Abstract
Although Pseudomonas aeruginosa is an opportunistic pathogen that does not often naturally infect alternate hosts, such as plants, the plant-P. aeruginosa model has become a widely recognized system for identifying new virulence determinants and studying the pathogenesis of the organism. Here, we examine how both host factors and P. aeruginosa PAO1 gene expression are affected in planta after infiltration into incompatible and compatible cultivars of tobacco (Nicotiana tabacum L.). N. tabacum has a resistance gene (N) against tobacco mosaic virus, and although resistance to PAO1 infection is correlated with the presence of a dominant N gene, our data suggest that it is not a factor in resistance against PAO1. We did observe that the resistant tobacco cultivar had higher basal levels of salicylic acid and a stronger salicylic acid response upon infiltration of PAO1. Salicylic acid acts as a signal to activate defense responses in plants, limiting the spread of the pathogen and preventing access to nutrients. It has also been shown to have direct virulence-modulating effects on P. aeruginosa. We also examined host effects on the pathogen by analyzing global gene expression profiles of bacteria removed from the intracellular fluid of the two plant hosts. We discovered that the availability of micronutrients, particularly sulfate and phosphates, is important for in planta pathogenesis and that the amounts of these nutrients made available to the bacteria may in turn have an effect on virulence gene expression. Indeed, there are several reports suggesting that P. aeruginosa virulence is influenced in mammalian hosts by the availability of micronutrients, such as iron and nitrogen, and by levels of O2.
There are many commonalities between the ways in which pathogens of plants and mammals infect their hosts and between the ways in which these hosts recognize and respond to pathogen attack. From the perspective of pathogen infection, the use of type III secretion systems (TTSS) to deliver effector molecules directly into host cells is one of the most notable similarities between plant and animal pathogens (4). On the host side, many of the mechanisms that plants and mammals use to identify and respond to these bacterial effectors are also similar (15, 35). For instance, the identification of pathogen-associated molecular patterns by pathogen recognition receptors in the host that activate common signaling pathways and induce core defense responses is conserved (18). Another parallel response is the production of reactive oxygen species and localized programmed cell death in response to pathogen attack. However, many of the signaling cascades and biosynthetic pathways leading to these responses differ or are only partially conserved (13, 22).
In the mid-1990s, it was established that plants could serve as useful alternatives to mammalian models for studying the pathogenesis of the human opportunistic pathogen Pseudomonas aeruginosa (27, 28, 38). These studies revealed that many overlapping factors are required for virulence in mammal and plant infections, providing the basis for an efficient, high-throughput screen for identifying new virulence factors. This plant-human pathogen model has been extended to include other human pathogens, such as Enterococcus faecalis (16) and Staphylococcus aureus (25). In nature, however, P. aeruginosa rarely infects plants despite the fact that it is a ubiquitous soil bacterium. Thus, we reasoned that by examining the global transcriptional response of P. aeruginosa in both compatible and incompatible plant models we could gain insight into what factors are necessary for successful host colonization and whether host defense responses may suppress these factors.
In recent years, improved molecular techniques have been applied to study the factors that determine successful colonization and infection by various pathogens in vivo. Plant-induced genes in Erwinia chrysanthemi were identified by comparing the global gene expression of bacteria recovered from a host plant to that of bacteria grown in culture medium (23). Global transcription changes in P. aeruginosa have been examined in the presence of human airway epithelial cells and in response to root exudates of Beta vulgaris L. and poplar rhizosphere (1, 11, 20). However, we are aware of only one published study that has examined the global gene expression of this bacterium growing in an intact host. Examination of P. aeruginosa isolated from the lungs of a naturally infected patient with cystic fibrosis provided important insights into in vivo expression of virulence factors, drug resistance, and nutrient acquisition and utilization (34). Here, we have assessed how basal defense responses and nutrient availability in a plant host affects P. aeruginosa global gene expression 24 h postinfiltration in two Nicotiana tabacum cultivars (Xanthi nc [NN] and Samsun nn) that differ in their susceptibilities to infection by the pathogen. By examining the interaction between a plant host and a human pathogen, we hope to gain new insights into how these pathogens behave in a human host.
MATERIALS AND METHODS
Plant material and bacterial strains.
Arabidopsis thaliana Col-0 seeds were obtained from Lehle Seed (Roundrock, TX), and A. thaliana Ag-O seeds were provided by Lawrence Rahme. All cultivars, transformants, and mutants of N. tabacum (Samsun nn, Xanthi nc [NN], Petite Havana SR-1::nn, pTG38, D46H, and G216E) were provided by Barbara Baker (University of California, Berkeley) and propagated in greenhouses at Colorado State University. P. aeruginosa PAO1 (selected for rifampin resistance) was generously provided by E. Peter Greenberg (University of Washington). Cultures were routinely grown in Luria-Bertani (LB) broth in a 37°C rotary shaker at 200 rpm, and rifampin (50 μg ml−1) was used for antibiotic selection for plate counts.
Determination of bacterial pathogenesis in N. tabacum plants.
Overnight cultures of PAO1 were diluted to 1 × 107 cells ml−1 in 10 mM MgSO4. Five hundred microliters of diluted culture was infiltrated into three leaves on each treated plant (the resulting initial inoculum was 5 × 106 cells leaf−1). The plants (N. tabacum cv. Samsun nn, Xanthi nc [NN], Petite Havana SR-1::nn, pTG38, D46H, and G216E) were injected with 500 μl of 10 mM MgSO4 per leaf for controls. The plants were incubated in temperature-controlled chambers at 28 to 30°C and high humidity. CFU were determined at 24-hour time points for 5 days by removing infected leaves and infiltrating them to saturation with sterile distilled H2O (dH2O). The saturated leaves were placed in a cotton-filtered syringe positioned in a centrifuge tube and spun at 5,000 rpm for 10 min to remove intracellular fluid. The amounts of intracellular fluid recovered from each leaf were adjusted to 1 ml with sterile dH2O, serially diluted, and drop-plated on LB solid medium containing 50 μg ml−1 of rifampin. The cultures were incubated at 37°C, and colonies were counted after 24 h. Three leaves from treated and control plants were plated for each time point, and each experiment was conducted at least two times. The figures represent average data from all experiments, and the error bars represent ±1 standard error (SE).
Determination of total SA content.
Total salicylic acid (SA) levels were measured for the tobacco cultivars (Xanthi and Samsun) using a modified version of previously described protocols (2, 24, 31). Briefly, The plants were infiltrated as described above and incubated overnight at 28 to 30°C. Approximately 1 g of fresh tissue per repetition was removed and stored at −80°C prior to extraction. The tissue was ground in liquid nitrogen and extracted at 5°C in 80% methanol. Samples were sonicated and centrifuged for 20 min at 13,000 rpm in a tabletop centrifuge. The supernatant was removed, and the pellet was reextracted as described above. The two supernatants were combined and evaporated to dryness. The sample was resuspended in 2.5 ml of 5% trichloroacetic acid and sonicated for 10 min. Free SA was separated from conjugated forms by extraction with 2 volumes of ethyl acetate-cyclopentane-isopropanol (50:50:1). The organic phase was evaporated to dryness and resuspended in 0.5 ml of MeOH. The aqueous phase was acidified to pH 1 with 1 N HCl, boiled for 30 min to release conjugated SA, and reextracted as described above. Samples were analyzed by high-performance liquid chromatography on a Dionex Acclaim Polar Advantage II C18 column (5 μm; 120Å; 4.6 by 150 mm) and separated using a 10 to 95% gradient of methanol-4 mM formic acid over 15 min at a flow rate of 0.5 ml min−1. Samples were quantified by comparison of peak areas using an SA standard (Sigma, St. Louis, MO).
RNA extraction and microarray analysis.
Infections and bacterial recovery for microarray analysis were carried out as described for the infection assays with N. tabacum, except the leaves were infiltrated to saturation. PAO1 was removed from the intracellular fluid of infected leaves 24 h after treatment. Bacterial pellets were flash frozen in liquid nitrogen, and RNA was isolated using a Ribopure RNA isolation kit (Ambion Inc., Austin, TX) according to the manufacturer's instructions with slight modification. Bacterial cells were vortexed three times for 10 min each time to increase the efficiency of mechanical cell lysis, resulting in a higher yield of extracted RNA. An additional DNase I treatment was included to minimize contamination by chromosomal DNA. Samples were concentrated using the RNeasy MiniElute Cleanup kit following the manufacturer's instructions (Qiagen Inc., Valencia, CA). The concentration and integrity of RNA were assessed using a Nanodrop system (Nanodrop, Wilmington, DE), by agarose gel electrophoresis, and by the 23S/16S ratio on an Agilent 2100 bioanalyzer (Agilent, Palo Alto, CA). Preparation of cDNA, labeling, and hybridization to Affymetrix P. aeruginosa GeneChips (Affymetrix Inc.) were conducted at Asuragen Inc., Austin, TX, using the following instruments: a GeneChip 640 hybridization oven, a GeneChip 450 Fluidics Station, and a high-resolution GeneChip 3000 scanner (GeneChip, Santa Clara, CA). Microarray hybridizations were performed in triplicate using RNA/cDNA obtained from concurrently conducted independent experiments. The data analysis was performed using a GeneSpring 7.2 (Silicon Genetics, Redwood City, CA) DNAChip Analyzer (dChip). The data were normalized per chip by dChip invariant set normalization, and each gene was normalized to the median measurement taken for that gene across all the samples. An average two-sample t test using a P value cutoff of ≤0.05, a present call in at least two replicates, and an average twofold or greater change compared to controls were applied to identify genes that were statistically differentially expressed. All gene annotations are from the Pseudomonas Genome Project website (http://www.pseudomonas.com).
Synthesis of cDNA for quantitative PCR.
Several of the genes involved in nutrient acquisition and transport that were significantly differentially expressed in host versus nonhost infection were chosen for validation using quantitative real-time PCR. RNA was isolated as described above, and cDNA was prepared using the procedure described by Affymetrix (Affymetrix, Santa Clara, CA). Briefly, P. aeruginosa RNA was incubated in a thermal cycler with random primers purchased from Invitrogen Life Technologies (Invitrogen, Carlsbad, CA) at 70°C and 25°C for 10 min each and cooled to 4°C. The incubated primer/RNA mixture was then added to cDNA synthesis components and incubated in a thermal cycler (25°C for 10 min, 37°C for 60 min, 42°C for 60 min, 70°C for 10 min, and chilling to 4°C). The RNA was removed by addition of 1 N NaOH and incubated at 65°C for 30 min, followed by addition of 1 N HCl to neutralize the reaction. MiniElute PCR purification columns (Qiagen Inc., Valencia, CA) were used to concentrate the samples, and the quantity of cDNA was determined using a Nanodrop system (Nanodrop, Wilmington, DE).
Validation of microarray experiments using real-time PCR quantification.
To validate the microarray analysis, prepared cDNAs were subjected to quantitative real-time PCR using seven primers for genes—primarily those involved in sulfur or phosphate transport and metabolism—that were shown to be significantly up- or downregulated (see Table S1 in the supplemental material). An iCycler IQ5 (Bio-Rad Laboratories, Hercules, CA) real-time PCR machine was used for the quantification of cDNA. For quantitative analysis of gene transcripts by quantitative reverse transcription-PCR (qRT-PCR), PCRs were performed using a Sybr greenER qPCR Supermix for iCycler instruments (Invitrogen Corp., Carlsbad, CA) according to the specifications of the supplier. qRT-PCRs were performed in 25-μl mixtures containing 21.5 μl of Master Mix, 2.5 μl of cDNA, and 0.5 μM (each) forward and reverse primers. External standard curves to determine primer efficiency were generated using five dilutions (101 to 105) of genomic DNA extracted from bacteria recovered 24 h after infiltration into N. tabacum cv. Samsun plants. PCRs were performed in triplicate for each gene and sample. The reaction mixtures were amplified using the following cycles: 1 cycle of 50°C for 2 min and 95°C for 10 min 30 s, 40 cycles of 95°C for 15 s and 60°C for 1 min, and 1 cycle of 95°C for 1 min and 55°C for 1 min. To correct for differences in the amounts of starting material, the ribosomal gene rpsL was chosen as a reference gene. Change ratios were determined by a variation of the Livak method using the following equations: relative expression (E) = 2CT (reference) − CT (target), where CT is the threshold cycle, and change ratio = E (reference)/E (target).
This modified method was used because the amplification efficiencies of the target and reference gene primers were similar but were not near enough to 100%.
Analysis of Pi and sulfate in intracellular fluid.
To determine the levels of inorganic phosphate (Pi) and sulfate in two tobacco cultivars, plants were infected as described above and incubated overnight at 28 to 30°C. Approximately 1 g of fresh tissue per repetition was removed and infiltrated with sterile dH2O, and the intracellular fluid was removed by centrifugation as described above. The intracellular fluid recovered from each repetition was centrifuged and filtered through a 0.2-μm filter to remove bacteria. The resulting fluid was placed in sterile containers, diluted to a total volume of 100 ml with sterile dH2O, and stored in the dark at 4°C until it was processed. Samples were processed at the Colorado State University's Soil, Water, and Plant Testing Laboratory by James Self. Briefly, sulfate from the intracellular fluid was quantified by ion chromatography using a Dionex 2000i/SP (Dionex Corporation, Sunnyvale, CA), and the Pi concentration was determined by a molybdate blue calorimetric method on a Milton Roy Spectronic-20 (Milton Roy Company, Ivyland, PA).
Microarray data accession number.
The gene expression data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (8) and are accessible through GEO series accession number GSE11544.
RESULTS
Determination of bacterial pathogenesis in N. tabacum plants.
To study the host-induced gene expression in P. aeruginosa, we first evaluated the pathogenicity of PAO1 on N. tabacum (cv. Xanthi nc [NN] and Samsun nn). Three leaves on each test plant were infiltrated with 5 × 106 cells leaf−1 of PAO1 and maintained at 28 to 30°C for the duration of the experiment. This high level of inoculum was used so that a sufficient number of cells could be recovered in subsequent experiments for RNA extraction 24 h postinfiltration. However, differences in PAO1 pathogenicity between the two tobacco cultivars were seen using initial inoculum levels as low as 2 × 105 cells leaf−1 (data not shown). The plants were monitored visually for symptom development, three leaves from each cultivar were removed daily over a period of 5 days, and the recovered bacteria were assessed for viability. Leaf infiltration in the two tobacco cultivars culminated in a successful infection in cv. Samsun, whose symptoms often included chlorosis and necrosis in the infected area, which eventually spread to other tissues of the plant. No visible symptoms were observed in cv. Xanthi aside from necrosis around the infiltration site, likely resulting from mechanical damage during leaf infiltration (Fig. 1A). The CFU counts of bacterial cells recovered from the intercellular space in the infiltrated tobacco leaves corroborate this phenotypic observation (Fig. 1B). After 5 days, the PAO1 population recovered from cv. Xanthi was only slightly higher (∼3-fold) than initial inoculum levels, suggesting that it is an incompatible host for these bacteria. Conversely, bacteria recovered from cv. Samsun continued to multiply and resulted in the recovery of approximately 50-fold more cells than initial inoculum levels 5 days postinfiltration.
FIG. 1.
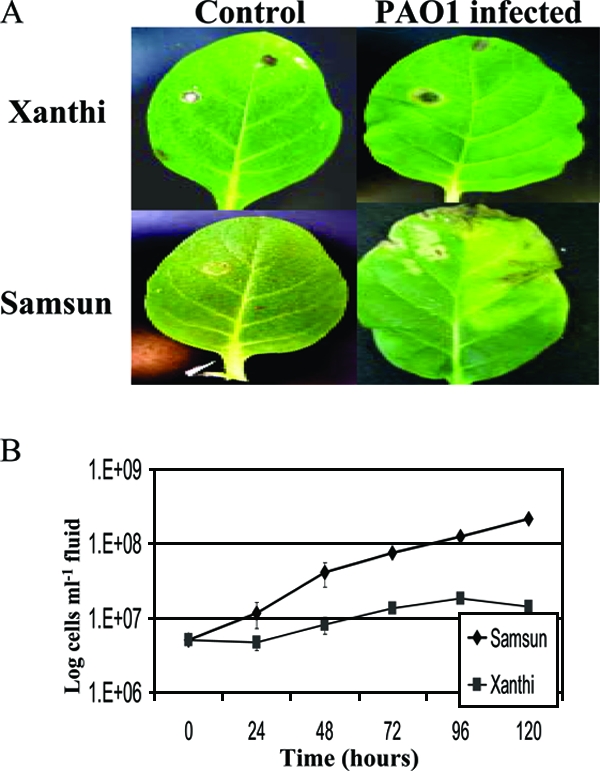
(A) Leaves of N. tabacum cv. Samsun (susceptible to PAO1) and cv. Xanthi (resistant to PAO1) 5 days after infiltration with 10 mM MgSO4 and PAO1. (B) Logarithmic graph of PAO1 CFU (CFU ml intracellular fluid−1) recovered from infiltrated plant tissue over a 5-day period. The data represent mean values from three replicates per experiment, with each experiment performed in triplicate. The error bars represent ±1 SE.
Determination of N-gene involvement in resistance to PAO1.
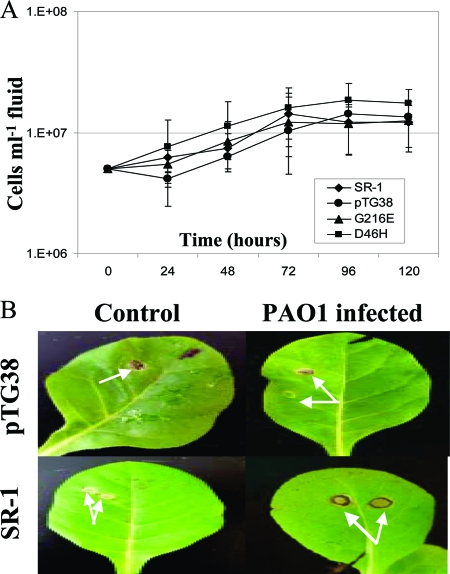
In some cases, the resistance of a plant to a particular pathogen is conveyed by the presence of a resistance (R) gene that has specifically evolved to overcome a virulence (avr) gene of that pathogen. In the case of N. tabacum, the presence of a dominant N gene triggers a hypersensitive response upon infection by tobacco mosaic virus (TMV), resulting in resistance to the virus (7). Although this “gene-for-gene” concept is thought to be a very specific interaction between one host gene and one pathogen gene, there have been reports of a single R gene conferring resistance to multiple attackers (12, 30). Because one tobacco cultivar that we tested had a dominant N gene (cv. Xanthi NN) and the other had a recessive n gene (cv. Samsun nn), and the presence of a dominant N gene was correlated with resistance to PAO1, we wanted to determine if PAO1 resistance was mediated by N-gene involvement or by other factors that differed between the two cultivars. To determine if the source of resistance to PAO1 was N gene related, we infected N. tabacum cv. Petite Havana SR-1 (SR-1::nn) and a transformant of this cultivar, pTG38, that contains a full-length N gene and shows a hypersensitive response to TMV infection (7). We also infiltrated PAO1 into the N-gene mutants D46H and G216E, which were transformed into an SR-1::nn background and crossed with wild-type Samsun NN (6). D46H carries a point mutation in the toll-interleukin 1 receptor region, and G216E carries a mutated nucleotide binding site region. None of these infiltrations resulted in a successful infection (Fig. 2A and B), suggesting that differences between the cultivars, independent of N-gene participation, are responsible for resistance against PAO1.
FIG. 2.
(A) The number of PAO1 CFU (CFU ml intracellular fluid−1) recovered from infiltrated tissue of N. tabacum cv. Petite Havana SR-1, which lacks a dominant N gene, did not differ significantly from the number recovered from a transformant carrying the dominant N gene or two N-gene mutants, confirming that this resistance gene plays no role in preventing infection by P. aeruginosa. The data represent mean values from three replicates per experiment, with each experiment performed at least twice. The error bars represent ±1 SE. (B) Infiltration of PAO1 did not result in symptom development in either N. tabacum cv. Petite Havana SR-1 or the N-gene transformant, pTG38. The arrows highlight areas of infiltration.
Evaluation of SA levels in N. tabacum cultivars.
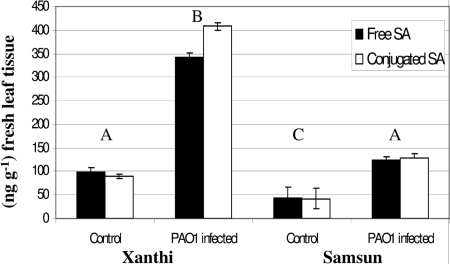
SA is an important defense signal in N-gene-mediated and N-gene-independent responses to pathogen attack in tobacco plants. In addition, as little as 0.1 mM of SA has been shown to have a direct effect on the virulence of P. aeruginosa strain PA14, resulting in the reduction of several virulence factors, including pyocyanin, elastase, and protease, and deterring biofilm formation (25). In fact, several of the same genes were up- or downregulated similarly in both studies, although these data sets were obtained at different time points (see Table S2 in the supplemental material). To determine if a stronger SA response might contribute to the resistance of cv. Xanthi against PAO1, we examined postinfiltration and mock-infiltration levels of SA in both the Samsun and Xanthi cultivars. The basal levels (mock inoculated) of total SA in leaves of cv. Samsun were significantly lower than the levels detected in cv. Xanthi (P = 0.021) (Fig. 3). In addition, there was a significant difference in the responses of the two cultivars 24 h after inoculation with PAO1.The levels of total SA detected in the PAO1-infected cv. Samsun (susceptible to PAO1 infection) were higher than the basal levels but did not differ significantly from the basal levels (P = 0.205) detected in cv. Xanthi and were approximately 2.5 times lower than the amount of total SA found in PAO1-infected cv. Xanthi (P < 0.0001) (Fig. 3). These data suggest that cv. Xanthi launches a more effective defense response against P. aeruginosa and that the amount of SA identified in the leaves may be sufficient to have direct negative effects on the virulence of PAO1.
FIG. 3.
Amounts of free and conjugated SA in 1 g of leaf tissue 24 hours after mock or PAO1 infiltration into N. tabacum cv. Samsun (susceptible to PAO1 infection) and Xanthi (resistant to PAO1). The data represent mean values of six individual replications from duplicated experiments, the error bars represent ±1 SE, and different letters denote significantly different means.
Gene expression analysis in PAO1 in response to compatible and incompatible hosts.
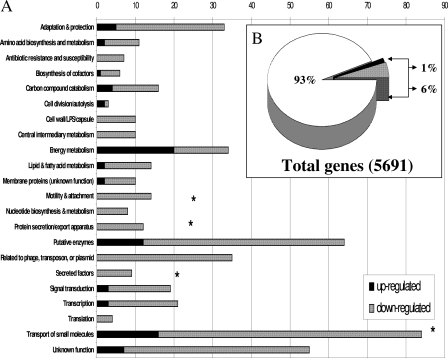
To examine how cultivar-specific factors affected PAO1 growth and virulence in planta, we examined global gene expression in bacteria recovered from the intracellular spaces of the infiltrated leaves of cv. Xanthi and Samsun. We chose to examine gene expression in P. aeruginosa 24 h postinfiltration because there was less difference in bacterial growth between the two cultivars at that time point and we wanted to unravel the determinants of early infection. Utilizing Affymetrix GeneChip microarray technology, we determined that approximately 7% of the genome was differentially expressed (P ≤ 0.05; change, ≥2-fold) with genes involved in adaptation and protection, transport, and energy metabolism being some of the largest functional categories affected (Fig. 4A; see Table S3 in the supplemental material). Using PAO1 gene expression from cv. Samsun (susceptible to PAO1 infection) as the calibrator data set, we determined that only 1% of the differentially expressed genes were upregulated and about 6% were downregulated (Fig. 4B). A large number of the genes that were upregulated are involved in sulfate transport and metabolism, and some of these genes, i.e., sbp and its associated operon, have been shown to be overexpressed in P. aeruginosa cells grown under conditions of sulfur stress (26). The upregulated genes included components of sulfate transporters, genes involved in transport and metabolism of alternate sulfur sources, and a gene thought to encode a protein-utilizing selenocysteine (Table 1). As expected, there were a number of genes thought to be related to virulence that were downregulated in PAO1 recovered from the resistant host plant (see Table S3 in the supplemental material). Among these genes was rpoS, a stationary-phase σ factor that positively regulates the expression of a number of exoproteins, including exotoxin A and alginate (36). Not surprisingly, genes involved in the biosynthesis of alginate (algE and alg44) and endotoxin A (toxA) were also downregulated. Additionally, 12 genes involved in protein export and secretion were downregulated, including several genes involved in the type II secretion system and TTSS. Six of these protein transport genes were probable components of the TTSS, including three psc genes that are thought to be components of the “injection needle” (41) and pcrV, which is involved in effector translocation (42). It was also interesting that a number of genes involved in the transport and metabolism of phosphorus, particularly phosphates, were also downregulated (Table 2). In addition to being an essential micronutrient for the bacteria, phosphorus is thought to be important in motility and biofilm formation in Pseudomonas spp. (21, 29). Downregulation in the resistant host plant of genes involved in phosphate transport and metabolism suggest that phosphorus is not a limiting micronutrient in that environment or that plant-specific factors could be modulating the expression of these genes.
FIG. 4.
(A) Functional categories of up- or downregulated genes in P. aeruginosa reisolated from the leaves of N. tabacum cv. Xanthi (resistant to PAO1) relative to bacteria reisolated from cv. Samsun (susceptible to PAO1). (B) Total percentage of genes differentially expressed more than twofold at a significance level where P is <0.05.
TABLE 1.
PAO1 genes involved in sulfur uptake and metabolism upregulated in the resistant host, cv. Xanthi, compared to the susceptible host, cv. Samsun
| Gene | Fold change | P value | Predicted function |
|---|---|---|---|
| cysA | 4.22 | 0.002 | Sulfate transport protein CysA |
| cysW | 3.86 | 0.023 | Sulfate transport protein CysW |
| cysT | 2.73 | 0.010 | Sulfate transport protein CysT |
| sbp | 3.20 | 0.005 | Sulfate-binding protein precursor |
| cysI | 3.36 | 0.019 | Sulfite reductase (cysteine biosynthesis) |
| PA3445 | 2.36 | 0.035 | Conserved sulfonate, nitrate, bicarbonate transporter |
| PA3446 | 3.44 | 0.007 | Conserved hypothetical protein (70% similar to Escherichia coli SSP4) |
| cysN | 3.64 | 0.028 | ATP sulfurylase GTP-binding subunit/APS kinase |
| cysD | 3.02 | 0.040 | ATP sulfurylase small subunit |
| PA5181 | 3.61 | 0.029 | Probable oxidoreductase (selenocysteine-containing protein) |
TABLE 2.
PAO1 genes involved in phosphorus uptake and metabolism downregulated in the resistant host, cv. Xanthi, compared to the susceptible host, cv. Samsun
| Gene | Fold change | P value | Predicted function |
|---|---|---|---|
| glpQ | −2.38 | 0.041 | Glycerophosphoryl diester phosphodiesterase |
| PA0450 | −6.03 | 0.011 | Probable phosphate transporter |
| phnX | −2.62 | 0.020 | 2-Phosphonoacetaldehyde hydrolase |
| pqqC | −2.15 | 0.014 | Pyrroloquinoline quinine biosynthesis protein C |
| oprP | −6.24 | 0.042 | Phosphate-specific outer membrane porin |
| oprO | −7.07 | <0.001 | Pyrophosphate-specific outer membrane porin |
| phoA | −3.93 | 0.004 | Alkaline phosphatase |
| plcN | −5.37 | 0.036 | Nonhemolytic phospholipase C precursor |
| PA3376 | −5.98 | 0.043 | Probable ATP-binding component of ABC phosphate transporter |
| PA3383 | −1.74 | 0.022 | Binding protein component of ABC phosphonate transporter |
| phnC | −5.17 | 0.010 | ATP-binding component of ABC phosphonate transporter |
| katA | −2.03 | 0.034 | Catalase |
| PA4861 | −5.92 | 0.030 | Probable ATP-binding component of ABC phosphate transporter |
| phoB | −2.33 | 0.009 | Two-component response regulator |
To validate these results, we examined gene expression in seven up- or downregulated genes using qRT-PCR and normalized it to the expression of the ribosomal gene rpsL. Trends in expression patterns for all genes examined confirmed the microarray data (Table 3). However, unlike the microarray experiments, where all of the biological replicates were collected from concurrently conducted experiments and processed together to minimize artificially introduced variability, the biological replications used for these analyses were collected from experiments that were conducted at different times and processed individually. This resulted in high variability of the biological and technical replications, and therefore, some of the qRT-PCR results were not statistically significant.
TABLE 3.
Comparison between qRT-PCR and microarray data
| Data source | Fold change in expressiona
|
||||||
|---|---|---|---|---|---|---|---|
| cysI | cysDb | sbpb | phoB | oprO | phnCb | algL | |
| qRT-PCR | 5.77 | 3.97 | 8.66 | −21.26 | −3.93 | −4.10 | −1.97 |
| Microarray | 3.36 | 3.02 | 3.20 | −2.33 | −7.07 | −5.17 | −4.56 |
Relative changes in gene expression of selected up- or downregulated genes in the resistant host, cv. Xanthi, relative to the susceptible host, cv. Samsun.
qRT-PCR results were not statistically different (P ≤ 0.05) between the two cultivars. However, trends in expression patterns were consistent between microarray and qRT-PCR data for these genes.
Determination of in planta levels of sulfate and inorganic phosphate.
As discussed above, some of the most obvious differences in gene expression between bacteria recovered from compatible versus incompatible plant material were the activation or repression of several genes involved in uptake and transport of sulfate and phosphates. To examine the cause of these differences, we mimicked the infiltration and collection protocol used for the microarray experiments to examine the available levels of sulfate and Pi present in the intercellular fluid. Mock-inoculated plants were used as controls. We discovered that the amounts of sulfate did not differ significantly between cultivars or treatments (P = 0.544) (Table 4). This suggests that there are no inherent differences in the levels of available sulfate between these cultivars that would account for the activation of sulfur stress pathways in PAO1 residing in the intracellular space of cv. Xanthi. However, it does not rule out the possibility that cv. Xanthi is able to physically confine the invading bacteria, creating a microenvironment where sulfate is more limited.
TABLE 4.
Amounts of sulfate and Pi detected in the intercellular fluid of N. tabacum cv. Xanthi and Samsun 24 h after infiltration with PAO1 or 10 mM MgSO4
| Cultivar | Treatment | Amt (mg liter−1) of:
|
|
|---|---|---|---|
| PO4 | SO4 | ||
| Samsun | MgSO4 | 0.325 ± 0.006 | 2.5 ± 0.05 |
| PAO1 | 0.228 ± 0.008 | 2.7 ± 0.13 | |
| Xanthi | MgSO4 | 0.426 ± 0.009 | 2.4 ± 0.05 |
| PAO1 | 0.368 ± 0.003 | 3.3 ± 0.01 | |
Assessments of Pi levels in the intracellular fluid could be interpreted in two ways. Statistical comparisons using Fisher's test (least significant difference) revealed that levels of Pi were significantly higher in the intracellular fluid of PAO1-infiltrated cv. Xanthi than in infected cv. Samsun (control, P = 0.106; treated, P = 0.039), suggesting that some differences in gene expression related to phosphate uptake and metabolism could be due to the availability of the nutrient in the different plant cultivars (Table 4). However, comparisons using the more stringent Tukey's test suggest that the amounts of Pi do not differ in either control or treated plants of these cultivars (control, P = 0.139; treated, P = 0.134), indicating that the difference is small compared to the variability between plants and may not be an important factor in determining gene expression related to phosphorus utilization.
DISCUSSION
The study of plant interactions with P. aeruginosa has revealed a great deal about individual factors that are necessary for the virulence of the bacterium (27, 28, 32, 43). In fact, the success of these studies has validated the use of plant models to examine key processes involved in the establishment of host-pathogen interactions (39). However, these studies were conducted using random or specific mutants of P. aeruginosa and reveal very little about the holistic interaction between P. aeruginosa and the model host. To address this deficiency, we chose to examine comprehensive transcriptional changes in PAO1 in the context of compatible and incompatible interactions with N. tabacum plants, as well as exploring some of the host factors that are involved in pathogen resistance.
We examined gene expression in the bacteria 24 h after infiltration into the different plant hosts. The rationale for choosing this time point was that we wanted to explore early determinants of infection, and at this time point, there was less difference in the in vivo growth of P. aeruginosa between the two tobacco cultivars. Despite this early time point, there were still roughly twice as many bacterial cells recovered from the susceptible host, cv. Samsun, than from cv. Xanthi. Because of the difference in bacterial growth between the two host environments, a number of genes downregulated in cv. Xanthi (the incompatible host) that were identified in this study are related to growth and primary metabolism of the bacteria, including many of the transport genes discussed, such as oprO and oprP. However, rather than confounding our attempts to identify early determinants of infection, we believe that fitness of the bacteria in a particular host is a determinant of whether the bacteria will successfully initiate an infection.
One of the most obvious differences identified in PAO1 gene expression was the induction of sulfur starvation pathways in bacteria removed from resistant host plant tissue. Sulfur starvation occurs in three primary stages. First, the bacteria upregulate genes involved in the acquisition of sulfate, the preferred sulfur source. Subsequently, they begin to upregulate genes for acquiring and utilizing secondary sources of sulfur, and ultimately to synthesize alternative forms of proteins containing reduced amounts of cysteine (26). We found that many genes that are required for sulfate acquisition (sbp and genes in the cysTWA operon) and one for a selenocysteine-containing protein were upregulated in the incompatible host environment, suggesting early stages of sulfur starvation. The levels of sulfate present in the intracellular fluids of both cultivars of N. tabacum did not differ significantly, indicating that only the amount of sulfate available to the bacteria was restricted. Although a hypersensitive reaction was rarely visible on PAO1-infected leaves of cv. Xanthi, the plant may create a physical barrier that restricts the growth of the bacteria and prevents them from accessing sulfur and other intercellular nutrients. Alternatively, sulfur-containing compounds, such as thionins, defensins, alliin, glutathione, and glucosinolates, have been both directly and indirectly linked to plant defense against microbial pathogens (14). One could speculate, then, that the resistant host, cv. Xanthi, is better defended than the susceptible cv. Samsun and actively utilizes sulfur sources to synthesize defense compounds. This may result in competition between the plant and PAO1 for available sulfur sources, accounting for the activation of sulfur stress pathways in PAO1 recovered from cv. Xanthi.
Another large group of differentially regulated genes were involved in the transport and metabolism of phosphates. Inorganic phosphate is an essential micronutrient required for bacterial growth, and it has also been implicated in the regulation of biofilm formation in Pseudomonas fluorescens via phosphate-dependent modulation of cyclic-di-GMP levels, which control the secretion of a surface adhesin (22). Relative to the calibrator data set (the compatible host, cv. Samsun), many genes required for acquisition and metabolism of phosphates were downregulated in incompatible host plants (cv. Xanthi). These results were contrary to what might be expected if resistant host plants prevent a flow of nutrients to PAO1. While it is enticing to speculate that defense-related factors produced by resistant host plants were able to modulate the expression of these genes, hindering the bacteria from acquiring phosphates, an examination of the Pi levels in the intercellular fluid offers a more mundane explanation. Using Fisher's least-significant-difference comparison, there was a significantly smaller amount of Pi present in the intracellular fluid of PAO1-infected samples from N. tabacum cv. Samsun than in the infected cv. Xanthi samples (P = 0.039). It is possible that the resulting differential expression of genes involved in phosphate uptake and metabolism was due to the activation of phosphate starvation pathways in cv. Samsun, since measured intracellular Pi levels were lower in these plants. The upregulation of oprO and oprP, which encode phosphate-specific membrane porins, supports this hypothesis. Additionally, the activation of pho genes, which are induced under low-Pi conditions and regulate genes such as phnC, suggests that the bacteria optimize their ability to utilize alternative sources of phosphorus (40). However, the basal levels of Pi in the two cultivars did not differ significantly (P = 0.106), and the more stringent Tukey's honestly significant difference statistical comparison suggests that there was no significant difference in the amounts of Pi between the two PAO1-infected cultivars (P = 0.134). Another possibility is that bacteria retrieved from host plants (cv. Samsun) had begun to utilize phosphorus in pathogenesis-related processes and needed to acquire additional Pi beyond the basic requirements for survival. It has been proposed that many Pi-regulated genes in P. aeruginosa function as a Pi-scavenging system that may be critical to pathogenesis (19, 40). Many of the phosphate acquisition and metabolism genes that were upregulated in bacteria retrieved from host plants relative to their resistant-host counterparts (plcN, phoA, glpQ, and oprO) were also shown to be upregulated after exposure to human airway epithelial cells (11), and one differentially regulated gene in this study, plcN, encodes a nonhemolytic phospholipase that has been associated with virulence in P. aeruginosa (5, 17).
An important virulence determinant of PAO1 in mammalian hosts is the secretion of exotoxins via the TTSS (10). The TTSS functions by facilitating the direct injection of bacterial effector proteins into host cells (3, 35). In P. syringae, a plant pathogen, some of these TTSS-mediated effectors are avr (avirulence) genes to which some hosts have developed resistance, often mediated by a single host gene that can recognize and neutralize a single avr gene in a “gene-for-gene” manner (9). However, in a susceptible plant host, these effectors can act on host protein substrates and interfere with host defenses and signaling by defense hormones, such as SA (37). In the resistant host (cv. Xanthi) plants, we saw that several genes related to TTSS were downregulated. This suggests that although a tobacco-PAO1 model has not been previously established, once PAO1 gains entry to susceptible tobacco plants, it can act as a true pathogen, utilizing TTSS and secreted virulence factors to compromise its host. The resistant N. tabacum cultivar, cv. Xanthi, contains a dominant N gene that can recognize an avirulence factor from TMV, resulting in an SA-mediated hypersensitive response, restricted spread of the virus, and systemic resistance to TMV, as well as other pathogens (7). Although the susceptible cultivar, Samsun, lacked a dominant N gene, subsequent experiments demonstrating PAO1 resistance in both cv. Petite Havana SR-1::nn and an N-gene-containing transformant of this cultivar, pTG38, suggest that the N gene does not play a role in resistance to PAO1. However, this elimination does not rule out the possibility that there are other, unidentified resistance genes that convey resistance to specific effectors of PAO1. It does appear that the plant defense signaling compound, SA, plays a role in the differential resistance that is visible between the two cultivars, as we detected substantially more SA in leaf tissue of the resistant cultivar in both infected and uninfected plants. SA may play a dual role in the defense of plants against P. aeruginosa and other human pathogens, as it is used by the plant to induce defense responses but also has direct negative effects on bacterial virulence (24, 25). This study demonstrates the importance of nutrient availability in disease development. For instance, the limitation of some nutrients, such as sulfate, appears to inhibit growth and subsequent pathogenicity. However, the limitation of other nutrients, like iron and phosphate, may stimulate the expression of virulence factors (19, 33). In summary, the results of this study suggest that the ability of plant defenses and innate immunity to modulate responses of the pathogen could be extrapolated to better understand pathogenesis in human hosts and further emphasize the importance of the use of plant models to study human pathogenesis.
Supplementary Material
Acknowledgments
This research was supported by a grant from the National Science Foundation to J.V. (MCB-0542642).
We also thank Barbara Baker and E. Peter Greenberg for supplying plant material and bacterial cultures and Emily Wortman-Wunder for helpful comments on the manuscript.
Footnotes
Published ahead of print on 18 July 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Attila, C., A. Ueda, S. L. G. Cirillo, J. D. Cirillo, W. Chen, and T. K. Wood. 2008. Pseudomonas aeruginosa PAO1 virulence factors and poplar tree response in the rhizosphere. Microb. Biotechnol. 1:17-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowling, S. A., A. Guo, H. Cao, A. S. Gordon, D. F. Klessig, and X. Dong. 1994. A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 6:1845-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chisholm, S. T., G. Coaker, B. Day, and B. J. Staskawicz. 2006. Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124:803-814. [DOI] [PubMed] [Google Scholar]
- 4.Cornelis, G. R., and F. Van Gijsegem. 2000. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54:735-774. [DOI] [PubMed] [Google Scholar]
- 5.Darby, C., C. L. Cosma, J. H. Thomas, and C. H. Manoil. 1999. Lethal paralysis of Caenorhabditis elegans by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:15202-15207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dinesh-Kumar, S. P., W. H. Tham, and B. J. Baker. 2000. Structure-function analysis of the tobacco mosaic virus resistance gene N. Proc. Natl. Acad. Sci. USA 97:14789-14794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dinesh-Kumar, S. P., S. Whitham, D. Choi, R. Hehl, C. Corr, and B. Baker. 1995. Transposon tagging of tobacco mosaic virus resistance gene N: its possible role in the TMV-N-mediated signal transduction pathway. Proc. Natl. Acad. Sci. USA 92:4175-4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edgar, R., M. Domrachev, and A. E. Lash. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flor, H. H. 1947. Inheritance of reaction to rust in flax. J. Agric. Res. 74:241-262. [Google Scholar]
- 10.Frank, D. W. 1997. The exoenzyme S regulon of Pseudomonas aeruginosa. Mol. Microbiol. 26:621-629. [DOI] [PubMed] [Google Scholar]
- 11.Frisk, A., J. R. Schurr, G. Wang, D. C. Bertucci, L. Marrero, S. H. Hwang, D. J. Hassett, and M. J. Schurr. 2004. Transcriptome analysis of Pseudomonas aeruginosa after interaction with human airway epithelial cells. Infect. Immun. 72:5433-5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grant, M. R., L. Godiard, E. Straube, T. Ashfield, J. Lewald, A. Sattler, R. W. Innes, and J. L. Dangl. 1995. Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 269:843-846. [DOI] [PubMed] [Google Scholar]
- 13.Hammond-Kosack, K. E., and J. E. Parker. 2003. Deciphering plant-pathogen communication: fresh perspectives for molecular resistance breeding. Curr. Opin. Biotechnol. 14:177-193. [DOI] [PubMed] [Google Scholar]
- 14.Hell, R. 1997. Molecular physiology of plant sulfur metabolism. Planta 202:138-148. [DOI] [PubMed] [Google Scholar]
- 15.Inohara, N., M. Chamaillard, C. McDonald, and G. Nuñez. 2005. NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annu. Rev. Biochem. 74:355-383. [DOI] [PubMed] [Google Scholar]
- 16.Jha, A. K., H. P. Bais, and J. M. Vivanco. 2005. Enterococcus faecalis uses mammalian virulence-related factors to exhibit potent pathogenicity in the Arabidopsis thaliana plant model. Infect. Immun. 73:464-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Konig, B., M. L. Vasil, and W. Konig. 1997. Role of hemolytic and nonhemolytic phospholipase C from Pseudomonas aeruginosa for inflammatory mediator release from human granulocytes. Int. Arch. Allergy Immunol. 112:115-124. [DOI] [PubMed] [Google Scholar]
- 18.Lee, S. W., S. W. Han, L. E. Bartley, and P. C. Ronald. 2006. Unique characteristics of Xanthomonas oryzae pv. oryzae AvrXa21 and implications for plant innate immunity. Proc. Natl. Acad. Sci. USA 103:18395-18400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, P. V. 1979. Toxins of Pseudomonas aeruginosa p. 63-88. In R. G. Doggett (ed.), Pseudomonas aeruginosa: clinical manifestations of infection and current therapy. Academic Press, Inc., New York, NY.
- 20.Mark, G. L., J. M. Dow, P. D. Kiely, H. Higgins, J. Haynes, C. Baysse, A. Abbas, T. Foley, A. Franks, J. Morrissey, and F. O'Gara. 2005. Transcriptome profiling of bacterial responses to root exudates identifies genes involved in microbe-plant interactions. Proc. Natl. Acad. Sci. USA 102:17454-17459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monds, R. D., P. D. Newell, R. H. Gross, and G. A. O'Toole. 2007. Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0-1 biofilm formation by controlling secretion of adhesion. Mol. Microbiol. 63:656-679. [DOI] [PubMed] [Google Scholar]
- 22.Nurnberger, T., and D. Scheel. 2001. Signal transmission in the plant immune response. Trends Plant Sci. 6:372-379. [DOI] [PubMed] [Google Scholar]
- 23.Okinaka, Y., C. H. Yang, N. T. Perna, and N. T. Keen. 2002. Microarray profiling of Erwinia chrysanthemi 3937 genes that are regulated during plant infection. Mol. Plant-Microbe Interact. 15:472-480. [DOI] [PubMed] [Google Scholar]
- 24.Prithiviraj, B., H. P. Bais, T. L. Weir, B. Suresh, E. H. Najarro, B. V. Dayakar, H. P. Schweizer, and J. M. Vivanco. 2005. Down regulation of virulence factors of Pseudomonas aeruginosa by salicylic acid attenuates its virulence on Arabidopsis thaliana and Caenorhabditis elegans. Infect. Immun. 73:5319-5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prithiviraj, B., H. P. Bais, A. K. Jha, and J. M. Vivanco. 2005. Staphylococcus aureus pathogenicity in Arabidopsis thaliana is mediated by either a direct effect of salicylic acid on the pathogen or by SA-dependent, NPR1-independent host responses. Plant J. 42:417-432. [DOI] [PubMed] [Google Scholar]
- 26.Quadroni, M., P. James, P. Dainese-Hatt, and M. A. Kertesz. 1999. Proteome mapping, mass spectrometric sequencing and reverse transcription-PCR for characterization of the sulfate starvation-induced response in Pseudomonas aeruginosa PAO1. Eur. J. Biochem. 266:986-996. [DOI] [PubMed] [Google Scholar]
- 27.Rahme, L. G., E. J. Stevens, S. F. Wolfort, J. Shao, R. G. Tompkins, and F. M. Ausubel. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 286:1899-1902. [DOI] [PubMed] [Google Scholar]
- 28.Rahme, L. G., M.-W. Tan, L. Le, S. M. Wong, R. G. Tompkins, S. B. Calderwood, and F. M. Ausubel. 1997. Use of model plant hosts to identify Pseudomonas aeruginosa virulence factors. Proc. Natl. Acad. Sci. USA 94:13245-13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rashid, M. H., K. Rumbaugh, L. Passador, D. G. Davies, A. N. Hamood, B. H. Iglewski, and A. Kornberg. 2000. Polyphosphate kinase is essential for biofilms development, quorum sensing, and virulence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 97:9636-9641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rossi, M., F. L. Goggin, S. B. Milligan, I. Kaloshian, D. E. Ullman, and V. M. Williamson. 1998. The nematode resistance gene Mi of tomato confers resistance against the potato aphid. Proc. Natl. Acad. Sci. USA 95:9750-9754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scott, I. M., S. M. Clarke, J. E. Wood, and L. A. J. Mur. 2004. Salicylate accumulation inhibits growth at chilling temperature in Arabidopsis. Plant Physiol. 135:1040-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silo-Suh, L., S. J. Suh, P. A. Sokol, and D. E. Ohman. 2002. A simple alfalfa seedling infection model for Pseudomonas aeruginosa strains associated with cystic fibrosis shows AlgT (sigma-22) and RhlR contribute to pathogenesis. Proc. Natl. Acad. Sci. USA 99:15699-15704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sokol, P. A., C. D. Cox, and B. H. Iglewski. 1982. Pseudomonas aeruginosa mutants altered in their sensitivity to the effect of iron on toxin A or elastase yields. J. Bacteriol. 151:783-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Son, M. S., W. R. Matthews, Jr., Y. Kang, D. T. Nguyen, and T. T. Hoang. 2007. In vivo evidence of Pseudomonas aeruginosa nutrient acquisition and pathogenesis in the lungs of cystic fibrosis patients. Infect. Immun. 75:5313-5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Staskawicz, B. J., M. B. Mudgett, J. L. Dangl, and J. E. Galan. 2001. Common and contrasting themes of plant and animal diseases. Science 292:2285-2289. [DOI] [PubMed] [Google Scholar]
- 36.Suh, S. J., L. Silo-Suh, D. E. Woods, D. J. Hassett, S. E. West, and D. E. Ohman. 1999. Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. J. Bacteriol. 181:3890-3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomma, B. P., I. A. Pennick, W. F. Broekaert, and B. P. Cammue. 2001. The complexity of disease signaling in Arabidopsis. Curr. Opin. Immunol. 13:63-68. [DOI] [PubMed] [Google Scholar]
- 38.van Baarlen, P., A. van Belkum, and B. P. H. J. Thomma. 2007. Disease induction by human microbial pathogens in plant-model systems: potential, problems, and prospects. Drug Disc. Today 12:167-173. [DOI] [PubMed] [Google Scholar]
- 39.Wanner, B. L., and J. A. Boline. 1990. Mapping and molecular cloning of the phn (psiD) locus for phosphonate utilization in Escherichia coli. J. Bacteriol. 172:1186-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weinberg, E. D. 1974. Iron and susceptibility to infectious disease. Science 134:952-955. [DOI] [PubMed] [Google Scholar]
- 41.Yahr, T. L., J. Goranson, and D. W. Frank. 1996. Exoenzyme S of Pseudomonas aeruginosa is secreted by a type III pathway. Mol. Microbiol. 22:991-1003. [DOI] [PubMed] [Google Scholar]
- 42.Yahr, T. L., L. M. Mende-Mueller, M. B. Friese, and D. W. Frank. 1997. Identification of type III secreted products of the Pseudomonas aeruginosa exoenzyme S regulon. J. Bacteriol. 179:7165-7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yorgey, P., L. G. Rahme, M. W. Tan, and F. M. Ausubel. 2001. The roles of mucD and alginate in the virulence of Pseudomonas aeruginosa in plants, nematodes, and mice. Mol. Microbiol. 41:1063-1076. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.