Abstract
The diversity of free-living protozoa in five meat-cutting plants was determined. Light microscopy after enrichment culturing was combined with sequencing of PCR-amplified, denaturing gradient gel electrophoresis (DGGE)-separated 18S rRNA gene fragments, which was used as a fast screening method. The general results of the survey showed that a protozoan community of amoebae, ciliates, and flagellates was present in all of the plants. Protozoa were detected mainly in floor drains, in standing water on the floor, on soiled bars of cutting tables, on plastic pallets, and in out-of-use hot water knife sanitizers, but they were also detected on surfaces which come into direct contact with meat, such as conveyer belts, working surfaces of cutting tables, and needles of a meat tenderizer. After 7 days of incubation at refrigerator temperature, protozoa were detected in about one-half of the enrichment cultures. Based on microscopic observations, 61 morphospecies were found, and Bodo saltans, Bodo spp., Epistylis spp., Glaucoma scintillans, Petalomonas spp., Prodiscophrya collini, and Vannella sp. were the most frequently encountered identified organisms. Sequencing of DGGE bands resulted in identification of a total of 49 phylotypes, including representatives of the Amoebozoa, Chromalveolata, Excavata, Opisthokonta, and Rhizaria. Sequences of small heterotrophic flagellates were affiliated mainly with the Alveolata (Apicomplexa), Stramenopiles (Chrysophyceae), and Rhizaria (Cercozoa). This survey showed that there is high protozoan species richness in meat-cutting plants and that the species included species related to known hosts of food-borne pathogens.
Protozoa are unicellular eukaryotic microorganisms which are ubiquitous in nature and anthropogenic environments. Protozoa feed on bacteria, microalgae, and particulate or dissolved matter. In turn, they serve as food for other protozoa and metazoa. Besides the prey-predator relationship, there is a particular association of bacteria with protozoa, namely, survival and/or replication of a bacterium within a protozoan. First, some bacteria commonly described as obligate endosymbionts live intracellularly in the cytoplasm or the macronucleus (29, 30, 32, 34) and often cannot be cultivated outside the protozoan host. Second, in the last two decades, more attention has been paid to bacteria which were not expected to have an intracellular protozoan life cycle. Internalization of human pathogens (Helicobacter pylori, Mycobacterium bovis) (60, 66) and food-borne pathogens (Campylobacter jejuni, Escherichia coli O157:H7, Listeria monocytogenes, Salmonella, Staphylococcus aureus) (6, 7, 14, 28, 35, 42, 59, 61, 68) in protozoa such as Acanthamoeba castellanii, Acanthamoeba polyphaga, Acanthamoeba rhysodes, and Tetrahymena pyriformis has been demonstrated. The mechanism of the bacterium-protozoan interaction has been intensively studied for Legionella pneumophila (1, 26). The facultative intracellular lifestyle of bacterial (food-borne) pathogens in protozoa is of special concern for several reasons: (i) some protozoa, such as A. castellanii, A. polyphaga, Glaucoma sp., and Tetrahymena spp., produce small vesicles which can contain living bacteria (10, 13, 14, 31), and these vesicles might be inhaled or can contaminate the environment; (ii) bacteria surviving within protozoa or protozoan cysts resist unfavorable conditions, such as desiccation and exposure to disinfectants (2, 27, 36, 37); and (iii) an increase in antimicrobial resistance and virulence of bacterial pathogens after passage through protozoa has been demonstrated (8, 17). The association of food-borne pathogens with free-living protozoa is of particular interest because it might explain how, besides well-known strategies such as biofilm formation, some food-borne pathogens persist in food-processing areas despite daily cleaning and disinfection.
Identification of protozoa is traditionally accomplished by using morphology (microscopy and ultrastructure analysis) and locomotion, although PCR-based techniques have increasingly been found to be valid. Morphological identification is often hampered by the small size of protozoa, limited or difficult diagnostic criteria for some taxa, and the time-consuming process necessary to obtain identification expertise. More recently, culture-independent techniques, such as denaturing gradient gel electrophoresis (DGGE), thermal gradient gel electrophoresis, single-strand conformation polymorphism, and terminal restriction fragment polymorphism, have been used to study protozoan communities in various ecosystems (19, 41, 43, 47). Molecular identification is obtained after sequencing of structural or functional genes. Moreover, several studies based on 18S rRNA genes revealed high phylogenetic diversity of uncultivable organisms (46, 48, 49), indicating that the protozoan biodiversity was much higher than that determined by traditional methods. In a few studies, traditional and molecular methods have been combined (4, 57). Compilation of the results obtained by two approaches leads to better species description for a particular environment.
In order to investigate the role of environmental free-living protozoa in the contamination of food by food-borne pathogens, an inventory of these unicellular eukaryotes in food-processing environments is a necessary first step. Although the bacterial flora of meat-processing environments has been described, to our knowledge, no studies of the environmental protozoan communities in these environments have been performed previously. The present study was designed to determine the diversity of free-living protozoa in meat-cutting plants using enrichment cultures and sequencing of PCR-amplified, DGGE-separated 18S rRNA gene fragments.
MATERIALS AND METHODS
Sample preparation and processing.
Five meat-cutting plants (one plant processing beef [plant A], two plants processing pork [plants B and C], and two plants processing beef, pork, and poultry [plants D and E; plant D also treated game]) were visited during the period from February to May 2007. Samples were taken after a waiting period of at least 2 h after cleaning and disinfection. A total of 105 samples were collected (Table 1). Liquid samples (n = 22) were taken with a sterile syringe or sterile pipette and transferred to sterile plastic tubes. Sterile cotton wool moistened with sterile demineralized water and dry cotton were used to sample dry and wet surfaces, respectively. Samples were collected from a surface area of 126 cm2 marked with a sterile template which was randomly placed on the sample point. Cotton wool samples (n = 71) were placed in numbered plastic bags, which were tightly closed to avoid drying of the cotton. Upon arrival in the lab (at most 2 h after sampling), 6 ml of sterile demineralized water was added to each cotton wool sample, which was subsequently gently massaged. The cotton wool was manually squeezed, and 5 ml of liquid was transferred to a sterile tube for further processing. For needles of tenderizers samples were obtained with sterile cotton swabs (n = 3), and the samples were further processed as described above. Meat residues (n = 5) which were found in corners or on the floor were transferred to sterile plastic tubes, and 5 ml of sterile demineralized water was added to each sample and vigorously shaken (vortexed). The supernatant was used for enrichment cultures. Tap water (n = 4) was collected in a sterile bottle.
TABLE 1.
Overview of protozoan status of sample points, as determined by microscopic observation and cumulative data from molecular sequence analyses
| Sample | Light microscopy
|
Sequence analysis
|
|||||
|---|---|---|---|---|---|---|---|
| Total no. positive/total no. of sample points | No. positive for:
|
Total no. of morphospecies | No. of sample points positive for protozoaa | No. of phylotypes | |||
| Amoebae | Flagellates | Ciliates | |||||
| No contact with meat | |||||||
| Air conditioning in cold storage | 0/2 | 0 | 0 | 0 | 0 | —b | — |
| Board tenderizer (underside) | 1/1 | 1 | 1 | 0 | 2 | 0 | 0 |
| Ceiling | 0/1 | 0 | 0 | 0 | 0 | — | — |
| Conveyer belt (underside) | 2/2 | 2 | 2 | 0 | 8 | 2 | 4 |
| Cutting table (underside) | 1/3 | 1 | 1 | 0 | 4 | 0 | 0 |
| Door | 0/1 | 0 | 0 | 0 | 0 | — | — |
| Floor | 3/6 | 1 | 3 | 0 | 3 | 1 | 1 |
| Floor drain | 13/18 | 6 | 11 | 4 | 19 | 11 | 13 |
| Hot water knife sanitizer (out of use) | 2/2 | 2 | 2 | 2 | 18 | 2 | 13 |
| Plastic pallet | 2/3 | 2 | 2 | 0 | 11 | 2 | 8 |
| Plastic strip | 2/4 | 0 | 2 | 2 | 7 | 2 | 4 |
| Rail | 2/7 | 1 | 2 | 2 | 8 | 2 | 7 |
| Soiled bar cutting table | 6/6 | 3 | 6 | 3 | 28 | 4 | 7 |
| Standing water on the floor | 5/6 | 5 | 4 | 3 | 17 | 4 | 12 |
| Wall | 3/10 | 1 | 3 | 1 | 7 | 3 | 6 |
| Wall truck (near air conditioning) | 0/2 | 0 | 0 | 0 | 0 | 1 | 1 |
| Direct contact with meat | |||||||
| Tenderizer | |||||||
| Needles | 1/3 | 1 | 1 | 0 | 2 | 1 | 3 |
| Board (top) | 1/3 | 1 | 1 | 0 | 3 | 1 | 3 |
| Conveyer belt (top) | 2/6 | 0 | 2 | 0 | 5 | 1 | 7 |
| Cutting table (working surface) | 2/10 | 2 | 2 | 0 | 5 | 1 | 2 |
| Others | |||||||
| Meat residues | 5/5 | 2 | 5 | 3 | 18 | 5 | 13 |
| Water supply | 0/4 | 0 | 0 | 0 | 0 | 0 | 0 |
A positive score was assigned when at least one band in the profile was affiliated with protozoan sequences (the results obtained with both primer sets for t0 and t7 are grouped together). The number of sample points analyzed was lower than the number analyzed for enrichment cultures (see Results).
—, sample point which was not analyzed or whose profile contained a prominent band(s) which was affiliated with eukaryotic sequences other than protozoan sequences.
Determination of microbiological hygiene status.
The general hygienic status of the plants was determined using the agar contact plate method as described in Commission Decision 2001/471/EC (21). Surface samples were collected from control points (conveyors, cutting tables, knives, mincing and packaging machines, saws, etc.) as described in the hazard analysis and critical control points plans of the meat processors. Plate count agar (Oxoid) was used as the cultivation medium, and the plates were aerobically incubated at 37 ± 1°C for 24 h.
Cultivation of protozoa and morphological identification.
One milliliter of an initial dilution or original liquid sample was transferred to a petri dish containing sterile Page's amoeba saline solution (Culture Collection of Algae and Protozoa recipe [www.ccap.ac.uk]) and heat-sterilized rice grain. All petri dishes were incubated for 7 days at refrigerator temperature (5 ± 1°C) in the dark and were examined microscopically (Olympus CKX41 inverted microscope) within 1 week for the presence of protozoa. Sample points were considered positive for protozoa when at least one representative of amoebae, ciliates, or flagellates was observed. Protozoa were identified morphologically using standard taxonomic sources for protozoan identification (22-25, 51, 52) and were classified as described by Adl et al. (3).
DNA extraction.
Three milliliters was removed from the original liquid sample or initial dilution (t0), and 2 ml was randomly withdrawn from each enrichment culture on day 7 (t7). The subsamples were centrifuged for 20 min at 20,800 × g at 4°C. The upper part was carefully withdrawn, and the pellet was suspended in the remaining 500 μl (final volume) of supernatant, which was subjected to DNA extraction. DNA extraction was performed with a ChargeSwitch genomic DNA micro tissue kit (Invitrogen) using a final eluent volume of 75 μl. The DNA extracts were stored at −20°C until analysis.
PCR amplification.
PCR amplification was performed with a PE Applied Biosystems 9700 temperature cycler. 18S rRNA gene primer sets Euk1A-Euk516r-GC (19) and F1427GC-R1616 (63) were used with the following modifications: 800 ng bovine serum albumin (Roche) was added to the PCR mixture, and 35 cycles of denaturation were used for both primer sets.
DGGE analysis.
Protozoan communities from t0 and t7 samples were analyzed by DGGE. t0 and t7 PCR products for the same sample points were placed on a gel next to each other. A molecular marker was not included on the gels. Approximately 42 μl of PCR product and 10 μl of loading dye (30% [vol/vol] glycerol, 0.125% bromophenol, 20 mM Tris-HCl) were applied to individual wells. A sample of bovine or porcine DNA was included on some gels which contained samples probably positive for the presence of this DNA (e.g., samples from conveyer belts and cutting tables). All the DGGE analyses were performed using the DCode universal mutation detection system (Bio-Rad). The polyacrylamide gels consisted of polyacrylamide in 1× Tris-acetate-EDTA (TAE) buffer diluted from a 50× TAE solution (2 M Tris base, 0.95 M glacial acetic acid, 50 mM EDTA). Two gels were used: a 30 to 50% gradient in a 6% (vol/vol) polyacrylamide gel and a 30 to 55% gradient in a 8% (vol/vol) polyacrylamide for primer sets Euk1A-Euk516r-GC and F1427GC-R1616, respectively (acrylamide/bisacrylamide ratio, 37.5:1; 100% denaturing polyacrylamide solution contained 7 M urea and 40% deionized formamide). The 24-ml gradient gels were cast using a gradient former and a pump set at a constant speed of 5 ml/min. The gels were run for 960 min at 50 V in 1× TAE buffer at 60°C and stained for 1 h in 1× TAE buffer containing 0.9 mg/liter ethidium bromide.
Recovery of DNA from DGGE gels.
The most prominent bands were sequenced, except for those whose positions were similar to the positions of bands for cows or pigs. The middle portions of selected DGGE bands were excised and transferred to 1.5-ml Eppendorf tubes containing 30 μl of 1× TE buffer (10 mM Tris-HCl, 1 mM EDTA; pH 8.0). The tubes were incubated overnight at 4°C. Four microliters was used for the next PCR, which was followed by DGGE to check the band position and purity. After the purity was ascertained, PCR was performed with the same primer set without a GC clamp.
Sequencing and analysis of excised gel bands.
Sequencing was performed using an Applied Biosystems ABI3130XL genetic analyzer. PCR products were purified for sequencing using shrimp alkaline phosphatase (1 U/μl; Amersham) and exonuclease I (20 U/μl; Epicentre Technologies) for 15 min at 37°C, followed by 15 min at 80°C. This material was subsequently used for cycle sequencing without any further purification using an ABI Prism BigDye V 3.1 terminator cycle sequencing kit (Applied Biosystems). Sequences were exported from BioEdit (33) as FASTA files and compared with the NCBI GenBank database using the nucleotide-nucleotide Basic Local Alignment Search Tool (BLAST) (5). All BLAST searches were performed in December 2007.
RESULTS
General hygiene status of the plants.
Only plant E fulfilled the legal requirement of a viable bacterial count of 0 to 10 CFU/cm2 for all surface samples tested (n = 16). For plants A to C, the limit was exceeded in only a minority of the samples (plant A, 2/10 samples; plant B, 1/20 samples; plant C, 2/20 samples). In plant D, seven of 10 samples were unacceptable, including samples from the balance, board meat tenderizer, cutting tables, and saws, which were heavily contaminated.
Recovery of eukaryotic organisms after enrichment culturing.
A total of 50.5% of 105 samples examined were positive for protozoa after enrichment culturing (t7) (Table 1), and 43.7% of 71 cotton samples resulted in positive cultures. About one-third of the protozoan-positive enrichment cultures were obtained from liquid samples (both samples from out-of-use hot water knife sanitizers, 13 of 18 samples from floor drains, and two samples from standing water on the floor). All samples from meat residues were positive after enrichment. In samples taken from air-conditioning systems, ceiling, doors, truck walls, and water supplies no protozoa were detected after 1 week. Flagellates, amoebae, and ciliates were observed in 94.3, 58.5, and 37.7% of the 53 positive enrichment cultures, respectively. Amoebae and/or flagellates were detected in cultures obtained from places which come in direct contact with meat, such as the working surfaces of two cutting tables, two top surfaces of conveyer belts, and the board and needles of one tenderizer. Metazoa, such as nematodes (present in five enrichment cultures) and rotifers (present in three enrichment cultures), were observed in two of the five plants but were not identified further. Fungi (mainly yeasts), either as pure cultures or in the presence of protozoa, were found in approximately one-quarter of the enrichment cultures.
The sample processing efficiency was checked by addition of 6 ml of sterile Page's amoeba saline and heat-sterilized rice grain to the squeezed cotton wool, followed by incubation for 7 days at refrigerator temperature and microscopic determination of the cultured protozoa after 1 week. The population obtained did not substantially differ from the t7 enrichment culture, although there were some species which were not found in the corresponding t7 cultures.
Morphological diversity of protozoa.
Representatives of amoebae, ciliates, and flagellates were found in each plant. Higher numbers of morphospecies were observed for cutting plants C (37 morphospecies, including 16 identified morphospecies), A (22 morphospecies, including 14 identified morphospecies), and B (22 morphospecies, including 9 identified morphospecies) than for plants D (18 morphospecies, including 7 identified morphospecies) and E (13 morphospecies, including 10 identified morphospecies) (Table 2). Enrichment cultures from six soiled table bars resulted in a total of 28 different morphospecies (Table 1). Other sample points showing high species diversity were floor drains, out-of-use hot water knife sanitizers, meat residues, and standing water on the floor. From a total of 61 different observed morphospecies (27 flagellates, 26 ciliates, and 8 amoebae), 27 protozoa were identified to the genus or species level (Table 2). The Chromalveolata comprised 15 identified ciliates. Epistylis sp., Glaucoma scintillans, and Prodiscophyra collini were ciliate species which were found in three of the five plants. Although flagellates were abundant in 94.3% of the positive enrichment cultures, the majority of these organisms could not be identified by light microscopy. Identified flagellates belonged to the Cercozoa and Euglenozoa. Bodo species 1 and Petalomonas species 2 were found in all meat-cutting plants. Amoebozoa were represented by Tubulinea and Flabellinea. Thecamoeba sp. was found in four of the five plants. Irregular, small (≤10 μm), dark amoebae were observed in some enrichment cultures, but their taxonomic positions could not be determined.
TABLE 2.
Morphologically identified protozoan taxa observed in enrichment cultures
| Supergroup | First rank | Second rank | Species | Meat-cutting planta
|
||||
|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | ||||
| Amoebozoa | Flabellinea | Cochliopodium | Cochliopodium actinophorum | + | ||||
| Thecamoebida | Thecamoeba sp. | + | + | + | + | |||
| Vannellida | Vannella sp. | + | + | |||||
| Tubulinea | Tubulinida | Hartmannella sp. | + | + | ||||
| Saccamoeba sp. | + | + | ||||||
| Chromalveolata | Alveolata | Ciliophora | Colpoda steinii | + | ||||
| Colpidium colpoda | + | |||||||
| Dexiostoma campylum | + | + | ||||||
| Glaucoma scintillans | + | + | + | |||||
| Glaucoma sp. | + | |||||||
| Tetrahymena pyriformis complex | + | + | ||||||
| Paramecium aurelia complex | + | |||||||
| Epistylis coronata | + | |||||||
| Epistylis sp. | + | + | + | |||||
| Vorticella sp. | + | |||||||
| Cinetochilum margaritaceum | + | |||||||
| Cyclidium glaucoma | + | + | ||||||
| Acineria incurvata | + | |||||||
| Chilodonella uncinata | + | |||||||
| Prodiscophrya collini | + | + | + | |||||
| Excavata | Euglenozoa | Euglenida | Petalomonas species 1 | + | + | + | ||
| Petalomonas species 2 | + | + | + | + | + | |||
| Kinetoplastea | Bodo saltans | + | + | + | + | |||
| Bodo species 1 | + | + | + | + | + | |||
| Bodo species 2 | + | + | ||||||
| Heterolobosea | Vahlkampfiidae | Vahlkampfia sp. | + | |||||
| Rhizaria | Cercozoa | Cercomonadida | Allantion tachyploon | + | ||||
+, present. The total numbers of identified morphospecies for plants A, B, C, D, and E were 14, 9, 16, 7, and 10, respectively.
DGGE profiles.
A total of 410 PCR products obtained from 100 t0 extracts (excluding four meat residue samples and one needle tenderizer sample) and 105 t7 extracts (each amplified with both primer sets) were subjected to DGGE. In 143 lanes, no bands were detected. The absence of profiles can be explained by an absence of eukaryotic DNA, breakdown of DNA due to chlorine residues originating from disinfection products, failure of the extraction method, or inhibition of the PCR. A subset of 109 profiles was not analyzed further for reasons such as weak bands, failure to amplify the excised bands, or dominance of fungi in the enrichment cultures. For the remaining 158 profiles, preliminary sequence analysis of prominent bands revealed that 68 profiles were affiliated with fungal and/or metazoan (Mammalia, Nematoda) sequences. However, not all bands of these 68 profiles were excised, and the possibility that some weak bands matched protozoan sequences cannot be excluded. Ninety profiles contained at least one band which was affiliated with protozoan DNA sequences. Also, not all bands could be excised, and visual selection was made. A number of DGGE profiles for t0 samples and the corresponding t7 samples revealed distinct changes in the presence of bands and/or the relative band intensities, suggesting that enrichment culturing changed the relative composition of the protozoan community (Fig. 1). Although not all sample points were included in sequence analyses (see above), the data confirmed the finding obtained by microscopy that out-of-use hot water knife sanitizers, floor drains, standing water on the floor, and meat residues showed the highest species diversity (Table 1).
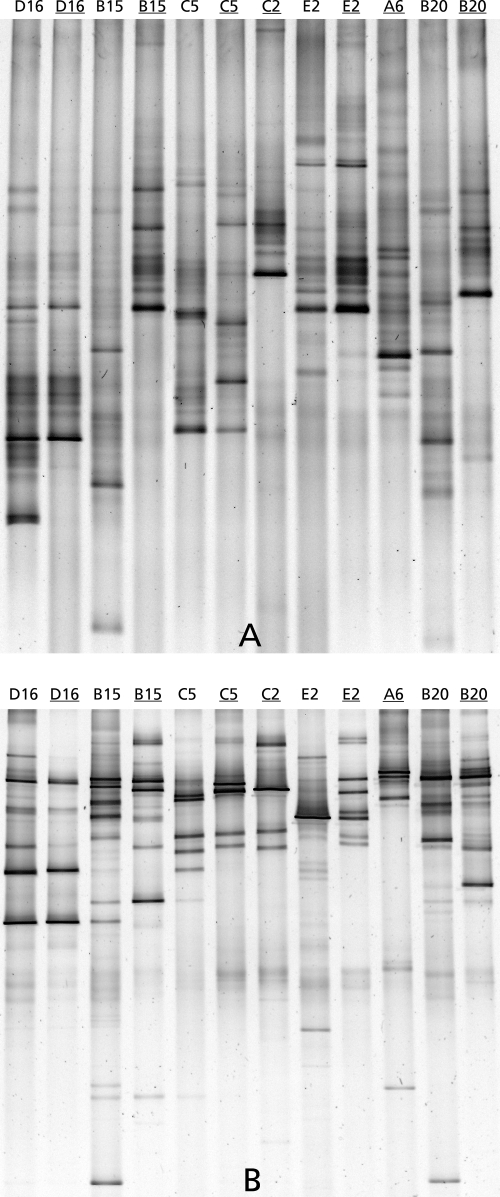
FIG. 1.
DGGE patterns obtained using primer set F1427GC-R1616 (A) and primer set Euk1A-Euk516r-GC (B) for sample points in five meat-cutting plants (plants A to E). Sample points: D16, needles of a meat tenderizer; B15, top of conveyer belt; C5, out-of-use hot water knife sanitizer; C2, floor drain; E2, standing water on the floor; A6, soiled table bar; B20, plastic pallet. Underlining indicates corresponding t7 enrichment cultures.
Molecular affiliation of protozoan DNA sequences.
All sequences were affiliated with eukaryotic organisms, which confirmed the specificity of the two eukaryotic primer sets and is in agreement with the original studies (19, 64). A total of 310 bands were sequenced, and 174, 102, and 17 of the sequences were affiliated with protozoa, fungi, and metazoa, respectively. Seventeen sequences were excluded due to sequence errors or ambiguous matches. Although the sequences with the highest BLAST scores often matched uncultured protozoan sequences, the most closely related organisms are shown in Table 3. Higher numbers of phylotypes were found for plants C (n = 21) and E (n = 20) than for plants A (n = 15), D (n = 13), and B (n = 12). The Chromalveolata was the largest group (25 phylotypes), followed by the Rhizaria (10 phylotypes), Amoebozoa (five phylotypes), Excavata (three phylotypes), Opisthokonta (three phylotypes), and Archaeplastida (two phylotypes). Within the Chromalveolata group, Chrysophyceae and Colpodellida were the flagellate groups which were encountered most often (Table 3). Spumella-like flagellate JBM/S11, which clusters with Spumella elongata (12), Colpodella tetrahymenae, and Telotrochidium matiense were found in four plants. The Rhizaria were mainly represented by Cercozoa spp. and the Heteromitidae. Bodomorpha (3, 50), Heteromita globosa, the “Costa Rica” flagellate (16), and soil flagellates AND21 and AND24 (39) belong to the latter group. Bodomorpha sp. and Lecythium sp. were the cercozoan phylotypes which were most frequently encountered in meat-cutting plants. Archaeplastida (Chlorophyta) were found in plants A and C. Representatives of the Excavata (Euglenozoa) and Opisthokonta (Choanomonada) were rarely found, and, with the exception of Parabodo caudatus, their BLAST scores were low (<92%). Remarkably, Petalomonas spp. were frequently observed in enrichment cultures, but they were detected only twice by the molecular tool. Amoebal DNA was seldom retrieved; only five species were found, and they belonged to the Flabellinea (n = 2), Tubulinea (n = 2), and incertae sedis Amoebozoa Spongomonadida (n = 1).
TABLE 3.
Protist community compositions at the different meat-cutting plants as determined by BLAST analysis of 18S rRNA gene sequences
| Supergroup | First rank | Second rank | Most closely related organism | Accession no. | Sequence similarity (%) | Meat-cutting planta
|
||||
|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | ||||||
| Amoebozoa | Flabellinea | Thecamoebida | Sappinia diploidea | DQ122380 | 97.2 | + | ||||
| Vannellida | Platyamoeba placida | AY294150 | 95.1 | + | ||||||
| Tubulinea | Lyptomyxida | Leptomyxa reticulata | AF293898 | 78.4 | + | |||||
| Incertae sedis Tubulinea | Echinamoeba exundans | AF293895 | 98.9 | + | ||||||
| Incertae sedis Amoebozoa | Spongomonadidae | Spongomonas minima | AF411280 | 97.6-100 | + | + | ||||
| Archaeplastida | Chloroplastida | Chlorophyta | Chloromonas sp. | AF514406 | 99.2 | + | ||||
| Polytoma uvella | U22942 | 99.2-100 | + | |||||||
| Chromalveolata | Alveolata | Apicomplexa | Ascogregarina culicis | DQ462457 | 92.8 | + | ||||
| Colpodella edax | AY234843 | 90.1-90.9 | + | + | ||||||
| Colpodella tetrahymenae | AF330214 | 88.2-98.8 | + | + | + | + | ||||
| Colpodella sp. | AY142075 | 95.8-96.9 | + | + | ||||||
| Cryptosporidium andersoni | AY954885 | 80.2-81.0 | + | + | ||||||
| Ciliophora | Colpidium campylum | X56532 | 99.1 | + | ||||||
| Discophrya collini | L26446 | 99.0 | + | |||||||
| Glaucomides bromelicola | AJ810077 | 99.4 | + | |||||||
| Loxophyllum rostratum | DQ190465 | 99.4 | + | |||||||
| Opisthonecta minima | EF417834 | 97.6-97.8 | + | |||||||
| Prodiscophrya sp. | AY331803 | 100 | + | |||||||
| Pseudocohnilembus persalinus | AY551906 | 99.8 | + | |||||||
| Pseudoplatyophrya nana | AF060452 | 99.5-99.6 | + | + | ||||||
| Sorogena stoianovitchae | AF300287 | 95.7-97.6 | + | + | ||||||
| Telotrochidium matiense | AY611065 | 96.2-100 | + | + | + | + | ||||
| Vorticella microstoma | DQ868347 | 97.9-99.6 | + | + | ||||||
| Dinozoa | Glenodinium inaequale | EF058237 | 80.9-85.0 | + | + | |||||
| Stramenopiles | Bicosoecida | Adriamonas peritocrescens | AF243501 | 76.0 | + | |||||
| Chrysophyceae | Ochromonas sp. | EF165126 | 98.6 | + | ||||||
| Ochromonas sp. | EF165144 | 99.4 | + | |||||||
| Paraphysomonas vestita | Z28335 | 98.7 | + | |||||||
| Spumella-like flagellate JBAS38 | DQ388538 | 99.3 | + | |||||||
| Spumella-like flagellate 1305 | DQ388568 | 99.4-100 | + | + | ||||||
| Spumella-like flagellate JBM/S11 | EF043285 | 99.4-100 | + | + | + | + | ||||
| Peronosporomycetes | Saprolegnia parasitica | AB086898 | 99.8 | + | ||||||
| Excavata | Euglenozoa | Euglenida | Notosolenus ostium | AF403159 | 75.1 | + | ||||
| Petalomonas cantuscygni | AF386635 | 78.2 | + | |||||||
| Kinetoplastea | Parabodo caudatus | DQ207590 | 100 | + | ||||||
| Opisthokonta | Choanomonada | Acanthoecidae | Diaphanoeca grandis | DQ059033 | 91.3 | + | ||||
| Stephanoeca diplocostata | AF084235 | 91.3-91.9 | + | + | ||||||
| Monosigidae | Monosiga ovata | AF084230 | 82.4 | + | ||||||
| Rhizaria | Cercozoa | Cercomonadida | Bodomorpha sp. | DQ211596 | 95.3-100 | + | + | + | + | |
| Cercomonas sp. | U42448 | 98.8 | + | |||||||
| Cercomonas sp. | DQ211598 | 99.4 | + | |||||||
| Cercomonas sp. | AY884338 | 98.9 | + | |||||||
| “Costa Rica” flagellate | AF411277 | 97.9 | + | |||||||
| Heteromita globosa | U42447 | 97.2-100 | + | + | ||||||
| Proleptomonas faecicola | AF411275 | 92.2 | + | + | ||||||
| Soil flagellate AND21 | AY965866 | 99.2-100 | + | + | + | |||||
| Soil flagellate AND24 | AY965867 | 99.8-100 | + | + | + | |||||
| Incertae sedis Cercozoa | Lecythium sp. | AJ514867 | 89.7-98.0 | + | + | + | + | + | ||
| Incertae sedis Eukaryotes | Breviata anathema | AF153206 | 96.7 | + | ||||||
+, present. The total numbers of phylotypes for plants A, B, C, D, and E were 15, 12, 21, 13, and 20, respectively.
Detection of eukaryotic DNA other than protozoan DNA.
A large proportion of the sequences (102/310 sequences) were affiliated with fungi, including the Ascomycota (n = 72), Basidiomycota (n = 23), Glomeromycota (n = 3), Urediniomycetes (n = 3), and Chytridiomycetes (n = 1). The yeast Yarrowia lipolytica was the species that was encountered most frequently (32 of the 72 ascomycete sequences, and in some DGGE lanes for a given sample point up to three bands were found at different migration positions), and it was found in all plants. Seven sequences matched sequences of Nematoda (genera Aduncospiculum, Choriorhabditis, Panagrolaimus, Pristionchus, and Rhabditis), one sequence matched an Arthropoda sequence, and one sequence matched an Oligochaeta sequence. Mammalian DNA was detected in eight cases, and the sequences were affiliated with cow, pig, or deer sequences.
DISCUSSION
The aim of this study was to determine the biodiversity of free-living protozoa in meat-cutting plants. Light microscopy after enrichment culture was combined with sequencing of PCR-amplified, DGGE-separated 18S rRNA gene fragments, which was used as a fast screening method. Preliminary microscopic observation of a few t0 samples revealed a low number of protozoa, and there were difficulties in analyzing fat- or protein-rich samples, which justified the decision to incorporate enrichment culturing. Moreover, the development of cryptic species (i.e., species inactive at the time of sampling) under favorable conditions (i.e., enrichment culture) indicated that they were present in the t0 samples. The enrichment cultures were incubated at a low temperature since the ambient processing temperature in the meat-cutting plants may not be more than 12°C. Two universal eukaryotic primer sets, those of Díez et al. (19) and van Hannen et al. (63), were used to capture a higher fraction of the eukaryotic communities. Because it was not practical to sequence all bands and analyze a subset of DGGE profiles, both of which inevitably resulted in underestimation of the species composition, it was not possible to make a detailed comparison of the two primer sets and the results obtained for t0 and t7 samples. Therefore, the survey resulted in a general overview of the protozoan community composition obtained by microscopy and sequencing.
A total of 105 samples were taken, and 53 of them resulted in protozoan-positive enrichment cultures, as determined microscopically. In most samples which yielded a protozoan-positive enrichment culture, residual organic material and/or water was present. For example, samples taken from floor drains, knife sanitizers, meat residues, soiled table bars, and standing water on the floor resulted in the highest number of protozoan-positive cultures. In addition, locations which were inadequately cleaned and disinfected because of ignorance or inaccessibility (e.g., holes in plastic pallets, undersides of cutting boards and conveyer belts, and upper sides of rails) harbored protozoa. Protein and fat residues provide an excellent nutrient source for bacteria, which in turn is favorable for the growth of protozoa. Detritus is rapidly colonized by small heterotrophic flagellates, whose concentrations can reach up to 105 organisms/ml or more (15). Of particular interest were the surfaces which come into direct contact with meat. Protozoa were found in enrichment cultures obtained from conveyer belts in plant B, and protozoan DNA was detected on the surface of a cutting table in plant E. In plant D, amoebae and flagellates were found in enrichment cultures obtained from two cutting tables and the board and needles of the tenderizer. The presence of protozoan DNA on needles was also observed at t0, and the DGGE pattern was similar to that at t7, suggesting that an active protozoan community was present at the moment of sampling (Fig. 1). The general hygiene status of some sample points as determined by microbial analysis in plant D was unacceptable according to Commission Decision 2001/471/EC and might explain the presence of protozoa on surfaces expected to have a low germ content. However, our results suggest that a good hygiene score (plant E) does not necessarily correlate with an absence of protozoa in the food-processing environment.
The overall species compositions (i.e., the occurrence of amoebae, flagellates, and ciliates) were quite similar in all plants, and our data do not suggest that process-related activity (i.e., cutting of cow carcasses versus cutting of pig carcasses) resulted in a specific protozoan community. Plant C showed the highest species richness, as determined by both methods. Little agreement was found between the light microscopy results and the sequencing results, although the analysis of a subset of bands limited our ability to make a reliable comparison. By using light microscopy, 27 morphospecies were identified, compared to 49 phylotypes. Higher ciliate diversity was observed when light microscopy was used than when the molecular approach was used (26 and 11 taxa, respectively). The taxa which were retrieved by both methods were P. collini and Vorticella spp. One species was morphologically identified as G. scintillans, while sequence analysis of the sample point indicated an affiliation with Glaucomides bromelicola. Sequence analysis showed that T. matiense was the most frequently encountered ciliate species, although it was not microscopically observed, in contrast to Epistylis spp., which were frequently observed in enrichment cultures. T. matiense is more closely related to Epistylis than to any other peritrichous genus and probably evolved from Peritrichia with a noncontractile stalk (44). Small heterotrophic flagellates were abundant in enrichment cultures but were difficult to identify microscopically in a reliable way. This was compensated for by sequence analyses of excised DGGE bands. Flagellates were represented mainly by Alveoloata (Apicomplexa), Stramenopiles (Chrysophyceae), and Rhizaria (Cercozoa). Bodo spp. and Petalomonas spp. were abundant in enrichment cultures but rarely detected by PCR. Amoeboid organisms were rarely identified by DGGE, although monopodial amoebae were frequently encountered in enrichment cultures. Our results agree with the finding that amoebae are rarely found in environmental molecular surveys (4, 11).
Although both culture methods and PCR-based methods have their specific biases (45, 58, 65) and it was difficult to identify all observed species morphologically and impossible to sequence all bands from the gels, the data obtained show that the two-method approach resulted in identification of a higher number of species than either method separately. Our results confirm the results of other studies (4, 45, 57) which recommended that a combination of methods should be used to study protozoan communities. The use of two universal eukaryotic primer sets resulted in detection of many eukaryotic organisms other than protozoa, including a high number of fungi. Amplification of fungal or metazoan DNA has also been found in other studies (19, 43) and can be attributed to several factors. First, yeasts were abundant in the enrichment cultures, leading to a higher fungal DNA content in the extract. Second, a high copy number of rRNA genes is present in the genomes of some yeasts, such as Y. lipolytica (18), which increases the probability that the genes are amplified during PCR. Third, protozoa form a paraphyletic group, and the use of universal eukaryotic primers inevitably leads to amplification of nonprotozoan DNA. The use of specific primer sets instead of universal primers can eliminate this problem and is strongly recommended when particular groups need to be studied (9, 20, 40, 53).
It is still unclear how protozoa are initially introduced and transported in food-processing areas. Air, drinking water, and human activities are the most likely transmission routes. Air can contain (cysts of) protozoa, including amoebae, flagellates, and ciliates, such as Colpoda steinii (38, 54). Protozoa are known to be common inhabitants of drinking water. The possibility that protozoa are spread by means of droplets formed by the aerosolization of water that is sprayed or splashed during cleaning and disinfection processes cannot be excluded. Daily cleaning and disinfection of food-processing areas creates an environment with a relative high moisture content, which favors the development of protozoa. In a study of moisture-damaged buildings, the occurrence of flagellates and ciliates on different building materials was demonstrated, including the presence of amoebae, which was positively correlated with the water content of the sampled material (67). In another study, 47% (23/49) of the swabs taken from moist areas (floor tiles, drains of sinks, water taps) in a hospital were positive for amoebae (56). In this study, amoebae were detected in 29.5% of the enrichment cultures. The amoebae identified by Rohr et al. (56) mainly belonged to the genera Hartmannella, Vahlfkampfia, and Vannella, which is in accordance with our data. Acanthamoeba spp. and Naegleria spp., however, were not found in the present survey. The protozoan community is subjected to different stress conditions, such as cold, desiccation, and cleaning and disinfection processes. Many, but not all, protozoa have the ability to encyst and can resist unfavorable conditions. Trophozoites and cysts have different susceptibilities to disinfectants, and cysts commonly show higher resistance (36). Our results suggest that protozoa in food-processing areas resist the daily cleaning and disinfection process, either due to excystment after the disinfection cycle, due to the inefficiency of disinfectants at the recommended user concentrations, due to inadequately cleaned surfaces, or due to uncharacterized cell response mechanisms. This suggestion is supported by the fact that protozoa have been found in other anthropogenic environments subjected to disinfection or having residual disinfectant concentrations, such as water systems and swimming pools (55, 62). However, susceptibility tests are necessary to determine the extent of resistance of free-living protozoa to disinfectants commonly applied in the food industry.
In conclusion, high protozoan species richness in meat-cutting plants was demonstrated. This diversity is preferably studied by a combination of microscopic observations and molecular techniques. As a recommendation for further studies, analysis of the composition of eukaryotic assemblages may be refined by designing specific primer sets to study particular groups of amoebae, ciliates, and flagellates. The ecological significance of free-living protozoa in food-processing areas remains unclear. Bacterivory by protozoa should influence the bacterial population in terms of number and species diversity. In this regard, protozoa might be considered “partners” in the control of bacterial levels. On the other hand, the diversity study showed that some of the protozoa are potential hosts for food-borne pathogens. Although an association of food-borne pathogens with protozoa such as T. pyriformis has been shown, whether food-borne pathogens really interact with protozoa isolated from food-processing environments remains to be determined, since most previous studies were carried out with culture collection strains under laboratory conditions. A recent study, however, showed that free-living protozoa isolated from vegetables were able to internalize and release food-borne pathogens (31). The detection of food-borne pathogens in environmental protozoa, as demonstrated by fluorescence in situ hybridization, is a logical next step. Furthermore, the survival of protozoa (and their internalized bacteria) under stress conditions (desiccation, disinfectants, extreme pH values, and heat), the increase in virulence of food-borne pathogens after passage through environmental protozoa, and the effect of grazing activities of protozoa on microbial communities in food-processing areas are all open fields of research.
Acknowledgments
This research was funded by doctoral fellowship O1J18206 from the Special Research Fund (BOF, Bijzonder Onderzoeksfonds) of Ghent University. Julie Baré is a doctoral fellow of the Flemish Institute for the Promotion of Innovation by Science and Technology in Flanders (grant IWT-SB/53141).
Johan Van Hende is acknowledged for technical assistance during sampling, Jeroen van Wichelen is acknowledged for checking microscopic determinations, and Andy Vierstraete is acknowledged for performing sequencing. We thank the companies involved for their cooperation.
Footnotes
Published ahead of print on 18 July 2008.
REFERENCES
- 1.Abu Kwaik, Y., L.-Y. Gao, B. J. Stone, C. Venkataraman, and O. S. Harb. 1998. Invasion of protozoa by Legionella pneumophila and its role in bacterial ecology and pathogenesis. Appl. Environ. Microbiol. 64:3127-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adékambi, T., S. Ben Salah, M. Khlif, D. Raoult, and M. Drancourt. 2006. Survival of environmental mycobacteria in Acanthamoeba polyphaga. Appl. Environ. Microbiol. 72:5974-5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adl, S. M., A. G. B. Simpson, M. A. Farmer, R. A. Andersen, O. R. Anderson, J. R. Barta, S. S. Bowser, G. Brugerolle, R. A. Fensome, S. Fredericq, T. Y. James, S. Karpov, P. Kugrens, J. Krug, C. E. Lane, L. A. Lewis, J. Lodge, D. H. Lynn, D. G. Mann, R. M. McCourt, L. Mendoza, Ø. Moestrup, S. E. Mozley-Standrigde, T. A. Nerad, C. A. Shearer, A. V. Smirnov, F. W. Spiegel, and M. F. J. R. Taylor. 2005. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol. 52:399-451. [DOI] [PubMed] [Google Scholar]
- 4.Aguilera, A., F. Gómez, E. Lospitao, and R. Amils. 2006. A molecular approach to the characterization of the eukaryotic communities of an extreme acidic environment: methods for DNA extraction and denaturing gradient gel electrophoresis analysis. Syst. Appl. Microbiol. 29:593-605. [DOI] [PubMed] [Google Scholar]
- 5.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Axelsson-Olsson, D., J. Waldenström, T. Broman, B. Olsen, and M. Holmberg. 2005. Protozoan Acanthamoeba polyphaga as a potential reservoir for Campylobacter jejuni. Appl. Environ. Microbiol. 71:987-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barker, J., T. J. Humphrey, and M. W. R. Brown. 1999. Survival of Escherichia coli O157 in a soil protozoan: implications for disease. FEMS Microbiol. Lett. 173:291-295. [DOI] [PubMed] [Google Scholar]
- 8.Barker, J., H. Scaife, and M. R. W. Brown. 1995. Intraphagocytic growth induces an antibiotic-resistant phenotype of Legionella pneumophila. Antimicrob. Agents Chemother. 39:2684-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berglund, J., K. Jürgens, I. Bruchmüller, M. Wedlin, and A. Andersson. 2005. Use of group-specific PCR primers for identification of chrysophytes by denaturing gradient gel electrophoresis. Aquat. Microb. Ecol. 39:171-182. [Google Scholar]
- 10.Berk, S. G., R. S. Ting, G. W. Turner, and R. J. Ashburn. 1998. Production of respirable vesicles containing live Legionella pneumophila cells by two Acanthamoeba spp. Appl. Environ. Microbiol. 64:279-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berney, C., J. Fahrni, and J. Pawlowski. 2004. How many novel eukaryotic ‘kingdoms’? Pitfalls and limitations of environmental DNA surveys. BMC Biol. 2:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boenigk, J., K. Pfandl, P. Stadler, and A. Chatzinotas. 2005. High diversity of the ‘Spumella-like’ flagellates: an investigation based on the SSU rRNA gene sequences of isolates from habitats located in six different geographic regions. Environ. Microbiol. 7:685-697. [DOI] [PubMed] [Google Scholar]
- 13.Bouyer, S., C. Imbert, M.-H. Rodier, and Y. Héchard. 2007. Long-term survival of Legionella pneumophila associated with Acanthamoeba castellanii vesicles. Environ. Microbiol. 9:1341-1344. [DOI] [PubMed] [Google Scholar]
- 14.Brandl, M. T., B. M. Rosenthal, A. F. Haxo, and S. G. Berk. 2005. Enhanced survival of Salmonella enterica in vesicles released by a soilborne Tetrahymena species. Appl. Environ. Microbiol. 71:1562-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caron, D. A. 1991. Heterotrophic flagellates associated with sedimenting detritus, p. 77-92. In D. J. Patterson and J. Larsen (ed.), The biology of free-living heterotrophic flagellates. Clarendon Press, Oxford, United Kingdom.
- 16.Cavalier-Smith, T., and E.-Y. Chao. 2003. Phylogeny and classification of phylum Cercozoa (Protozoa). Protist 154:341-358. [DOI] [PubMed] [Google Scholar]
- 17.Cirillo, J. D., S. L. G. Cirillo, L. Yan, L. E. Bermudez, S. Falkow, and L. S. Tompkins. 1999. Intracellular growth in Acanthamoeba castellanii affects monocyte entry mechanisms and enhances virulence of Legionella pneumophila. Infect. Immun. 67:4427-4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clare, J. J., L. S. Davidow, D. C. Gardner, and S. G. Oliver. 1986. Cloning and characterization of the ribosomal RNA genes of the dimorphic yeast, Yarrowia lipolytica. Curr. Genet. 10:449-452. [DOI] [PubMed] [Google Scholar]
- 19.Díez, B., C. Pedrós-Alió, T. L. Marsh, and R. Massana. 2001. Application of denaturing gradient gel electrophoresis (DGGE) to study the diversity of marine picoeukaryotic assemblages and comparison of DGGE with other molecular techniques. Appl. Environ. Microbiol. 67:2942-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dopheide, A., G. Lear, R. Stott, and G. Lewis. 2008. Molecular characterization of ciliate diversity in stream biofilms. Appl. Environ. Microbiol. 74:1740-1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.European Union. 2001. Commission decision of 8 June 2001 laying down rules for the regular checks on the general hygiene carried out by the operators in establishments according to Directive 64/433/EEC on health conditions for the production and marketing of fresh meat and Directive 71/118/EEC on health problems affecting the production and placing on the market of fresh poultry meat (2001/471/EC). Off. J. Eur. Commun. L 165:48-53. [Google Scholar]
- 22.Foissner, W., H. Berger, H. Blatterer, and F. Kohmann. 1995. Taxonomische und ökologische Revision der Ciliaten des Saprobiensystems. Band IV: Gymnostomatea, Loxodes, Suctoria. Informationsberichte des Bayer. Landesamtes für Wasserwirtschaft, Heft 1/95. Bartels und Wernitz Druck, München, Germany.
- 23.Foissner, W., H. Berger, and F. Kohmann. 1992. Taxonomische und ökologische Revision der Ciliaten des Saprobiensystems. Band II: Peritrichia, Heterotrichida, Odontostomatida. Informationsberichte des Bayer. Landesamtes für Wasserwirtschaft, Heft 5/92. Bartels und Wernitz Druck, München, Germany.
- 24.Foissner, W., H. Berger, and F. Kohmann. 1994. Taxonomische und ökologische Revision der Ciliaten des Saprobiensystems. Band III: Hymenostomata, Protomatida, Nassulida. Informationsberichte des Bayer. Landesamtes für Wasserwirtschaft, Heft 1/94. Bartels und Wernitz Druck, München, Germany.
- 25.Foissner, W., H. Blatterer, H. Berger, and F. Kohmann. 1991. Taxonomische und ökologische Revision der Ciliaten des Saprobiensystems. Band I: Cyrtophorida, Oligotrichida, Hypotrichia, Colpodea. Informationsberichte des Bayer. Landesamtes für Wasserwirtschaft, Heft 1/91. Bartels und Wernitz Druck, München, Germany.
- 26.Gao, L.-Y., and Y. Abu Kwaik. 2000. The mechanism of killing and exiting the protozoan host Acanthamoeba polyphaga by Legionella pneumophila. Environ. Microbiol. 2:79-90. [DOI] [PubMed] [Google Scholar]
- 27.García, M. T., S. Jones, C. Pelaz, R. D. Millar, and Y. Abu Kwaik. 2007. Acanthamoeba polyphaga resuscitates viable non-culturable Legionella pneumophila after disinfection. Environ. Microbiol. 9:1267-1277. [DOI] [PubMed] [Google Scholar]
- 28.Gaze, W. H., N. Burroughs, M. P. Gallagher, and E. M. H. Wellington. 2003. Interactions between Salmonella typhimurium and Acanthamoeba polyphaga, and observation of a new mode of intracellular growth within contractile vacuoles. Microb. Ecol. 46:358-369. [DOI] [PubMed] [Google Scholar]
- 29.Görtz, H.-D. 2001. Intracellular bacteria in ciliates. Int. Microbiol. 4:143-150. [DOI] [PubMed] [Google Scholar]
- 30.Görtz, H.-D., and T. Brigge. 1998. Intracellular bacteria in protozoa. Naturwissenschaften 85:359-368. [DOI] [PubMed] [Google Scholar]
- 31.Gourabathini, P., M. T. Brandl, K. S. Redding, J. H. Gunderson, and S. G. Berk. 2008. Interactions between food-borne pathogens and protozoa isolated from lettuce and spinach. Appl. Environ. Microbiol. 74:2518-2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greub, G., and D. Raoult. 2004. Microorganisms resistant to free-living amoebae. Clin. Microbiol. Rev. 17:413-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 34.Horn, M., and M. Wagner. 2004. Bacterial endosymbionts of free-living amoebae. J. Eukaryot. Microbiol. 51:509-514. [DOI] [PubMed] [Google Scholar]
- 35.Huws, S. A., R. J. Morley, M. V. Jones, M. R. W. Brown, and A. W. Smith. 2008. Interactions of some common pathogenic bacteria with Acanthamoeba polyphaga. FEMS Microbiol. Lett. 282:258-265. [DOI] [PubMed] [Google Scholar]
- 36.Kilvington, S., and J. Price. 1990. Survival of L. pneumophila within cysts of Acanthamoeba polyphaga following chlorine exposure. J. Appl. Bacteriol. 68:519-525. [DOI] [PubMed] [Google Scholar]
- 37.King, C. H., E. B. Shotts, Jr., R. E. Wooley, and K. G. Porter. 1988. Survival of coliforms and bacterial pathogens within protozoa during chlorination. Appl. Environ. Microbiol. 54:3023-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kingston, D., and D. C. Warhurst. 1969. Isolation of amoebae from the air. J. Med. Microbiol. 2:27-36. [DOI] [PubMed] [Google Scholar]
- 39.Lara, E., C. Berney, F. Ekelund, H. Harms, and A. Chatzinotas. 2007. Molecular comparison of cultivable protozoa from a pristine and a polycyclic aromatic hydrocarbon polluted site. Soil Biol. Biochem. 39:139-148. [Google Scholar]
- 40.Lara, E., C. Berney, H. Harms, and A. Chatzinotas. 2007. Cultivation-independent analysis reveals a shift in ciliate 18S rRNA gene diversity in a polycyclic aromatic hydrocarbon-polluted soil. FEMS Microbiol. Ecol. 62:365-373. [DOI] [PubMed] [Google Scholar]
- 41.Lepère, C., D. Boucher, L. Jardillier, I. Domaizon, and D. Debroas. 2006. Succession and regulation of factors of small eukaryote community composition in a lacustrine ecosystem (Lake Pavin). Appl. Environ. Microbiol. 72:2971-2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ly, T. M. C., and H. E. Müller. 1990. Ingested Listeria monocytogenes survive and multiply in protozoa. J. Med. Microbiol. 33:51-54. [DOI] [PubMed] [Google Scholar]
- 43.Marie, D., F. Zhu, V. Balagué, J. Ras, and D. Vaulot. 2006. Eukaryotic picoplankton communities of the Mediterranean Sea in summer assessed by molecular approaches (DGGE, TGGE, QPCR). FEMS Microbiol. Ecol. 55:403-415. [DOI] [PubMed] [Google Scholar]
- 44.Martín-Cereceda, M., A. Guinea, E. Bonaccorso, P. Dyal, G. Novarino, and W. Foissner. 2007. Classification of the peritrich ciliate Opisthonecta matiensis (Martín-Cereceda et al. 1999) as Telotrochidium matiense nov. comb., based on new observations and SSU rDNA phylogeny. Eur. J. Protistol. 43:265-279. [DOI] [PubMed] [Google Scholar]
- 45.Massana, R., V. Balagué, L. Guillou, and C. Pedrós-Alió. 2004. Picoeukaryotic diversity in an oligotrophic coastal site studied by molecular and culturing approaches. FEMS Microbiol. Ecol. 50:231-243. [DOI] [PubMed] [Google Scholar]
- 46.Massana, R., and C. Pedrós-Alió. 2008. Unveiling new microbial eukaryotes in the surface ocean. Curr. Opin. Microbiol. 11:213-218. [DOI] [PubMed] [Google Scholar]
- 47.Medlin, L. K., K. Metfies, H. Mehl, K. Wiltshire, and K. Valentin. 2006. Picoeukaryotic plankton diversity at the Helgoland time series site as assessed by three molecular methods. Microb. Ecol. 52:53-71. [DOI] [PubMed] [Google Scholar]
- 48.Moon-van der Staay, S. Y., R. De Wachter, and D. Vaulot. 2001. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature 409:607-610. [DOI] [PubMed] [Google Scholar]
- 49.Moreira, D., and P. López-García. 2002. The molecular ecology of microbial eukaryotes unveils a hidden world. Trends Microbiol. 10:31-38. [DOI] [PubMed] [Google Scholar]
- 50.Myl'nikov, A. P., and S. A. Karpov. 2004. Review of diversity and taxonomy of cercomonads. Protistology 3:201-217. [Google Scholar]
- 51.Page, F. C. 1988. A new key to freshwater and soil Gymnamoebae. Freshwater Biological Association, Ambleside, United Kingdom.
- 52.Patterson, D. J. 1998. Free-living freshwater protozoa: a colour guide. Manson Publishing Ltd., London, United Kingdom.
- 53.Rasmussen, L. D., F. Ekelund, L. H. Hansen, S. J. Sørensen, and K. Johnsen. 2001. Group-specific PCR primers to amplify 24S α-subunit rRNA genes from Kinetoplastida (Protozoa) used in denaturing gradient gel electrophoresis. Microb. Ecol. 42:109-115. [DOI] [PubMed] [Google Scholar]
- 54.Rivera, F., A. Lugo, E. Ramírez, P. Bonilla, A. Calderón, S. Rodríguez, R. Ortiz, E. Gallegos, A. Labastida, and M. P. Chavez. 1992. Seasonal distribution of air-borne protozoa in Mexico City and its suburbs. Water Air Soil Pollut. 61:17-36. [Google Scholar]
- 55.Rivera, F., E. Ramírez, P. Bonilla, A. Calderón, E. Gallegos, S. Rodríguez, R. Ortiz, B. Zaldívar, P. Ramírez, and A. Durán. 1993. Pathogenic and free-living amoebae isolated from swimming pools and physiotherapy tubs in Mexico. Environ. Res. 62:43-52. [DOI] [PubMed] [Google Scholar]
- 56.Rohr, U., S. Weber, R. Michel, F. Selenka, and M. Wilhelm. 1998. Comparison of free-living amoebae in hot water systems of hospitals with isolates from moist sanitary areas by identifying genera and determining temperature tolerance. Appl. Environ. Microbiol. 64:1822-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Savin, M. C., J. L. Martin, M. LeGresley, M. Giewat, and J. Rooney-Varga. 2004. Plankton diversity in the Bay of Fundy as measured by morphological and molecular methods. Microb. Ecol. 48:51-65. [DOI] [PubMed] [Google Scholar]
- 58.Smirnov, A. V. 2003. Optimizing methods of the recovery of gymnamoebae from environmental samples: a test of ten popular enrichment media, with some observations on the development of cultures. Protistology 3:47-57. [Google Scholar]
- 59.Snelling, W. J., J. P. McKenna, D. M. Lecky, and J. S. G. Dooley. 2005. Survival of Campylobacter jejuni in waterborne protozoa. Appl. Environ. Microbiol. 71:5560-5571. (Author's correction, 71:7631.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taylor, S. J., L. J. Ahonen, F. A. A. M. de Leij, and J. W. Dale. 2003. Infection of Acanthamoeba castellanii with Mycobacterium bovis and M. bovis BCG and survival of M. bovis within the amoebae. Appl. Environ. Microbiol. 69:4316-4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tezcan-Merdol, D., M. Ljungström, J. Winiecka-Krusnell, E. Linder, L. Engstrand, and M. Rhen. 2004. Uptake and replication of Salmonella enterica in Acanthamoeba rhysodes. Appl. Environ. Microbiol. 70:3706-3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thomas, V., T. Bouchez, V. Nicolas, S. Robert, J. F. Loret, and Y. Lévi. 2004. Amoebae in domestic water systems: resistance to disinfection treatments and implications in Legionella persistence. J. Appl. Microbiol. 97:950-963. [DOI] [PubMed] [Google Scholar]
- 63.van Hannen, E. J., W. Mooij, M. P. van Agterveld, H. J. Gons, and H. J. Laanbroek. 1999. Detritus-dependent development of the microbial community in an experimental system: qualitative analysis by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 65:2478-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van Hannen, E. J., M. P. van Agterveld, H. J. Gons, and H. J. Laanbroek. 1998. Revealing genetic diversity of eukaryotic microorganisms in aquatic environments by denaturing gradient gel electrophoresis. J. Phycol. 34:206-213. [Google Scholar]
- 65.von Wintzingerode, F., U. B. Göbel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 66.Winiecka-Krusnell, J., K. Wreiber, A. von Euler, L. Engstrand, and E. Linder. 2002. Free-living amoebae promote growth and survival in Helicobacter pylori. Scand. J. Infect. Dis. 34:253-256. [DOI] [PubMed] [Google Scholar]
- 67.Yli-Pirilä, T., J. Kusnetsov, S. Haatainen, M. Hänninen, P. Jalava, M. Reiman, M. Seuri, M.-R. Hirvonen, and A. Nevalainen. 2004. Amoebae and other protozoa in material samples from moisture-damaged buildings. Environ. Res. 96:250-256. [DOI] [PubMed] [Google Scholar]
- 68.Zhou, X., J. Elmose, and D. R. Call. 2007. Interactions between the environmental pathogen Listeria monocytogenes and a free-living protozoan (Acanthamoeba castellanii). Environ. Microbiol. 9:913-922. [DOI] [PubMed] [Google Scholar]