Abstract
Flux balance analysis and phenotypic data were used to provide clues to the relationships between the activities of gene products and the phenotypes resulting from the deletion of genes involved in respiratory function in Saccharomyces cerevisiae. The effect of partial or complete respiratory deficiency on the ethanol production and growth characteristics of hap4Δ/hap4Δ, mig1Δ/mig1Δ, qdr3Δ/qdr3Δ, pdr3Δ/pdr3Δ, qcr7Δ/qcr7Δ, cyt1Δ/cyt1Δ, and rip1Δ/rip1Δ mutants grown in microaerated chemostats was investigated. The study provided additional evidence for the importance of the selection of a physiologically relevant objective function, and it may improve quantitative predictions of exchange fluxes, as well as qualitative estimations of changes in intracellular fluxes. Ethanol production was successfully predicted by flux balance analysis in the case of the qdr3Δ/qdr3Δ mutant, with maximization of ethanol production as the objective function, suggesting an additional role for Qdr3p in respiration. The absence of similar changes in estimated intracellular fluxes in the qcr7Δ/qcr7Δ mutant compared to the rip1Δ/rip1Δ and cyt1Δ/cyt1Δ mutants indicated that the effect of the deletion of this subunit of complex III was somehow compensated for. Analysis of predicted flux distributions indicated self-organization of intracellular fluxes to avoid NAD+/NADH imbalance in rip1Δ/rip1Δ and cyt1Δ/cyt1Δ mutants, but not the qcr7Δ/qcr7Δ mutant. The flux through the glycerol efflux channel, Fps1p, was estimated to be zero in all strains under the investigated conditions. This indicates that previous strategies for improving ethanol production, such as the overexpression of the glutamate synthase gene GLT1 in a GDH1 deletion background or deletion of the glycerol efflux channel gene FPS1 and overexpression of GLT1, are unnecessary in a respiration-deficient background.
The quantitative description of biological systems for accurate predictions of genotype-phenotype relationships is a major goal of postgenomic biology. While the completion of genome sequences for many species has triggered a phase of systematic analysis of gene function (21, 28, 29, 39), the Human Genome Project and sequencing of model organisms resulted in the annotation of many genes. Knowledge of the specific functions of many genes at the biochemical level does not, however, enable accurate prediction of the phenotypes resulting from the loss or mutation of any particular gene. This is because of the interdependency of the activities of gene products within complex biological systems. For instance, many proteins catalyze reactions that form part of, or are involved in the regulation of, several pathways. Although genome scale models of the metabolic networks of model organisms, such as Escherichia coli (10) or Saccharomyces cerevisiae (13), have been shown to have remarkable predictive power (9), new methods are still required to reduce the dimensionality of the large parametric space encompassed. Metabolomics is the level of functional genomic analysis that is closest to function, and therefore, a practical approach to overcome system complexity may be the use of metabolic “snapshots” of gene deletion mutants to provide clues to the relationships between the activities of gene products and their resultant phenotypes.
Flux balance analysis (FBA) is used to predict metabolic phenotypes under different conditions, such as substrate and oxygen availability, by simply constraining the appropriate fluxes (15, 20). FBA can be applied to genome scale constraint-based models of the metabolic network to predict a particular flux distribution using linear optimization (5, 20). The predicted growth or by-product secretion rates were found to be consistent with the experimental data in cases where E. coli was grown on acetate or yeast was grown on glucose (9, 11). However, in other cases, FBA predictions may be inconsistent with experimental data, even after adaptation to a particular environment, as in the cases of some E. coli strains bearing deletions in metabolic genes (12). Identification of a physiologically relevant objective function is important, and methods have been developed for constraint-based models to identify such objective functions (7, 34).
There is renewed interest in bioethanol as a fuel, and its production is considered to be a good model system for the optimization of flux through central carbon metabolism. In order to increase the overall conversion yield, several strategies to redirect the flow of carbon going to biomass or glycerol toward ethanol have been adopted. Manipulation of the redox pathways by the deletion of the genes encoding NADPH- and NADH-dependent glutamate dehydrogenases (GDH1 and GDH2, respectively) reduced glycerol production by 30%, while overexpression of both the glutamate synthase (GLT1) and the glutamine synthetase (GLN1) genes in a gdh1 deletion background resulted in a 38% reduction in the glycerol yield and an increase in the ethanol yield under anaerobic conditions (27). Bro et al. (6) have used genome scale models of the metabolic network of S. cerevisiae to evaluate a number of different strategies for metabolic engineering of redox metabolism to decrease glycerol production and increase ethanol yields on glucose under anaerobic conditions. Kong et al. (22) have constructed an ethanol-overproducing S. cerevisiae strain by deleting the gene for a glycerol efflux channel, FPS1, and overexpressing GLT1. Hutter and Oliver (18) suggested that 100% respiration-deficient nuclear petites may be used in the commercial production of ethanol under circumstances in which the oxygen supply cannot be tightly controlled. Indeed, the use of respiration-deficient S. cerevisiae strains as hosts for the construction of recombinant strains increased the ethanol yield from starch (38).
In the present study, FBA and metabolic “snapshots” were used to provide clues to the relationships between the activities of gene products and the resultant phenotypes of partially or completely respiration-deficient deletion strains of S. cerevisiae. Fermentation characteristics and the results of FBA of completely respiration-deficient nuclear petite diploids generated by the deletion of both copies of QCR7, RIP1, or CYT1 were investigated; each of these genes encodes a different subunit of the respiratory chain complex III. The partially respiration-deficient qdr3Δ/qdr3Δ strain was also investigated and compared to the others. The predictive power of FBA was tested on deletion mutants of two genes, MIG1 and HAP4, which encode transcription factors involved in glucose repression and the control of respiration, respectively.
The genome scale metabolic reconstruction and metabolic snapshots were used to model the behavior of the deletion mutants by setting the appropriate objective function. Predicted fluxes were analyzed and compared to elucidate the molecular mechanisms leading to an increase in the ethanol yield. Principal-component analysis (PCA) was used to reveal the relative phenotypic similarity of deletion mutants.
MATERIALS AND METHODS
Strains and media.
Eight deletion strains of S. cerevisiae with the genetic background of BY4743 (MATa/MATΔ his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 lys2Δ0/+ met15Δ0/+ ura3Δ0/ura3Δ0), namely, hoΔ/hoΔ, hap4Δ/hap4Δ, mig1Δ/mig1Δ, qdr3Δ/qdr3Δ, pdr3Δ/pdr3Δ, qcr7Δ/qcr7Δ, cyt1Δ/cyt1Δ, and rip1Δ/rip1Δ strains from the European Saccharomyces cerevisiae Archive for Functional Analysis, were used in this study. The absence of the deleted genes was verified using PCR-based methods.
Precultures were inoculated with a single colony taken from yeast extract-peptone-dextrose (YPD) agar plates and incubated in YPD medium (2% [wt/vol] d-glucose, 2% [wt/vol] peptone, 1% [wt/vol] yeast extract) up to an optical density at 600 nm of 1.2 ± 0.1 at 30°C and 180 rpm in an orbital shaker.
Chemostat cultures.
Microaerated 1.5-liter chemostat cultures, with a dilution rate of 0.1 h−1, were conducted in 3-liter Bioflo3000 New Brunswick fermentors with agitation at 400 rpm, pH 5.5 to 6.5, and the temperature at 30°C. The experiments were performed under low-dissolved-oxygen conditions with no external control of the oxygen supply. A 1% (vol/vol) preculture was used to inoculate the fermentor, and the cells were grown in YPD for 30 h to allow three residence times in the reactor. The culture was demonstrated to be nitrogen starved in its stationary phase. All experiments were carried out in duplicate.
One-milliliter samples taken from the fermentor at regular intervals were centrifuged at 8,000 rpm for 6 min (Eppendorf 5415C; Germany) to determine substrate utilization, extracellular-product formation, and metabolite concentrations.
Quantification of biomass, glucose, and extracellular metabolites.
For dry-weight determination, triplicate samples were collected during the exponential phase of growth. The cell dry weights and the corresponding optical-density values were used to prepare individual calibration curves for the seven strains.
Extracellular glucose, ethanol, and succinate concentrations were determined enzymatically using Boehringer-Mannheim kits, and the concentration of pyruvate was determined using an enzyme kit purchased from Sigma.
The accepted variance in biomass-dry-weight calibrations and metabolite measurements was <10%.
Student's t test.
In order to identify observed significant differences from the wild type, the probabilities associated with Student's t test were calculated using a two-tailed probability distribution and assuming that the two samples had equal variances. P values lower than 0.05 denoted a significant difference between the performance of the mutants and that of the reference strains.
PCA.
PCA was carried out with the PLS Toolbox of MATLAB 7.0 by using the measured metabolites as variables and the strains as the objects, as described by Wold et al. (41).
FBA.
The comprehensive genome scale model described by Förster et al. (13) was employed, using two different objective functions—the maximization of ethanol production and the optimization of oxygen uptake, upper bounded by biomass production. Glucose consumption, biomass, and succinate and pyruvate production fluxes were introduced as constraints. For the three mutants of the cytochrome bc1 complex, the model was modified to constrain the flux through complex III of the respiratory chain to zero. The simulations were carried out with TOMLAB run under MATLAB 7.0. Alternate optima and the related flux distributions were eliminated to obtain a unique set of flux distributions for each strain, using the method of Mahadevan and Schilling (24).
Clustering and functional analysis.
The unique predicted fluxes were clustered using GeneCluster2 (35).
RESULTS
The effects of partial or complete respiratory deficiency on the ethanol production and growth characteristics of eight deletion strains of S. cerevisiae were investigated and compared using microaerated chemostats. The hoΔ/hoΔ deletant was included in order to demonstrate that the insertion of a deletion cassette does not create a difference in respiratory capacity, i.e., there is no marker effect (2). Nuclear petites generated by the deletion of QCR7, RIP1, and CYT1, encoding different subunits of respiratory chain complex III, and a homozygous deletion strain of QDR3 displaying a partial respiratory deficiency, as well as deletion strains of MIG1 and HAP4, encoding proteins which are involved in glucose repression and control of respiration, respectively, were included in this study. QDR3 encodes a multidrug transporter (37), and strains with this gene deleted display partial respiratory deficiency (data not shown); PDR3 encodes a protein whose sole function is drug/chemical transport and regulation of these transport processes (8, 25, 26, 31). The respiration-sufficient mutant pdr3Δ/pdr3Δ was included as a control strain with respect to the partially respiration-deficient qdr3Δ/qdr3Δ mutant.
The fermentation characteristics of the nine strains exhibited either respirofermentative or completely fermentative behavior on glucose. The concentrations of biomass, ethanol, pyruvate, glucose, and succinic acid at steady state were measured and are shown in Table 1, together with the yields of biomass on glucose (Yxs) and of ethanol on glucose (Yes) for each strain (Table 2).
TABLE 1.
Comparison of growth characteristics of S. cerevisiae in chemostats as duplicates
| Strain | Amt (g/liter) (%)a
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Biomass | P | Glucose | P | Pyruvate | P | Ethanol | P | Succinate | P | |
| BY4743 | 2.70 ± 0.22 (100) | 1.66 ± 0.35 (100) | 0.04 ± 0.01 (100) | 6.88 ± 0.33 (100) | 0.15 ± 0.04 (100) | |||||
| hoΔ/hoΔ | 3.03 ± 0.24 (112) | 0.41 ± 0.19 (25) | 0.02 ± 0.01 (50) | 7.23 ± 0.24 (105) | 0.11 ± 0.01 (73) | |||||
| mig1Δ/mig1Δ | 2.04 ± 0.40 (76) | 7.6 × 10−2 | 0.21 ± 0.09 (13) | 2.4 × 10−1 | 0.02 ± 0.00 (50) | 4.9 × 10−1 | 7.75 ± 0.61 (113) | 2.2 × 10−1 | 0.29 ± 0.03 (193) | 1.1 × 10−2 |
| hap4Δ/hap4Δ | 1.37 ± 0.05 (51) | 3.8 × 10−3 | 0.46 ± 0.09 (28) | 4.0 × 10−1 | 0.01 ± 0.00 (25) | 4.4 × 10−1 | 8.97 ± 0.23 (131) | 4.1 × 10−3 | 0.29 ± 0.04 (193) | 4.4 × 10−1 |
| qdr3Δ/qdr3Δ | 2.22 ± 0.16 (82) | 7.1 × 10−2 | 1.39 ± 0.33 (80) | 6.0 × 10−1 | 0.04 ± 0.01 (100) | 6.0 × 10−1 | 8.78 ± 0.50 (127) | 1.5 × 10−2 | 0.19 ± 0.08 (127) | 3.6 × 10−1 |
| pdr3Δ/pdr3Δ | 2.72 ± 0.08 (101) | 5.9 × 10−1 | 0.94 ± 0.21 (57) | 8.8 × 10−1 | 0.04 ± 0.00 (100) | 6.0 × 10−1 | 7.64 ± 1.07 (111) | 4.6 × 10−1 | 0.16 ± 0.02 (107) | 7.7 × 10−1 |
| qcr7Δ/qcr7Δ | 2.49 ± 0.19 (92) | 2.4 × 10−1 | 2.14 ± 0.41 (129) | 1.6 × 10−1 | 0.04 ± 0.00 (100) | 6.0 × 10−1 | 8.07 ± 0.63 (117) | 1.0 × 10−1 | 0.10 ± 0.01 (67) | 3.9 × 10−1 |
| cyt1Δ/cyt1Δ | 1.34 ± 0.02 (50) | 3.4 × 10−3 | 0.16 ± 0.02 (10) | 2.2 × 10−1 | 0.01 ± 0.00 (25) | 4.4 × 10−1 | 9.81 ± 0.61 (143) | 4.4 × 10−3 | 0.06 ± 0.04 (40) | 1.5 × 10−1 |
| rip1Δ/rip1Δ | 0.60 ± 0.01 (22) | 7.7 × 10−4 | 3.11 ± 0.19 (187) | 2.7 × 10−2 | 0.00 ± 0.00 (00) | 3.9 × 10−1 | 8.25 ± 0.91 (120) | 1.3 × 10−1 | 0.16 ± 0.09 (107) | 6.6 × 10−1 |
The percentages are in comparison to the wild-type strain, and the P values indicate the significance of observed differences from the wild type.
TABLE 2.
Comparison of yields of biomass and ethanol on glucose in chemostats as duplicates
| Strain | Yxs (cells/g glucose) | Yes (g/g glucose) |
|---|---|---|
| BY4743 | 0.15 ± 0.02 | 0.38 ± 0.03 |
| hoΔ/hoΔ | 0.15 ± 0.02 | 0.37 ± 0.02 |
| mig1Δ/mig1Δ | 0.10 ± 0.02 | 0.39 ± 0.03 |
| hap4Δ/hap4Δ | 0.07 ± 0.00 | 0.46 ± 0.01 |
| qdr3Δ/qdr3Δ | 0.12 ± 0.01 | 0.47 ± 0.04 |
| pdr3Δ/pdr3Δ | 0.14 ± 0.01 | 0.40 ± 0.05 |
| qcr7Δ/qcr7Δ | 0.14 ± 0.01 | 0.45 ± 0.03 |
| cyt1Δ/cyt1Δ | 0.07 ± 0.00 | 0.49 ± 0.03 |
| rip1Δ/rip1Δ | 0.04 ± 0.01 | 0.49 ± 0.06 |
The growth patterns, steady-state glucose consumption, and ethanol production levels (as well as the yields of biomass and ethanol on glucose) varied considerably for the deletion mutants of S. cerevisiae BY4743 in the microaerated chemostat cultures. For the wild type (BY4743) and the hoΔ/hoΔ deletant, the difference between their yields of ethanol and biomass on glucose was insignificant, with P values of 0.85 and 0.65, respectively.
As expected, the respiration-deficient complex III mutants had reduced biomass yields and increased ethanol yields. However, while the cyt1Δ/cyt1Δ and rip1Δ/rip1Δ mutants had similar yields for ethanol (P = 0.94), the qcr7Δ/qcr7Δ mutant showed higher levels of biomass and lower levels of ethanol than the other two.
Mutants with the regulatory genes deleted, mig1Δ/mig1Δ and hap4Δ/hap4Δ, displayed different fermentation characteristics. The ethanol and biomass yields of the completely respiration-deficient hap4Δ/hap4Δ strain were typical of those for completely respiration-deficient strains, while the respiration-competent mig1Δ/mig1Δ mutant gave values similar to those of the wild type.
The ethanol and biomass production performances of the strains lacking the two drug resistance proteins Pdr3p and Qdr3p were very different from each other. The ethanol yield of the qdr3Δ/qdr3Δ mutant on glucose was 18% higher than that of the pdr3Δ/pdr3Δ mutant, and its biomass yield on glucose was only 86% of that of the pdr3Δ/pdr3Δ mutant. Both the ethanol and biomass production yields of the pdr3Δ/pdr3Δ mutant on glucose were similar to those of the wild-type BY4743 strain. However, the ethanol yield of the qdr3Δ/qdr3Δ mutant was 24% higher than that of the wild type. The difference between the two deletants could also be observed statistically for both the yield of ethanol on glucose (P = 0.04) and the yield of biomass on glucose (P = 0.03).
The phenotypic similarities of the strains included in this study were assessed through PCA. Principal components were determined for a total of nine strains with five measured metabolites, with the first two principal components covering >70% of the variance. BY4743 and the hoΔ/hoΔ mutant, the wild-type and reference strains, as well as the two respiration-deficient mutants of complex III, cyt1Δ/cyt1Δ and rip1Δ/rip1Δ, were separately clustered together, excluding the other respiration-deficient strain of the same complex, qcr7Δ/qcr7Δ. The two mutants lacking genes involved in multiple drug resistance, qdr3Δ/qdr3Δ and pdr3Δ/pdr3Δ, were clearly separated by the PCA, as were the two regulatory mutants, mig1Δ/mig1Δ and hap4Δ/hap4Δ.
FBA.
Variations in the flux distributions of strains having a range of respiratory capacities from respiratory competence to partial or complete respiratory deficiency were investigated by FBA, using a genome scale metabolic model constructed by Förster et al. (13), and were compared in order to gain further insight into their metabolic differences. In our analyses, the measured fluxes were used as equality constraints, and ethanol was kept unconstrained. The distribution of fluxes was predicted for each of the eight deletion mutants, as well as the wild-type strain, under two different objective functions, and changes in fluxes were identified. The optimization of oxygen uptake (upper bounded by biomass production) and the maximization of ethanol production were used as separate objective functions. The first objective function is suitable for respiring strains, while the second is more suitable for the fermenting strains. The alternate optima and the related flux distributions were eliminated to obtain a unique set of flux distributions for each strain.
The genome scale metabolic network modeled the behavior of the organism satisfactorily when the appropriate objective function was set according to the physiology of the organism. When oxygen uptake (upper bounded by biomass) was used as the objective function, the flux of ethanol could be accurately predicted for the hoΔ/hoΔ, BY4743, and pdr3Δ/pdr3Δ strains (with a difference between predicted and experimental results of <9%). The behavior of the respiration-deficient hap4Δ/hap4Δ, cyt1Δ/cyt1Δ, and rip1Δ/rip1Δ strains could be described more accurately when the maximization of ethanol production was selected as the objective function (the difference between the experimental and the predicted ethanol yields was <5%). The predicted ethanol fluxes of the qcr7Δ/qcr7Δ mutant, as well as that of the qdr3Δ/qdr3Δ mutant, were found to be within ca. 8% of the experimentally measured values (Table 3). In the case of the regulatory mig1Δ/mig1Δ mutant, neither objective function could predict its ethanol production to within 10% of the experimental value, indicating that they were unable to account for the function of Mig1p in glucose repression.
TABLE 3.
Comparison of experimental and predicted fluxes, by FBA, for ethanol production using a suitable objective functiona
| Strain | Flux (mol mol glucose−1 h−1)
|
|
|---|---|---|
| Experimental | FBA | |
| BY4743 | 1.47 | 1.59 |
| hoΔ/hoΔ | 1.45 | 1.56 |
| hap4Δ/hap4Δ | 1.80 | 1.82 |
| mig1Δ/mig1Δ | 1.53 | 1.69/1.74b |
| qdr3Δ/qdr3Δ | 1.85 | 1.70 |
| pdr3Δ/pdr3Δ | 1.57 | 1.55 |
| rip1Δ/rip1Δ | 1.91 | 1.92 |
| cyt1Δ/cyt1Δ | 1.93 | 1.85 |
| qcr7Δ/qcr7Δ | 1.77 | 1.64 |
Suitable objective functions were the maximization of ethanol production for hap4Δ/hap4Δ, qdr3Δ/qdr3Δ, rip1Δ/rip1Δ, qcr7Δ/qcr7Δ, and cyt1Δ/cyt1Δ strains and the optimization of oxygen uptake upper bounded by biomass for BY4743, hoΔ/hoΔ, and pdr3Δ/pdr3Δ strains.
The values give the optimization of oxygen uptake upper bounded by biomass as the objective function and the maximization of ethanol production as the objective function, respectively.
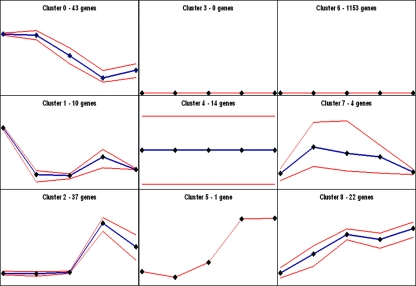
The predicted fluxes through the enzymes that catalyze the reactions in the metabolic pathways included in the genome scale model were clustered to investigate their coresponses to changes in three completely respiration-deficient mutants of complex III (Fig. 1). The five data points represented in the figure correspond to the behavior of fluxes in the wild-type, hoΔ/hoΔ, qcr7Δ/qcr7Δ, cyt1Δ/cyt1Δ, and rip1Δ/rip1Δ strains.
FIG. 1.
Clustering of in silico flux distributions via self-organizing maps (SOM) in wild-type, hoΔ/hoΔ, qcr7Δ/qcr7Δ, cyt1Δ/cyt1Δ, and rip1Δ/rip1Δ strains. The SOM centers of each strain are represented by black diamonds, and the number of fluxes falling into each cluster is stated in the upper middle region of each cell.
Fluxes (43) in cluster 0 (c0) were decreased or eliminated in respiration-deficient strains (see Table S2 in the supplemental material). The members of the COX gene family forming complex IV and ATP1, which encodes the alpha subunit of the FoF1 ATPase complex, were completely inactive in the rip1Δ/rip1Δ and cyt1Δ/cyt1Δ mutants, and a decrease in these fluxes was observed in the qcr7Δ/qcr7Δ mutant.
The reactions through enzymes Sdh3p and Fum1p and the uncharacterized product of the open reading frame YEL047c of complex II were inactivated in all complex III mutants, suggesting that disruption of complex III not only renders the downstream reactions in the respiratory chain zero, but also affects the upstream processes involving complex II. Transport of pyruvate across the mitochondrial membrane was eliminated in rip1Δ/rip1Δ mutants and reduced in cyt1Δ/cyt1Δ and qcr7Δ/qcr7Δ deletants.
The fluxes through the gene products of GDH3, YDR11c, SER1, SER2, and SER3 (c0) were eliminated in rip1Δ/rip1Δ and cyt1Δ/cyt1Δ strains, whereas they remained unchanged in the qcr7Δ/qcr7Δ mutant. The fluxes through Aac1p (transport of mitochondrial ATP to the cytoplasm and of molecular oxygen to the mitochondria) were also zero in these two respiration-deficient strains. These transport fluxes were predicted to decrease in qcr7Δ/qcr7Δ deletants.
The fluxes catalyzed by the products of the genes GPM2 and ERR1 (both glycolysis/gluconeogenesis), MDH1 (a complex II member), NDI1 (oxidative phosphorylation), PHO84 (plasma membrane transport), and MIR1 and DIC1 (mitochondrial-membrane transporters) were clustered together (c1) and were predicted to decrease in all deletion strains in comparison to BY4743.
The 37 fluxes clustered together in c2 were shown to display similar changes in wild-type, hoΔ/hoΔ, and qcr7Δ/qcr7Δ strains but were elevated in rip1Δ/rip1Δ and cyt1Δ/cyt1Δ deletants. Fluxes through Gln1p and Glt1p were found to be increased in rip1Δ/rip1Δ and cyt1Δ/cyt1Δ mutants, whereas they remained unchanged in qcr7Δ/qcr7Δ deletants. The flux through Acs1p, which is involved in pyruvate metabolism, was increased only in rip1Δ/rip1Δ and cyt1Δ/cyt1Δ deletants.
Fluxes through c2 reactions, involved in arginine metabolism and catalyzed by Cpa2p, Arg1p, Arg3p, Arg4p, and Car1p, were increased only in rip1Δ/rip1Δ mutants but remained unchanged in cyt1Δ/cyt1Δ and qcr7Δ/qcr7Δ deletants. These proteins catalyze the production of glutamine, arginosuccinate, citrate, fumarate, and urea, respectively. Reactions involving several members of the alanine and aspartate, glycine, serine, and threonine pathways (Aat2p, Yat1p, Cat2p, Gcv1p, Hom2p, Hom3p and Hom6p, and Thr1p and Thr4p) were elevated in two of the deletion mutants of complex III, rip1Δ/rip1Δ and cyt1Δ/cyt1Δ, but not in the qcr7Δ/qcr7Δ mutant.
The 14 genes in c4 displayed no change in fluxes in any of the strains.
c6 contained 1,153 enzymes through which either the flux value was zero or no change was observed between the strains. Gene products involved in glycerol/glycerolipid metabolism, NADPH-dependent glutamate dehydrogenase (Gdh1p), and NADH-dependent glutamate synthase (Gdh2p) were clustered there. The reactions catalyzed by these gene products have been predicted to have zero fluxes in all of the strains included in this study. The flux through the channel Fps1p, which is responsible for changes in glycerol permeability during osmotic adaptation, was also predicted to be zero in all strains under the conditions used in this study.
Four enzymes through which fluxes displayed different behaviors in wild-type and deletion strains constitute c7. The flux through Ndh2p, a mitochondrial NADH dehydrogenase, was slightly increased in cyt1Δ/cyt1Δ and qcr7Δ/qcr7Δ mutants, whereas it remained zero in the rip1Δ/rip1Δ mutant. Mitochondrial transport of glutamate was rendered zero in cyt1Δ/cyt1Δ and qcr7Δ/qcr7Δ mutants but remained unchanged in rip1Δ/rip1Δ deletants.
c8 comprises 22 enzymes. The fluxes through the enzymes involved in glycolysis/gluconeogenesis, Tdh1p, Gpm1p, and Eno1p, together with Pgm1p, involved in the pentose-phosphate cycle, increased in all respiration-deficient strains. The fluxes through Lsc2p of the tricarboxylic acid (TCA) cycle and Adh5p, which catalyze the conversion of ethanol to acetaldehyde, were eliminated in all these respiration-deficient strains. The flux through the reaction catalyzed by Shm2p, converting serine into glycine, switched its direction and was reduced 10-fold in the absence of Rip1p and Cyt1p, but not in the absence of Qcr7p.
DISCUSSION
We used the genome scale metabolic network of S. cerevisiae to predict in vivo cell behavior in order to evaluate its usefulness for the rational design of yeast strains for high-level ethanol production. The ethanol production performances of three nuclear petites of S. cerevisiae resulting from the deletion of different components of the cytochrome bc1 complex and the deletion mutants of two key regulatory genes mediating glucose repression (hap4Δ/hap4Δ and mig1Δ/mig1Δ) were investigated, together with a deletion mutant of a drug resistance gene, qdr3Δ/qdr3Δ. Deletion of QDR3 results in partial respiratory deficiency, so the respiration-sufficient deletion mutant of another drug resistance gene, PDR3, was included to control for any correlation between multidrug resistance and ethanol productivity.
Our studies confirmed that the BY4743 and hoΔ/hoΔ strains can safely be used interchangeably as control strains, because they gave very similar ethanol and biomass yields. This is in agreement with our previous findings that the ho::kanMX4/ho::kanMX deletion had only a small, but measurable, effect on the growth rate under aerobic conditions (2) and no measurable effect on the metabolome (30, 32).
The hap4Δ/hap4Δ, rip1Δ/rip1Δ, and cyt1Δ/cyt1Δ deletants all displayed reduced biomass yields, with that of the rip1Δ/rip1Δ mutant the lowest, followed by those of the cyt1Δ/cyt1Δ and hap4Δ/hap4Δ mutants. The maximum ethanol yield was observed in the cyt1Δ/cyt1Δ and rip1Δ/rip1Δ mutants, followed by the qdr3Δ/qdr3Δ and hap4Δ/hap4Δ strains. The observation of an increased ethanol production capacity, 21% higher than that of the wild type, in the case of the hap4 deletant was in good agreement with previous studies. In contrast, a strain overexpressing HAP4 has been reported to display a 17% reduction in ethanol production and a 10% increase in biomass production compared to the wild type (33, 40). These results indicate that the manipulation of the regulators of respiration may also lead to an improvement in ethanol production. Experiments in other laboratories (17, 33) have demonstrated that higher ethanol yields are obtained in batch than in chemostat culture. Our own batch experiments (not shown) demonstrated that Yes is equal to 0.44 g/g glucose for the wild type, 0.49 g/g glucose for the qdr3Δ/qdr3Δ mutant, and 0.48 g/g glucose for the qcr7Δ/qcr7Δ mutant.
PCA revealed that the cyt1Δ/cyt1Δ and rip1Δ/rip1Δ deletants were clustered together, confirming that their corresponding proteins are functionally linked. However, the PCA separated these two deletion mutants from that for the gene encoding a third member of the cytochrome bc1 complex, qcr7Δ/qcr7Δ. This result suggests that the requirement for Qcr7p in respiration may be bypassed or that it has a distinct mechanism of action, regulation, or assembly.
The mig1Δ/mig1Δ and hap4Δ/hap4Δ strains were located far from the other strains in the PCA plot and quite far from each other, although both were found in the same quadrant. This result is consistent with the fact that Mig1p and Hap4p are transcription factors with distinct, but overlapping, roles (3, 14, 23, 33). The pdr3Δ/pdr3Δ and qdr3Δ/qdr3Δ mutants were positioned far from each other in the PCA projection. This indicates that Qdr3p may play a role in respiration that is quite distinct from its activity in multidrug resistance, which it shares with Pdr3p. The elucidation of the respiratory role of Qdr3p merits further investigation.
Flux distributions, calculated using whole-genome models with the optimization of oxygen uptake upper bounded by biomass as the objective function, led to successful predictions of ethanol production in respiration-sufficient strains. The experimental ethanol flux of 1.47 mol/mol glucose for the wild-type strain, BY4743, and the computed flux of 1.59 mol/mol glucose are comparable to the value of 1.59 mol/mol glucose reported by Gombert et al. (16). When maximization of ethanol production was used as the objective function, better predictions were obtained for the respiration-deficient strains, with almost no difference between the experimental and computational values in the case of the rip1Δ/rip1Δ mutant. The respiration-deficient deletion mutant of the regulatory gene HAP4 showed similar behavior through FBA using the same objective function; predictions, again, were within 10% of the measured values. Furthermore, ethanol production was successfully predicted by FBA in the case of the qdr3Δ/qdr3Δ mutant when maximization of ethanol production was chosen as the objective function. This result supports experimental findings suggesting a role for Qdr3p in respiration, in addition to drug resistance.
Ethanol production could not be predicted accurately for the mig1Δ/mig1Δ deletant. This may be because the metabolic model did not include regulatory pathways, such as glucose repression, or because neither of the chosen objective functions reflects the physiological impact of this regulatory gene properly. FBA could not reveal the effect of the deletion of PDR3 either, since we were unable to devise objective functions suitable for describing drug transport, chemical resistance, and its regulatory function.
A reconstructed genome scale model and a metabolic snapshot were used to predict metabolic fluxes in strains carrying deletions of genes encoding components of respiratory complex III in S. cerevisiae. Estimated changes in metabolic fluxes were used to evaluate, at least qualitatively, the impact on S. cerevisiae grown under microaerobic conditions in a chemostat.
The fluxes through the enzymes catalyzing the different steps in glycolysis and gluconeogenesis (c0, c1, c2, and c8) were, except for Pda1p, affected similarly in all respiration-deficient strains. The flux through Pda1p, which has a role in mitochondrial conversion of pyruvate to acetyl-coenzyme A (CoA), was predicted to be zero in rip1Δ/rip1Δ and cyt1Δ/cyt1Δ mutants but remained unchanged in qcr7Δ/qcr7Δ deletants.
The fluxes through reactions catalyzed by Idp1p, Kgd1p, Sdh3p, the protein encoded by YEL047c, Fum1p, and Lsc2p in the TCA cycle were eliminated in all completely respiration-deficient strains. In rip1Δ/rip1Δ and cyt1Δ/cyt1Δ mutants, the flux through mitochondrial Cit1p was lost while that through cytoplasmic Cit2p increased. The same fluxes remained unchanged in the qcr7Δ/qcr7Δ mutant. It has been reported that cells lacking a mitochondrial genome trigger expression of CIT2, which encodes the cytoplasmic citrate synthase, in order to maintain the levels of TCA cycle intermediates to ensure the synthesis of glutamate (19). Deletion of both copies of QCR7 seemed to be sufficient to cause respiratory deficiency but may not have been sufficient to induce retrograde regulation of Cit2p levels (36). Therefore, these results indicate the functional disruption of mitochondria in rip1Δ/rip1Δ and cyt1Δ/cyt1Δ mutants, but not in qcr7Δ/qcr7Δ deletants.
It has also been reported recently that a different set of genes of the PDR pathway, involved in multidrug resistance in S. cerevisiae, is strongly induced in the absence of mitochondrial function. The fluxes through the mitochondrial Cit1p disappeared in the absence of either Qdr3p or Pdr3p, but that through the cytoplasmic Cit2p remained unchanged in both strains, as was the case for the qcr7Δ/qcr7Δ mutant. Partial respiratory deficiency caused by the loss of QDR3 may also be related to mitochondrial dysfunction in this strain. However, the role of Qdr3p in mitochondrial function or the coordination of the mitochondria and cytoplasm remains to be elucidated.
The fluxes through the steps catalyzed by Zwf1p, Tkl1p, and Gnd2p of the pentose phosphate pathway were also reduced in rip1Δ/rip1Δ and cyt1Δ/cyt1Δ mutants but remained unchanged in qcr7Δ/qcr7Δ deletants (4, 14). However, extracellular pyruvate levels were found to decrease dramatically in all strains with elevated ethanol production, namely, rip1Δ/rip1Δ, cyt1Δ/cyt1Δ, and hap4Δ/hap4Δ mutants, indicating a rapid conversion of pyruvate into other metabolites before it could be released. Indeed, the reactions catalyzed by Pdc6p, which mediates the conversion of pyruvate to acetaldehyde, were predicted to be elevated in all respiration-deficient strains. The flux through Acs1p leading to the production of acetyl-CoA from CoA, in the same pathway, was increased in rip1Δ/rip1Δ and cyt1Δ/cyt1Δ mutants. The flux through the anapleurotic reaction catalyzed by Pyc1p, converting pyruvate and CO2 into oxaloacetate in the cytoplasm, was also elevated in rip1Δ/rip1Δ and cyt1Δ/cyt1Δ mutants but remained unchanged in qcr7Δ/qcr7Δ deletants.
The reactions catalyzed by Gdh3p and Gln1p had zero and increased fluxes, respectively, in rip1Δ/rip1Δ and cyt1Δ/cyt1Δ deletants but were unchanged in qcr7Δ/qcr7Δ mutants. Fluxes through Glt1p were found to be elevated in rip1Δ/rip1Δ and cyt1Δ/cyt1Δ strains, whereas they remained unchanged in qcr7Δ/qcr7Δ mutants. Reactions catalyzed by the NADPH-dependent glutamate dehydrogenase, encoded by GDH1, and the NADH-dependent glutamate synthase, encoded by GDH2, have been predicted to have zero fluxes in all strains under the investigated conditions. It has been reported that the overexpression of both the glutamate synthase (Glt1p) and the glutamine synthetase (Gln1p) genes in the gdh1 deletion background resulted in a 38% reduction in the glycerol yield and an increase in the ethanol yield under anaerobic conditions (27). Therefore, the predicted flux distributions indicate that intracellular fluxes were reorganized to avoid imbalance in NAD+/NADH in the two respiration-deficient strains, rip1Δ/rip1Δ and cyt1Δ/cyt1Δ, but not in the qcr7Δ/qcr7Δ deletant.
The flux through the glycerol efflux channel Fps1p was estimated to be zero in all strains. Deletion of the FPS1 gene and overexpression of GLT1 in S. cerevisiae were also reported to be associated with high ethanol production (22). Therefore, we may conclude that further genetic manipulations, such as the overexpression of the glutamate synthase gene GLT1 in a gdh1 deletion background or deletion of the glycerol efflux channel gene FPS1 and overexpression of GLT1, may not provide improvements in the ethanol production properties of rip1Δ/rip1Δ and cyt1Δ/cyt1Δ mutants.
In this study, we demonstrated that genome scale metabolic networks may be useful tools in the rational design of S. cerevisiae strains for increased ethanol production. This study has also provided additional evidence for the importance of the selection of an objective function in FBA. The selection of a physiologically relevant objective function may improve the quantitative predictions of exchange fluxes, as well as qualitative estimates of changes in intracellular fluxes. Differences in both the performance and the estimated intracellular fluxes between qcr7Δ/qcr7Δ mutants and those of rip1Δ/rip1Δ and cyt1Δ/cyt1Δ deletants indicated that the effect of the loss of the Qcr7 subunit of complex III may somehow be compensated for. The molecular mechanism underlying this phenomenon requires further examination. Elucidation of the molecular mechanism of regulatory elements, such as Mig1p, and the incorporation of the regulatory information into genome-wide models through the integration of transcriptome data, as proposed by Akesson et al. (1), may improve the predictions and contribute to the rational design of strains with higher ethanol yields.
Supplementary Material
Acknowledgments
This work was financially supported by the Bogaziçi University Research Fund through projects 04A503 and 04HA503D and by the State Planning Organization of the Turkish Government through project DPT-03K120250. The Ph.D. studies of Duygu Dikicioglu are supported by BDP-TÜBITAK. Work on systems biology in the laboratory of S.G.O. is supported by BBSRC.
Footnotes
Published ahead of print on 27 June 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Akesson, M., J. Forster, and J. Nielsen. 2004. Integration of gene expression data into genome-scale metabolic models. Metab. Eng. 6:285-293. [DOI] [PubMed] [Google Scholar]
- 2.Baganz, F., A. Hayes, D. Marren, D. C. J. Gardner, and S. G. Oliver. 1997. Suitability of replacement markers for functional analysis studies in Saccharomyces cerevisiae. Yeast 13:1563-1573. [DOI] [PubMed] [Google Scholar]
- 3.Blom, J., M. J. Teixeira de Mattos, and L. A. Grivell. 2000. Redirection of the respiro-fermentative flux distribution in Saccharomyces cerevisiae by overexpression of the transcription factor Hap4p. Appl. Environ. Microbiol. 66:1970-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boles, E., and F. K. Zimmerman. 1993. Induction of pyruvate decarboxylase in glycolysis mutants of Saccharomyces cerevisiae correlates with the concentrations of 3-carbon glycolytic intermediates. Arch. Microbiol. 160:324-328. [DOI] [PubMed] [Google Scholar]
- 5.Bonarius, H. P. J., G. Schmid, and J. Tramper. 1997. Flux analysis of underdetermined metabolic networks: the quest for the missing constraints. Trends Biotechnol. 15:308-314. [Google Scholar]
- 6.Bro, C., B. Regenberg, J. Forster, and J. Nielsen. 2006. In silico aided metabolic engineering of Saccharomyces cerevisiae for improved bioethanol production. Metab. Eng. 8:102-111. [DOI] [PubMed] [Google Scholar]
- 7.Burgard, A. P., and C. D. Maranas. 2003. Optimization-based framework for inferring and testing hypothesized metabolic objective functions. Biotechnol. Bioeng. 82:670-677. [DOI] [PubMed] [Google Scholar]
- 8.Cernicka, J., Z. Kozovska, M. Hnatova, M. Valachovic, I. Hapala, Z. Riedl, G. Hajos, and J. Subik. 2007. Chemosensitisation of drug-resistant and drug-sensitive yeast cells to antifungals. Int. J. Antimicrob. Agents 29:170-178. [DOI] [PubMed] [Google Scholar]
- 9.Edwards, J. S., R. U. Ibarra, and B. O. Palsson. 2001. In silico predictions of Escherichia coli metabolic capabilities are consistent with experimental data. Nat. Biotechnol. 19:125-130. [DOI] [PubMed] [Google Scholar]
- 10.Edwards, J. S., and B. O. Palsson. 2000. The Escherichia coli MG1655 in silico metabolic genotype: Its definition, characteristics, and capabilities. Proc. Natl. Acad. Sci. USA 97:5528-5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Famili, I., J. Forster, J. Nielsen, and B. O. Palsson. 2003. Saccharomyces cerevisiae phenotypes can be predicted by using constraint-based analysis of a genome-scale reconstructed metabolic network. Proc. Natl. Acad. Sci. USA 100:13134-13139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fong, S. S., A. Nanchen, B. O. Palsson, and U. Sauer. 2006. Latent pathway activation and increased pathway capacity enable Escherichia coli adaptation to loss of key metabolic enzymes. J. Biol. Chem. 281:8024-8033. [DOI] [PubMed] [Google Scholar]
- 13.Förster, J., I. Famili, P. Fu, B. O. Palsson, and J. Nielsen. 2003. Genome-scale reconstruction of the Saccharomyces cerevsiae metabolic network. Genome Res. 13:244-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gancedo, J. M. 1998. Yeast carbon catabolite depression. Microbiol. Mol. Biol. Rev. 62:334-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gombert, A. K., and J. Nielsen. 2000. Mathematical modeling of metabolism. Curr. Opin. Biotechnol. 11:180-186. [DOI] [PubMed] [Google Scholar]
- 16.Gombert, A. K., M. Moreira dos Santos, B. Christensen, and J. Nielsen. 2001. Network identification and flux quantification in the central metabolism of Saccharomyces cerevisiae under different conditions of glucose repression. J. Bacteriol. 183:1441-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heux, S., J. M. Sablayrolles, R. Cachon, and S. Dequin. 2006. Engineering a Saccharomyces cerevisiae wine yeast that exhibits reduced ethanol production during fermentation under controlled microoxygenation conditions. Appl. Environ. Microbiol. 72:5822-5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hutter, A., and S. G. Oliver. 1998. Ethanol production using nuclear petite yeast mutants. Appl. Microbiol. Biotechnol. 49:511-516. [DOI] [PubMed] [Google Scholar]
- 19.Jia, Y. K., A. M. Becam, and C. J. Herbert. 1997. The CIT3 gene of Saccharomyces cerevisiae encodes a second mitochondrial isoform of citrate synthase. Mol. Microbiol. 24:53-59. [DOI] [PubMed] [Google Scholar]
- 20.Kauffman, K. J., P. Prakash, and J. S. Edwards. 2003. Advances in flux balance analysis. Curr. Opin. Biotechnol. 14:491-496. [DOI] [PubMed] [Google Scholar]
- 21.Kemmeren, P., T. T. J. P. Kockelkorn, T. Bijma, R. Donders, and F. C. P. Holstege. 2004. Predicting gene function through systematic analysis and quality assessment of high-throughput data. Bioinformatics 21:1644-1652. [DOI] [PubMed] [Google Scholar]
- 22.Kong, Q. X., J. G. Gu, L. M. Cao, A. L. Zhang, X. Chen, and X. Zhao. 2006. Improved production of ethanol by deleting FPS1 and over-expressing GLT1 in Saccharomyces cerevisiae. Biotechnol. Lett. 28:2033-2038. [DOI] [PubMed] [Google Scholar]
- 23.Lascaris, R., J. Piwowarsk, H. van der Spek, J. Teixeira de Mattos, L. Grivell, and J. Blom. 2004. Overexpression of HAP4 in glucose-derepressed yeast cells reveals respiratory control of glucose-regulated genes. Microbiology 150:929-934. [DOI] [PubMed] [Google Scholar]
- 24.Mahadevan, R., and C. H. Schilling. 2003. The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab. Eng. 5:264-276. [DOI] [PubMed] [Google Scholar]
- 25.Mamnun, Y. M., C. Schuller, and K. Kuchler. 2004. Expression regulation of the yeast PDR5 ATP-binding cassette (ABC) transporter suggests a role in cellular detoxification during the exponential growth phase. FEBS Lett. 554:111-117. [DOI] [PubMed] [Google Scholar]
- 26.Moye-Rowley, M. S. 2005. Retrograde regulation of multidrug resistance in Saccharomyces cerevisiae. Gene 354:15-21. [DOI] [PubMed] [Google Scholar]
- 27.Nissen, T. L., M. C. Kielland-Brandt, J. Nielsen, and J. Villadsen. 2000. Optimization of ethanol production in Saccharomyces cerevisiae by metabolic engineering of the ammonium assimilation. Metab. Eng. 2:69-77. [DOI] [PubMed] [Google Scholar]
- 28.Oliver, S. G. 1996. From DNA sequence to biological function. Nature 379:597-600. [DOI] [PubMed] [Google Scholar]
- 29.Oliver, S. G. 1996. A network approach to the systematic analysis of yeast gene function. Trends Genet. 12:241-242. [DOI] [PubMed] [Google Scholar]
- 30.Oliver, S. G., M. K. Wilson, D. B. Kell, and F. Baganz. 1998. Systematic functional analysis of the yeast genome. Trends Biotechnol. 16:373-378. [DOI] [PubMed] [Google Scholar]
- 31.Onda, M., K. Ota, T. Chiba, Y. Sakaki, and I. Takashi. 2004. Analysis of gene network regulating yeast multidrug resistance by artificial activation of transcription factors: involvement of Pdr3 in salt tolerance. Gene 332:51-59. [DOI] [PubMed] [Google Scholar]
- 32.Raamsdonk, L. M., B. Teusink, D. Broadhurst, N. Zhang, A. Hayes, M. C. Walsh, J. A. Berden, K. M. Brindle, D. B. Kell, J. J. Rowland, H. V. Westerhoff, K. van Dam, and S. G. Oliver. 2001. A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations. Nat. Biotechnol. 19:45-50. [DOI] [PubMed] [Google Scholar]
- 33.Raghevendran, V., K. R. Patil, L. Olsson, and J. Nielsen. 2006. Hap4 is not essential for activation of respiration at low specific growth rates in Saccharomyces cerevisiae. J. Biol. Chem. 281:12308-12314. [DOI] [PubMed] [Google Scholar]
- 34.Raghunathan, A. U., J. R. Perez-Correa, and L. T. Bieger. 2003. Data reconciliation and parameter estimation in flux-balance analysis. Biotechnol. Bioeng. 84:700-709. [DOI] [PubMed] [Google Scholar]
- 35.Reich, M., K. Ohm, P. Tamayo, M. Angelo, and J. P. Mesirov. 2004. GeneCluster 2.0: an advanced toolset for bioarray analysis. Bioinformatics 20:1797-1798. [DOI] [PubMed] [Google Scholar]
- 36.Small, W. C., R. D. Brodeur, A. Sandor, N. Fedorova, G. Li, R. A. Butow, and P. A. Srere. 1995. Enzymatic and metabolic studies on retrograde regulation mutants of yeast. Biochemistry 34:5569-5576. [DOI] [PubMed] [Google Scholar]
- 37.Tenreiro, S., R. Vargas, M. C. Teixeira, C. Magnani, and I. Sa-Correia. 2005. The yeast multidrug transporter Qdr3 (Ybr043c): localization and role as a determinant of resistance to quinidine, barban, cisplatin, and bleomycin. Biochem. Biophys. Res. Commun. 327:952-959. [DOI] [PubMed] [Google Scholar]
- 38.Toksoy-Oner, E., S. G. Oliver, and B. Kirdar. 2005. Production of ethanol from starch by respiration-deficient recombinant Saccharomyces cerevisiae. Appl. Environ. Microbiol. 71:6443-6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tong, A. H. Y., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader, N. Page, M. Robinson, S. Raghibizadeh, C. W. V. Hogue, H. Bussey, B. Andrews, M. Tyers, and C. Boone. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294:2364. [DOI] [PubMed] [Google Scholar]
- 40.van Maris, A. J. A., B. M. Bakker, M. Brandt, A. Boorsma, M. J. T. de Mattos, L. A. Grivell, J. T. Pronk, and J. Blom. 2001. Modulating the distribution of fluxes among respiration and fermentation by overepression of HAP4 in Saccharomyces cerevisiae. FEMS Yeast Res. 1:139-149. [DOI] [PubMed] [Google Scholar]
- 41.Wold, S., K. Esbensen, and P. Geladi. 1987. Principal component analysis. Chemometrics Intell. Lab. Syst. 2:37-52. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.