Abstract
Sequence analysis of the five genes (gutRMCBA) downstream from the previously described sorbitol-6-phosphate dehydrogenase-encoding Lactobacillus casei gutF gene revealed that they constitute a sorbitol (glucitol) utilization operon. The gutRM genes encode putative regulators, while the gutCBA genes encode the EIIC, EIIBC, and EIIA proteins of a phosphoenolpyruvate-dependent sorbitol phosphotransferase system (PTSGut). The gut operon is transcribed as a polycistronic gutFRMCBA messenger, the expression of which is induced by sorbitol and repressed by glucose. gutR encodes a transcriptional regulator with two PTS-regulated domains, a galactitol-specific EIIB-like domain (EIIBGat domain) and a mannitol/fructose-specific EIIA-like domain (EIIAMtl domain). Its inactivation abolished gut operon transcription and sorbitol uptake, indicating that it acts as a transcriptional activator. In contrast, cells carrying a gutB mutation expressed the gut operon constitutively, but they failed to transport sorbitol, indicating that EIIBCGut negatively regulates GutR. A footprint analysis showed that GutR binds to a 35-bp sequence upstream from the gut promoter. A sequence comparison with the presumed promoter region of gut operons from various firmicutes revealed a GutR consensus motif that includes an inverted repeat. The regulation mechanism of the L. casei gut operon is therefore likely to be operative in other firmicutes. Finally, gutM codes for a conserved protein of unknown function present in all sequenced gut operons. A gutM mutant, the first constructed in a firmicute, showed drastically reduced gut operon expression and sorbitol uptake, indicating a regulatory role also for GutM.
Sorbitol, also called glucitol, is found in many fruits in nature and is widely used in the food industry as a sweetener, humectant, texturizer, and softener (25). Besides, a probiotic effect has been demonstrated for this hexitol (22), which would be important for the development of functional foods. Utilization of sorbitol as a carbon source has been described for some bacteria, including Escherichia coli (30), Erwinia amylovora (1), Clostridium beijerinckii (27), Streptococcus mutans (2), and Lactobacillus casei (32). All these organisms rely on the phosphoenolpyruvate:sugar phosphotransferase system (PTS) for the transport of sorbitol into the cell. The PTS consists of the general phosphotransferase proteins enzyme I (EI) and HPr and carbohydrate-specific transporters (EII). The EI autophosphorylates at a histidine residue by using phosphoenolpyruvate and then transfers the phosphoryl group to HPr, which becomes phosphorylated at the conserved histidine-15 residue. P∼His-HPr (where the P∼ prefix stands for phospho) functions as a phosphoryl donor for the different PTS transporters, which usually consist of three different proteins or domains: the cytoplasmic domains EIIA and EIIB and the transmembrane transporter EIIC (20). The EIIs responsible for sorbitol uptake (EIIGut) have an unusual domain structure, consisting of EIIC, EIIBC, and EIIA, where EIIC corresponds only to the amino-terminal part of the EIIC domain and the carboxy-terminal half is fused to the EIIB domain (1, 2, 27).
The enzymes responsible for sorbitol transport and metabolism are encoded by the gut operon. This operon includes the genes encoding the EII domains of the PTS involved in the transport and phosphorylation of sorbitol to sorbitol-6-phosphate, the gene encoding sorbitol-6-phosphate dehydrogenase, which converts sorbitol-6-phosphate to fructose-6-phosphate, and two genes encoding regulatory proteins (gutM and gutR). The role of these regulators in E. coli has been studied, and a complex but yet unknown regulatory mechanism, where GutM is an activator and GutR a repressor, has been suggested (30). In E. coli, gutM and gutR are located downstream from the EII and sorbitol-6-phosphate dehydrogenase-encoding genes. Constitutive transcription of gutR occurs from its own promoter (monocistronic mRNA), whereas inducible transcription starts at the gut promoter upstream from the EIIC-encoding gene (polycistronic mRNA). It has been suggested that the ratio of GutR and GutM might determine the extent of gut operon expression (30). In firmicutes, the gut operons analyzed so far also contain a gutM homologue, but the putative regulator GutR differs from E. coli GutR. The activity of E. coli GutR is controlled by a sorbitol-6-phosphate-binding domain, but GutR from firmicutes contains PTS regulated domains (PRDs) and a mannitol/fructose-specific EIIA-like domain (EIIAMtl domain) (8, 26). GutRs of firmicutes were therefore thought to be controlled via PTS-mediated phosphorylation, an assumption which so far has not been experimentally confirmed (2, 27). An exception is the presumed gut operon of listeriae, which contains no gutR but which contains gutM (lmo0545/lin0549) that is neither of the E. coli nor of the firmicute type.
The L. casei gutF gene encoding the sorbitol-6-phosphate dehydrogenase involved in sorbitol metabolism has previously been cloned and characterized (32). Sequence analyses of the region downstream from gutF revealed the presence of five genes, gutR, gutM, gutC, gutB, and gutA. The genes gutR, gutM, and gutB were disrupted, and their role in gut operon transcriptional regulation and sorbitol uptake was studied. Additionally, GutR and GutM were overproduced and purified from E. coli, allowing for the first time the identification of the DNA-binding site of a PRD-containing GutR regulator within the gut promoter region.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and plasmids.
The strains and plasmids used in this study are listed in Table 1. The L. casei strains (ATCC 334 and BL23 and its various gut mutant derivatives) were grown at 37°C under static conditions on MRS medium (Oxoid) or MRS fermentation medium (Adsa-Micro) supplemented with 0.5% of the appropriate sugar, as indicated in the figure legends and text. E. coli and Lactococcus lactis strains, which were used as hosts in cloning experiments, were grown in Luria-Bertani medium at 37°C and M17 medium (Oxoid) plus 0.5% glucose at 30°C, respectively. The corresponding solid media were prepared by adding 1.5% agar. E. coli transformants were selected with ampicillin (50 μg/ml), and L. lactis and L. casei transformants were selected with erythromycin (5 μg/ml).
TABLE 1.
Strains and plasmids used in this studya
| Strain or plasmid | Genotype or properties | Source or reference |
|---|---|---|
| L. casei | ||
| BL23 | Wild type | CECT 5275 |
| BL282 | BL23 gutB (480-bp deletion in gutB) | This work |
| BL285 | BL23 gutR (frameshift at PstI restriction site) | This work |
| BL286 | BL23 gutM (frameshift at PstI restriction site) | This work |
| BL291 | BL286 containing pT1NgutM; Ermr | This work |
| BL307 | BL285 containing pT1NgutR; Ermr | This work |
| ATCC 334 | Wild type; carries an insertion element in gutR | ATCC |
| L. lactis MG1363 | Plasmid- and phage-free derivative of NCDO712 | 9 |
| E. coli | ||
| DH5a | F−endA1 hsdR17 gyrA96 thi-1 recA1 relA1 supE44 DlacU169 (F80 lacZ DM15) | Stratagene |
| M15(pREP4) | Nals, Strs, Rifs, Thi−, Lac−, Ara+, Gal+, Mtl−, F−, RecA+, Uvr+, Lon+ | Qiagen |
| Plasmids | ||
| pRV300 | Suicide vector carrying Ermr from pAMb1 | 15 |
| pRVB1 | pRV300 with a fragment carrying a deletion in gutB | This work |
| pRVR1 | pRV300 with a frameshift at PstI site in gutR | This work |
| pRVM4 | pRV300 with a frameshift at PstI site in gutM | This work |
| pT1NX | Contains the P1 constitutive promoter; Ermr | 23 |
| pT1NgutM | pT1NX containing gutM-encoding region | This work |
| pT1NgutR | pT1NX containing gutR-encoding region | This work |
| pQE30 | Ampr | Qiagen |
| pQEgutR | pQE30 containing gutR-encoding region | This work |
| pQEgutM | pQE30 containing gutM-encoding region | This work |
CECT, Colección Española de Cultivos Tipo; ATCC, American Type Culture Collection; Ampr, ampicillin resistant; Ermr, erythromycin resistant; Nals, nalidixic acid sensitive; Strs, streptomycin sensitive; Rifs, rifampin sensitive.
Vector pRV300 (15) was used for cloning experiments with E. coli and for insertional inactivation of genes in L. casei. Vector pT1NX (23) was used for cloning experiments with L. lactis and for expression of GutM and GutR in L. casei. E. coli M15(pREP4) was used for the synthesis of His-tagged GutR and His-tagged GutM. E. coli and L. lactis strains were transformed by electroporation with a Gene Pulser apparatus (Bio-Rad Laboratories) as recommended by the manufacturer (E. coli) or as described earlier (L. lactis) (11). L. casei strains were transformed as previously described (19).
DNA manipulation, oligonucleotides, and sequencing.
The oligonucleotides used in this study are listed in Table 2. Total DNA was isolated from L. casei BL23 as described before (19). Recombinant DNA techniques were performed by following standard procedures (21). All PCRs were performed with an Expand High Fidelity PCR system (Roche), which contains an enzyme mix of Taq and Tgo DNA polymerases. DNA sequencing was carried out by the Central Service of Research Support of the University of Valencia (Spain) by using the dideoxynucleotide DNA chain termination method. M13 universal and reverse primers or custom primers hybridizing within the appropriate DNA fragments were used for sequencing. Sequence analyses were carried out with DNAMAN 4.0 for Windows (Lynnon BioSoft), and sequence similarities were analyzed with the BLAST program.
TABLE 2.
Oligonucleotides used in this study
| Function and name | Sequencea | Locationb | Utilization |
|---|---|---|---|
| Construction of mutants | |||
| gutbc1 | CTGCCATACCAGCAATGGC | −285 | Cloning gutB with an internal 480-bp deletion |
| gutbc2 | TGCCAGCGGACCAGACTTC | +315 | |
| gutbc3 | TATCGTCATCGGCTTTATCTG | +796 | |
| gutbc4 | ATCCTGCGAGCTCAGGTG | +1455 | |
| gutF1 | GCAAGTAAAGGCATC | −350 | Cloning part of gutR to create a frameshift at |
| gutR1 | TGGTGAATCGTCCATCAAG | +689 | PstI restriction site (codon 48) |
| gutR2 | TGATGCTGGCAGAATCATC | −451 | Cloning gutM to create a frameshift at PstI |
| gutC1 | TGGAACAGCTTCATGAAGC | +574 | restriction site (codon 16) |
| Complementation of mutants | |||
| gutRBglII | CATAAGATCTGGGAGGTAACCTG | −24 | Cloning gutR in pT1NX |
| gutRSpeI | TATCAACTAGTTCATTTAAAGTCCCTC | +1884 | |
| gutMBglII | CCTTAACAGATCTCACGGAGGGAC | −29 | Cloning gutM in pT1NX |
| gutMSpeI | CATCTACTAGTTTTAAATTGTATTC | +522 | |
| Transcription analysis | |||
| ribu1 | TATTATGGGCAGGATAATGC | −302 | DNA probe (the primer location is indicated |
| gutF3 | GCCGTTAACAACCGTAGCG | +117 | with respect to guF) |
| gut3 | GACTTTAAGGACCCTAA | +133 | RT-PCR analysis (the primer location is |
| gutbc4 | ATCCTGCGAGCTCAGGTG | +5277 | indicated with respect to guF) |
| gut5 | TTTTCAACTTCGTCAGGATC | +197 | Identification of the transcription start site of |
| gutF3 | GCCGTTAACAACCGTAGCG | +117 | the gut operon (the primer location is indicated with respect to guF) |
| Overexpression of genes | |||
| gutRBamHI | GGGAGGGATCCGGATTGAATAG | −14 | Cloning gutR in pQE30 |
| gutRSalI | CAAAACGTCGACTTAAAGTCCC | +1881 | |
| gutMBamHI | CGGAGGGATCCAAAATGAACG | −14 | Cloning gutM in pQE30 |
| gutMHindIII | CTCCTAAGCTTAAATTGTATTC | +519 | |
| Gel mobility shift and DNase I footprinting | |||
| gut8D | GTCAAGCACCGCTAACC | −200 | Amplified 179-bp DNA fragment of the gut |
| gutF4 | TTTCTGCTCTATGGTAGC | −22 | promoter region (the primer location is indicated with respect to guF) |
Letters in italics indicate newly created restriction sites.
The numbers indicate the position of the 5′ end of the primer with respect to the start codon of the gene specified in the “Utilization” column.
Construction of a gutB mutant strain.
A DNA fragment containing 253 bp of the 3′ end of gutC and 314 bp of the 5′ end of gutB was amplified by PCR using L. casei BL23 chromosomal DNA and primers gutbc1 and gutbc2. Another DNA fragment, containing 321 bp of the 3′ end of gutB and 310 bp of the 5′ end of gutA, was amplified with primers gutbc3 and gutbc4. The two DNA fragments with blunt ends were cloned successively in the appropriate orientation in the integrative vector pRV300. The resulting plasmid, pRVB1, was used for single integration into L. casei BL23. Subsequently, one of the integrants was selected and grown without erythromycin for approximately 200 generations to facilitate the second recombination. One of the mutants with a deleted gutB region was selected and named BL282. The deletion was confirmed by PCR analysis and DNA sequencing.
Construction of gutR and gutM mutant strains.
DNA fragments containing part of gutR or gutM were obtained by PCR using L. casei BL23 chromosomal DNA. A gutR fragment was obtained with the primers gutF1 and gutR1; the gutM fragment was obtained with primers gutR2 and gutC1. These DNA fragments were cloned in the integrative vector pRV300 digested with SmaI and EcoRV, giving pRVR1 and pRVM4, respectively. These plasmids contain a unique PstI restriction site present in the gutR and gutM coding regions. pRVR1 and pRVM4 were digested with PstI, and the 3′-protruding ends were removed with the exonuclease activity of the Klenow fragment of DNA polymerase I. The plasmids were subsequently self-ligated to introduce a frameshift into each gene, which was confirmed by DNA sequencing. These constructs were used to transform L. casei BL23. One integrant of each gene was grown for 200 generations on medium without antibiotics to facilitate the second recombination, which would excise the vector. Two mutant strains were obtained, one with a frameshift in gutR (named BL285) and the other with a frameshift in gutM (named BL286). The two frameshifts were confirmed by PCR analysis, using as a template L. casei BL285 and BL286 chromosomal DNA, respectively, followed by DNA sequencing.
Complementation of gutR and gutM mutants.
The coding regions of gutR and gutM were amplified by PCR using L. casei BL23 chromosomal DNA as a template. The gutR gene was obtained with the primers gutRBglII and gutRSpeI, and the gutM gene was obtained with primers gutMBglII and gutMSpeI. The amplified DNA fragments were digested with BglII and SpeI and cloned into the replicative vector pT1NX previously digested with the same enzymes, giving pT1NgutR and pT1NgutM. These plasmids were used to transform L. casei strains BL285 and BL286, respectively. One transformant of each strain was selected, and the transformants were named BL307 (BL285[pT1NgutR]) and BL291 (BL286[pT1NgutM]).
RNA isolation and Northern blot analysis.
RNA was isolated from L. casei cells grown to an optical density at 550 nm (OD550) of 0.8 in MRS fermentation medium containing 0.5% of the appropriate sugars. Cells extracts were prepared as previously described (31), and total RNA was isolated according to the protocol of the Trizol manufacturer. Sample preparation, denaturing agarose gel electrophoresis, and RNA transfer were performed by standard methods (21). The DNA probe for the gut operon was synthesized by PCR using L. casei BL23 chromosomal DNA as a template, primers ribu1 and gutF3, and digoxigenin DNA labeling mix (Roche).
RT-PCR and RACE.
Reverse transcriptase PCR (RT-PCR) was carried out as previously described (31). RT reactions were performed with 2 μg total RNA isolated from L. casei BL23 cells grown on sorbitol, avian myeloblastosis virus RT (AMV-RT) (Sigma), and gutbc4 oligonucleotide. The subsequent PCR amplifications were performed with purified cDNA and primers gut3 and gutbc4. The transcription initiation of the gut operon was determined with a 5′/3′ rapid amplification of cDNA ends (RACE) kit (Roche) by following the manufacturer's instructions. Total RNA isolated from L. casei BL23 cells grown on sorbitol and the primer gut5 were used for cDNA synthesis. The cDNA was dA tailed and then amplified by PCR using the primer oligonucleotide dT anchor supplied in the kit and the primer gutF3. The resulting PCR product was used in a second PCR with primer PCR anchor (supplied with the kit) and primer gutF3. The amplified DNA fragment of about 0.2 kb was purified and sequenced.
Sorbitol uptake assays.
d-[14C]sorbitol uptake by whole cells of L. casei was performed according to the method described by Chassy and Thompson (5, 6). BL23 (wild type), BL282 (gutB), BL285 (gutR), BL286 (gutM), BL291 (BL286[pT1NgutM]), and BL307 (BL285[pT1NgutR]) were grown in 10 ml of MRS fermentation medium supplemented with a mixture of 0.5% sorbitol and 0.5% ribose to an OD550 of 0.8. Cells were prepared and incubated as previously described (31) and d-[14C]sorbitol (125 μM final concentration; 0.6 mCi/mmol) was added. The uptake rate was determined in the first 2 min of the reaction. One-milliliter aliquots were withdrawn and filtered through Millipore membranes (pore size, 0.45 μm). Filters were washed with cold buffer, and radioactivity was quantified by scintillation counting.
Expression and purification of His-tagged GutR and GutM.
The coding regions of gutR and gutM were amplified by PCR using chromosomal DNA from L. casei BL23 as a template and appropriate primers (Table 2), which added restriction sites to the 5′ and 3′ ends. The PCR fragments of 1,881 bp and 509 bp, respectively, were cleaved with the restriction enzymes recognizing the sites added by the PCR and cloned into pQE30. The resulting plasmids, pQEgutR and pQEgutM, were used to transform E. coli M15(pREP4), and the correct sequence of the inserts was confirmed by DNA sequencing. One clone of each was grown in 0.5 liter of Luria-Bertani medium with ampicillin (100 μg/ml) and kanamycin (25 μg/ml) at 30°C with agitation. When the cultures reached an OD550 of 0.5, isopropyl-β-d-thiogalactopyranoside (1 mM) was added and incubation was continued for 1 h. Cells were harvested by centrifugation and resuspended in ice-cold lysis buffer A (50 mM NaH2PO4 [pH 7.5], 300 mM NaCl, and 10 mM imidazole) and buffer B (50 mM NaH2PO4 [pH 7.5], 800 mM NaCl, 2.5 mM imidazole, 0.1% Tween 20, and 10 mM β-mercaptoethanol) for GutR and GutM purification, respectively. The suspensions were treated with 1 mg/ml lysozyme on ice for 10 min, quickly frozen in a dry ice/ethanol bath, thawed at 37°C, and then returned to ice. Cells were then sonicated, and the cell debris was removed by centrifugation at 12,000 × g for 20 min at 4°C. The cleared extracts were loaded onto Ni-nitrilotriacetic acid agarose columns (Qiagen). After the columns were washed with buffer A containing 20 mM imidazole and buffer B containing 15 mM imidazole for GutR and GutM purification, respectively, GutR was eluted with 250 mM imidazole and GutM was eluted with a 50 to 250 mM imidazole gradient. Fractions containing the proteins of interest were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels, pooled, dialyzed against 10 mM NaH2PO4 (pH 7.5) containing 45% glycerol and 10 mM β-mercaptoethanol, and kept frozen at −80°C. Protein concentrations were determined with a Bio-Rad dye-binding assay.
Gel mobility shift and DNase I footprinting assays.
The upstream region of the gutF gene was amplified by PCR with L. casei BL23 chromosomal DNA and primers gutF4 and gut8D. The amplified 179-bp DNA fragment was used in electrophoretic mobility shift assays with purified His-tagged GutR or purified His-tagged GutM. The binding assay was carried out in binding buffer (20 mM Tris-HCl [pH 6.5], 40 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol, and 10% glycerol) with 0.1 μg of target DNA and different amounts of His-tagged GutR or His-tagged GutM. The binding mixtures were incubated for 30 min at room temperature and separated on 5% nondenaturing polyacrylamide gels in 100 mM Tris-HCl [pH 7.5[-1 mM EDTA buffer at 60 V for 1 h. The DNA was stained with ethidium bromide. To perform footprinting assays, two single-end 32P-labeled DNA probes of the sorbitol promoter region were synthesized in two separated PCRs. The 179-bp DNA fragment described above was used as a template, and the oligonucleotides gutF4 or gut8D, 5′ labeled with T4 polynucleotide kinase and [γ-32P]ATP, were used as primers with the second oligonucleotide gut8D or gutF4 unlabeled. The reaction mixture contained binding buffer, 30,000 dpm of the labeled PCR fragments, 0.5 μg of herring sperm DNA, and different amounts of His-tagged GutR, in a final volume of 25 μl. After incubation for 30 min at room temperature, the DNA was digested with 0.0125 U of DNase I for 1 min at room temperature. The digestion was stopped by heating the samples for 10 min to 75°C in the presence of 5 mM EDTA. The reaction mixtures were passed through MicroSpin G-50 columns (GE Healthcare) to remove glycerol and subsequently separated on a 5% sequencing gel. A reference sequence ladder was generated using a Sequenase version 2.0 kit (GE Healthcare) with the same labeled oligonucleotide.
Nucleotide sequence accession number.
The nucleotide sequence reported in this work has been deposited in GenBank under accession number EU315692.
RESULTS
Characterization of the L. casei sorbitol operon.
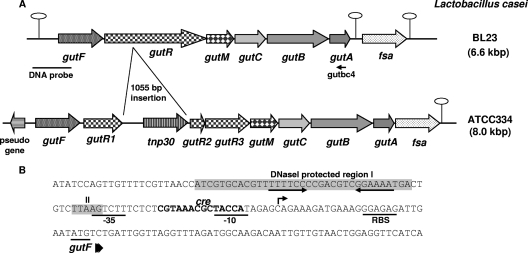
The gene encoding sorbitol-6-phosphate dehydrogenase (GutF), involved in sorbitol metabolism in L. casei BL23, has previously been isolated (32). Analysis of the chromosomal region of this gene using the genomic sequence of L. casei BL23 (GenBank accession no. FM177140) (13), showed that, downstream from gutF, five genes (gutRMCBA) were present which probably constitute an operon (Fig. 1). gutR encodes a protein homologous to putative sorbitol operon transcriptional regulators, showing the highest similarity to GutR from Lactobacillus salivarius (28% identity, 49% similarity) (7). The gutM gene encodes a protein homologous to putative sorbitol operon activators, e.g., SrlM1 from Lactobacillus plantarum (45% identity, 72% similarity) (14). Following gutM, three genes (gutCBA) encode proteins with significant homology to membrane-bound (EIICGut and EIIBCGut) and soluble (EIIAGut) components of the sorbitol-specific PTS. This transport system has the singularity that the EIIC domain is divided into two parts; one corresponds only to the amino-terminal half (189 residues) and is encoded by gutC, whereas the carboxy-terminal end is fused to the EIIB domain and the fused protein (372 residues) is encoded by gutB. Two putative rho-independent terminators were identified downstream from gutA, one between gutA and the following gene (fsa) (ΔG, −19.4 kcal/mol) and another located in the fsa 3′ region (ΔG, −12.3 kcal/mol). The deduced amino acid sequence of Fsa shows significant homology to transaldolase-like fructose-6-phosphate aldolases (24). Genes encoding transaldolase-like proteins have been found associated with catabolic operons in other organisms, for example, the gut operon from C. beijerinckii (27) and the sorbose operon from L. casei (33).
FIG. 1.
(A) Genetic organization of the chromosomal locus containing the gut operon in L. casei strains BL23 and ATCC 334. Hairpin loops indicate putative rho-independent transcriptional terminators. fsa refers to fructose-6-phosphate aldolase. The positions of the DNA probe used in Northern blot experiments and primer gutbc4 used in RT-PCR analysis are indicated. (B) DNA sequence of the gut promoter region and the 5′ end of gutF. The GutR-protected regions (I and II) are shown in shaded boxes. The inverted repeat motif is shown by convergent arrows. The transcriptional start site of the gut operon is indicated by an arrow. The putative ribosome-binding site (RBS) and −10 and −35 promoter sequences are underlined. A cre-like sequence is shown in boldface type.
When comparing the gut operon of L. casei strain BL23 with the equivalent region in L. casei strain ATCC 334 (GenBank accession no. CP000423) (16), we observed the same gene organization, except that in the latter strain, gutR contains a 1,055-bp DNA insertion. The inserted DNA encodes a putative transposase of the IS30 family (Fig. 1). In addition, a frameshift was found in the 3′ half of the ATCC 334 gutR homologue; therefore, the mutated gutR in the ATCC 334 genome is annotated as three different open reading frames: LSEI_2728, LSEI_2726, and LSEI_2725 (gutR1, gutR2, and gutR3 in Fig. 1).
The gut genes are induced by sorbitol and repressed by glucose.
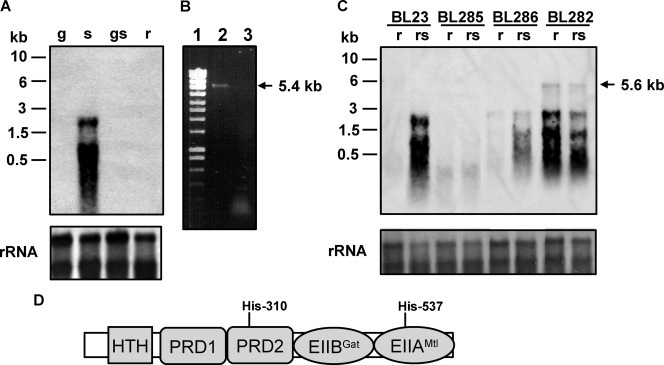
Northern blot experiments were performed with RNA isolated from L. casei BL23 (wild type) grown in media containing glucose, sorbitol, glucose plus sorbitol, or ribose (Fig. 2A). The results showed that the sorbitol operon was induced by sorbitol and repressed by glucose. This was in agreement with the presence of a DNA sequence matching the consensus motif of a cre (catabolite responsive element) gene (12), the target site for carbon catabolite repression in firmicutes, in the promoter region of the gut operon (Fig. 1B). The transcription signal was at the level of ribosomal RNAs (Fig. 2A), likely due to degradation or comigration of the RNA, a common problem in the analysis of long transcripts (33). However, RT-PCR analysis using total RNA isolated from strain BL23 grown on sorbitol showed a DNA band of about 5.4 kb, which confirmed that the gutFRMCBA genes are transcribed as a single mRNA (Fig. 2B). The transcription initiation site was determined by 5′ RACE and shown to be the cytosine located 27 bp upstream from the gutF start codon (Fig. 1B).
FIG. 2.
(A) Northern blot analysis performed with RNA (1 μg) isolated from L. casei BL23 (wild type) grown in MRS fermentation medium with 0.5% glucose (g), 0.5% sorbitol (s), 0.5% glucose plus 0.5% sorbitol (gs), or 0.5% ribose (r). The DNA probe spanned the promoter region of the gut operon and the 5′ end of gutF. Positions of size standard markers are indicated on the left. Stained rRNAs (23S and 16S) are shown as loading controls. (B) Agarose gel showing a RT-PCR band obtained with RNA isolated from BL23 grown on 0.5% sorbitol. Total RNA was used in RT reactions with primer gutbc4 and AMV-RT (lane 2) or without AMV-RT (lane 3). The cDNAs obtained were used in PCRs with primers gut3 and gutbc4. Ecoladder 1 (Ecogen) was used as a size standard marker (lane 1). The size of the fragment obtained is marked on the right. (C) Northern blot analysis using RNA (1 μg) isolated from L. casei BL23 (wild-type), BL282 (gutB), BL285 (gutR), and BL286 (gutM) strains. The conditions of cell cultures (0.5% ribose or 0.5% ribose plus 0.5% sorbitol [rs]) and the probe are the same as in panel A. A transcript of 5.6 kb is indicated with an arrow. Positions of size standard markers are indicated on the left. Stained rRNAs (23S and 16S) are shown as loading controls. (D) Domain organization of GutR. HTH, helix-turn-helix. Potential PTS phosphorylation sites (histidyl residues) are indicated.
gutR codes for a transcriptional activator of gut genes.
To determine the participation of GutR in the regulation of sorbitol utilization in L. casei BL23, a mutant with a frameshift in gutR was constructed (strain BL285). This mutant lost the ability to ferment sorbitol, and transcription of the gut operon in cells grown with a mixture of ribose (a sugar that does not trigger catabolite repression in L. casei) plus sorbitol was abolished (Fig. 2C). In addition, the sorbitol uptake rate in strain BL285 was 88-fold lower than in the wild type (30.76 ± 0.89 and 0.35 ± 0.21 nmol mg of dry weight−1 min−1 for wild-type and BL285 strains, respectively). gutR precedes gutM and the genes coding for the putative transport proteins of PTSGut. Although it is likely that the absence of GutR or GutM (see below) from strains BL285 and BL286, respectively, is responsible for the loss of sorbitol utilization capacity, we cannot exclude the possibility that the frameshift mutations might have a polar effect on the downstream gut genes. To test this possibility, BL285 was transformed with pT1NgutR (Table 1), which constitutively expresses gutR in trans. In the complemented strain (BL307), sorbitol uptake was restored (23.79 ± 1.5 nmol mg of dry weight−1 min−1), and the strain was able to ferment sorbitol, demonstrating that the phenotype of BL285 was due to an impaired gutR. These results indicate that GutR acts as a transcriptional activator and that no other sorbitol utilization system is present in BL23. In accordance with these data, L. casei ATCC 334, which contains a gutR inactivated by an insertion element, was also found to be unable to utilize sorbitol.
A sequence analysis of GutR showed a multidomain structure of the protein (Fig. 2D). In its N-terminal region, GutR contains a helix-turn-helix domain (type 11), located at positions 88 to 146. GutR exhibits significant similarity (42%) to LicR from Bacillus subtilis, which controls the expression of a cellobiose-specific PTS. The helix-turn-helix domain in LicR is followed by two PRDs, a galactitol-specific EIIB-like domain (EIIBGat domain) and, finally, an EIIA domain of a mannitol/fructose (EIIAMtl/Fru) class PTS (8). The four phosphorylatable histidyl residues in the two PRDs of LicR were identified as the sites of positive regulation by P∼His-HPr, whereas the conserved histidine in the EIIAMtl domain represents the site of negative control by P∼EIIBCel; where Cel is cellobiose (28). The regulation of L. casei GutR must be slightly different, as it contains only one of the four phosphorylatable histidyl residues in the two PRDs (His-310, the first conserved His in PRD2) (Fig. 2D). A phosphorylatable histidine in PRD1 is present only in GutR of L. salivarius, and PRD1 is generally poorly conserved among GutR proteins. Sequence comparisons between L. casei GutR and other members of this transcriptional regulator family showed that the L. casei protein also contains an EIIBGat domain with a length of approximately 80 amino acids. The potentially phosphorylatable cysteyl residue in the EIIBGat domain is lacking in GutR of L. casei and other lactobacilli, but it is present in GutR of, for example, clostridia, enterococci, and Streptococcus mutans. In contrast, the phosphorylatable histidyl residue in the EIIAMtl domain is conserved in GutR of all known gut operon-containing firmicutes (His-537 in GutR of L. casei). In fact, the EIIAMtl/Fru domain is characteristic of DeoR-type PRD- containing transcriptional activators in firmicutes (8). In analogy with B. subtilis LicR, it was likely that L. casei GutR would be positively controlled by P∼His-HPr-mediated phosphorylation at His-310 and negatively regulated by P∼EIIBCGut-mediated phosphorylation at His-537 (Fig. 2D).
In order to demonstrate the presumed effect of the PTSGut on the activity of GutR, we constructed a strain (BL282) with a mutated gutB gene, which encodes the sorbitol-specific EIIBCGut. As expected, this strain displayed a sorbitol-negative phenotype and showed a sorbitol uptake rate (0.54 ± 0.20 nmol mg of dry weight−1 min−1) 57-fold lower than that of the wild type. However, in contrast to BL285, Northern blot analysis showed that transcription of the gut operon in BL282 occurred independently of the presence of sorbitol (Fig. 2C). This indicated that the product of gutB is necessary for sorbitol uptake and that it has a negative effect on the expression of the gut operon, which it probably exerts via GutR phosphorylation at His-537. A transcript of about 5.6 kb was observed for strain BL282, which would correspond to an mRNA starting at the gutF gene and extending to the first putative rho-independent terminator located downstream from gutA, thus supporting the above RT-PCR results.
GutM is necessary for transcription of the gut genes.
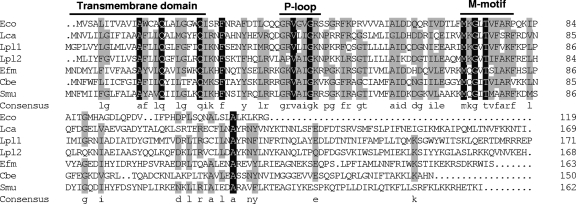
Sequence analysis showed that GutM proteins encoded within different sorbitol operons in both proteobacteria and firmicutes share relatively high conservation in their N-terminal part, with less homology in the C terminus (Fig. 3). The TMHMM program (server version 2.0; CBS, Denmark) for the prediction of transmembrane helices in proteins detected a potential transmembrane segment of 20 or 23 amino acids at the N terminus of GutM (positions 2 to 4 to positions 21 to 26). The alignment of GutM proteins also showed a presumed Walker motif A (G-R-V-A-I-G-K-S/N/R/V), which differs in one residue from the P-loop pattern A/G-X(4)-G-K-S/T (Fig. 3). Some protein kinases that differ in one residue at the beginning or at the end of the sequence pattern without losing their functionality have also been described (3). Downstream from the P-loop domain, there is another amino acid sequence motif (M-K-G-hydrophobic-T-V-F-A-R-F; boldface indicates conservation in all the GutM proteins) of unknown function but well conserved in all GutM proteins, which we dubbed M-motif (Fig. 3).
FIG. 3.
Multiple amino acid sequence alignment of GutM from L. casei (Lca, this work), E. coli (Eco, GenBank accession no. NP_417186), L. plantarum (Lpl1; GenBank accession no. NP_786817), L. plantarum (Lpl2; GenBank accession no. NP_786847), Enterococcus faecium (Efm; GenBank accession no. ZP_00604866), C. beijerinckii (Cbe; GenBank accession no. YP_001307479), and S. mutans (Smu, GenBank accession no. AAD33520). The residue number of each protein is indicated on the right. Residues conserved in all sequences are shown against a black background. Residues conserved among at least four of the seven sequences appear against a grey background. The consensus sequence (at least four residues conserved) is shown in lowercase letters. The predicted transmembrane domain and the P-loop motif are shown. M-motif indicates a highly conserved region of unknown function.
According to these data, GutM might be a membrane-bound protein with a nucleotide-binding activity, but no clear function has been reported for it. In order to shed some light on GutM function, a strain with a frameshift in this gene was constructed (BL286). This strain lost the ability to ferment sorbitol, indicating that the gene is essential for sorbitol metabolism. In addition, Northern blot experiments showed that transcription of the gut genes was strongly reduced in this strain (Fig. 2C) and that the sorbitol uptake rate was 70-fold lower than in the wild type (30.76 ± 0.89 and 0.44 ± 0.05 nmol mg of dry weight−1 min−1 for wild-type and BL286 strains, respectively), suggesting that GutM plays an important regulatory role in gut operon expression. gutM precedes the genes coding for the putative transport proteins of the PTSGut. To test the possibility if a frameshift in gutM leads to polar effects on the downstream genes, BL286 was transformed with pT1NgutM (Table 1), which constitutively expresses gutM in trans. In the complemented strain (BL291), sorbitol uptake was restored (9.41 ± 0.23 nmol mg of dry weight−1 min−1), albeit to a lower rate than that of the wild-type, and the strain was able to ferment sorbitol, demonstrating that the phenotype of BL286 was due to an impaired gutM.
Binding of GutR to the gutFRMCBA promoter.
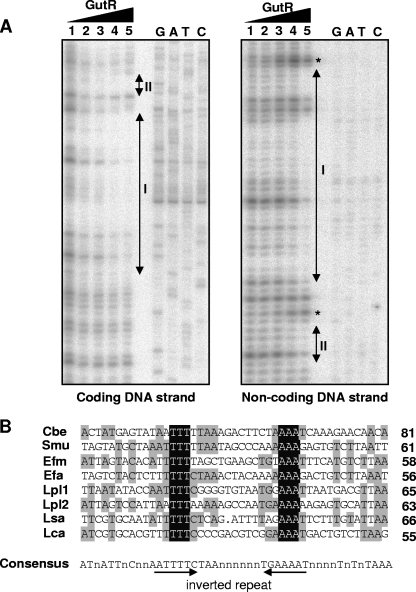
To address the question of whether the regulatory effect of the products of the gutR and gutM genes required a direct interaction with the promoter region of the gut operon, His-tagged GutR and GutM were produced in E. coli and purified. Both proteins were used in gel mobility shift assays with a 179-bp DNA fragment from positions −173 to +6 relative to the translation initiation site. Under the tested conditions, His-tagged GutR retarded the DNA fragment, while His-tagged GutM did not (data not shown). The inclusion of GutM in binding assays containing GutR did not influence its binding activity under the tested conditions (data not shown). To identify the exact DNA-binding site within the gut promoter recognized by GutR, DNase I footprinting experiments with the purified His-tagged GutR were performed. A radiolabeled DNA fragment from position −173 to +6 was incubated with different amounts of GutR, and the footprint analysis showed a protected region (I) spanning 35 bases, from positions −73 to −39 in the coding strand. The equivalent region was protected on the noncoding strand (Fig. 4). Additionally, a short region (II) extending from positions −33 to −29 just downstream from region I was protected in both strands. Hypersensitivity to DNase I digestion, induced by the presence of GutR, was observed at positions −35 and −75 in the noncoding strand (Fig. 4A). The 35-bp protected sequence contains an inverted repeat that could be the recognition motif for GutR (Fig. 1B). An alignment of the DNA regions upstream from gut operons from various firmicutes with the DNA-binding sequence determined for L. casei GutR revealed for all bacteria a consensus inverted repeat containing a 3T/3A conserved core (Fig. 4B). This motif could be the putative gut transcriptional activator recognition sequence in firmicutes.
FIG. 4.
(A) DNase I footprinting analysis of GutR binding to the sorbitol promoter region. Protection of the coding and noncoding DNA strands in the presence of 0.00, 0.02, 0.17, 0.68, and 1.36 μg of His-tagged GutR (lanes 1 to 5, respectively). The GutR-protected regions (I and II) in the coding and noncoding DNA strands are indicated by double-arrowhead vertical bars. Asterisks indicate the DNA positions with hypersensitivity to DNase I digestion. Binding positions were determined by the sequence ladder (GATC). (B) Alignment of the GutR-protected region of L. casei (Lca; see panel A) with the hypothetical promoter regions (45 bases) of gut operons from C. beijerinckii (Cbe; GenBank accession no. NC_009617.1), S. mutans (Smu; accession no. AF132127.1), E. faecium (Efm; accession no. NZ_AAAK03000093.1), E. faecalis (Dfa; accession no. NC_004668.1), (Lpl) L. plantarum (accession no. NC_004567.1.1), and L. salivarius (Lsa; accession no. NC_007930.1). At the right end of each sequence lane, we indicated the distance (in bases) to the start codon of the first gene of the corresponding gut operon. Bases conserved in all sequences are shown against a dark background. Bases conserved in at least four of the eight sequences appear against a shaded background. A consensus sequence (50% conservation) is shown below the alignment and contains an inverted repeat marked with arrows.
DISCUSSION
The L. casei gutFRMCBA operon involved in sorbitol utilization has been characterized. The gutF gene encodes a sorbitol-6-phosphate dehydrogenase enzyme that converts sorbitol-6-phosphate to fructose-6-phosphate with the associated reduction of NAD+ to NADH (32). The gutR and gutM genes encode proteins involved in regulation of the gut operon, and gutC, gutB, and gutA encode the EII domains of a sorbitol PTS. The gene organization in the L. casei gut operon is also found in L. plantarum (14) and in L. salivarius (7). This organization is typical of firmicutes, with the exception of C. beijerinckii, in which the gutF gene is located downstream from gutA (27), and listeriae, which lack the gutR gene. Firmicutes therefore differ from proteobacteria, where the regulatory genes are located downstream from the transporter and enzyme genes, leading to the organization gutCBAFMR (1, 30). Curiously, one of the regulatory proteins (GutM) is conserved in both proteobacteria and firmicutes, whereas the other (GutR) belongs to different families of regulators. GutRs from proteobacteria are repressors of the DeoR family controlled by small metabolites, whereas GutRs from firmicutes are PRD-containing transcriptional activators controlled via phosphorylation (1, 2, 27, 30). Since the EIIGut are highly conserved in both bacterial groups, it is possible that gutCBA and gutM could have a common origin and the different gutR genes might have been acquired later.
An L. casei gutB mutant strain (BL282) constitutively expressed the gut operon but was impaired in sorbitol uptake. These results strongly suggest that EIIBCGut participates in sorbitol transport and that this PTS probably represents the only route for sorbitol uptake in L. casei. Furthermore, a mutant defective in the general PTS protein EI failed to ferment sorbitol (29). The fact that transcription of the gut operon in BL282 occurs independently of the presence of sorbitol also indicates that the product of gutB has a negative effect on the expression of the operon. The target for this regulatory effect is most probably the GutR regulator, which is required for gut operon transcription. Previous results have shown that mutations in levB, encoding the EIIBLev of a PTSLev, abolished the uptake of fructose via this PTS in B. subtilis and led to constitutive expression of the lev operon in B. subtilis and L. casei (4, 18). For both bacteria, it was demonstrated that phosphorylation of the levanase operon regulator, LevR, by EIIBLev at a histidine in PRD2 inhibits its transcriptional activator function (17, 18). Similarly, regulation of the L. casei esu operon depended on a LevR-like activator (EsuR) that was negatively regulated by EIIBEsu (34). However, GutR of L. casei more resembles LicR- and MtlR-like regulators, which are controlled via P∼EIIB-mediated phosphorylation at the C-terminal EIIAMtl-like domain (8). Because L. casei GutR also contains an EIIAMtl domain, its transcriptional activator function might be inhibited by EIIBCGut-mediated phosphorylation at the conserved His-537. In accordance with LicR and MtlR, the conserved His-310 in PRD2 of GutR might become phosphorylated via P∼His-HPr. In the case of B. stearothermophilus MtlR, phosphorylation at PRD2 by P∼His-HPr increases its affinity for its DNA-binding site (10). The absence of P∼His-HPr-mediated phosphorylation during the uptake of a rapidly metabolizable PTS sugar serves as a secondary carbon catabolite repression mechanism (8). However, other possibilities cannot be excluded at present, and an understanding of the detailed regulation mechanism of GutR will require additional studies.
We have shown that GutR from L. casei binds to a region of the gut promoter, which is conserved within firmicutes, suggesting that this group shares a common mechanism of regulation. Unlike MtlR, which protected five small and discontinuous DNA regions, GutR binds upstream from the promoter and protects a 35-base continuous DNA stretch, which contains an inverted repeat with a 3T/3A conserved core and thus resembles the DNA-binding site determined for LicR of B. subtilis (28). Additionally, a short region of five bases that partially overlaps the −35 box was also protected.
Transcription analysis of the gut operon indicated that it is transcribed as a polycistronic mRNA comprising the six gut genes, their expression being induced by sorbitol. This result supports the earlier biochemical studies that indicated that sorbitol-fermenting L. casei possess an inducible sorbitol-6-phosphate dehydrogenase (32). Sorbitol availability must be sensed by the phosphorylation state of EIIGut proteins which in turn leads to changes in the phosphorylation state of the transcriptional activator GutR. The fact that L. casei gutR was transcribed from the same GutR-regulated promoter adds an autoregulation mechanism to the system. This regulation differs from that described for E. coli (30), although both E. coli and firmicutes share a common regulator (GutM). An L. casei gutM mutant (BL286) showed lower levels of gutFRMCBA transcription than the wild-type strain, denoting that GutM also exhibits an activation effect on gut operon expression. Despite the different nature of E. coli GutR, transcription of the gut genes in this organism is also activated by GutM (30). Compared to GutM from E. coli, the L. casei protein contains 50 additional residues at the carboxy-terminal end. Nevertheless, the two proteins exhibit significant similarity (22% identity). GutM from E. coli was postulated to be a DNA-binding protein. However, this has never been proven, and L. casei GutM did not bind to the promoter region of the gut operon or a DNA fragment comprising the intergenic region between gutF and gutR (data not shown). A sequence analysis of GutM proteins revealed that they contain a potential transmembrane domain at the N-terminal end and a presumed nucleotide-binding domain (Walker motif A), suggesting that they might be membrane-located sensors. The presence of a gutM gene in the PTSGut-encoding operons and the results obtained with E. coli and L. casei favor the hypothesis that GutM plays a role in the transcriptional regulation of the gut operon.
In this study, functional and regulatory molecular and genetic analyses of the sorbitol operon from L. casei have been carried out. Although several sorbitol utilization operons have been described, information about them is very limited. We have characterized for the first time the binding site for a PRD-containing GutR regulator, appearing to be a PTS-controlled transcriptional activator, which binds to a DNA site conserved in all sorbitol operons of firmicutes. We began to study the function of GutM in this group of bacteria and showed that it probably plays a regulatory role. Further work will be necessary to elucidate the exact function of GutM in the regulation of the gut operon.
Acknowledgments
This work was financed by funds of the Spanish Ministry for Science and Technology (project AGL2004-03886). Genome sequencing of L. casei BL23 was supported by funds from the Région Basse Normandie, CNRS, INRA, AgroParisTech, and IATA (CSIC).
Footnotes
Published ahead of print on 1 August 2008.
REFERENCES
- 1.Aldridge, P., M. Metzger, and K. Geider. 1997. Genetics of sorbitol metabolism in Erwinia amylovora and its influence on bacterial virulence. Mol. Gen. Genet. 256:611-619. [DOI] [PubMed] [Google Scholar]
- 2.Boyd, D. A., T. Thevenot, M. Gumbmann, A. L. Honeyman, and I. R. Hamilton. 2000. Identification of the operon for the sorbitol (glucitol) phosphoenolpyruvate:sugar phosphotransferase system in Streptococcus mutans. Infect. Immun. 68:925-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brakoulias, A., and R. M. Jackson. 2004. Towards a structural classification of phosphate binding sites in protein-nucleotide complexes: an automated all-against-all structural comparison using geometric matching. Proteins 56:250-260. [DOI] [PubMed] [Google Scholar]
- 4.Charrier, V., J. Deutscher, A. Galinier, and I. Martin-Verstraete. 1997. Protein phosphorylation chain of a Bacillus subtilis fructose-specific phosphotransferase system and its participation in regulation of the expression of the lev operon. Biochemistry 36:1163-1172. [DOI] [PubMed] [Google Scholar]
- 5.Chassy, B. M., and J. Thompson. 1983. Regulation of lactose-phosphoenolpyruvate-dependent phosphotransferase system and β-d-phosphogalactoside galactohydrolase activities in Lactobacillus casei. J. Bacteriol. 154:1195-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chassy, B. M., and J. Thompson. 1983. Regulation and characterization of the galactose-phosphoenolpyruvate-dependent phosphotransferase system in Lactobacillus casei. J. Bacteriol. 154:1204-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Claesson, M. J., Y. Li, S. Leahy, C. Canchaya, J. P. van Pijkeren, A. M. Cerdeno-Tarraga, J. Parkhill, S. Flynn, G. C. O'Sullivan, J. K. Collins, D. Higgins, F. Shanahan, G. F. Fitzgerald, D. van Sinderen, and P. W. O'Toole. 2006. Multireplicon genome architecture of Lactobacillus salivarius. Proc. Natl. Acad. Sci. USA 103:6718-6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deutscher, J., C. Francke, and P. W. Postma. 2006. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70:939-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henstra, S. A., R. H. Duurkens, and G. T. Robillard. 2000. Multiple phosphorylation events regulate the activity of the mannitol transcriptional regulator MtlR of the Bacillus stearothermophilus phosphoenolpyruvate-dependent mannitol phosphotransferase system. J. Biol. Chem. 275:7037-7044. [DOI] [PubMed] [Google Scholar]
- 11.Holo, H., and I. F. Nes. 1989. High-frequency transformation by electroporation of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hueck, C. J., W. Hillen, and M. H. Saier, Jr. 1994. Analysis of a cis-active sequence mediating catabolite repression in Gram-positive bacteria. Res. Microbiol. 145:503-518. [DOI] [PubMed] [Google Scholar]
- 13.Klaenhammer, T., E. Altermann, F. Arigoni, A. Bolotin, F. Breidt, J. Broadbent, R. Cano, S. Chaillou, J. Deutscher, M. Gasson, M. van de Guchte, J. Guzzo, A. Hartke, T. Hawkins, P. Hols, R. Hutkins, M. Kleerebezem, J. Kok, O. Kuipers, M. Lubbers, E. Maguin, L. McKay, D. Mills, A. Nauta, R. Overbeek, H. Pel, D. Pridmore, M. Saier, D. van Sinderen, A. Sorokin, J. Steele, D. O'Sullivan, W. de Vos, B. Weimer, M. Zagorec, and R. Siezen. 2002. Discovering lactic acid bacteria by genomics. Antonie van Leeuwenhoek 82:29-58. [DOI] [PubMed] [Google Scholar]
- 14.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. Fiers, W. Stiekema, R. M. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leloup, L., S. D. Ehrlich, M. Zagorec, and F. Morel-Deville. 1997. Single-crossover integration in the Lactobacillus sake chromosome and insertional inactivation of the ptsI and lacL genes. Appl. Environ. Microbiol. 63:2117-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makarova, K., A. Slesarev, Y. Wolf, A. Sorokin, B. Mirkin, E. Koonin, A. Pavlov, N. Pavlova, V. Karamychev, N. Polouchine, V. Shakhova, I. Grigoriev, Y. Lou, D. Rohksar, S. Lucas, K. Huang, D. M. Goodstein, T. Hawkins, V. Plengvidhya, D. Welker, J. Hughes, Y. Goh, A. Benson, K. Baldwin, J.-H. Lee, I. Díaz-Muñiz, B. Dosti, V. Smeianov, W. Wechter, R. Barabote, G. Lorca, E. Altermann, R. Barrangou, B. Ganesan, Y. Xie, H. Rawsthorne, D. Tamir, C. Parker, F. Breidt, J. Broadbent, R. Hutkins, D. O'Sullivan, J. Steele, G. Unlu, M. Saier, T. Klaenhammer, P. Richardson, S. Kozyavkin, B. Weimer, and D. Mills. 2006. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. USA 103:15611-15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin-Verstraete, I., V. Charrier, J. Stülke, A. Galinier, B. Erni, G. Rapoport, and J. Deutscher. 1998. Antagonistic effects of dual PTS-catalysed phosphorylation on the Bacillus subtilis transcriptional activator LevR. Mol. Microbiol. 28:293-303. [DOI] [PubMed] [Google Scholar]
- 18.Mazé, A., G. Boël, S. Poncet, I. Mijakovic, Y. L. Breton, A. Benachour, V. Monedero, J. Deutscher, and A. Hartke. 2004. The Lactobacillus casei ptsHI47T mutation leads to overexpression of a LevR-regulated, but RpoN-independent operon encoding a mannose class phosphotransfrase system. J. Bacteriol. 186:4543-4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Posno, M., R. J. Leer, N. van Luijk, M. J. F. van Giezen, P. T. Heuvelmans, B. C. Lokman, and P. H. Pouwels. 1991. Incompatibility of Lactobacillus vectors with replicons derived from small cryptic Lactobacillus plasmids and segregational instability of the introduced vectors. Appl. Environ. Microbiol. 57:1822-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Postma, P. W., J. W. Lengeler, and G. R. Jacobson. 1993. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57:543-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 22.Sarmiento-Rubiano, L. A., M. Zúñiga, G. Pérez-Martínez, and M. J. Yebra. 2007. Dietary supplementation with sorbitol results in selective enrichment of lactobacilli in rat intestine. Res. Microbiol. 158:694-701. [DOI] [PubMed] [Google Scholar]
- 23.Schotte, L., L. Steidler, J. Vandekerckhove, and E. Remaut. 2000. Secretion of biologically active murine interleukin-10 by Lactococcus lactis. Enzyme Microb. Technol. 27:761-765. [DOI] [PubMed] [Google Scholar]
- 24.Schürmann, M., and G. A. Sprenger. 2001. Fructose-6-phosphate aldolase is a novel class I aldolase from Escherichia coli and is related to a novel group of bacterial transaldolases. J. Biol. Chem. 276:11055-11061. [DOI] [PubMed] [Google Scholar]
- 25.Silveira, M. M., and R. Jonas. 2002. The biotechnological production of sorbitol. Appl. Microbiol. Biotechnol. 59:400-408. [DOI] [PubMed] [Google Scholar]
- 26.Stülke, J., M. Arnaud, G. Rapaport, and I. Martin-Verstraete. 1998. PRD—a protein domain involved in PTS-dependent induction and carbon catabolite repression of catabolic operons in bacteria. Mol. Microbiol. 28:865-874. [DOI] [PubMed] [Google Scholar]
- 27.Tangney, M., J. K. Brehm, N. P. Minton, and W. J. Mitchell. 1998. A gene system for glucitol transport and metabolism in Clostridium beijerinckii NCIMB 8052. Appl. Environ. Microbiol. 64:1612-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tobisch, S., J. Stülke, and M. Hecker. 1999. Regulation of the lic operon of Bacillus subtilis and characterization of potential phosphorylation sites of the LicR regulator protein by site-directed mutagenesis. J. Bacteriol. 181:4995-5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Viana, R., V. Monedero, V. Dossonnet, C. Vadeboncoeur, G. Pérez-Martínez, and J. Deutscher. 2000. Enzyme I and HPr from Lactobacillus casei: their role in sugar transport, carbon catabolite repression and inducer exclusion. Mol. Microbiol. 36:570-584. [DOI] [PubMed] [Google Scholar]
- 30.Yamada, M., and M. H. Saier, Jr. 1988. Positive and negative regulators for glucitol (gut) operon expression in Escherichia coli. J. Mol. Biol. 203:569-583. [DOI] [PubMed] [Google Scholar]
- 31.Yebra, M. J., V. Monedero, M. Zúñiga, J. Deutscher, and G. Pérez-Martínez. 2006. Molecular analysis of the glucose-specific phosphoenolpyruvate: sugar phosphotransferase system from Lactobacillus casei and its links with the control of sugar metabolism. Microbiology 152:95-104. [DOI] [PubMed] [Google Scholar]
- 32.Yebra, M. J., and G. Pérez-Martínez. 2002. Cross-talk between the l-sorbose and d-sorbitol (d-glucitol) metabolic pathways in Lactobacillus casei. Microbiology 148:2351-2359. [DOI] [PubMed] [Google Scholar]
- 33.Yebra, M. J., A. Veyrat, M. A. Santos, and G. Pérez-Martínez. 2000. Genetics of l-sorbose transport and metabolism in Lactobacillus casei. J. Bacteriol. 182:155-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yebra, M. J., R. Viana, V. Monedero, J. Deutscher, and G. Pérez-Martínez. 2004. An esterase gene from Lactobacillus casei cotranscribed with genes encoding a phosphoenolpyruvate:sugar phosphotransferase system and regulated by a LevR-like activator and σ54 factor. J. Mol. Microbiol. Biotechnol. 8:117-128. [DOI] [PubMed] [Google Scholar]