Abstract
Vibrio cholerae non-O1, non-O139 was isolated from natural surface waters from different sites sampled in diarrhea endemic zones in Kolkata, India. Twenty-one of these isolates were randomly selected and included in the characterization. The multiserogroup isolates were compared by their virulence traits with a group of clinical non-O1, non-O139 isolates from the same geographic area. Of the 21 environmental isolates, 6 and 14 strains belonged to Heiberg groups I and II, respectively. Three of the environmental isolates showed resistance to 2,2-diamine-6,7-diisopropylpteridine phosphate. All of the non-O1, non-O139 strains were positive for toxR, and except for one environmental isolate, none of them were positive for tcpA in the PCR assay. None of the isolates were positive for genes encoding cholera toxin (ctxA), heat-stable toxin (est), heat-labile toxin (elt), and Shiga toxin variants (stx) of Escherichia coli. Additionally, except for one environmental isolate (PC32), all were positive for the gene encoding El Tor hemolysin (hly). The culture supernatants of 86% (18 of 21) of the environmental isolates showed a distinct cytotoxic effect on HeLa cells, and some of these strains also produced cell-rounding factor. The lipase, protease, and cell-associated hemagglutination activities and serum resistance properties of the environmental and clinical isolates did not differ much. However, seven environmental isolates exhibited very high hemolytic activities (80 to 100%), while none of the clinical strains belonged to this group. The environmental isolates manifested three adherence patterns, namely, carpet-like, diffuse, and aggregative adherence, and the clinical isolates showed diffuse adherence on HeLa cells. Of the 11 environmental isolates tested for enteropathogenic potential, 8 (73%) induced positive fluid accumulation (≥100) in a mouse model, and the reactivities of these isolates were comparable to those of clinical strains of non-O1, non-O139 and toxigenic O139 V. cholerae. Comparison of the counts of the colonized environmental and clinical strains in the mouse intestine showed that the organisms of both groups had similar colonizing efficiencies. These findings indicate the presence of potentially pathogenic V. cholerae non-O1, non-O139 strains in surface waters of the studied sites in Kolkata.
Vibrio cholerae non-serogroup O1 strains are ubiquitous in aquatic environments (35) and have been recognized as the causative agents of sporadic cholera-like disease and outbreaks (5, 34, 39, 43). In addition, environmental nontoxigenic, non-O1 strains play an important role in the evolution of toxigenic V. cholerae (19). However, only a minority of the strains of V. cholerae non-O1 seem to be enteropathogenic. The pathogenicity markers of vibrios are lacking in many species, including V. cholerae non-O1, non-O139 strains. The nature of virulence and the infective doses need to be determined for the establishment of guidelines for risk assessment of each species in surface water and food material earmarked for human consumption (18). Cholera surveillance is in progress in many countries, based primarily on detection of V. cholerae O1 and O139 and determining the presence of cholera toxin (CT) by using biological and molecular methods (37). However, virulence-associated factors in V. cholerae isolates from environmental sources are of concern (37). The emergence of serogroup O139 as a second etiologic agent of cholera epidemics, along with the possible conversion of non-O1 to O1 serotype (9) and the emergence of V. cholerae O10, O12, O31, O37, and O53 as bacterial serogroups associated with cholera-like epidemics (2, 11, 12, 22, 24, 38, 43), has caused further interest in the non-O1, non-O139 V. cholerae strains.
The pathogenic mechanisms by which these enteropathogens cause diarrhea are not yet well established. Monitoring existing environmental strains and undertaking detailed studies of how pathogenic strains evolved from them are essential to our understanding of human disease. The present study reports on the isolation of V. cholerae non-O1, non-O139 strains from natural surface waters and whether these isolates possess the ability to produce virulence-associated factors compared with some clinical isolates from our culture collection. The virulence-associated factors studied here include CT; Shiga toxin variants, heat-labile toxin, and heat-stable toxin of Escherichia coli; cytotonic toxin; cytotoxin; hemolysin; lipase; protease; cell-associated hemagglutinin (HA); toxin-coregulated pilus subunit A (TcpA) (classical and El Tor); toxin regulatory protein (ToxR); serum sensitivity; adherence to HeLa cells; and antibiotic resistance. We also investigated the enteropathogenic potentials of some of the isolates in an animal model and compared them with clinical isolates.
MATERIALS AND METHODS
Study area and sample collection.
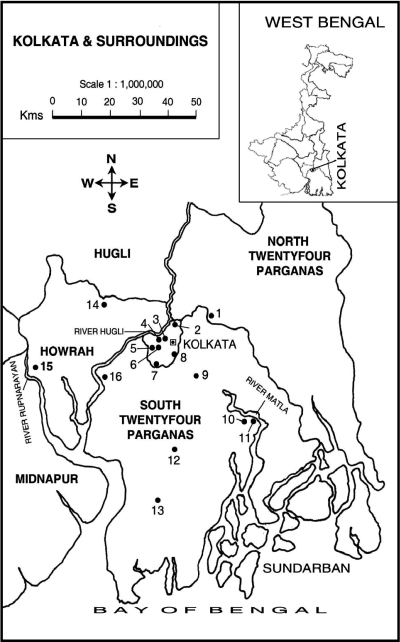
The study sites in Kolkata and surrounding areas, West Bengal, India, are shown in Fig. 1. The main water sources in these rural and urban communities were identified, sampled, and included in this study. Diarrheal diseases are endemic in these selected areas. The ponds and canals are used for domestic purposes (bathing, washing of cloths and utensils, and cooking) by the community and are probably linked to infections by enteropathogens. The collection of water samples from the ponds and canals (5 cm below the water surface) in different areas was done biweekly between April 2001 and March 2003. The samples were collected in sterile 250-ml glass bottles. Inoculations into selective medium were made within 24 h after the collection of water samples.
FIG. 1.
Sample collection sites. 1, Rajarhat; 2, Dum Dum; 3, Kalighat; 4, Chetla; 5, Mudialy; 6, Taratala; 7, Behala; 8, Jadavpur; 9, Sonarpur; 10, Taldi; 11, Canning; 12, South Barasat; 13, Laksmikantapur; 14, Bally; 15, Deulti; and 16, Budge Budge.
Isolation and identification of bacteria.
A water sample (10 ml) was added to 10 ml of double-strength alkaline peptone-water (pH 8.6) and incubated at 37°C for 24 h (35). A sample from this enrichment culture was transferred with a loop to thiosulfate-citrate-bile salts-sucrose (TCBS) agar (Eiken, Tokyo, Japan) and incubated at 37°C for 24 h. Four of the yellow colonies suspected to be Vibrio spp. per sample were picked up from the TCBS agar plates. These colonies were first examined with a multitest medium (25). The isolates that gave a typical alkaline slant-acid butt reaction in the multitest medium were tested for oxidase reaction (Kovacs method). The oxidase-positive isolates were also tested for string reaction (47) and sensitivity to the vibriostatic agent 2,2-diamine-6,7-diisopropylpteridine phosphate (O/129) with 150-μg disks. Organisms showing oxidase-positive reactions were identified by methods described previously (13, 15, 49) and by use of biochemical tests by conventional methods (8, 13). Their shapes and motilities were determined with a phase-contrast microscope (model BX51/B52; Olympus, Japan). Salt tolerance was determined by growth of the isolates at 37°C in 1% peptone broth without NaCl or supplemented with either 1 or 7% NaCl. The isolates were further examined by V. cholerae-specific ompW PCR (27). Serological confirmation of V. cholerae strains was performed by an agglutination test with polyvalent O1-specific antiserum and with O139-specific monoclonal antibody (MAb). Isolates that did not agglutinate with either O1 antiserum or O139 MAb were further serogrouped by the somatic O antigen serogrouping scheme for V. cholerae developed at the National Institute of Infectious Diseases, Tokyo, Japan (44). The strains were maintained in nutrient agar as stabs at room temperature.
Clinical strains.
Five non-O1, non-O139 strains (PL2, PL72, PG5, PG109, and AS67) and one O139 strain (SG24) of V. cholerae isolated from hospitalized patients with acute diarrhea in Kolkata, India (7, 24), were included in this study.
PCR.
Amplification of the target gene was carried out by PCR assay using bacterial cell lysate as the source of template DNA (27, 43). Briefly, bacterial cells were grown overnight at 37°C on Luria agar plates. Isolated colonies were picked up and mixed with 100 μl of normal saline, and the bacterial cells were pelleted by centrifugation. The cell pellet was resuspended in 100 μl of double-distilled water and boiled for 10 min. Cell debris was removed by centrifugation, and the supernatant containing the template DNA was placed in a fresh microcentrifuge tube for PCR assay. Amplification was performed in a thermal cycler (Mastercycler Personal; Eppendorf, Germany) using 200-μl PCR tubes with a reaction mixture volume of 25 μl. Each of the reaction mixtures contained 3 μl of template DNA (lysate), 2.5 μl of each primer (10 pmol/μl), 2.5 μl of 2.5 mM deoxynucleoside triphosphates, 0.3 μl (5 U/μl) of Taq DNA polymerase (Takara Shuzo, Japan), 2.5 μl of 10× reaction buffer containing 20 mM MgCl2 (Extaq; Takara), and 11.8 μl of distilled water.
PCR for detecting the genes representing CT (primers, ctxA-F [5′-CTCAGACGGGATTTGTTAGGCACG-3′] and ctxA-R [5′-TCTATCTCTGTAGCCCCTATTACG-3′]) (20), E. coli heat-labile toxin (primers, elt-F [5′-GGCGACAGATTATACCGTGC-3′] and elt-R [5′-CCGAATTCTGTTATATATGTC-3′]) (21), E. coli heat-stable toxin (primers, est-F [5′-TTAATAGCACCCGGTACAAGCAGG-3′] and est-R [5′-CCTGACTCTTCAAAAGAGAAAATTAC-3′]) (21), Shiga toxin variants of E. coli (primers, stx1-F [5′-CAACACTGGATGATCTCAG-3′] and stx1-R [5′-CCCCCTCAACTGCTAATA-3′], and stx2-F [5′-ATCAGTCGTCACTCACTGGT-3′] and stx2-F [5′-CTGCTGTCACAGTGACAAA-3′]) (21), V. cholerae outer membrane protein (primers, VcompW-F [5′-CACCAAGAAGGTGACTTTATTGTG-3′] and VcompW-R [5′-GAACTTATAACCACCCGCG-3′]) (27), toxin regulatory protein for V. cholerae (primers, toxR-F [5′-CGGGATCCATGTTCGGATTAGGACAC-3′] and toxR-R [5′-CGGGATCCTACTCACACACTTTGATGGC-3′]) (14), and El Tor hemolysin (primers, hlyA-F [5′-GGCAAACAGCGAAACAAATACC-3′] and hlyA-R [5′-CTCAGCGGGCTAATACGGTTTA-3′]) (37) was done as described elsewhere (6, 14, 20, 21, 27, 37).
Previous reports identified several variant TcpA sequences among non-O1, non-O139 isolates (6, 14, 28, 31, 41), and a PCR assay was done for the detection of those variant tcpA genes using primers (tcpA, classical and El Tor variants) designed by Keasler and Hall (20). In the present study, a PCR assay was performed (6, 14, 20, 28, 31, 41, 43) for the detection of the tcpA genes of V. cholerae strains using primers (classical variant, tcpA-F [5′-CACGATAAGAAAACCGGTCAAGAG-3′] and tcpA-R [5′-ACCAAATGCAACGCCGAATGGAGC-3′]; El Tor variant, tcpA-F [5′-GAAGAAGTTTGTAAAAGAAGAACAC-3′] and tcpA-R [5′-GAAAGCACCTTCTTTCACGTTG-3′]) (20). The amplification program began with denaturation at 94°C for 5 min, followed by 30 cycles consisting of denaturation at 94°C for 1.5 min, annealing at 60°C for 1.5 min, and extension at 72°C for 1.5 min, and a final extension step of 72°C for 7 min.
The PCR products were electrophoresed through a 1.5% (wt/vol) agarose gel to resolve the amplified products, which were visualized under UV light after ethidium bromide staining.
Preparation of cell-free culture supernatants.
Trypticase soy broth (TSB) (HiMedia) and AKI medium (peptone, 1.5%; yeast extract, 0.4%; NaCl, 0.5%; NaHCO3, 0.3%) were used for assessing the production of various toxins by the strains. The test strains were grown in the above-mentioned media at 37°C with shaking (200 rpm) for 18 h. After centrifugation (15,000 × g for 20 min at 4°C), the culture supernatant was filtered using a 0.22-μm filter (Millipore Corp.) and used in the tissue culture and hemolysin assays.
Tissue culture assay.
HeLa cells were grown as monolayers in tissue culture flasks (25 cm2) using Dulbecco's modified essential medium (DMEM) (Gibco Laboratories) supplemented with 10% (vol/vol) horse serum (Gibco) at 37°C in a humidified 5% CO2 incubator (model HF; Shanghai Lishen Scientific Equipment Co. Ltd., Shanghai, China). Freshly trypsinized cells were resuspended in DMEM supplemented with 1% horse serum at a final concentration of 2 × 104 cells per ml, and 0.2 ml was added to each well of the 96-well tissue culture plate. The cell-free culture filtrates of the test strains were serially diluted with sterile 10 mM phosphate-buffered saline (PBS) (pH 7.2), and aliquots (50 μl) of each dilution were added in duplicate to the assay plate and incubated as described above. Morphological changes and cytotoxic effects were recorded after 24 h of incubation using an inverted microscope (Olympus).
Adherence assay.
The isolates were examined for their adherence to HeLa cells following methods described previously (10). The HeLa cells were grown in DMEM containing 10% fetal calf serum to 50 to 70% confluence on glass coverslips in a 24-well flat-bottom tissue culture plate (Tarson, Mumbai, India). The isolates were grown in TSB at 37°C without shaking for 18 h and incubated (20 μl of TSB culture per ml of tissue culture medium) with HeLa cell monolayers at 37°C for 3 h. The monolayers were washed three times with PBS to remove nonadherent bacteria, fixed with 70% methanol, and stained with 10% Giemsa for 15 min. E. coli strain EDL933 was used as a positive control in the adherence assay. The adherence patterns were examined under ×400 magnification using a light microscope (Olympus). The adhesion index was determined as the percentage of epithelial cells with adhering bacteria; if at least 40% of the HeLa cells had adhering bacteria, the strain was considered positive (10).
Assay of hemolysin and cell-associated hemagglutinating activity.
The hemolytic and cell-associated hemagglutinating activities of the strains with human erythrocytes were determined as described previously (34). The amount of released hemoglobin in the supernatant was measured spectrophotometrically (U-3210; Hitachi, Japan) at 540 nm. An optical density of ≥0.1 was considered positive for hemolysin. Finally, the results were expressed as the percentage of lysis by comparing these optical density values with that of an identical erythrocyte suspension lysed (100%) with an equal volume of Triton X-100 solution. For cell-associated hemagglutinating activity, strains were recorded as having a reaction that was immediate and complete or as a response that was incomplete or that was not instantaneous but occurred within 5 min. PBS was included for each assay as the negative control.
Autoagglutination.
For the autoagglutination test, strains were grown in LB (pH 6.5) supplemented with 1% (wt/vol) NaCl and incubated at 37°C with aeration for 18 h. A visible clumping of bacteria indicated a positive result (6, 46).
Detection of extracellular enzymes.
Proteolytic (gelatinase and HA/protease) and lipolytic (lipase) activities were evaluated by the plate assay method (48). A clear zone around the bacterial colony indicated a positive result. A known strain of V. cholerae O1 for gelatinase and HA/protease and Pseudomonas aeruginosa for lipase were included as the positive controls.
Serum resistance test.
The susceptibility of bacteria to human serum was determined by the method of Hughes et al. (16). Twenty-five microliters of bacterial suspension (ca. 106 CFU/ml) in PBS and 75 μl of undiluted normal human serum were kept in a sterile microtiter tray and incubated at 37°C. Responses were graded as highly sensitive, intermediately sensitive, or resistant according to the established system (16). A control experiment was done by replacing the 75 μl of normal human serum with 75 μl of PBS, with all other conditions remaining the same as for the test experiment. Each test was repeated three times.
Antimicrobial susceptibility test.
Antimicrobial susceptibility testing was performed by the disk diffusion method (4) using commercially available disks (HiMedia) with 11 antimicrobial drugs on Mueller-Hinton agar (HiMedia). Isolates were considered susceptible, reduced susceptible, or resistant to a particular antimicrobial agent on the basis of the diameters of the inhibitory zones that matched the criteria of the manufacturer's interpretive table, which followed the recommendations of the National Committee for Clinical Laboratory Standards (29). E. coli ATCC 25922 was used for quality control.
Enteropathogenicity assay.
The enteropathogenicity of the strains was examined in the sealed-adult-mouse model of Richardson et al. (36) using BALB/c mice weighing ca. 10 to 20 g. The animals, kept in wire-mesh polycarbonate cages with autoclaved bedding, were acclimated to laboratory conditions (12-h dark/12-h light cycles; 24 ± 1°C) and had free access to food and water ad libitum. The mice fasted for 24 h before administration of bacterial inocula. For the preparation of the bacterial inocula, V. cholerae strains were grown in TSB at 37°C with shaking. After being harvested by centrifugation, the cells were resuspended in PBS. The mice were given 100 μl of 5% sodium bicarbonate (wt/vol) intragastrically with a ball-tipped inoculating needle. After 15 min, the bacterial inocula (2 × 1010 CFU/ml) in 200 μl of PBS were given to the test mouse. At 5 h postinoculation, the animals were sacrificed, and the fluid accumulation (FA) ratios were determined (36). FA ratios of ≥100 were considered positive (23). For the colonization assay, infections were allowed to proceed for 18 h. The mice were sacrificed, and the intestines were aseptically removed. Sections from the intestine were washed with PBS to remove unbound bacteria, weighed, and homogenized in PBS. Various dilutions were plated on TCBS agar and incubated at 37°C for 24 h. CFU counts were taken, and the colonization ability was expressed as log10 CFU per g of tissue.
RESULTS
One hundred and thirty-nine samples of natural surface waters were collected from 16 sites located in different diarrhea endemic zones in and around Kolkata. Sixty-one (44%) of the samples contained presumptive V. cholerae, and the species was isolated from surface waters in 6 of 16 sites sampled (sites, 1, 6, 8, 11, 14, and 16) (Fig. 1). All the isolates were gram negative, rod shaped, positive for oxidase, and motile. All the isolates grew in TCBS and MacConkey agars. A total of 250 isolates of V. cholerae were tested for serotyping using O1 antiserum and O139 MAb, and none of the isolates was agglutinated with these antibodies. These strains of V. cholerae were inferred to be non-O1, non-O139 serogroups. Twenty-one of these V. cholerae isolates were randomly selected, including at least two strains from each site, for further characterization. Serotyping of the 21 non-O1, non-O139 strains revealed that they belonged to serogroups O5, O9, O14, O20, O26, O28, O34, O48, O64, O65, O119, O121, O144, O149/O151, and O201. Six isolates could not be typed with available 206 O antisera (Table 1). In addition, these strains were positive for ompWVc, which is specific to V. cholerae (27).
TABLE 1.
Biochemical characteristics of isolates from surface waters in different regions of Kolkata, India
| Biochemical testa | Result in strain (serogroup)b
|
||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SG24 (O139) | NB2 (O1) | PC1 (O65) | PC2 (O144) | PC3 (O119) | PC4 (O26) | PC5 (O64) | PC6 (OUT) | PC7 (OUT) | PC8 (O9) | PC9 (O14) | PC10 (OUT) | PC11 (OUT) | PC13 (OUT) | PC14 (O20) | PC15 (OUT) | PC32 (O149/O151) | PC57 (O5) | PC65 (O48) | PC89 (O201) | PC90 (O121) | PC91 (O34) | PC96 (O28) | |
| Oxidase | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| ONPG | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| DNase | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Motility | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| ADH | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| LDC | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + |
| ODC | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Citrate | + | + | + | + | − | + | + | + | − | + | + | + | + | + | + | + | − | + | − | − | + | + | − |
| dl-Lactate | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Urease | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| TDA | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Indole | + | + | − | + | + | + | − | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + |
| VP reaction | − | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | − | + | + | + | + | − | + |
| Gelatinase | + | + | − | + | + | + | − | + | + | + | + | + | + | + | + | − | − | + | + | + | + | − | + |
| Glucose (gas) | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Acid from: | |||||||||||||||||||||||
| Glucose | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| d-Mannitol | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | − | − | + | + | + | + |
| m-Inositol | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| d-Sorbitol | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − |
| l-Rhamnose | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − |
| Sucrose | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Melibiose | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − |
| Amygdalin | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| l-Arabinose | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − |
| Mannose | − | − | + | − | − | − | + | − | − | − | − | + | + | − | + | − | + | − | − | + | − | + | − |
| Salicin | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − |
| Lactose | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | + | − | − | − | + | − | − |
| String test | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + |
| Catalase | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Growth in: | |||||||||||||||||||||||
| 0% NaCl | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 1% NaCl | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 7% NaCl | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| TCBS | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| H2S | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| KIA | K/ANG | K/ANG | K/ANG | K/ANG | K/ANG | K/ANG | K/ANG | K/ANG | K/ANG | K/ANG | K/ANG | K/ANG | K/ANG | K/ANG | K/ANG | K/ANG | K/ANG | K/ANG | K/ANG | K/ANG | K/ANG | K/ANG | K/ANG |
| Nitrate reduction | + | + | + | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − |
| Sensitivity to O/129 (150 μg) | R | S | S | S | S | S | S | S | S | S | S | S | R | S | S | S | R | S | S | R | S | S | S |
ONPG, orthonitrophenyl galactoside; ADH, arginine dihydrolase; LDC, lysine decarboxylase; ODC, ornithine decarboxylase; TDA, tryptophan deaminase; KIA, Kligler iron agar; VP, Voges-Proskauer.
OUT, O untypeable; Y, yellow colony; K, alkaline slant; A, acid butt; NG, no gas; S, susceptible; R, resistant; +, plus; −, negative.
The biochemical characteristics of the isolates are shown in Table 1, and for comparison, the results of biochemical tests of standard toxigenic V. cholerae O1 El Tor (strain NB2) and O139 (SG24) were included. All the environmental isolates were invariably positive in nine tests (orthonitrophenyl galactoside, ornithine decarboxylase, DNase, catalase, growth in 0 and 1% NaCl, dl-lactate, and acid production from glucose and sucrose) and invariably negative for another eight tests (arginine dihydrolase, gas production from glucose, tryptophan deaminase, acid production from amygdalin and m-inositol, urease, H2S production, and growth in 7% NaCl). Variable results were obtained in the following tests: ≥90% of the isolates were positive for the Voges-Proskauer test, lysine decarboxylase, acid from d-mannitol, and nitrate reduction, and 40 to 80% of the isolates were positive for utilization of citrate, indole, gelatinase, and acid from mannose.
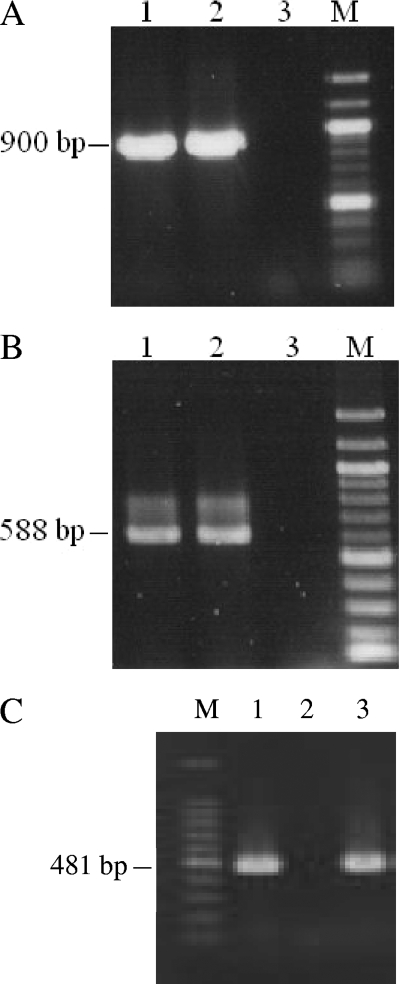
In the PCR assay, all of the non-O1, non-O139 isolates yielded positive results for the toxR gene, and except for one (PC65), all of the isolates yielded negative results for the tcpA genes (classical and El Tor alleles). The isolate PC65 gave a positive result for the classical variant of tcpA. Furthermore, the clinical non-O1, non-O139 strains were determined to be negative for the tcpA (classical and El Tor alleles) and ctxA genes. In addition, none of the isolates was positive for ctxA. All of the isolates were negative for the genes representing the Shiga toxin variants, heat-stable toxin, and heat-labile toxin of Escherichia coli. Additionally, except for one strain (PC32), all of the non-O1, non-O139 strains were positive for the gene encoding El Tor hemolysin. PCR results for the toxR, ompWVc, and hlyA genes of the representative strains are shown in Fig. 2.
FIG. 2.
Amplification of toxR (900 bp) (A), ompWVc (588 bp) (B), and hlyA (El Tor; 481 bp) (C) by PCR. (A and B) Lanes: 1, V. cholerae O139 strain SG24 (positive control); 2, non-O1, non-O139 strain PC1; and 3, Aeromonas hydrophila strain PC16 (negative control). (C) Lanes 1 and 3, V. cholerae strains NB2 (O1; El Tor; positive control) and PC1; lane 2, A. hydrophila strain PC16 (negative control); lane M, 100-bp ladder.
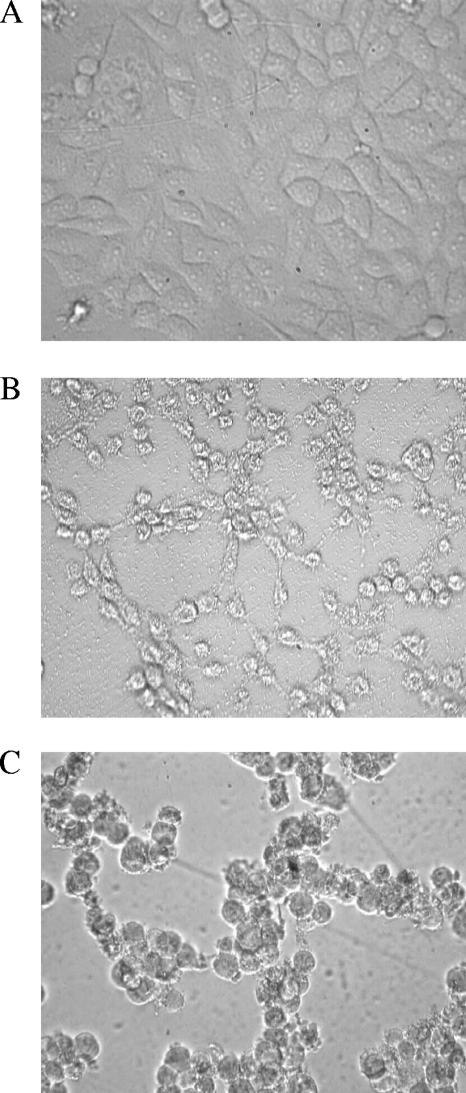
The effects of cell-free culture filtrates of the test isolates on HeLa cells are shown in Fig. 3. The cytotoxic responses of the test isolates could be categorized into two types: type 1, which was associated with complete membrane damage (Fig. 3B), and type 2, which was associated with cell rounding (cell-rounding factor) (Fig. 3C). Cell culture supernatants of 86% (18 of 21) of the environmental isolates grown in TSB medium evoked a distinct cytotoxic effect on HeLa cells with an end point titer of 4 to 128 (the reciprocal of the dilution that showed a cytotoxic effect on ≥50% of the cells), while culture supernatants of 8 (38%) isolates also produced a cell-rounding effect when they were cultured in AKI medium (Table 2). Four and one (AS67) clinical non-O1, non-O139 isolates exhibited cytotoxic and cell-rounding effects on HeLa cells, respectively, as detected in a previous study (43).
FIG. 3.
Effects of culture filtrates of the environmental isolates on HeLa cells. Confluent growth of HeLa cells (A), cytotoxic effect (B), and cytotoxic effect associated with cell rounding (C). Magnification, ×400.
TABLE 2.
Virulence phenotypes of environmental and clinical strains of V. cholerae non-O1, non-O139
| Source | Strain | Serotype | Cytotoxic activity (end point titer)a | Hemolytic activity (OD540)b | Lipase activityc | Protease activityc | Hemagglutinating activityd | Serum sensitivity gradee | Adherencef | Resistant/reduced susceptible (reference)g |
|---|---|---|---|---|---|---|---|---|---|---|
| Environment | PC1 | O65 | Cr (32) | 100 (1.7) | + | − | − | 1-2 | Diffuse | −/Fz A |
| PC2 | O144 | Ct (32) | 100 (1.7) | + | + | + | 5-6 | − | A/T S N Fz | |
| PC3 | O119 | Cr (4) | 12 (0.2) | + | + | ++ | 5-6 | Diffuse | −/T S N Fz | |
| PC4 | O26 | Ct (64) | 12 (0.2) | + | + | ++ | 5-6 | Carpet-like | A Fz/T S G | |
| PC5 | O64 | Cr (128) | 94 (1.6) | + | − | + | 1-2 | Diffuse | A/N | |
| PC6 | OUT | Cr (4) | 24 (0.4) | + | − | ++ | 1-2 | − | −/A T N Fz | |
| PC7 | OUT | Ct (32) | 12 (0.2) | + | − | ++ | 5-6 | Diffuse | A Fz/T S N Na Nx | |
| PC8 | O9 | Ct (32) | 94 (1.6) | + | + | + | 1-2 | Diffuse | −/A T Fz | |
| PC9 | O14 | Ct (128) | 59 (1.0) | + | + | + | 3-4 | Aggregative | −/A T Fz N | |
| PC10 | OUT | Cr (32) | 59 (1.0) | + | + | ++ | 3-4 | Carpet-like | −/A T N | |
| PC11 | OUT | 0 (0.0) | + | − | − | 5-6 | Aggregative | A/Co | ||
| PC13 | OUT | 0 (0.0) | + | − | − | NDh | ND | ND | ||
| PC14 | O20 | Cr (4) | 6 (0.1) | + | − | ++ | 5-6 | Diffuse | A T/N Fz | |
| PC15 | OUT | Ct (16) | 94 (1.6) | + | − | ++ | 3-4 | Diffuse | A T Co/S N Fz | |
| PC32 | O149/O151 | 0 (0.0) | − | − | + | 3-4 | ND | ND | ||
| PC57 | O5 | Ct (64) | 59 (1.0) | + | + | ++ | 3-4 | Diffuse | A Fz/T S N G Co Cf | |
| PC65 | O48 | Ct (32) | 94 (1.6) | + | − | + | 3-4 | − | ND | |
| PC89 | O201 | Ct (64) | 12 (0.2) | + | + | ++ | 5-6 | Carpet-like | ND | |
| PC90 | O121 | Ct (32) | 59 (1.0) | + | + | + | 3-4 | Diffuse | ND | |
| PC91 | O34 | Cr (32) | 88 (1.5) | + | − | − | 1-2 | Diffuse | ND | |
| PC96 | O28 | Cr (4) | 12 (0.2) | + | + | ++ | 5-6 | Diffuse | −/T S N Fz | |
| Clinical | PL2 | O10 | Ct (4) | 6 (0.1) | + | + | ++ | 5-6 | Diffuse | A Fz N/− (6) |
| PL71 | O7 | Ct (16) | 35 (0.6) | + | + | − | 3-4 | ND | AFz T/− | |
| PG5 | O12 | Ct (32) | 41 (0.7) | + | + | ++ | 5-6 | Diffuse | A Fz/ (6) | |
| PG109 | O27 | Ct (16) | 24 (0.4) | + | − | − | 1-2 | ND | A Cf Fz N/− (6) | |
| AS67 | O190 | Cr (32) | 35 (0.6) | + | + | + | ND | ND | A Fz N S/− (38) |
Reciprocal of dilution of TSB/AKI culture filtrate producing cytotoxic effect on ≥50% of HeLa cells. Ct, cytotoxic; Cr, cell rounding.
TSB culture filtrate was mixed 1:1 with human erythrocytes suspended in PBS and incubated at 37°C for 1 h. The released hemoglobin was measured spectrophotometrically. OD540, optical density at 540 nm.
+, positive; −, negative.
Cell-associated hemagglutinating activity was determined using pooled group O human blood cells. ++, reaction was immediate; +, reaction was incomplete or not instantaneous but occurred within 5 min; −, no agglutination.
Following Hughes et al. (16). Grades 5-6, resistant; grades 3-4, intermediate (delayed) sensitive; grades 1-2, completely sensitive to normal human serum.
−, negative (<40% of HeLa cells had adhering bacteria).
A, ampicillin (10 μg); C, chloramphenicol (30 μg); T, tetracycline (30 μg); S, streptomycin (10 μg); N, neomycin (30 μg); G, gentamicin (10 μg); Na, nalidixic acid (30 μg); Fz, furazolidone (100 μg); Nx, norfloxacin (10 μg); Co, cotrimoxazole (25 μg); Cf, ciprofloxacin (5 μg); −, negative (i.e., susceptible to the rest of the antibiotics tested).
ND, not determined.
Twenty-five (20 environmental and 5 clinical) non-O1, non-O139 isolates were positive for lipase production, and 14 isolates (10 environmental and 4 clinical) showed protease activity (Table 2). Seventeen (81%) environmental and four clinical isolates showed hemolytic activity against human erythrocytes (above 10%). However, seven of the environmental isolates had very high hemolytic activities (80 to 100%), while none of the clinical strains belonged to this group (Table 2). Twenty isolates (17 environmental and 3 clinical) showed moderate to strong cell-associated hemagglutination (Table 2).
The incidences of serum resistance of the isolates belonging to both groups were found to vary considerably (Table 2).
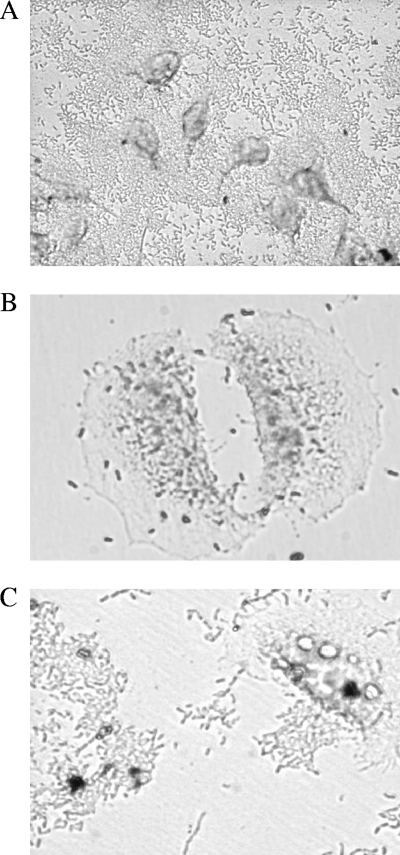
Three adherence patterns were manifested by the isolates; carpet-like adherence (Fig. 4A), diffuse adherence (Fig. 4B), and aggregative adherence (Fig. 4C). Eighteen (86%) of the isolates adhered to HeLa cells with an index of ≥40% (Table 2).
FIG. 4.
Adherence patterns of the environmental isolates on HeLa cells by Giemsa stain. (A) Carpet-like adherence to HeLa cells and a glass surface. (B) Diffuse adherence; bacteria are dispersed over the cell surface and the cell matrix. (C) Aggregative adherence; bacteria adhere to each other away from the cells, as well as to the cell surface. Magnification, ×400.
Multidrug resistance was exhibited by both clinical and environmental isolates (Table 2). Resistance and reduced susceptibility to ampicillin, furazolidone, neomycin, streptomycin, tetracycline, and ciprofloxacin were observed among these isolates. In addition, environmental isolates showed reduced susceptibility to cotrimoxazole, gentamicin, nalidixic acid, and norfloxacin.
Eleven environmental and two clinical isolates of non-O1, non-O139 V. cholerae were tested for their enteropathogenic potentials in the sealed-adult-mouse model using live bacterial cells. A toxigenic strain of V. cholerae O139 (SG24) was used as a positive control. Of the 11 environmental isolates, eight induced positive FA (FA ≥ 100), and the reactivities of these isolates were comparable to those of clinical isolates of non-O1, non-O139 and O139 V. cholerae (Table 3). In addition, two isolates (PC6 and PC9) showed FAs significantly higher than that in the PBS control (P < 0.05). A comparison of the counts of the colonized environmental and clinical strains in mouse intestines showed that the isolates of both groups had almost the same colonizing potential. Colonization, in mean log10 CFU ± standard deviation per gram of tissue, ranged between 5.31 ± 1.25 and 9.90 ± 1.40 (Table 3).
TABLE 3.
Enteropathogenicities of environmental and clinical isolates of V. cholerae in the mouse modela
| Source | Strain | Virulence phenotypeb | Colonization (log10 CFU/g of tissue [mean ± SD])c | FA ratio (mean ± SD)c |
|---|---|---|---|---|
| Environment | PC1 | (Cyt Hly Lip Ad)+ (Hae Pro SR)− | 6.51 ± 1.86 | 100.5 ± 4.50 |
| PC2 | (Cyt Hly Hae Lip Pro SR)+ Ad− | 7.47 ± 0.62 | 63.67 ± 6.13 | |
| PC4 | (Cyt Hly Hae Lip Pro SR Ad)+ | 5.40 ± 0.80 | 118.65 ± 1.82 | |
| PC5 | (Cyt Hly Hae Lip Ad)+ (Pro SR) − | 5.36 ± 0.59 | 104.38 ± 8.68 | |
| PC6 | (Cyt Hly Hae Lip)+ (Pro SR Ad)− | 6.00 ± 2.05 | 86.15 ± 3.15 | |
| PC8 | (Cyt Hly Hae Lip Pro Ad)+ SR− | 6.44 ± 0.29 | 138.25 ± 0.75 | |
| PC9 | (Cyt Hly Hae Lip Pro SR Ad)+ | 8.60 ± 0.43 | 94.26 ± 10.66 | |
| PC10 | (Cyt Hly Hae Lip Pro SR Ad)+ | 8.19 ± 0.15 | 126.70 ± 35.30 | |
| PC11 | (Lip SR Ad)+ (Cyt Hly Hae Pro)− | 5.45 ± 0.87 | 129.54 ± 27.28 | |
| PC15 | (Cyt Hly Lip SR Ad)+ (Hae Pro)− | 8.99 ± 1.65 | 176.00 ± 23.00 | |
| PC32 | (Hae SR)+ AdND (Cyt Hly Lip Pro)− | 5.38 ± 1.76 | 150.00 ± 40.82 | |
| Clinical | PG5 | (Cyt Hly Hae Lip Pro SR Ad)+ | 5.31 ± 1.25 | 155.33 ± 39.50 |
| PL2 | (Cyt Hly Hae Lip Pro SR Ad)+ | 7.94 ± 0.18 | 126.67 ± 20.50 | |
| SG24 | 9.90 ± 1.40 | 133.00 ± 27.00 | ||
| PBS control | 62.00 ± 2.80 |
All the non-O1, non-O139 strains were negative for the ctxA, elt, stx1, and stx2 genes, and the O139 strain (SG24) was positive for ctxA.
+, presence, and −, absence of listed phenotype; ND, not determined; Cyt, cytotoxic (HeLa cells); Hly, hemolysin (active against human erythrocytes); Hae, hemagglutination (human O-group blood); Pro, protease; Lip, lipase; SR, serum resistance; Ad, adherence (HeLa cells).
The values represent the average of results in three to five adult BALB/c mice.
DISCUSSION
In the present study, the biochemical and virulence traits of V. cholerae non-O1, non-O139 isolates from surface waters from different sites sampled in Kolkata and the surrounding area were extensively characterized to evaluate their pathogenic potentials. Members of the genus Vibrio are fermentative, facultatively anaerobic, gram-negative, straight or curved motile rods. The growth of these bacteria is stimulated by Na+, which, except for V. cholerae and Vibrio mimicus, is an absolute requirement (18). V. cholerae and V. mimicus are biochemically similar species and can be differentiated by sucrose fermentation. V. mimicus cannot ferment sucrose. All of the environmental isolates of non-O1, non-O139 V. cholerae examined showed growth in 1% peptone broth without NaCl and were able to ferment sucrose. V. cholerae gives a positive string test (47). All of the environmental strains except strain PC32 showed a positive string reaction. The oxidase reaction differentiates Vibrio metschnikovii, which is oxidase negative, from other sucrose-fermenting Vibrio species (18). The lysine and ornithine decarboxylase and arginine dihydrolase activities of the pathogenic Vibrios are useful for separating them into groups (18). V. cholerae is arginine dihydrolase negative and lysine and ornithine decarboxylase positive (15). All of the environmental isolates included in this study were arginine dihydrolase negative and ornithine decarboxylase positive, and all of these strains except strain PC32 were positive for lysine decarboxylase. Of the 21 environmental isolates, 6 and 14 belonged to Heiberg groups I (mannose and sucrose fermenting and arabinose nonfermenting) and II (sucrose fermenting and mannose and arabinose nonfermenting), respectively. There did not appear to be any correlation between the sites sampled and the Heiberg fermentation patterns of the isolates. V. cholerae gives negative reactions for d-sorbitol, l-rhamnose, melibiose, l-arabinose, and salicin fermentation (15). However, all of the environmental isolates except PC32 were negative for sorbitol, rhamnose, melibiose, arabinose, and salicin fermentation. The positive activities for lactate and DNase and negative reaction for H2S production differentiate V. cholerae from Aeromonas sobria, which is sucrose fermenting, arginine dihydrolase negative, and lysine decarboxylase positive (14). The susceptibility of vibrios to the vibriostatic agent O/129 is an important taxonomical trait to differentiate between vibrios and Aeromonas spp. However, all the V. cholerae O139 and several V. cholerae O1 strains were resistant to this vibriostatic agent (23). In the present study, three (14%) of the environmental isolates showed resistance to O/129.
Several extracellular products of V. cholerae play important roles in the disease process, including CT, a heat-stable toxin of E. coli, a Shiga-like toxin, hemolysin, and HAs (2, 18, 34, 44). The non-O1, non-O139 isolates tested in the present study belonged to different serogroups and did not possess the genes encoding heat-stable toxin, heat-labile toxin, or Shiga toxins of E. coli and CT. However, from the present study, it appears that the majority of the environmental and clinical isolates produced a cytotoxic effect on HeLa cells. It has previously been reported that the cytotoxic effect of non-O1 V. cholerae is probably related to the production of hemolysin (17). However, some of our isolates with weak hemolytic activity showed strong cytotoxic activity. It could be that cytotoxic activity against HeLa cells and hemolytic activity against human erythrocytes are elicited by two different factors produced by these vibrios. It has been demonstrated that the purified non-membrane-damaging cytotoxin has enterotoxic activity (40), and a cell-rounding effect was associated with human diarrhea with sole infection by non-O1 V. cholerae (34, 43). Interestingly, some of our isolates evoked a cell-rounding effect, followed by cell death, on HeLa cells. These isolates also showed significant enterotoxic activity in a mouse model (FA ≥ 100).
The non-O1, non-O139 isolates showed strong adherence to HeLa cells. For many bacterial enteropathogens, the ability to adhere to the intestinal mucosa is a first step in colonization and is a prerequisite for causing diarrhea (22). In a previous study, occurrences of diffuse, as well as clustered, adherence to HEp-2 cells were observed among the vibrios recovered from estuarine waters (3). Of three of the HeLa cell adherence patterns observed among the isolates, diffuse adherence was manifested by most of the V. cholerae isolates in our study. In addition, two environmental strains (PC9 and PC11) exhibited aggregative adherence, identical to the patterns shown by enteroaggregative E. coli, and these strains were also able to colonize the mouse intestine and showed FA reaction. It is interesting that a carpet-like adherence by three environmental isolates (serogroup O26, OUT [O untypeable], and O201) to HeLa cells similar to the reported adherence pattern of a clinical strain of V. cholerae VIG1587 (serogroup O12) to HEp-2 cells and a glass surface (12) was observed in the present study. Except for PC65, all of our non-O1, non-O139 isolates were negative for the genes representing TcpA (classical and El Tor alleles). Previously, it was also reported that the majority of strains belonging to non-O1, non-O139 serogroups do not contain genes for CT and/or TCP (7, 22, 43). Although TCP protein is one of the colonization factors in V. cholerae, some of our isolates showed adherence to HeLa cells and colonized mouse intestine, and 14 (12 environmental and 2 clinical) showed an autoagglutination phenotype that was previously shown to be correlated with the expression of TcpA (6). However, pilins, as well as outer membrane proteins, of the enteropathogenic bacteria have been demonstrated to be autoagglutinins (6, 46). Furthermore, V. cholerae non-O1, non-O139 strains possess other fimbrial antigens, surface polysaccharide, hemagglutinating function, and outer membrane proteins, and the involvement of these cell surface antigens in adherence and intestinal colonization has been demonstrated (26, 30, 42).
Proteasic, hemagglutinating, and hemolytic activities have been related to virulence in Vibrio spp. in several findings (1, 17, 18, 42, 45). Although our non-O1, non-O139 strains were producers of these putative virulence factors, the precise roles of the factors in vivo are not yet clear. However, it has previously been suggested that hemagglutinins of non-O1 strains are important for their survival in aquatic environments because they govern the ability to attach to different substrates (1). In addition, a previous study demonstrated that HA/protease (HapA) of V. cholerae El Tor was necessary for full expression of enterotoxicity, and hapA (encoding HapA) mutants expressed less CT and TcpA (45). However, further study is needed to establish the relationship between HA production and full expression of enterotoxicity for non-O1, non-O139 strains. Although the majority of our strains were producers of lipase, the precise role of this protein in pathogenesis has yet to be determined. It has been reported that lipase (LipA) could potentially damage host cells, but mutation of the lipA gene did not affect colonization potential or virulence in an infant mouse cholera model (32). Since LipA is encoded in the vicinity of hlyA, and HlyA toxin is capable of host tissue damage, which would release a variety of cellular components, including membrane lipids that could be degraded by lipases, it has been proposed that lipases are involved in the acquisition of nutrients (32). V. cholerae is known to be a noninvasive organism that colonizes the extracellular mucosal surface in the gut. It has been postulated that the bactericidal activity of antibody and the “complement-like” bactericidal activity are operative in the intestinal mucosa, contributing to the colonization properties of a variety of bacterial pathogens (33). If this is true, then serum resistance properties of V. cholerae could play an essential role in intestinal colonization, and it is interesting that some of our isolates showed serum resistance properties.
An earlier study revealed that the drug resistance patterns of the clinical and environmental strains showed remarkable differences, with the clinical strains being resistant to more drugs and exhibiting multidrug resistance along with their environmental counterparts (7). However, in the present study, multidrug resistance that included resistance/reduced susceptibility to ampicillin, tetracycline, cotrimoxazole, furazolidone, streptomycin, or neomycin was exhibited by the majority of the strains studied.
Previously, it was reported that V. cholerae non-O1 strains that produced hemolysin, that were cytotoxic, and that produced HAs were isolated from hospitalized patients most often as the sole pathogen (34). Our findings in this study revealed that strains of non-O1, non-O139 V. cholerae isolated from surface waters were capable of expressing the putative virulence traits that were associated with the clinical isolates from the same geographical area. It was also interesting that some of the environmental isolates showed enterotoxic activity and colonization in mouse intestines that were comparable to those of toxigenic strains of V. cholerae O139. Studies with the O10 and O12 strains of V. cholerae isolated from an outbreak in Peru showed that none of the strains produced enterotoxin but a majority of the strains produced a cytotoxin, as assessed in Y1 and HeLa cells (12). Three of our environmental isolates (PC11, PC13, and PC32) were nonhemolytic and did not evoke a cytotoxic response to HeLa cells. In addition, strains PC11 and PC13 were positive and strain PC32 was negative for hlyA (El Tor) as determined by PCR. It was interesting that two of these strains (PC11 and PC32) showed significantly high enterotoxic activities. Collectively, the results presented here failed to show any absolute correlation between serogroups and hemagglutination, hemolytic, protease, or cytotoxic activities and enterotoxic activities among the strains of both groups tested (Table 3). Thus, our findings agree with the previous findings that the enteropathogenicity of non-O1, non-O139 V. cholerae is multifactorial, and the presence of a single factor should not be construed as the cause of pathogenicity (34). It still remains unclear which factor(s) is specific for the determination of the enteropathogenic potential of non-O1, non-O139 V. cholerae. The results of the present investigation indicate that further epidemiological studies and continuous monitoring are necessary to elucidate the ecology and the public health significance of V. cholerae in the aquatic environment.
Acknowledgments
This work was partially supported by the University Grants Commission (UGC), New Delhi, India, and the Department of Science and Technology, Government of West Bengal, India. The fellowships of T. K. Hajra and P. Bhowmik were provided by UGC.
Footnotes
Published ahead of print on 18 July 2008.
REFERENCES
- 1.Amaro, C., A. E. Toranzo, E. A. González, J. Blanco, M. J. Pujalte, R. Aznar, and E. Garay. 1990. Surface and virulence properties of environmental Vibrio cholerae non-O1 from Albufera Lake (Valencia, Spain). Appl. Environ. Microbiol. 56:1140-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagchi, K., P. Echeverria, J. D. Arthur, O. Scthabutr, O. Serichantalergs, and C. W. Hoge. 1993. Epidemic diarrhoea caused by Vibrio cholerae non-O1 that produced heat-stable toxin among Khmers in a camp in Thailand. J. Clin. Microbiol. 31:1315-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbieri, E., L. Falzano, C. Fiorentini, A. Pianetti, W. Baffone, A. Fabbri, P. Matarrese, A. Casiere, M. Katouli, I. Kühn, R. Möllby, F. Bruscolini, and G. Donelli. 1999. Occurrence, diversity, and pathogenicity of halophilic Vibrio spp. and non-O1 Vibrio cholerae from estuarine waters along the Italian Adriatic coast. Appl. Environ. Microbiol. 65:2748-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauer, A. W., W. M. Kirby, J. C. Sherris, and M. Jurck. 1996. Antibiotic susceptibility testing by a standardized single disc method. Am. J. Pathol. 45:493-496. [PubMed] [Google Scholar]
- 5.Bhattacharya, S. K., M. K. Bhattacharya, T. Ramamurthy, A. Pal, P. K. Bag, T. Takeda, S. Chakraborti, P. Dutta, A. Debnath, S. C. Pal, and G. B. Nair. 1992. Acute secretory diarrhoea caused by V. cholerae non-O1 which does not produce cholera like and heat stable enterotoxins. J. Diar. Dis. Res. 10:161-163. [PubMed] [Google Scholar]
- 6.Chakraborty, S., A. K. Mukhopadhyay, R. K. Bhadra, A. N. Ghosh, R. Mitra, T. Shimada, S. Yamasaki, S. M. Faruque, Y. Takeda, R. R. Colwell, and G. B. Nair. 2000. Virulence genes in environmental strains of Vibrio cholerae. Appl. Environ. Microbiol. 66:4022-4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakraborty, S., P. Garg, T. Ramamurthy, M. Thungapathra, J. K. Gautam, C. Kumar, S. Maity, S. Yamasaki, T. Shimada, Y. Takeda, A. Ghosh, and G. B. Nair. 2001. Comparison of antibiogram, virulence genes, ribotypes and DNA fingerprints of Vibrio cholerae of matching serogroups isolated from hospitalised diarrhoea cases and from the environment during 1997-1998 in Calcutta, India. J. Med. Microbiol. 50:879-888. [DOI] [PubMed] [Google Scholar]
- 8.Collee, J. G., A. G. Fraser, B. P. Marmion, and A. Simmons (ed.). 1996. Mackie and McCartney practical medical microbiology, 14th ed. Churchill Livingstone, New York, NY.
- 9.Colwell, R. R., A. Huq, M. A. R. Chowdhury, P. R. Brayton, and B. Xu. 1995. Serogroup conversion of Vibrio cholerae. Can. J. Microbiol. 41:946-950. [DOI] [PubMed] [Google Scholar]
- 10.Cravioto, A., R. J. Gross, S. M. Scotland, and B. Rowe. 1979. An adhesive factor in strains of Escherichia coli belonging to the traditional infantile enteropathogenic serotypes. Curr. Microbiol. 3:95-99. [Google Scholar]
- 11.Dakin, S. P. H., D. J. Howel, R. G. A. Sutton, M. E. O'Keffe, and P. Thoms. 1994. Gastroenteritis due to non-agglutinable (non-cholera) vibrios. Med. J. 2:490. [DOI] [PubMed] [Google Scholar]
- 12.Dalsgaard, A., N. J. Albert, D. N. Taylor, T. Shimada, R. Meza, O. Serichantalergs, and P. Echeverria. 1995. Characterization of Vibrio cholerae non-O1 serogroups obtained from an outbreak of diarrhea in Lima, Peru. J. Clin. Microbiol. 33:2715-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ewing, W. H., and B. R. Davis. 1981. Medium and tests for differentiation of Enterobacteriaceae. U.S. Department of Health and Human Services publication no. CDC 81-8236. U.S. Department of Health and Human Services, Washington, DC.
- 14.Ghosh, C., R. K. Nandy, S. K. Dasgupta, G. B. Nair, R. H. Hall, and A. C. Ghose. 1997. A search for cholera toxin (CT), toxin coregulated pilus (TCP), the regulatory element ToxR and other virulence factors in non-O1/non-O139 Vibrio cholerae. Microb. Pathog. 22:199-208. [DOI] [PubMed] [Google Scholar]
- 15.Holt, J. G., N. R. Krieg, P. H. Sneath, J. T. Staley, and S. T. Williams. 1994. Bergey's manual of determinative bacteriology, 9th ed. Williams and Wilkins, Baltimore, MD.
- 16.Hughes, C., R. Phillips, and A. P. Robert. 1982. Serum resistance among Escherichia coli strains causing urinary tract infection in relation to O type and the carriage of hemolysin, colicin, and antibiotic resistance determinants. Infect. Immun. 35:270-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ichinose, Y., K. Yamamoto, N. Nakasone, M. J. Tanabe, T. Takeda, T. Miwatani, and M. Iwanaga. 1987. Enterotoxicity of ElTor-like haemolysin of non-O1 Vibrio cholerae. Infect. Immun. 55:1090-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janda, J. M., C. Powers, R. G. Bryant, and S. L. Abbott. 1988. Current perspectives on the epidemiology and pathogenesis of clinically significant Vibrio spp. Clin. Microbiol. Rev. 1:245-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karaolis, D. K. R., R. Lan, and P. R. Reeves. 1995. The sixth and seventh cholera pandemics are due to independent clones separately derived from environmental, nontoxigenic, non-O1 Vibrio cholerae. J. Bacteriol. 177:3191-3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keasler, S. P., and R. H. Hall. 1993. Detecting and biotyping Vibrio cholerae O1 with multiplex chain reaction. Lancet 341:1661. [DOI] [PubMed] [Google Scholar]
- 21.Khan, A., S. Yamasaki, T. Santo, T. Ramamurthy, A. Pal, S. Datta, N. R. Chowdhury, S. C. Das, A. Sikdar, T. Tsukamoto, S. K. Bhattacharya, Y. Takeda, and G. B. Nair. 2002. Prevalence and genetic profiling of virulence determinants of non-O157 Shiga toxin producing Escherichia coli isolated from cattle, beef and human cases in Calcutta, India. Emerg. Infect. Dis. 8:54-62. [PubMed] [Google Scholar]
- 22.Morris, J. G., Jr., T. Takeda, B. D. Tall, G. A. Losonsky, S. K. Bhattacharya, B. D. Forrest, B. A. Kay, and M. Nishibuchi. 1990. Experimental non-O group 1 Vibrio cholerae gastroenteritis in humans. J. Clin. Investig. 85:697-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moyenuddin, M., K. Wachsmuth, S. H. Richardson, and W. L. Cook. 1992. Enteropathogenicity of non-toxigenic Vibrio cholerae O1 for adult mice. Microb. Pathog. 12:451-458. [DOI] [PubMed] [Google Scholar]
- 24.Mukhopadhyay, A. K., P. K. Saha, S. Garg, S. K. Bhattacharya, T. Shimada, T. Takeda, Y. Takeda, and G. B. Nair. 1995. Distribution and virulence of Vibrio cholerae belonging to serogroups other than O1 and O139: a nation survey. Epidemiol. Infect. 114:65-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nair, G. B., S. Misra, R. K. Bhadra, and S. C. Pal. 1987. Evaluation of the multitest medium for rapid presumptive identification of Vibrio cholerae from environmental sources. Appl. Environ. Microbiol. 53:1203-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakasone, N., and M. Iwanaga. 1990. Pili of V. cholerae non-O1. Infect. Immun. 58:1640-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nandi, B., R. K. Nandy, S. Mukhopadhyay, G. B. Nair, T. Shimada, and A. C. Ghose. 2000. Rapid method for species-specific identification of Vibrio cholerae using primers targeted to the gene of outer membrane protein OmpW. J. Clin. Microbiol. 38:4145-4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nandi, B., R. K. Nandy, A. C. Vicente, and A. C. Ghose. 2000. Molecular characterization of a new variant of toxin-coregulated pilus protein (TcpA) in a toxigenic non-O1/non-O139 strain of Vibrio cholerae. Infect. Immun. 68:948-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.NCCLS. 2002. Performance standards for antimicrobial susceptibility testing, 12th informational supplement. NCCLS document M100-S12. NCCLS, Wayne, PA.
- 30.Nesper, J., S. Schild, C. M. Lauriano, A. Kraiss, K. E. Klose, and J. Reidl. 2002. Role of Vibrio cholerae O139 surface polysaccharides in intestinal colonization. Infect. Immun. 70:5990-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Novais, R. C., A. Coelho, C. A. Salles, and A. C. Vicente. 1999. Toxin-co-regulated pilus cluster in non-O1, non-toxigenic Vibrio cholerae: evidence of a third allele of pilin gene. FEMS Microbiol. Lett. 171:49-55. [DOI] [PubMed] [Google Scholar]
- 32.Ogierman, M. A., A. Fallrino, T. Riess, S. G. Williams, S. R. Attridge, and P. A. Manning. 1997. Characterization of the Vibrio cholerae El Tor lipase operon lipAB and a protease gene downstream of the hly region. J. Bacteriol. 179:7072-7080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parsot, C., E. Taxman, and J. J. Mekalanos. 1991. ToxR regulates the production of lipoproteins and the expression of serum resistance in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 88:1641-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramamurthy, T., P. K. Bag, A. Pal, S. K. Bhattacharya, M. K. Bhattacharya, D. Sen, T. Shimada, T. Takeda, and G. B. Nair. 1993. Virulence patterns of V. cholerae Non-O1 isolated from hospitalized patients with acute diarrhoea in Calcutta, India. J. Med. Microbiol. 39:310-317. [DOI] [PubMed] [Google Scholar]
- 35.Rhodes, J. B., H. L. Smith, Jr., and J. E. Ogg. 1986. Isolation of non-O1 Vibrio cholerae serovars from surface waters in western Colorado. Appl. Environ. Microbiol. 51:1216-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richardson, S. H., J. C. Giles, and K. S. Kruger. 1984. Sealed adult mice: new model for enterotoxin evaluation. Infect. Immun. 43:482-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rivera, I. N. G., J. Chun, A. Huq, R. B. Sack, and R. R. Colwell. 2001. Genotypes associated with virulence in environmental isolates of Vibrio cholerae. Appl. Environ. Microbiol. 67:2421-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rudra, S., R. Mahajan, M. Mathur, K. Kathuria, and V. Talwar. 1996. Cluster of cases of clinical cholera due to Vibrio cholerae O10 in east Delhi, India. J. Med. Res. 103:71-73. [PubMed] [Google Scholar]
- 39.Russel, R. G., B. D. Tall, and J. G. Morris, Jr. 1992. Non-O1 Vibrio cholerae intestinal pathology and invasion in removable intestinal tie adult rabbit diarrhea model. Infect. Immun. 60:435-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saha, P. K., H. Koley, and G. B. Nair. 1996. Purification and characterization of an extracellular secretogenic non-membrane-damaging cytotoxin produced by clinical strains of Vibrio cholerae non-O1. Infect. Immun. 64:3101-3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sarkar, A., R. K. Nandy, G. B. Nair, and A. C. Ghose. 2002. Vibrio pathogenicity island and cholera toxin genetic element-associated virulence genes and their expression in non-O1 non-O139 strains of Vibrio cholerae. Infect. Immun. 70:4735-4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sengupta, D. K., T. K. Sengupta, and A. C. Ghose. 1992. Major outer membrane proteins of Vibrio cholerae and their role in induction of protective immunity through inhibition of intestinal colonization. Infect. Immun. 60:4848-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharma, C., M. Thungapathra, A. Ghosh, A. K. Mukhopadhyay, A. Basu, R. Mitra, I. Basu, S. K. Bhattacharya, T. Shimada, T. Ramamurthy, T. Takeda, S. Jamasaki, Y. Takeda, and G. B. Nair. 1998. Molecular analysis of non-O1, non-O139 Vibrio cholerae associated with an unusual upsurge in the incidence of cholera-like disease in Calcutta, India. J. Clin. Microbiol. 36:756-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shimada, T., E. Arakawa, K. Itoh, T. Okitsu, A. Matsushima, Y. Asai, S. Yamai, T. Nakazato, G. B. Nair, M. J. Albert, and Y. Takeda. 1994. Extended serotyping scheme for Vibrio cholerae. Curr. Microbiol. 28:175-178. [Google Scholar]
- 45.Silva, A. J., G. J. Leitch, A. Camilli, and J. A. Benitez. 2006. Contribution of hemagglutinin/protease and motility to the pathogenesis of El Tor biotype cholera. Infect. Immun. 74:2072-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skurnik, M., I. Bolin, H. Heikkine, S. Piha, and H. Wolf-Watz. 1984. Virulence plasmid-associated autoagglutination in Yersinia spp. J. Bacteriol. 158:1033-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith, H. L., Jr. 1970. A presumptive test for vibrios: the “string” test. Bull. W. H. O. 42:817-818. [PMC free article] [PubMed] [Google Scholar]
- 48.West, P. A., and R. R. Colwell. 1984. Identification and characterization of Vibrionaceae. An overview, p. 285-363. In R. Colwell (ed.), Vibrios in the environment. John Wiley & Sons, Inc., New York, NY.
- 49.World Health Organization. 1987. Manual for laboratory investigations of acute enteric infections, CDD/83.3. World Health Organization, Geneva, Switzerland.