 4-6, 8-10, 12, 13, 15, 17, 18, 20), the extraction of RNA is still a challenge. Randazzo et al. (16) studied the bacterial flora of an artisanal cheese by performing denaturing gradient gel electrophoresis analysis after reverse transcription of rRNA. Bonaïti et al. (2) reverse transcribed rRNA from bacteria and yeasts after extraction of RNA from experimental cheeses. To our knowledge, there has been only one study that described analysis of mRNA transcripts in cheese samples (22). In that study, the RNA was extracted from cheese after separation of the bacterial cells from the cheese matrix.
The objective of the present study was to determine whether RNA samples compatible with quantitative mRNA transcript analysis can be obtained without prior separation of the cells from the cheese matrix. The main advantage of such a method would be that the production and degradation of mRNA transcripts could be stopped immediately after a cheese is sampled.
MATERIALS AND METHODS
Strain and growth conditions.
Lactococcus lactis subsp. lactis LD61 was obtained from SOREDAB (La Boissière-Ecole, France) and was routinely grown under static conditions in M17 lactose broth (21) at 30°C.
Cheese production.
Ultrafiltered milk was produced as described by Hannon et al. (7) and stored at −20°C. Its final fat concentration was 5.5%. To produce cheese, strain LD61 was inoculated into 100 ml of reconstituted skim milk (100 g/liter; Difco Laboratories, Detroit, MI) that was previously heated for 10 min at 110°C and incubated for 15 h at 30°C. Ultrafiltered milk was inoculated using a concentration of 107 CFU/ml, and rennet (chymosin with an activity of 180 international milk-clotting units/ml; Maxiren 180; DSM Food Specialities, Delft, The Netherlands) was added at a final concentration of 60 μl/liter. After incubation for 7 h at 30°C, the curd was transferred onto a grid in a sterile crystallizing basin. Ripening was then performed either at 12°C or at 30°C. The latter temperature is far above the temperature generally used during cheese ripening. It was chosen in order to provide conditions under which the RNA transcripts would be less stable.
Measurement of culturable bacterial concentration.
One gram of cheese was mixed with 9 ml of physiological saline (9 g/liter NaCl). After dispersion with a mechanical blender (Ultra-Turrax model T25; Ika Labortechnik, Staufen, Germany) for 1 min at 11,500 rpm, 10-fold serial dilutions in physiological saline were prepared and plated on M17 agar. Colonies were enumerated after the plates were incubated for 2 days at 30°C.
Extraction of DNA.
DNA was extracted from cheese as previously described (11).
Extraction of RNA.
During the purification procedure, only RNase-free reagents and plastic utensils were used, and care was taken to avoid any contamination by RNases. Approximately 125 mg of cheese was placed into a 2-ml tube containing 800 mg of zirconium beads (diameter, 0.1 mm; BioSpec Products, Bartlesville, OK), and this was followed by immediate addition of 1.25 ml TRIzol reagent (Invitrogen, Cergy Pontoise, France). The tubes were vigorously shaken in a bead beater (FastPrep-24 system; MP Biomedicals, Illkirch, France) by using three 60-s mixing sequences at a speed of 6.5 m/s. The tubes were cooled on ice for 5 min before each mixing sequence. After centrifugation for 10 min at 12,000 × g and 4°C, each supernatant (approximately 1,100 μl) was transferred into a 2-ml tube containing 300 μl of a gel that improved separation of the aqueous and organic phases (Phase Lock Gel Heavy; Eppendorf, Hamburg, Germany). In some cases, an approximately 3-mm red layer formed just above the beads. This layer was also transferred. However, the fat layer, which was at the top the liquid phase, was not transferred. The tubes were incubated for 5 min at room temperature before addition of 230 μl chloroform. They were then shaken for 15 s, incubated for 3 min at room temperature and for 2 min on ice, and centrifuged for 15 min at 12,000 × g and 4°C. Each aqueous phase (approximately 700 μl) was recovered in a 2-ml tube, and 700 μl of phenol-chloroform-isoamyl alcohol (125:24:1, pH 4.7) was added. The tubes were then shaken for 15 s and centrifuged for 10 min at 12,000 × g and 4°C. Each aqueous phase (approximately 550 μl) was recovered, taking care not to recover any part of the organic phase. In order to increase the final RNA concentration in the samples, the contents of tubes from three replications were pooled and a volume of ethanol (100%) corresponding to 55% of the volume of the aqueous phase was added. Seven hundred microliters of the sample was loaded on an RNeasy spin column (Qiagen, Courtaboeuf, France), which was then centrifuged for a few seconds at 12,000 × g and room temperature. After elimination of the flowthrough, the remainder of the sample was loaded on the column and treated in the same way, until the entire sample was used. When a large number of samples was pooled (for example, 10 samples), the procedure was considered time-consuming due the numerous successive centrifugations required. In such a case, the samples were loaded on the column using a vacuum manifold (QIAvac 24; Qiagen). Then 350 μl of RW1 buffer (Qiagen) was loaded on the column, and following incubation for 5 min at room temperature, the tube was centrifuged for a few seconds at 12,000 × g. The flowthrough was discarded, and a second washing step with 350 μl of RW1 buffer was performed. Two washing steps were then performed with 500 μl of RPE buffer (Qiagen), and the tube was centrifuged for 1 min in order to eliminate all traces of RPE buffer. The RNA was recovered after addition of 30 μl of RNase-free water, incubation for 2 min at room temperature, and centrifugation for 1 min.
Quantification of RNA and quality control.
Purified RNA was quantified at 260 nm using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). The quality of the RNA was analyzed with a 2100 bioanalyzer (Agilent, Palo Alto, CA) using RNA 6000 NANO chips according to the manufacturer's instructions.
DNase treatment and reverse transcription.
When DNase treatment was performed, the reaction was conducted with 800 ng of RNA in a 20-μl reaction mixture, using a TURBO DNA-free kit (Ambion, Austin, TX). cDNA was synthesized from RNA or DNase-treated RNA using the SuperScript III First-Strand synthesis system (Invitrogen, Cergy Pontoise, France) according to the manufacturer's recommendations. The concentrations of RNA used in the reaction mixtures are indicated below. Priming was performed using random hexamers. The resulting cDNA samples were stored at −20°C.
Real-time PCR conditions.
SYBR green I PCR amplification was performed using a LightCycler instrument (Roche Applied Science, Mannheim, Germany). Amplification was carried out in a 20-μl (final volume) mixture containing 5 μl of DNA or cDNA sample, 4 mM MgCl2, each primer at a concentration of 0.3 μM, and 2 μl of LightCycler FastStart DNA Master SYBR green I (Roche). The primers were synthesized by Eurogentec (Seraing, Belgium). Amplification involved incubation at 95°C for 8 min for the initial denaturation, followed by 45 cycles of (i) denaturation at 95°C for 10 s, (ii) annealing at 60°C for 7 s, (iii) extension at 72°C for 6 s, and (iv) fluorescence acquisition (530 nm) at the end of extension. The temperature transition rate was 20°C/s for each step. After real-time PCR, a melting curve analysis was performed by continuously measuring fluorescence during heating from 65 to 95°C at a transition rate of 0.1°C/s. Threshold cycle (CT) values were determined with LightCycler software (version 3.3), using the second derivative method. Standard curves were generated by plotting the CT values as a function of the log of the DNA concentration (analysis of genomic DNA) or of the dilution of the cDNA sample (analysis of reverse-transcribed RNA). PCR efficiency (E) was then calculated using the following formula: E = 10−1/slope (14). The genes investigated in the present study and the corresponding primer pairs are shown in Table 1. The primers were designed using LightCycler probe design software (v1.0; Roche Applied Science) and a melting temperature of 65°C. The expression of selected genes in cheese was measured using the relative standard curve method (ABI Prism 7700 sequence detection system user bulletin 2, 1997). Reverse transcription results were standardized using an RNA concentration of 2.5 ng/μl, and real-time PCRs were performed after dilution of the samples in order to avoid PCR inhibition and to obtain a linear relationship between the CT value and the logarithm of the amount of RNA. Twenty-five-fold dilutions were used for analysis of the tuf gene, and 125-fold dilutions were used for analysis of the 16S rRNA and 23S rRNA genes. The amount of target at different sampling times was divided by the amount of target at 3 h, which was chosen as the calibrator sample. Thus, data were expressed as differences relative to the results for the sample at 3 h.
TABLE 1.
Primers used in this study
| Gene | Gene product | Forward primer (5′→3′) | Reverse primer (5′→3′) | Reference |
|---|---|---|---|---|
| adhE | Alcohol-acetaldehyde dehydrogenase | ATC TAT ACT GAT GCA ATG CGT CC | ACG AAC CCA TTG TGG GC | This study |
| aldB | Alpha-acetolactate decarboxylase | AAT GGC AGG CCT TTA CGA G | AGC TTG GTA GGC TTT ACC G | This study |
| als | Alpha-acetolactate synthase | GCA CGT CAT TTC AAA TCA TAC GAA C | CCT CCA TCA CCA GAG TGT GAA TA | This study |
| busAA | Betaine ABC transporter ATP binding protein | CGA TTA AGC AAA CGC AAC AA | AGG TGA GCC CAA AAA TGA AA | This study |
| butB | 2,3-Butanediol dehydrogenase | AGT AGG TGA CCA TGT CGT TG | ACC TCC ACC ATT TCC GC | This study |
| citD | Citrate lyase acyl-carrier protein | GTC TTC AGA TGT GCA AAT CAT G | CGC ATT TTC CAA TGG CAT AAG C | 22 |
| citF | Citrate lyase alpha chain | TCG CTG GAA GAA GTG GTT TT | AGA AGA CGG AGC AAT CCT CA | This study |
| codY | Transcriptional repressor | CAG AAG AAA GCC TTG GCG A | CGT CTT CAC GCC ACA TGA T | This study |
| cspD | Cold shock protein D | TGG CAA ATG GAA CAG TAA AAT G | GCT GAG AAG TGA GCG AAC AA | This study |
| cspE | Cold shock protein E | TAA CAG CTT GAG GTC CAC GA | ACG ACG TTT TCG CTC ACT TC | This study |
| cysS | Cysteinyl-tRNA synthetase | TGG ATG AGC AAG CTG AGT CT | TTC TTT GAC CTC AGG CCC AC | This study |
| cysK | Cysteine synthase | GAG CGT CGT CAG ATT ATT CAA G | GCA TAA ACC AAC CAT TTT CTT C | 22 |
| deoB | Phosphopentomutase | CGA AAG TCC GCT CAA AAC G | TTC CCA GTG CCC AGT CAT T | This study |
| dnaK | Molecular chaperone | GCA GCT CTT GCT TAT GGT CTT G | CGG CAA CCA TCC AGT CAA TG | This study |
| gapB | Glyceraldehyde 3-phosphate dehydrogenase | GTT GTT ATC ACT GCA CCT GG | CAT TGG AGC AAG ACA GTT AGT TG | 22 |
| glnQ | Glutamine ABC transporter ATP-binding protein | AAA GAT GCA ATG CCT GAA ATG | CGT CAA AGA GCA TAA CGT CAG | This study |
| groEL | Chaperonin | TGTA GCT GAT GAT GTT GAT GGA GA | CTT TAC GAC GAT CAC CAA ATC CT | This study |
| gyrA | DNA gyrase subunit A | CAC GGG CAC TTC CTG ATG TA | CCA TAA CTT CAC CGA CAA TA | 22 |
| ldh | l-Lactate dehydrogenase | GTC GCT GTA GCT CTT GCT CG | GTT GAC CAA GGT AGC AGT CG | 22 |
| ldhB | l-Lactate dehydrogenase | AGG TTT TCC ACC TAC CGT CGT | TCT CGA ATT GTC GGA ACT GGT | This study |
| metA | Homoserine O- succinyltransferase | CCT TGA TAA ACC GCA TAA TTC A | GAA GGA CTG AAA CCC AAT CTT C | 19 |
| metB1 | Cystathionine gamma-synthase | GTT GCT AGA GCA AAT TGT CCT G | CAA CGA CCT ATC AAC ATC CAG A | 19 |
| mleS | Malolactic enzyme | ACC ACC CTG AAA ACA TTA CTG AA | CTT GAG TTC CCC AGT CTC CA | This study |
| pfl | Pyruvate-formate lyase | ATG AGC CAC AAG CAT TCT TCT AT | AAG TGG TGA GAC ACA ACA AGA G | This study |
| purM | Phosphoribosylaminoimidazole synthetase | GAT TGC GTA GCC ATG TGC GTC | GCC ACT CCA GCC ACA ACT TG | 22 |
| rpiA | Ribose-5-phosphate isomerase A | TCG AAG AAT TGG GTC GAA GAA | AATTTCATCTGCGCCATCAAC | This study |
| tuf | Elongation factor Tu | CTC TAA AGT ATT GTC TGA CA | GTG TTG ATT GTG ATA CCA CG | 22 |
| 16S rRNA | 16S rRNA | GCT CAC CAA GGC GAT GAT ACA TA | ACC AAC GTT CTT CTC TAC CAA CA | This study |
| 23S rRNA | 23S rRNA | ACA GGA TAG GTA GGA GCC ATT | CGA TTA TGC CAG CGG GTT AG | This study |
Statistical analyses.
The means of CT values were compared using a Student test. Three repetitions (separate RNA extracts) were performed for each extraction method, using the same initial cheese sample.
RESULTS
Development of the RNA extraction method.
In order to establish an efficient method for extracting RNA from cheese without prior separation of bacterial cells, both the quality and extraction yield of the purified RNA were taken into account. Experiments were performed with a model semifat cheese produced with L. lactis strain LD61. After numerous initial attempts, the procedure described in Materials and Methods was chosen. The most critical factor is the ratio of the amount of cheese to the volume of TRIzol reagent. This ratio should not exceed 150 mg of cheese per ml of reagent, as a higher ratio affects the quality and quantity of the purified RNA. Consequently, the amount of RNA that can be recovered from cheese with this protocol is limited by the amount of cheese that can be processed in the first step. To minimize this problem, several tubes containing the same sample were processed in parallel and pooled later in the protocol (for example, before the purification on silica membrane-based columns). The samples could also be pooled after the bead-beating step, but this required the use of larger tubes in subsequent steps. The presence of fat in cheese does not interfere with the purification of RNA since the fat is eliminated after the first centrifugation step. The amount of beads and the time and intensity parameters of the bead-beating treatment were optimized for the destruction of L. lactis cells. No further improvement was obtained when additional bead-beating procedures were performed, indicating that the destruction of the cells was probably complete. TRIzol-based purification methods do not usually require any further extraction of the samples with phenol. However, we observed that slight degradation of RNA sometimes occurred, except when an acid phenol treatment was included. Under these conditions, the level of contamination of the RNA samples with genomic DNA was also lower. An acid phenol extraction step was therefore included in the final purification protocol.
Validation of the conditions for DNase treatment, reverse transcription, and real-time PCR.
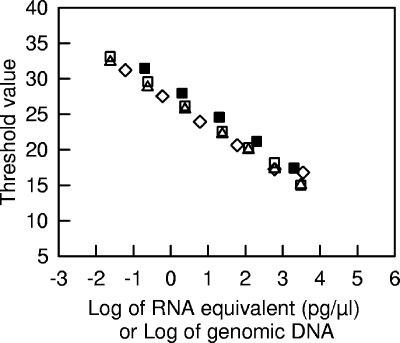
Different standard curves for real-time PCR amplification of genomic DNA or of reverse-transcribed RNA are shown in Fig. 1. For the tuf gene, the amplification efficiency with genomic DNA was constant between 0.2 and 2,000 pg/μl, and the corresponding PCR efficiency was 1.93 (97%). RNA extracted from cheese after 15 h of incubation at 30°C was treated with DNase and reverse transcribed using an RNA concentration equivalent to 15 ng/μl. When dilutions of the corresponding cDNA were analyzed, the PCR efficiency (1.96) was similar to that obtained with genomic DNA. This indicated that the reverse transcription reaction mixture did not inhibit the real-time PCR. However, this type of inhibition was detected when the reverse-transcribed RNA was not diluted before real-time PCR analysis (results not shown). Furthermore, omission of the DNase treatment did not change the threshold values of the reverse-transcribed RNA (both types of measurements were performed during the same PCR run). This means that the RNA samples did not contain significant amounts of genomic DNA, that the DNase treatment did not degrade the RNA, and that the DNase mixture did not change the reverse transcription efficiency. In addition, when different initial amounts of RNA were reverse transcribed and analyzed using a constant dilution factor (25-fold), the relationship between the threshold value and the concentration of RNA was similar to the relationship obtained by analysis of various dilutions of a cDNA which was produced by reverse transcription of 15 ng/μl of RNA. This means that under our conditions, the efficiency of the reverse transcription was constant. We also verified that the reproducibility of the reverse transcription and real-time PCR steps was good. For example, when the same reverse-transcribed RNA sample (15 ng/μl of RNA) was analyzed during the same PCR run, the standard deviation of the threshold value was as low as 0.15 (n = 4). Taken together, these results show that the conditions for DNase treatment, reverse transcription, and real-time PCR that were used are compatible with quantitative analysis of RNA transcripts from cheese. For the measurements described below, we included assays that ensured that no inhibition of reverse transcription or of real-time PCR occurred and that the amount of genomic DNA in the RNA samples was negligible.
FIG. 1.
Standard curve analyses of tuf amplification for genomic DNA (▪), RNA (⋄), cDNA (▵), and cDNA from DNase-treated RNA (□) dilution series. RNA and genomic DNA were extracted from cheese incubated for 15 h at 30°C. For the DNA standard curve, various dilutions of the DNA sample were directly analyzed by real-time PCR. For the RNA standard curve, various amounts of RNA were reverse transcribed and analyzed by real-time PCR after 25-fold dilution. For the two cDNA standard curves, reverse transcription was done using an RNA concentration of 15 ng/μl, and various dilutions were then analyzed by real-time PCR.
Extraction and analysis of RNA from cheese after different incubation times.
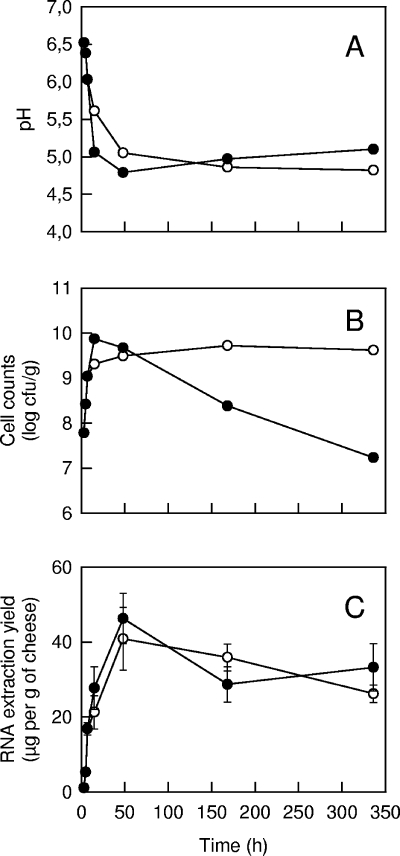
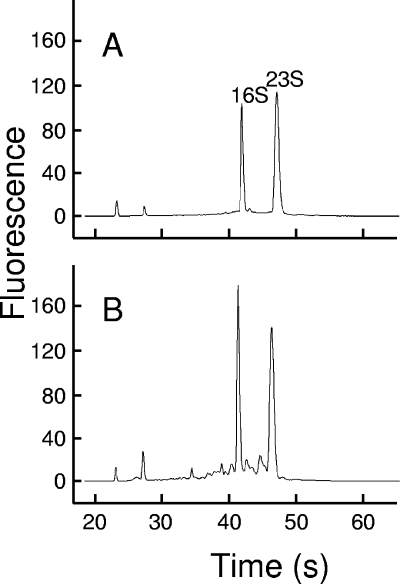
During the production of cheese with strain LD61, samples were taken after 3, 5, and 7 h (one cheese at each sampling time). After 7 h, some cheeses were incubated on a grid at 30°C, while others were incubated at 12°C, and samples were taken at 15 h and 2, 7, and 14 days. The pH of the cheeses decreased at the beginning of production (Fig. 2) due to the acidifying activity of L. lactis. For the cheeses incubated at 30°C, the culturable cell concentration reached the maximum value at 15 h and decreased thereafter. After 14 days, only 0.2% of the cells survived. However, no significant decrease was observed for the cheeses incubated at 12°C. The maximum RNA extraction yield was obtained after 2 days, and the value was close to 40 μg per g of cheese. It is interesting that there was only a slight decrease in the extraction yield between 2 and 14 days, even for the cheeses incubated at 30°C, in which cell viability decreased substantially. When the extraction of RNA was repeated (three separate RNA samples, obtained from the same initial cheese), the RNA extraction yield was relatively constant. Indeed, the mean coefficient of variation calculated from the data shown in Fig. 2C was 15%. Analysis of RNA samples with a 2100 bioanalyzer is based on capillary electrophoresis, followed by calculation of a value, known as the RIN value, which is representative of the integrity of the RNA. A RIN value of 10 corresponds to apparently intact material. Examples of electropherograms are shown in Fig. 3. The RNA extracted after 5 h of incubation at 30°C showed very good integrity (RIN value, 9.6). A few small peaks were present in the electropherogram of RNA extracted after 14 days of incubation at 30°C. The corresponding RIN value was 8.3, which is still good for most types of RNA analyses. For all the other samples, the RNA integrity was very good since the RIN values were between 9.2 and 9.6.
FIG. 2.
Changes in pH (A), culturable cell concentration (B), and RNA extraction yield (mean ± standard deviation) (C) during cheese production. After incubation for 7 h at 30°C, the curd was transferred onto a grid and incubated further at 30°C (•) or 12°C (○).
FIG. 3.
RNA quality assessment with the Agilent bioanalyzer: electropherograms of RNA preparations from cheese after 5 h (A) or 14 days (B) of incubation at 30°C.
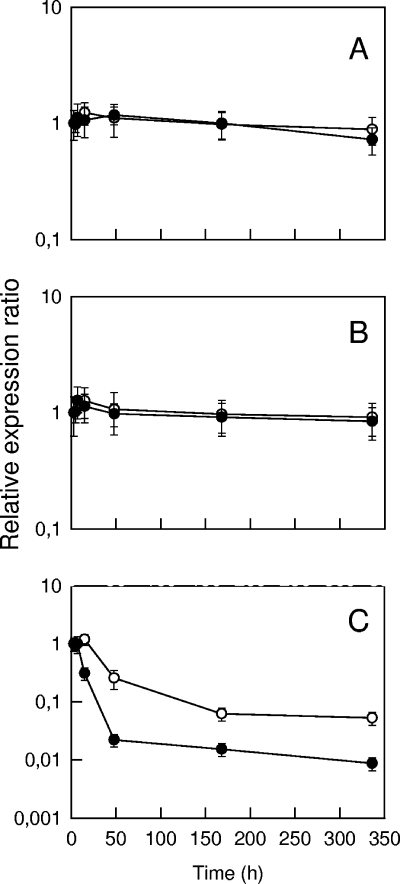
The results of some quantitative analyses of RNA transcripts in cheese samples are shown in Fig. 4. For each cheese sample, three separate RNA extractions were performed. The error bars thus represent the combined variability of the extraction, reverse transcription, and real-time PCR steps. However, the amount of RNA was standardized in all the reverse transcription reactions, and the real-time PCR results were expressed relative to a calibrator (the sample after 3 h), which was included in all the PCR runs. Since the samples contained no significant amount of genomic DNA, DNase treatment was not performed. There was no significant variation over time of the 16S rRNA and 23S rRNA transcript abundance. Furthermore, there were only limited differences between repeated samples. For example, after 48 h of incubation at 30°C, the means ± standard deviations of the variation corresponding to the three samples resulting from separate RNA extractions were 1.18 ± 0.20 and 0.98 ± 0.22 for 16S rRNA and 23S rRNA, respectively. The expression ratio of the tuf gene was constant from 3 to 7 h at 30°C and even at 15 h when the cheese was transferred at 12°C. The ratio decreased substantially after this, especially for the cheeses incubated at 30°C. After 14 days, the abundance of tuf transcripts was approximately 1 and 10% of the abundance after 3 h for the cheeses incubated at 30 and 12°C, respectively.
FIG. 4.
Changes in the relative expression ratios of 16S rRNA (A), 23S rRNA (B), and tuf (C) genes during the production of cheese. After incubation for 7 h at 30°C, the curd was transferred onto a grid and incubated further at 30°C (•) or 12°C (○). The levels of expression are the levels relative to the expression of the sample at 3 h (calibrator), and the error bars indicate standard deviations. Reverse transcription was done using an RNA concentration of 2.5 ng/μl, and real-time PCR were performed using a 25-fold dilution for the tuf gene and a 125-fold dilution for the 16S rRNA and 23S rRNA genes.
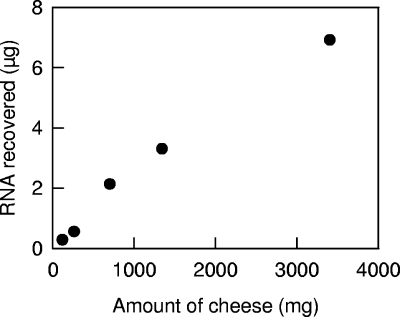
Effect of the amount of cheese on the recovery of RNA.
Since it was not possible to increase the amount of cheese processed in the first step of the protocol, the possibility of parallel processing of several samples and pooling after the bead-beating step was assessed. In all these experiments, the amount of RNA loaded on the silica membrane-based column was less than the column capacity (100 μg). The recovery of RNA was linear, and there were only small differences in the extraction yields between the different samples (Fig. 5). No significant difference in RNA quality or in 16S rRNA, 23S rRNA, and tuf transcript abundance was detected in these samples (results not shown). The concentration of RNA in samples can thus be increased by pooling samples after the bead-beating step. However, this considerably increases the cost and duration of the purification method. Since the amount of cheese recovered in each tube is approximately 125 mg, an adequate sampling procedure (e.g., the number of tubes that are subsequently pooled and the spatial distribution of the samples) has to be considered.
FIG. 5.
Recovery of RNA from various amounts of cheese. Samples (approximately 125 mg) of cheese produced after 5 h of incubation at 30°C were treated with TRIzol reagent and subjected to bead beating as described in Materials and Methods. Various numbers of samples (1 to 24 samples) were then pooled, and RNA was recovered using a single silica membrane-based purification column.
Comparison with the RNA extraction method based on prior separation of cells from cheese.
The extraction method developed by Ulvé et al. (22) consists of purification of RNA after separation of the bacterial cells from the cheese matrix. In order to compare this method with the method described in the present study, the abundance of transcripts from 29 genes from the same cheese after 15 h of incubation at 30°C was measured (Table 2). Several of the selected genes are involved in the response to various stresses. No significant difference in transcript abundance could be detected for 17 of these genes, including the 16S rRNA and 23S rRNA genes. Interestingly enough, for the other 12 genes, there was always a higher abundance in the RNA samples extracted after separation of cells from cheese. For most of these genes (busAA, cspD, cspE, glnQ, metA, metB1, and mleS), previous studies showed that their expression was modified by heat, acid, or osmotic stresses (23, 24). The largest difference was the difference for busAA, which encodes a betaine ABC transporter ATP-binding protein and for which the abundance of transcripts was about 50 times higher with the RNA samples extracted after separation of cells from cheese. Similar results were obtained when the RNA samples were treated with a DNase before reverse transcription (results not shown), even for the genes for which the highest threshold values were obtained (for example, metA and metB1). This shows that the contamination of the samples with genomic DNA was negligible.
TABLE 2.
Measurement of gene expression in cheese incubated for 15 h at 30°C using RNA directly extracted from cheese or after separation of cells
| Gene | Gene product | Threshold valuea
|
Gene expression ratiob | ||
|---|---|---|---|---|---|
| Direct RNA extraction | RNA extraction after separation of cells | Difference | |||
| adhE | Alcohol-acetaldehyde dehydrogenase | 22.70 ± 0.30 | 23.00 ± 0.12 | 0.30 | 0.82 |
| aldB | Alpha-acetolactate decarboxylase | 22.04 ± 0.22 | 21.53 ± 0.23 | 0.51 | 1.40 |
| als | Alpha-acetolactate synthase | 19.44 ± 0.34 | 19.66 ± 0.06 | −0.22 | 0.87 |
| busAA | Betaine ABC transporter ATP-binding protein | 25.58 ± 0.37 | 19.69 ± 0.33 | 5.89c | 51.51 |
| butB | 2,3-Butanediol dehydrogenase | 23.33 ± 0.38 | 23.81 ± 0.19 | −0.48 | 0.73 |
| citD | Citrate lyase acyl-carrier protein | 19.10 ± 0.20 | 19.73 ± 0.26 | −0.63 | 0.66 |
| citF | Citrate lyase alpha chain | 16.78 ± 0.10 | 16.92 ± 0.35 | −0.14 | 0.91 |
| codY | Transcriptional repressor | 24.56 ± 0.24 | 22.31 ± 0.30 | 2.25c | 4.46 |
| cspD | Cold shock protein D | 19.20 ± 0.35 | 16.14 ± 0.28 | 3.06c | 6.71 |
| cspE | Cold shock protein E | 19.92 ± 0.07 | 18.06 ± 0.35 | 1.87c | 3.30 |
| cysS | Cysteinyl-tRNA synthetase | 26.43 ± 0.11 | 23.97 ± 0.34 | 2.46c | 9.77 |
| cysK | Cysteine synthase | 18.92 ± 0.17 | 18.73 ± 0.07 | 0.19 | 1.13 |
| deoB | Phosphopentomutase | 18.73 ± 0.12 | 19.31 ± 0.37 | −0.58 | 0.68 |
| dnaK | Molecular chaperone | 16.85 ± 0.21 | 16.98 ± 0.32 | −0.13 | 0.91 |
| gapB | Glyceraldehyde 3-phosphate dehydrogenase | 15.70 ± 0.12 | 16.40 ± 0.39 | −0.70 | 0.64 |
| glnQ | Glutamine ABC transporter ATP-binding protein | 25.89 ± 0.47 | 21.62 ± 0.37 | 4.28c | 16.13 |
| groEL | Chaperonin | 20.58 ± 0.22 | 20.58 ± 0.11 | 0.00 | 1.00 |
| gyrA | DNA gyrase subunit A | 20.83 ± 0.06 | 19.67 ± 0.06 | 1.17c | 2.10 |
| ldh | l-Lactate dehydrogenase | 18.33 ± 0.06 | 18.27 ± 0.26 | 0.07 | 1.04 |
| ldhB | l-Lactate dehydrogenase | 24.36 ± 0.10 | 25.07 ± 0.57 | −0.71 | 0.64 |
| metA | Homoserine O-succinyltransferase | 30.72 ± 0.21 | 27.71 ± 0.27 | 3.01c | 6.23 |
| metB1 | Cystathionine gamma-synthase | 29.97 ± 0.17 | 26.77 ± 0.16 | 3.20c | 7.67 |
| mleS | Malolactic enzyme | 27.50 ± 0.28 | 25.54 ± 0.18 | 1.96c | 3.51 |
| pfl | Pyruvate-formate lyase | 25.10 ± 0.14 | 23.99 ± 0.25 | 1.12c | 2.03 |
| purM | Phosphoribosylaminoimidazole synthetase | 24.99 ± 0.04 | 22.75 ± 0.03 | 2.24c | 4.85 |
| rpiA | Ribose-5-phosphate isomerase A | 24.02 ± 0.23 | 24.03 ± 0.18 | −0.01 | 1.00 |
| tuf | Elongation factor Tu | 17.44 ± 0.12 | 17.89 ± 0.21 | −0.45 | 0.75 |
| 16S rRNA | 16S rRNA | 9.60 ± 0.19 | 9.92 ± 0.12 | −0.32 | 0.81 |
| 23S rRNA | 23S rRNA | 10.83 ± 0.15 | 11.10 ± 0.22 | −0.27 | 0.84 |
The values are means ± standard deviations for three repetitions (separate RNA extractions) using the same cheese sample. Reverse transcription was standardized using an RNA concentration of 15 ng/μl, and real-time PCR were done using a 25-fold dilution, except for 16S rRNA and 23S rRNA, for which a 125-fold dilution was used.
Ratio of the gene abundance measured using RNA extracted after separation of cells to the abundance measured using RNA extracted directly from cheese.
The threshold values of the two protocols were significantly different (P < 0.05).
DISCUSSION
The RNA extraction method that we propose in the current study does not require separation of cells from the cheese matrix. In this procedure, the first step is addition of TRIzol reagent to a sample. This reagent is appropriate for isolating RNA using the method of Chomczynski and Sacchi (3). Since it contains phenol and guanidine isothiocyanate, it has the ability to rapidly stop the cellular processes and to inactivate RNases. Hence, the main advantage of this procedure is that the cellular processes are stopped at the very beginning of the extraction. When cheeses were produced with L. lactis LD61, the integrity of the purified RNA was excellent, even for 2-week-old cheeses. In addition, the RNA samples did not contain any traces of RNases, and the amount of genomic DNA was negligible. A good level of reproducibility could also be achieved. However, because it is impossible to increase the ratio of cheese weight to the volume of TRIzol reagent, the amount of RNA that can be recovered in each extraction tube is limited. This problem can be reduced by processing several identical samples in parallel and pooling them after the bead-beating step. However, it is clear that such a procedure is more time-consuming than the procedure based on separation of cells from cheese (22), which allows a large amount of cheese to be processed without difficulty.
Quantitative data for the abundance of rRNA and mRNA transcripts during the manufacturing of cheese are reported here. Several factors support the hypothesis that these data may reflect the “true” quantitative evolution of the transcripts. For example, the reverse transcription and real-time PCR steps were reproducible, and their efficiency was constant. In addition, similar results were obtained with RNA samples from repeated extractions. Furthermore, because the RNA contained mainly rRNA, because the amount of RNA subjected to reverse transcription was standardized, and because the integrity of the RNA was good, the abundance of 16S and 23S rRNA transcripts should have been constant in our experiments. This was verified for all the samples that we tested, even for 2-week-old cheeses incubated at 30°C. It has often been stated that rRNA is characteristic of living and active cells. No such conclusion can be drawn from our results, since the abundance of 16S and 23S rRNA remained constant, even in samples in which the culturable cell concentration decreased substantially.
In a previous study, the tuf gene was used as a reference gene for normalization of real-time PCR results for the sulfur amino acid metabolism of L. lactis (19). In the current study the abundance of the tuf transcript was observed to be constant at the beginning of cheese production but to be substantially decreased after 7 h (30°C) or 15 h (12°C). The main function of a reference gene is to normalize for differences in sample extraction and cDNA synthesis efficiencies. Our measurements were normalized using total cellular RNA, which took possible differences in sample extraction efficiency into account, and we checked that the reverse transcription and real-time PCR efficiencies were constant. However, normalization with total RNA would be inappropriate for cheeses in which several microbial species are present. In such a case, one could normalize the results with a reference gene. However, it is likely that, as observed for tuf, the abundance of transcripts from most genes is not constant during cheese production. A careful interpretation of the results would then be required. Another possibility would be to normalize the results with 16S or 23S rRNA from the target species. Indeed, we showed that real-time PCR measurements of L. lactis 16S or 23S rRNA are representative of the total RNA of this species. A drawback of this approach is that real-time PCR analysis of rRNA and mRNA transcripts cannot be performed with the same dilution of the reverse-transcribed RNA due to large differences in abundance. Since the abundance of tuf transcripts decreased together with the concentration of culturable cells during ripening, it is tempting to use the tuf gene as a marker of cell viability. This would require additional validation experiments.
For most genes investigated, the transcript abundance was the same for RNA samples obtained using the method proposed in this study and for samples obtained after separation of the cells from the cheese matrix. Differences were observed mainly for genes in which expression has previously been shown to be modified by heat, acid, or osmotic stresses. It is thus likely that the treatments used for separating the cells from the cheese matrix (mixing in a mechanical blender, centrifugation) activate the transcription of several genes. As a consequence, the extraction method that we developed may be appropriate for the study of the genes involved in stress response. However, since the abundance of transcripts was never lower than the value reported for the method based on separation of cells, there is no evidence that degradation of some transcripts occurs during the separation of cells. It may be interesting to devise an RNA extraction method that is based on separation of cells but in which the transcription of genes is totally inhibited during cell separation steps. The performance of potential transcription inhibitors could be evaluated by comparing the abundance of transcripts from genes such as busAA and glnQ (Table 2) with the abundance obtained with the direct RNA extraction method.
In conclusion, in the present work, RNA was successfully extracted from cheeses manufactured with L. lactis, and rRNA and mRNA transcripts were quantified by real-time PCR. The extraction method could probably be used or adapted for cheeses in which other microbial species are present.
Acknowledgments
This work was carried out with support from a grant from PNRA (French National Agency of Research, GENOFERMENT project).
We thank Charlotte Beltramo (SOREDAB) for designing real-time PCR primers and John Hannon for his critical review of the manuscript.
Footnotes
Published ahead of print on 25 July 2008.
REFERENCES
- 1.Baruzzi, F., A. Matarante, L. Caputo, and M. Morea. 2005. Development of a culture-independent polymerase chain reaction-based assay for the detection of lactobacilli in stretched cheese. J. Rapid Methods Automat. Microbiol. 13:177-192. [Google Scholar]
- 2.Bonaïti, C., S. Parayre, and F. Irlinger. 2006. Novel extraction strategy of ribosomal RNA and genomic DNA from cheese for PCR-based investigations. Int. J. Food Microbiol. 107:171-179. [DOI] [PubMed] [Google Scholar]
- 3.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 4.Coppola, S., G. Blaiotta, D. Ercolini, and G. Moschetti. 2001. Molecular evaluation of microbial diversity occurring in different types of Mozzarella cheese. J. Appl. Microbiol. 90:414-420. [DOI] [PubMed] [Google Scholar]
- 5.Fernandez, M., B. del Rio, D. M. Linares, M. C. Martin, and M. A. Alvarez. 2006. Real-time polymerase chain reaction for quantitative detection of histamine-producing bacteria: use in cheese production. J. Dairy Sci. 89:3763-3769. [DOI] [PubMed] [Google Scholar]
- 6.Fukushima, H., K. Katsube, Y. Hata, R. Kishi, and S. Fujiwara. 2007. Rapid separation and concentration of food-borne pathogens in food samples prior to quantification by viable-cell counting and real-time PCR. Appl. Environ. Microbiol. 73:92-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hannon, J. A., S. M. Deutsch, M. N. Madec, J. Y. Gassi, M. P. Chapot-Chartier, and S. Lortal. 2006. Lysis of starters in UF cheeses: behaviour of mesophilic lactococci and thermophilic lactobacilli. Int. Dairy J. 16:324-334. [Google Scholar]
- 8.Hein, I., A. Lehner, P. Rieck, K. Klein, E. Brandl, and M. Wagner. 2001. Comparison of different approaches to quantify Staphylococcus aureus cells by real-time quantitative PCR and application of this technique for examination of cheese. Appl. Environ. Microbiol. 67:3122-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henri-Dubernet, S., N. Desmasures, and M. Guéguen. 2004. Culture-dependent and culture-independent methods for molecular analysis of the diversity of lactobacilli in “Camembert de Normandie” cheese. Lait 84:179-189. [Google Scholar]
- 10.McKillip, J. L., L. A. Jaykus, and M. A. Drake. 2000. A comparison of methods for the detection of Escherichia coli O157:H7 from artificially-contaminated dairy products using PCR. J. Appl. Microbiol. 89:49-55. [DOI] [PubMed] [Google Scholar]
- 11.Monnet, C., K. Correia, A.-S. Sarthou, and F. Irlinger. 2006. Quantitative detection of Corynebacterium casei in cheese by real-time PCR. Appl. Environ. Microbiol. 72:6972-6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ogier, J.-C., V. Lafarge, V. Girard, A. Rault, V. Maladen, A. Gruss, J.-Y. Leveau, and A. Delacroix-Buchet. 2004. Molecular fingerprinting of dairy microbial ecosystems by use of temporal temperature and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 70:5628-5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ogier, J.-C., O. Son, A. Gruss, P. Tailliez, and A. Delacroix-Buchet. 2002. Identification of the bacterial microflora in dairy products by temporal temperature gradient gel electrophoresis. Appl. Environ. Microbiol. 68:3691-3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pfaffl, M. W. 2004. Quantification strategies in real-time PCR, p. 87-112. In S. A. Bustin (ed.), A-Z of quantitative PCR. International University Line, La Jolla, CA.
- 15.Rademaker, J. L. W., J. D. Hoolwerf, A. A. Wagendorp, and M. C. te Giffel. 2006. Assessment of microbial population dynamics during yoghurt and hard cheese fermentation and ripening by DNA population fingerprinting. Int. Dairy J. 16:457-466. [Google Scholar]
- 16.Randazzo, C. L., S. Torriani, A. D. L. Akkermans, W. M. de Vos, and E. E. Vaughan. 2002. Diversity, dynamics, and activity of bacterial communities during production of an artisanal Sicilian cheese as evaluated by 16S rRNA analysis. Appl. Environ. Microbiol. 68:1882-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rantsiou, K., G. Comi, and L. Cocolin. 2004. The rpoB gene as a target for PCR-DGGE analysis to follow lactic acid bacterial population dynamics during food fermentations. Food Microbiol. 21:481-487. [Google Scholar]
- 18.Rudi, K., K. Naterstad, S. M. Dromtorp, and H. Holo. 2005. Detection of viable and dead Listeria monocytogenes on gouda-like cheeses by real-time PCR. Lett. Appl. Microbiol. 40:301-306. [DOI] [PubMed] [Google Scholar]
- 19.Sperandio, B., P. Polard, D. S. Ehrlich, P. Renault, and E. Guedon. 2005. Sulfur amino acid metabolism and its control in Lactococcus lactis IL1403. J. Bacteriol. 187:3762-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stevens, K. A., and L. A. Jaykus. 2004. Direct detection of bacterial pathogens in representative dairy products using a combined bacterial concentration-PCR approach. J. Appl. Microbiol. 97:1115-1122. [DOI] [PubMed] [Google Scholar]
- 21.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ulvé, V. M., C. Monnet, F. Valence, J. Fauquant, H. Falentin, and S. Lortal. RNA extraction from cheese for analysis of in situ gene expression of Lactococcus lactis. J. Appl. Microbiol., in press. [DOI] [PubMed]
- 23.Wouters, J. A., H. Frenkiel, W. M. de Vos, O. P. Kuipers, and T. Abee. 2001. Cold shock proteins of Lactococcus lactis MG1363 are involved in cryoprotection and in the production of cold-induced proteins. Appl. Environ. Microbiol. 67:5171-5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie, Y., L.-S. Chou, A. Cutler, and B. Weimer. 2004. DNA macroarray profiling of Lactococcus lactis subsp. lactis IL1403 gene expression during environmental stresses. Appl. Environ. Microbiol. 70:6738-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]