Abstract
To develop a piglet model for studying diarrheal disease and developing vaccines, we challenged gnotobiotic piglets with isogenic Escherichia coli strains constructed to express porcine 987P(F6) fimbriae and a heat-labile or a heat-stable enterotoxin to examine clinical outcomes. Piglets developed identical diarrheal diseases when inoculated with constructs expressing human or porcine enterotoxins.
Enterotoxigenic Escherichia coli (ETEC) strains that colonize the small intestines and produce enterotoxins are the major cause of diarrheal disease in humans and animals (15, 29, 35, 36). The key virulence factors of ETEC in diarrhea include enterotoxins and colonization factor antigens or fimbriae. These colonization factor antigens or fimbriae mediate attachment of bacteria to host epithelium cells and facilitate bacterial colonization. Enterotoxins stimulate fluid secretion in the intestinal lumens, which results in diarrhea. The pathogenesis of ETEC-associated diarrhea has been extensively studied. However, significant progress toward disease prevention is still lacking, partially because most investigators lack a suitable animal model with which to study host-pathogen interactions and to develop prevention strategies.
Human subjects, especially highly susceptible young children, have to be excluded from diarrheal studies due to a higher level of risks. Challenge studies with adult human volunteers have been limited and have provided insufficient data to critically assess the contribution of each ETEC virulence determinant (9, 18, 31). It is therefore necessary to employ animal models to study ETEC pathogenicity and to develop prevention strategies. Mouse and rabbit models have been used to assess the pathology of ETEC enterotoxins (6, 14, 17, 20, 22, 27). However, mice are not naturally susceptible to ETEC, and fundamental differences in the pathogenicities of ETEC in mice and in humans exist, including the ability to sufficiently adhere to human but not mouse intestinal epithelium. In addition, after being orally inoculated with a human diarrheagenic ETEC strain, mice did not develop diarrhea or become dehydrated and had a significantly low colonization of the ETEC strain in their small intestines (1). A mouse model may have limited value for the study of ETEC, particularly with regard to the study of immune development. Similar to mice, rabbits are not susceptible to ETEC strains. Thus, a rabbit model has the same limitation as a mouse model and is not suitable for studying ETEC diarrhea.
In contrast to mice and rabbits, young pigs that express receptors of ETEC adhesins are naturally susceptible to diarrheagenic porcine ETEC strains (11, 13, 39). Infected young pigs develop typical diarrhea and may become dehydrated (3, 32, 38), similar to clinical outcomes of human diarrheic patients. The similarity between porcine and human ETEC infections in pathogenesis and clinical outcomes suggests that young pigs would be a good model to study human ETEC diarrhea. However, the heat-labile (LT) and heat-stable (ST) enterotoxins produced by porcine and human ETEC strains are different. Although the nucleotide sequences of the eltAB genes (coding for porcine LT [pLT] and human LT [hLT]) and the estA genes (coding for porcine STa [pSTa] and human STa [hSTa] toxins) are highly homologous (5, 10, 21, 33), the genes coding for hLT and hSTa toxins are not found in porcine ETEC strains that cause diarrhea, and the porcine and human toxins differ in epitopes and thus antigenicities (16, 37). Differences in hLT or hSTa in biological activity from pLT and pSTa will be relevant to the development of a porcine model of human ETEC disease. In this study, we constructed isogenic ETEC strains expressing 987P(F6) fimbria and an enterotoxin (pLT, pSTa, hLT, or hSTa) and conducted comparative challenge studies in gnotobiotic piglets to evaluate clinical disease in the piglet infection model.
We used a nonpathogenic porcine E. coli field isolate, G58-1 (12), as the parental strain to construct isogenic strains that expressed 987P fimbria and either a human or a porcine LT or STa toxin. The G58-1 strain does not carry genes for any known enterotoxins (LT, STa, STb, EAST1, and Stx2e) or fimbrial or nonfimbrial adhesins [K88(F4), F18, K99(F5), F41, 987P, Paa, AIDA-I and EAE]. It does not stimulate fluid accumulation in porcine ligated gut loops, nor does it cause diarrhea in young gnotobiotic piglets. In our previous study, we used nonpathogenic E. coli isolate 1836-2 as the parental strain to assess contributions of LT and STb to diarrheal disease (38). However, in a subsequent study we found that strain 1836-2 carries two potential virulence genes: the astA gene coding for EAST1 toxin and the paa adhesin gene. To eliminate any influences from these potential virulence factors, we used G58-1 as the parental strain for the present study. G58-1 cells were first made competent by following standard protocols (26) and transformed with plasmid pDMS167, which expressed the fimbria 987P (a gift from D. M. Schifferli, University of Pennsylvania) (28) to produce a 987P-positive E. coli construct, 8293 (G58/987P [Table 1]). Expression of the 987P fimbriae in the 8293 strain was confirmed in a Western blot using anti-987P polyclonal antibodies (data not shown). The enterocyte brush border binding activity of the 987P fimbria was verified in a modified porcine brush border adherence assay (2, 4, 25, 30). We first identified 987P-receptor-positive pigs by using a 987P+ ETEC wild-type 1194 strain in the adherence assay and then used the 987P-receptor-positive brush borders to test binding of strain 8293. The adherence assay showed that on average 15 8293 cells bound to a single brush border, equivalent to the level of binding of wild-type strain 1194. This indicated that 987P fimbriae were expressed properly on the bacterium surface in strain 8293. Next, we used strain 8293 as host cells to construct isogenic strains expressing a porcine or a human enterotoxin. All constructs were grown in Luria-Bertani (LB) medium supplemented with chloramphenicol (20 μg/ml) overnight at 37°C.
TABLE 1.
Strains and plasmids used for this study
| Strain or plasmid | Virulence determinant(s) | Source or reference |
|---|---|---|
| Strains | ||
| G58-1 | Nonpathogenic porcine isolate | 12 |
| 3030-2 | Porcine ETEC strain | 12 |
| 8293 | 987P | This study |
| 8331 | 987P/pACYC184, as negative control | This study |
| 8330 | 987P/pSTa | This study |
| 8351 | 987P/hSTa | This study |
| 8484 | 987P/hLT | This study |
| 8561 | 987P/pLT | This study |
| H10407 | hLT/hSTa | Human ETEC strain |
| 04-21018 | F18/STa/STb | Porcine field isolate |
| 04-10779 | STa | Porcine field isolate |
| 1194 | 987P/pSTa/pSTb | Porcine field isolate |
| Plasmids | ||
| pDMS167 | 987P fimbrial gene cluster in pBR322 derivative | 28 |
| pACYC184 | GenBank accession no. X06403 | Promega, Madison, WI |
| p8297 | Porcine estA gene in pACYC184 at SfcI/EagI | This study |
| p8299 | Human estA gene in pACYC184 at SfcI/EagI | This study |
| p8477 | Human eltAB gene in pACYC184 at EcoRV | This study |
| p8458 | Porcine eltAB gene in pACYC184 at NheI/EagI | This study |
To express porcine or human enterotoxins, we isolated the human eltAB (LT) and estA (STa) genes from a human ETEC strain H10407 (kindly provided by P. Hardwidge), and the porcine eltAB gene from a porcine ETEC wild-type strain, 3030-2 (12). The porcine estA was isolated from porcine STa+ ETEC field strain 04-21018. Strains 3030-2 and 04-21018 were isolated from weaned pigs with diarrhea submitted to the Animal Disease Research and Diagnostic Laboratory at the Veterinary Science Department, South Dakota State University. Total genomic DNA prepared with a DNeasy tissue kit (Qiagen, Valencia, CA) was used as the templates to amplify the human or porcine eltAB or estA gene with the specific primers listed in Table 2. PCRs were performed in an MJ PT-100 thermocycler (Bio-Rad, Hercules, CA) in a reaction of 50 μl containing 1× Pfu DNA polymerase buffer (with Mg2+), 0.2 mM deoxynucleoside triphosphate (dNTP), 0.5 μM each forward and reverse primers, 100 ng of DNA template, and 1 U of Pfu DNA polymerase (Strategene, La Jolla, CA). The PCR program contained one cycle of 2 min at 94°C and 30 cycles of 45 s at 94°C, 45 s at 52°C, and 2 min at 72°C, followed by an extension of 6 min at 72°C. Amplified PCR products were separated in 1% agarose (FMC Bioproducts, Rockland, MA) gels by electrophoresis and purified using a QIAquick gel extraction kit according to the manufacturer's instructions (Qiagen).
TABLE 2.
PCR primers used to amplify porcine and human eltAB and estA genes
| Gene and primer | Sequence |
|---|---|
| eltAB | |
| Human | |
| hLT-F | 5′-ATGATTGACATCATGTTGCATATAGG-3′ |
| hLT-R | 5′-CCCCTCCAGCCTAGCTTAGTT-3′ |
| Porcine | |
| pLT-F | 5′-ATCCTCGCTAGCATGTTTTAT-3′ |
| pLT-R | 5′-CCCCTCCGGCCGAGCTTAGTT-3′ |
| estA | |
| Human | |
| hSTaSfcI-F2 | 5′-GATTAACCCCCACAACTATAGTCA-3′ |
| STaEagI-R | 5′-GTGGAGCCGGCCGAAACA-3′ |
| Porcine | |
| pSTaSfcI-F2 | 5′-GTTTATTGATGTTCTGCAGACA-3′ |
| STaEagI-R | 5′-GTGGAGCCGGCCGAAACA-3′ |
Purified PCR products were digested with restriction enzymes and then were cloned into vector pACYC184 (New England Biolab, Beverly, MA). Vector pACYC184 was selected because of its compatibility with vector pBR322, which was used for cloning and expressing the 987P fimbriae in pDMS167. PCR products of the human and porcine estA genes were digested with SfcI and EagI, and PCR products of the porcine eltAB gene were digested with NheI and EagI restriction enzymes, respectively. PCR products of the human eltAB gene were used for blunt end ligation. Vector pACYC184 was also digested with SfcI and EagI, NheI and EagI, and EcoRV for cloning of the porcine and human estA genes, porcine eltAB gene, and the human eltAB gene, respectively. Digested products from the inserts and the vector were separated by gel electrophoresis, purified with the QIAquick gel extraction kit, and ligated with T4 DNA ligase (Promega, Madison, WI) at 14°C overnight.
Two microliters of each T4 ligated product was introduced into 25 μl of TOPO cells (Invitrogen, CA) in a standard electroporation. Antibiotic-selected colonies were initially screened by PCR, and positive colonies were sequenced using an ABI DigDye terminator sequencing kit (Applied Biosystem, CA) to verify the insertion of each gene and to ensure that the inserted gene was in the reading frame. Four plasmids, p8458 (pLT), p8477 (hLT), p8297 (pSTa), and p8299 (hSTa), were produced (Table 1). Transformation of competent 8293 cells (G58/987P) with each plasmid resulted in four isogenic E. coli strains 8330 (987P/pSTa), 8351 (987P/hSTa), 8484 (987P/hLT), and 8561 (987P/pLT), respectively. Empty pACYC184 vectors were introduced into strain 8293 to produce the negative control strain 8331 (Table 1). A colony passage study indicated that over 95% of isogenic cells retained the 987P and enterotoxin plasmids after five passages.
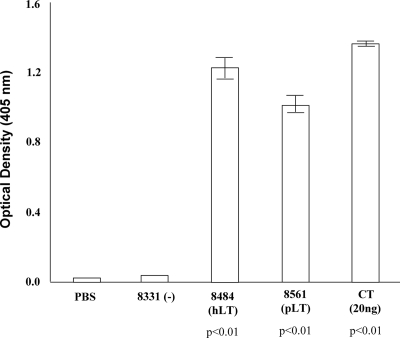
Expressed LT and STa proteins were detected by GM1 ganglioside enzyme-linked immunosorbent assay (ELISA) and STa competitive ELISA, respectively. The LT constructs were grown in 20 ml of LB broth overnight at 37°C in the presence of chloramphenicol (20 μg/ml), and the overnight-grown culture was used to prepare total proteins by using B-PER reagent (in phosphate buffer) (Pierce, Rockford, IL). Total protein samples from the LT constructs were used for GM1 ELISA with anti-cholera toxin (anti-CT) rabbit serum (Sigma) as the primary antibody as described previously (3, 24, 38). We included purified CT (20 ng) as the positive control, with phosphate-buffered saline as a background check, and tested triplets of total protein samples from each construct in the ELISA. Results from the GM1 ELISA indicated that LT proteins were expressed in strains 8484 and 8561 at approximately 150 to 180 ng per ml of culture suspension, equivalent to that in a wild-type ETEC strain 3030-2 (200 ng/ml) (38). LT was not produced by strain 8331 (Fig. 1). Statistical analysis using Student's t test indicated that the LT expression in both the pLT and hLT constructs was significantly different from that in strain 8331 (P < 0.01).
FIG. 1.
GM1 ELISA to detect expression of LT proteins from hLT and pLT constructs. Pellets from an overnight-grown culture were used for total protein preparation using B-PER. Each sample was assayed in triplicate. Anti-CT serum (1:5,000) was used as the primary antibody, and goat anti-rabbit HRP-conjugated IgG (1:5,000) was used as the secondary antibody. Optical densities were measured at a wavelength of 405 nm. PBS, phosphate-buffered saline.
An assessment of STa expression was conducted in a competitive ELISA following a protocol modified from Lockwood and Robertson (19). The hSTa and the pSTa constructs were grown in LB broth at 37°C for 8 h, and then similar amounts of cells (based on optical density measurements) were transferred to 4AA medium (19) and grown at 37°C overnight. Overnight-grown cultures were pelleted, and 75 μl of supernatant from each strain was used in the STa competitive ELISA. ELISA microtiter plates (Costar 2595; Corning, Inc., Corning, NY) were coated with 1.25 ng of STa-ovalbumin conjugate in ELISA dilution buffer (128 mM NaCl, 2.68 mM KCl, 1.47 mM KH2PO4, 8.10 mM Na2HPO4, 0.05% ovalbumin [pH 7.0 to 7.2]) per well. Coated plates were blocked with casein blocking buffer (2.5% casein in 0.3 N NaOH [pH 7.0]) and washed with STa washing buffer (ELISA dilution buffer with addition of 0.05% Tween 20). Seventy-five microliters of culture supernatant and 75 μl of anti-STa serum (1:10,000 in dilution) were added to each well, followed by incubation at 37°C for 2 h on a shaker (180 rpm). A series of control samples containing synthetic STa peptide at 0, 0.01, 0.05, 0.10, 0.50, 1.0, 5.0, and 10.0 ng in 75 μl ELISA buffer were also mixed with 75 μl of anti-STa serum (1:10,000) to generate a standard reference curve. After three washes, plates were blotted dry and incubated with goat anti-rabbit horseradish peroxidase (HRP)-conjugated IgG (1:10,000) at 37 C for 1 h. The optical densities from the reaction of bound IgG with ABTS [2,2′-azinobis(3-ethylbenzthiasoline sulfonic acid)] substrate were measured in a plate reader at 405 nm.
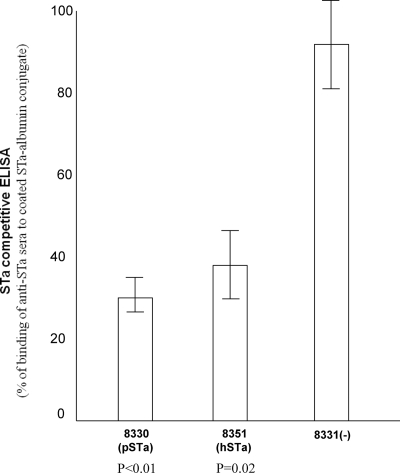
The STa competitive ELISA showed that STa proteins were detected in the hSTa and the pSTa isogenic strains. Protein samples from the pSTa and hSTa strains blocked the binding of the anti-STa serum to the STa-ovalbumin conjugate immobilized on the ELISA plate. By reference to a standard curve, the binding of the anti-STa serum to the coated STa-ovalbumin in wells mixed with STa proteins from strains 8330 and 8351 were only 30% ± 4.6% and 37.6% ± 8.7%, respectively, while binding in wells mixed with protein samples from negative control strain 8331 was 92.5% ± 10.4% (Fig. 2). Differences in STa expression between pSTa and hSTa constructs likely were caused by differences in recognition of two STa proteins by the anti-STa antibody, since pSTa and hSTa strains were constructed using the same host strain and the same expression vector. A porcine STa+ wild-type ETEC strain, 04-10779, and a human ETEC strain, H10407, were also included in the ELISA, and the levels of binding in wells mixed with protein samples from 04-10779 and H10407 were 19.6% ± 0.7% and 26.1% ± 3.5%, respectively. These results suggested that the STa proteins expressed in strains 8330 and 8351 blocked two-thirds of the binding from the anti-STa serum to the immobilized STa-ovalbumin conjugates, which was significantly higher than the blocking by the negative control strain 8331 (P < 0.01 and P = 0.02, respectively).
FIG. 2.
STa competitive ELISA to detect expression of STa proteins from hSTa and pSTa constructs. Supernatants from overnight-grown culture (4AA medium) were used as protein samples for ELISA. Microtiter plates were coated with STa-ovalbumin conjugate, and anti-STa serum (1:10,000) was used as the primary antibody. Goat anti-rabbit HRP-conjugated IgG (1:10,000) was used as the secondary antibody. Optical densities were measured at a wavelength of 405 nm. Blocking of STa ELISA was calculated based on the standard reference curve (using a series of synthetic STa peptide samples) generated in the same assay.
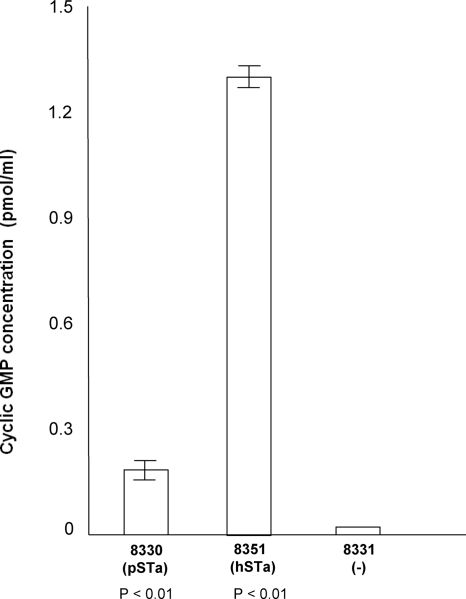
The ability of STa to stimulate cyclic GMP (cGMP) in T-84 human colon cells was measured using a direct cGMP enzyme immunoassay (EIA) kit according to the manufacturer's instructions (acetylated version) (Correlate EIA; Assay Designs, Ann Arbor, MI) and using 75 μl of the same culture supernatant as in STa competitive ELISA (in triplicate). Results from the cGMP ELISA showed a significant increase of cGMP levels in cells incubated with culture supernatant from hSTa- and pSTa-positive strains (Fig. 3), suggesting the STa proteins from the hSTa and pSTa strains were enterotoxic. A greater increase of cGMP level in cells incubated with the hSTa protein sample than in cells incubated with the pSTa proteins was observed, although the pSTa strain showed a slightly higher expression of STa based on results from the STa competitive ELISA. Cells incubated with the H10407 protein sample showed a larger increase in cGMP (2.8 ± 0.6 pmol/ml), but cells incubated with protein samples from porcine STa strains had a smaller increase in cGMP (<0.2 pmol/ml). It is not clear whether the GC-C receptor on human T84 cells is more specific for the hSTa toxin than it is for the pSTa toxin, thus causing differences in cGMP stimulation. However, it was reported that human diarrheal patients suffered more severely from infection with ETEC strains that expressed hSTa than from those that expressed pSTa enterotoxin (34). We attempted to use the porcine cell line IPEC-J2 to test STa stimulation of cGMP but were unable to optimize the assay. Further studies are needed to examine whether hSTa and pSTa stimulate similar increases in cGMP in porcine intestinal epithelial cells.
FIG. 3.
cGMP ELISA to detect biological activity of STa proteins expressed from the hSTa and pSTa constructs through stimulation of cGMP in T-84 cells. The effect of stimulation on increasing cGMP levels was measured with a direct cGMP EIA kit (Correlate EIA; Assay Designs).
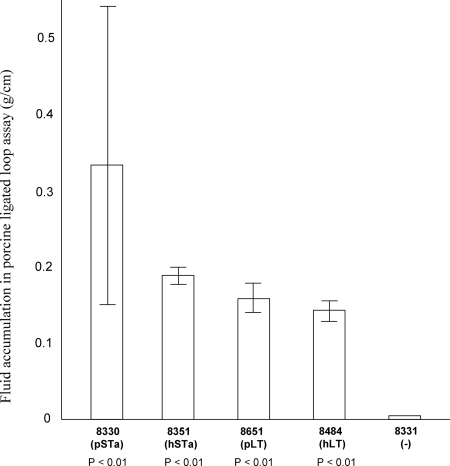
Biological activity of the expressed LT and STa toxins in stimulation of fluid secretion in vivo was tested in a porcine ligated loop assay as described previously (38). A 5-day-old 987P receptor-positive piglet was used in a gut loop assay. Results from the assay showed that the LT and STa proteins from the constructed isogenic strains were biologically functional (Fig. 4). After 8 h of incubation with 2 × 109 CFU of overnight-grown culture in ligated loops (in duplicate), fluid accumulated in loops incubated with the 8330, 8351, 8651, 8484, and 8331 strains was measured as 0.33 ± 0.21, 0.19 ± 0.06, 0.16 ± 0.08, 0.15 ± 0.06, and 0.01 g per cm of loop, respectively. Statistical analysis showed that fluid accumulation in loops containing the LT and STa constructs was significantly higher than that in loops containing the negative control strain (P < 0.01). However, analysis showed no significant differences in fluid stimulation between strains expressing LT and STa (P = 0.21), between strains expressing hSTa and pSTa (P = 0.46), or between strains expressing hLT and pLT (P = 0.95).
FIG. 4.
Porcine ligated gut loop assay to determine biological activity of the LT and STa toxins from the pSTa, hSTa, pLT, and hLT constructs. Overnight-grown culture (2 × 109 CFU) from each strain was injected into a ligated loop. After 8 h of incubation, the fluid accumulated in each loop was measured in grams per centimeter of loop length.
After verifying the enterotoxicity of LT and STa constructs, we conducted animal challenge studies. All animal studies complied with the Animal Welfare Act, followed the Guide for the Care and Use of Laboratory Animals (21a), and were under the supervision of South Dakota State University's Institutional Animal Care and Use committee. Groups of five randomly assigned gnotobiotic piglets, delivered by cesarean section, were inoculated with each construct to compare clinical disease outcomes. At the age of 3 days, the pigs were orally inoculated with 3 × 109 CFU of each construct in 50 ml of sterile milk replacer. Piglets were observed to consume the inoculum and monitored every 2 to 3 h for clinical signs, including vomiting, diarrhea, dehydration, and lethargy. At 24 h postinoculation, piglets were euthanized and subjected to necropsy. Small intestinal ileum and jejunum samples from each pig were collected to assess bacterial colonization by methods previously described (38). Additional jejunum samples from each piglet were collected to prepare brush borders as also described previously (2, 38). In porcine brush border adherence assays, large numbers of 987P-positive bacteria were observed to adhere to enterocyte brush borders prepared from each pig used in this study. Results indicated that each pig contained receptors for the 987P fimbriae (data not shown).
Results from challenge studies showed that 987P fimbrial E. coli strains expressing hLT or hSTa were equally diarrheagenic to gnotobiotic piglets to those expressing pLT or pSTa. All pigs in the groups challenged with the pSTa or the hSTa construct developed diarrhea within 6 to 8 h. Similarly, all pigs in the groups challenged with a pLT or hLT construct developed diarrhea, but clinical disease first appeared at about 12 h postinoculation. None of the pigs challenged with the negative control strain exhibited signs of diarrheal disease. Differences in the incubation times required by strains expressing heat-labile or heat-stable enterotoxins for developing diarrhea were noted. Pigs challenged with either the hSTa or pSTa strain developed diarrhea 4 to 6 h earlier than those inoculated with the hLT or pLT construct, which may explain why STa+ ETEC strains have been observed to cause a more severe disease (23, 34). The time course difference in developing diarrhea in the present study seemed to correlate with the observation that gut loops incubated with either an hSTa strain or a pSTa strain had relatively more (although not significantly) fluid accumulated than those incubated with the LT strains in 8 h of incubation. However, the mechanism responsible for the time differences in onset of disease is unclear.
In this study, we selected to use 987P-receptor-positive piglets because young pigs appear to have receptors to 987P (7, 8). Other porcine ETEC strains that express either K88 or F18 fimbriae exhibit virulence in only a portion of the swine population. However, unlike K88 fimbrial ETEC strains, which are diarrheagenic to K88-receptor-positive pigs up to 8 weeks of age, 987P+ ETEC strains cause diarrhea only in piglets less than 6 days old (7, 8). Therefore, we used 3-day-old piglets for animal challenge studies to avoid the influence of age-related resistance to 987P fimbrial ETEC colonization. Quantitative culturing indicated that all 987P constructs colonized well in the small intestines from the challenged piglets, with 1.0 × 109, 1.5 × 109, 1.85 × 109, 1.55 × 109, and 1.3 × 109 CFU per g of ileum tissue for the pLT, hLT, pSTa, hSTa, and negative constructs, respectively. It seems that 987P fimbrial strains expressing LT or STa did not colonize significantly higher than the 987P-positive but enterotoxin-negative control strain, which is different from the observation that LT enhanced colonization of the K88 fimbrial strain in porcine small intestines (3, 38). Further studies are needed to determine whether associations between different fimbriae and enterotoxins affect bacterial colonization.
Identical disease outcomes and equivalent colonizations by E. coli constructs that expressed human or porcine enterotoxin in pigs make this pig model suitable for studying human ETEC-associated diarrhea. A suitable animal model is critical to facilitate strategies for development of effective treatments for preventing or controlling this disease and improving our knowledge regarding interactions between the host and ETEC. Gnotobiotic piglets may also be a useful model for studying other enteric diseases.
Acknowledgments
We thank D. M. Schifferli for providing the pDMS167 plasmid and Philip Hardwidge for sharing the H10407 strain with us, as well as Kristina Mateo and Diane Baker for assistance in animal surgery and animal challenge studies.
Financial support for this study was provided by the South Dakota 2010 Initiative grant 01-05 (D. Francis), NIH AI068766 (W. Zhang), National Pork Board (NPB) grant 07-006 (W. Zhang), and the South Dakota Agricultural Experiment Station.
Footnotes
Published ahead of print on 25 July 2008.
REFERENCES
- 1.Allen, K. P., M. M. Randolph, and J. M. Fleckenstein. 2006. Importance of heat-labile enterotoxin in colonization of the adult mouse small intestine by human enterotoxigenic Escherichia coli strains. Infect. Immun. 74:869-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, D. R., L. O. Billey, and D. H. Francis. 1996. Distribution of K88 Escherichia coli-adhesive and nonadhesive phenotypes among pigs of four breeds. Vet. Microbiol. 54:123-132. [DOI] [PubMed] [Google Scholar]
- 3.Berberov, E. M., Y. Zhou, D. H. Francis, M. A. Scott, S. D. Kachman, and R. A. Moxley. 2004. Relative importance of heat-labile enterotoxin in the causation of severe diarrheal disease in the gnotobiotic piglet model by a strain of enterotoxigenic Escherichia coli that produces multiple enterotoxins. Infect. Immun. 72:3914-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bijlsma, I. G. W., A. de Nijs, C. van der Meer, and J. F. Frik. 1982. Different pig phenotypes affect adherence of Escherichia coli to jejunal brush borders by K88ab, K88ac, or K88ad antigen. Infect. Immun. 37:891-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dallas, W. S. 1983. Conformity between heat-labile toxin genes from human and porcine enterotoxigenic Escherichia coli. Infect. Immun. 40:647-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dean, A., Y. C. Ching, R. Williams, and L. Harden. 1972. Test for Escherichia coli enterotoxin using infant mice: application in a study of diarrhea in children in Honolulu. J. Infect. Dis. 125:407-411. [DOI] [PubMed] [Google Scholar]
- 7.Dean, E. A., S. C. Whipp, and H. W. Moon. 1989. Age-specific colonization of porcine intestinal epithelium by 987P-piliated enterotoxigenic Escherichia coli. Infect. Immun. 57:82-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dean-Nystrom, E. A., and J. E. Samuel. 1994. Age-related resistance to 987P fimbria-mediated colonization correlates with specific glycolipid receptors in intestinal mucus in swine. Infect. Immun. 62:4789-4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DuPont, H. L., S. B. Formal, R. B. Hornick, M. J. Synder, J. P. Libonati, D. G. Sheahan, E. H. Labrec, and J. P. Kalas. 1971. Pathogenesis of Escherichia coli diarrhea. N. Engl. J. Med. 285:1-9. [DOI] [PubMed] [Google Scholar]
- 10.Finkelstein, R. A. 1988. Cholera, the cholera enterotoxins, and the cholera enterotoxin-related enterotoxin family, p. 85-102. In P. Owen and T. J. Foster (ed.), Immunochemical and molecular genetic analysis of bacterial pathogens. Elsevier, New York, NY.
- 11.Francis, D. H. 2002. Enterotoxigenic Escherichia coli infection in pigs and its diagnosis. J. Swine Health Prod. 10:171-175. [Google Scholar]
- 12.Francis, D. H., and J. A. Willgohs. 1991. Evaluation of a live avirulent Escherichia coli vaccine for K88+, LT+ enterotoxigenic colibacillosis in weaned pigs. Am. J. Vet. Res. 52:1051-1055. [PubMed] [Google Scholar]
- 13.Frydendahl, K. 2002. Prevalence of serogroups and virulence genes in Escherichia coli associated with postweaning diarrhoea and edema disease in pigs and a comparison of diagnostic approaches. Vet. Microbiol. 85:169-182. [DOI] [PubMed] [Google Scholar]
- 14.Giannella, R. A. 1976. Suckling mouse model for detection of heat-stable Escherichia coli enterotoxin: characteristics of the model. Infect. Immun. 14:95-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilligan, P. H. 1999. Escherichia coli, EAEC, EHEC, EIEC, ETEC. Clin. Lab. Med. 19:505-521. [PubMed] [Google Scholar]
- 16.Hirayama, T. 1995. Heat-stable enterotoxin of Escherichia coli, p. 281-296. In J. Moss, B. Iglewski, M. Vanghan, and A. T. Tu (ed.), Bacterial toxins and virulence factors in disease. Marcel Dekker, Inc., New York, NY.
- 17.Lariviere, S., C. L. Gyles, and D. A. Barnum. 1972. A comparative study of the rabbit and pig gut loop system for the assay of Escherichia coli enterotoxin. Can. J. Comp. Med. 36:319-328. [PMC free article] [PubMed] [Google Scholar]
- 18.Levine, M. M., J. B. Kaper, R. E. Black, and M. L. Clements. 1983. New knowledge on pathogenesis of bacterial enteric infections as applied to vaccine development. Microbiol. Rev. 47:510-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lockwood, D. E., and D. C. Robertson. 1984. Development of a competitive enzyme-linked immunosorbent assay (ELISA) for Escherichia coli heat-stable enterotoxin (STa). J. Immunol. Methods 75:293-307. [DOI] [PubMed] [Google Scholar]
- 20.Moon, H. W., S. C. Whipp, and A. L. Baetz. 1971. Comparative effects of enterotoxins from Escherichia coli and Vibrio cholerae on rabbit and swine small intestine. Lab. Investig. 25:133-140. [PubMed] [Google Scholar]
- 21.Moseley, S. L., J. W. Hardy, M. I. Huq, P. Echeverria, and S. Falkow. 1983. Isolation and nucleotide sequence determination of a gene encoding a heat-stable enterotoxin of Escherichia coli. Infect. Immun. 39:1167-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21a.National Research Council. 1996. Guide for the care and use of laboratory animals. National Academy Press, Washington, DC.
- 22.Olsson, E., and O. Söderlind. 1980. Comparison of different assays for definition of heat-stable enterotoxigenicity of Escherichia coli porcine strains. J. Clin. Microbiol. 11:6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qadri, F., S. K. Das, A. S. G. Faruque, G. J. Fuchs, M. J. Albert, R. B. Sack, and A. M. Svennerholm. 2000. Prevalence of toxin types and colonization factors in enterotoxigenic Escherichia coli isolated during a 2-year period from diarrheal patients in Bangladesh. J. Clin. Microbiol. 38:27-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ristaino, P. A., M. M. Levine, and C. R. Young. 1983. Improved GM1-enzyme-linked immunosorbent assay for detection of Escherichia coli heat-labile enterotoxin. J. Clin. Microbiol. 18:808-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rutter, J. M., M. R. Burrows, R. Sellwood, and R. A. Gibbons. 1975. A genetic basis for resistance to enteric disease caused by Escherichia coli. Nature 257:135-136. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 27.Savarino, S. J., A. Fassno, D. C. Robertson, and M. M. Levine. 1991. Enteroaggregative Escherichia coli elaborate a heat stable enterotoxin demonstrable in an in vitro rabbit intestinal model. J. Clin. Investig. 87:1450-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schifferli, D. M., and M. A. Alrutz. 1994. Permissive linker insertion sites in the outer membrane protein of 987P fimbriae of Escherichia coli. J. Bacteriol. 176:1099-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sears, C. L., and J. B. Kaper. 1996. Enteric bacterial toxins: mechanisms of action and linkage to intestinal secretion. Microbiol. Rev. 60:167-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sellwood, R., R. A. Gibbons, G. W. Jones, and J. M. Rutter. 1975. Adhesion of enteropathogenic Escherichia coli to pig intestinal brush borders: the existence of two pig phenotypes. J. Med. Microbiol. 8:405-411. [DOI] [PubMed] [Google Scholar]
- 31.Skjelkvale, R., and T. Uemura. 1977. Experimental diarrhoea in human volunteers following oral administration of Clostridium perfringens enterotoxin. J. Appl. Bacteriol. 43:281-286. [DOI] [PubMed] [Google Scholar]
- 32.Smith, H. W., and M. A. Linggood. 1971. Observation on the pathogenic properties of the K88, HIY and ENT plasmids of Escherichia coli with particular reference to porcine diarrhea. J. Med. Microbiol. 4:467-485. [DOI] [PubMed] [Google Scholar]
- 33.So, M., and B. J. McCarthy. 1980. Nucleotide sequence of transposon Tn 1681 encoding a heat-stable toxin (ST) and its identification in enterotoxigenic Escherichia coli strains. Proc. Natl. Acad. Sci. USA 77:4011-4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steinland, H., P. Valentiner-Branth, M. Perch, F. Dias, T. K. Fischer, P. Aaby, K. Molbak, and H. Sommerfelt. 2002. Enterotoxigenic Escherichia coli infections and diarrhea in a cohort of young children in Guinea-Bissau. J. Infect. Dis. 186:1740-1747. [DOI] [PubMed] [Google Scholar]
- 35.Svennerholm, A. M., and D. Steels. 2004. Progress in enteric vaccine development. Best Pract. Res. Clin. Gastroenterol. 18:421-445. [DOI] [PubMed] [Google Scholar]
- 36.Walker, R. I. 2005. Considerations for development of whole cell bacterial vaccines to prevent diarrheal diseases in children in developing countries. Vaccine 23:3369-3385. [DOI] [PubMed] [Google Scholar]
- 37.Yamamoto, T., T. Gojobori, and T. Yokota. 1987. Evolutionary origin of pathogenic determinants in enterotoxigenic Escherichia coli and Vibrio cholerae O1. J. Bacteriol. 169:1352-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang, W., E. M. Berberov, J. Freeling, D. He, R. A. Moxley, and D. H. Francis. 2006. Significance of heat-stable and heat-labile enterotoxins in porcine colibacillosis in an additive model for pathogenicity studies. Infect. Immun. 76:3107-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, W., M. Zhao, L. Ruesch, A. Omot, and D. H. Francis. 2007. Prevalence of virulence genes in Escherichia coli strains recently isolated from young pigs with diarrhea in the US. Vet. Microbiol. 123:145-152. [DOI] [PubMed] [Google Scholar]