Abstract
Microorganisms associated with the stems and roots of nonnodulated (Nod−), wild-type nodulated (Nod+), and hypernodulated (Nod++) soybeans [Glycine max (L.) Merril] were analyzed by ribosomal intergenic transcribed spacer analysis (RISA) and automated RISA (ARISA). RISA of stem samples detected no bands specific to the nodulation phenotype, whereas RISA of root samples revealed differential bands for the nodulation phenotypes. Pseudomonas fluorescens was exclusively associated with Nod+ soybean roots. Fusarium solani was stably associated with nodulated (Nod+ and Nod++) roots and less abundant in Nod− soybeans, whereas the abundance of basidiomycetes was just the opposite. The phylogenetic analyses suggested that these basidiomycetous fungi might represent a root-associated group in the Auriculariales. Principal-component analysis of the ARISA results showed that there was no clear relationship between nodulation phenotype and bacterial community structure in the stem. In contrast, both the bacterial and fungal community structures in the roots were related to nodulation phenotype. The principal-component analysis further suggested that bacterial community structure in roots could be classified into three groups according to the nodulation phenotype (Nod−, Nod+, or Nod++). The analysis of root samples indicated that the microbial community in Nod− soybeans was more similar to that in Nod++ soybeans than to that in Nod+ soybeans.
The genetic requirements for rhizobial and mycorrhizal interactions in plants overlap in a common signaling pathway, leading to successful symbioses (21, 23). Plants are also known to control the degrees of nodulation and mycorrhization of roots by rhizobia and mycorrhizae, respectively (4, 16). This autoregulatory mechanism occurs through long-distance signaling between the shoot and the root (20). Plants deficient in autoregulation of nodulation develop hypernodulated roots. In the case of soybean [Glycine max (L.) Merril], hypernodulated mutants differ in their ability to autoregulate root colonization by arbuscular mycorrhizal fungi (16). However, the degree to which plants use similar or identical systems, such as a common signaling pathway and autoregulation, for interactions with other microorganisms remains unclear (23). Recently, it was shown that the wild type and symbiosis-defective mutants of the model legume Medicago truncatula have different bacterial community structures (19), and certain bacteria preferentially associate with roots with mycorrhizae. These examples indicate that genetic alteration in the nodulation/mycorrhization signaling pathways can in turn alter the accompanying plant microflora, aside from rhizobia and mycorrhizae.
Characterization of soybean-associated microbial communities has been based solely on culture-dependent methods (8, 14, 25, 30), and only a few studies have used culture-independent techniques (12). Moreover, the impact of nodulation on the microbial community in soybean is unknown.
The aim of the present study was to determine the effect of nodulation phenotypes on microbes associated with soybeans grown under field conditions. A parental line (nodulating) and derived nonnodulating and hypernodulating soybean mutants were used for microbial community analyses. The microfloras associated with soybeans with different nodulation phenotypes were surveyed by using ribosomal intergenic spacer analysis (RISA) and automated RISA (ARISA). The differential bands for the nodulation phenotypes were cloned and sequenced from RISA, and ARISA profiles were subjected to principal-component analysis (PCA) to resolve differences in microbial community structures.
MATERIALS AND METHODS
Plant materials and soil characteristics.
The experimental field used in this work has been cultivated with a rotation of rice (paddy field conditions) and soybean (upland field conditions) every year since 1997. Soybean seeds with five genotypes were obtained from the collection of Makie Kokubun (Tohoku University) and Kazunori Sakamoto (Chiba University). The plants included parental cultivar Enrei (wild-type nodulating cultivar, Nod+), cultivars En1314 and En1282 (nonnodulating mutants derived from Enrei, Nod−) (6), and cultivars En6500 and Sakukei 4 (hypernodulating mutants derived from Enrei, Nod++) (1, 15). Mutations in En1314 and En1282 soybeans were found in the nfr1 gene, which is known to be responsible for a LysM-type receptor kinase for Nod factor recognition (26; Masaki Hayashi, personal communication). A mutation in En6500 was found in the har1 gene, which mediates systemic autoregulation of nodulation (18), and Sakukei 4 is a descendant with a hypernodulation phenotype that was derived from En6500 (15).
The seeds were planted on 31 May 2006 in an experimental field at Tohoku University (Kashimadai, Miyagi, Japan). The field soil was classified as gray lowland soil (pH[H2O], 5.9; pH[KCl], 4.3; total carbon content, 1.21%; total nitrogen content, 0.11%; Truog phosphorus content, 69 mg P2O5 kg−1).
Sampling and DNA extraction.
Soybean plants (five plants per genotype) were harvested on 7 September 2006 and immediately transported on ice to the laboratory. The plants were washed well with tap water. Leaves and nodules were removed manually. The roots were then separated from the stem. Tissues from each plant sample were separately stored at −80°C until they were used. Plant tissues were ground to a powder in liquid nitrogen with a mortar and pestle. DNA was extracted from 200 to 300 mg of powdered tissue by the DNA extraction method developed by Ikeda et al. (11).
RISA.
RISA was performed as previously described (29), using three primer sets: bacterial primer set ITSF/ITSReub (3) and fungal primer sets ITS1F/ITS4 and ITS1F/ITS4-B specific for basidiomycetes (7). After electrophoresis, digital fingerprinting images were obtained with a fluorescent scanner (FLA-2000; Fujifilm, Tokyo, Japan), and the bands that were stably present in a nodulation phenotype were identified and were cut out from the gel for cloning and sequencing.
Molecular cloning and sequencing.
The protocol of Ikeda et al. (10) was used for molecular cloning and sequencing. Briefly, DNA was extracted from the bands and reamplified by PCR under the conditions that were used for RISA, except that a nonfluorescent labeled primer was used. Purified DNA fragments were then ligated into a pGEM-T Easy vector and subsequently transformed into competent cells of Escherichia coli by using a pGEM-T Easy vector system I cloning kit (Promega, Madison, WI) according to the manufacturer's instructions. Transformants were selected, and plasmid DNA was extracted by using a QIAprep spin miniprep kit (Qiagen, Hilden, Germany). To select clones corresponding to each band in RISA profiles, the RISA procedure described above was performed with 1 μl of plasmid extract as the template DNA. Sequencing reactions were performed for selected plasmid clones by using an ABI Prism BigDye terminator cycle sequencing ready reaction kit (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. Using a Perkin-Elmer 9600 thermal cycler and an ABI Prism 377 DNA sequencer (Applied Biosystems), both ends of each PCR clone were sequenced with the T7 and SP6 primers. Sequences were manually edited to remove the vector backbone, primer regions, and ambiguous sequences, and contiguous sequences were generated by using overlapping regions of sequence data for both ends of the fragments. The resulting data were compared with public database entries by using the BLASTN subroutine search protocol (2), and sequence matches were considered to be significant when the score was >50 (17).
ARISA and PCA.
PCA was used in conjunction with ARISA to evaluate similarities of microbial communities for plant samples. The samples used for ARISA were prepared like those used for RISA, as described above. PCR products (1 μl) were mixed with 1 μl of the LIZ1200 internal size standard (Applied Biosystems), and 20 μl of deionized formamide was added. The mixture was denatured at 95°C for 5 min and cooled on ice. Next, the PCR product was placed in an ABI Prism 310 genetic analyzer (Applied Biosystems) with a 47-cm, 50-μm capillary filled with POP-4 polymer (Applied Biosystems). The samples were run under standard ABI Prism 310 denaturing electrophoresis conditions for 40 min. The profile data (up to 1,200 bases) obtained by ARISA were initially analyzed with ABI GeneMapper software (Applied Biosystems) and were processed further with the RIBOSORT program (32) for assigning fragment size and calculating relative abundance for each ribotype. The fragments with fewer than 100 fluorescence units were eliminated from the analyses. Finally, PCA was performed by using CANOCO (version 4.5 for Windows; Microcomputer Power, Ithaca, NY) with default parameters (except that intersample scaling was used) to generate ordination plots based on the scores of the first two principal components.
Phylogenetic analysis.
For the phylogenetic analysis, alignment was performed with the CLUSTAL W program (34). The neighbor-joining method was used to construct the phylogenetic trees (31). The PHYLIP format tree output was obtained by the bootstrap procedure (5), and 1,000 bootstrap trials were used. The trees were constructed with Tree View software (22).
Nucleotide sequence accession numbers.
Nucleotide sequences of the PCR clones isolated from RISA profiles have been deposited in the DDBJ database under accession numbers AB432939 to AB432941 and AB432943 to AB432945.
RESULTS
RISA profiles for stem- and root-associated microbial communities.
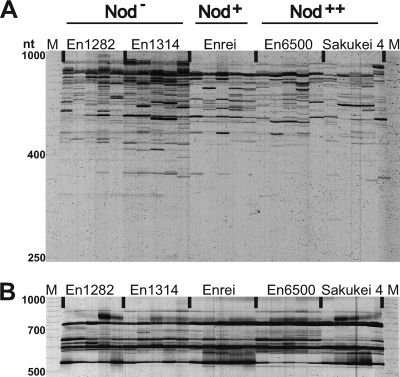
The RISA profiles of bacteria from stem samples were examined (Fig. 1A). No DNA band was clearly associated with “a nodulation phenotype (Nod−, Nod+, or Nod++),” although some polymorphic DNA bands were specifically present or more abundant in a plant genotype. The RISA profiles of fungi from stem samples contained only a few noticeable bands, and the patterns were essentially identical for all plant genotypes (Fig. 1B).
FIG. 1.
RISA profiles of microbial communities in soybean stems. (A) Bacterial RISA with primers ITSF and ITSReub. (B) Fungal RISA with primers ITS1F and ITS4. For each treatment, five independent samples were used. Each lane represents an individual plant. Lanes M, size marker (50-1000 MapMarker; Bio Ventures); lanes En1282 and En1314, nonnodulating cultivars En1282 and En1314, respectively; lane Enrei, wild-type nodulating cultivar Enrei; lanes En6500 and Sakukei 4, hypernodulating soybean cultivars En6500 and Sakukei 4, respectively.
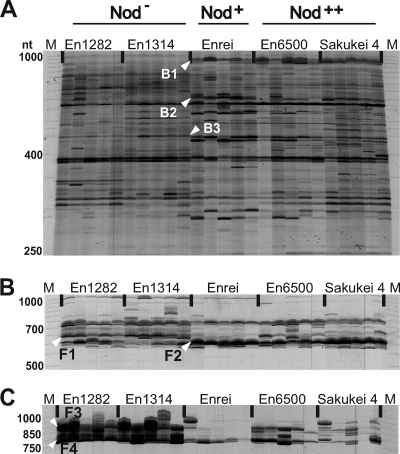
As expected, both the bacterial and fungal RISA profiles for root samples were more complex than those for stem samples (Fig. 2). Several polymorphic DNA bands for bacteria and fungi specific to or abundant in a nodulation phenotype were observed, as reported in other cases (27). For the bacterial RISA, sequence analysis of these DNA bands revealed that the closest relatives among known bacterial species were Bradyrhizobium japonicum (clone B1, for Nod+ and Nod++), Pseudomonas fluorescens (clone B2, for Nod+), and Micromonospora echinospora (clone B3, specific for Nod−) (Fig. 2A and Table 1). The absence of the band corresponding to band B1 in nonnodulated plants suggests that the habitat of B. japonicum may be restricted to the rhizosphere of nodulated soybeans or that this species is present as a minor population whose size is less than the detection limit of RISA. For fungal RISA of root samples, two noticeable DNA bands (bands F1 and F2) were observed when the ITS1F/ITS4 primer set was used (Fig. 2B). Band F1 was stably detected in all Nod− soybean roots and weakly present in the nodulated (Nod+ and Nod++) roots. In contrast, the signal pattern for band F2 was just the opposite. Sequence analysis revealed that the closest relatives of these fungi among known species were Exidia saccharina for band F1 and Fusarium solani for F2 (Fig. 2B and Table 1). However, when the ITS1F/ITS4-B primer set was used for fungal RISA of the same root samples, drastic differences among the nodulation phenotypes were observed in the profile patterns and band intensities (Fig. 2C). Two distinct bands (bands F3 and F4) with very strong signals were detected in all Nod− soybean roots, whereas the corresponding bands in the nodulated (Nod+) roots were very weak or faint (Fig. 2C). The profile for Nod++ soybean roots was similar to that for Nod− soybean roots, but the band signals were less intense. A BLAST search for clone F4 showed that it was closely related to Exidiopsis sp., a member of the Exidia/Exidiopsis group in the Auriculariales (Table 1), while band F3 could not be cloned in the present study.
FIG. 2.
RISA profiles of microbial communities in soybean roots. (A) Bacterial RISA with primers ITSF and ITSReub. (B) Fungal RISA with primers ITS1F and ITS4. (C) Basidiomycete-specific fungal RISA with primers ITS1F and ITS4-B. For each treatment, five independent samples were used. Each lane represents an individual plant. Lanes M, size marker (50-1000 MapMarker; Bio Ventures); lanes En1282 and En1314, nonnodulating cultivars En1282 and En1314, respectively; lane Enrei, wild-type nodulating cultivar Enrei; lanes En6500 and Sakukei 4, hypernodulating soybean cultivars En6500 and Sakukei 4, respectively.
TABLE 1.
Sequence similarities of RISA clones to known microbes
| Type of organism | Clonea | Accession no.b | Length (nucleotides) | Score | % Similarity | Closest BLAST match (accession no.b) |
|---|---|---|---|---|---|---|
| Bacterium | B1 | AB432939 | 860 | 1528 | 99 | Bradyrhizobium japonicum (AF293381) |
| B2 | AB432940 | 559 | 994 | 99 | Pseudomonas fluorescens PfO-1 (CP000094) | |
| B3 | AB432941 | 403 | 147 | 74 | Micromonospora echinospora (AY524043) | |
| Fungus | F1 | AB432944 | 639 | 639 | 87 | Exidia saccharina (AF291277) |
| F2 | AB432943 | 562 | 1014 | 100 | Fusarium solani (AB369907) | |
| F4 | AB432945 | 720 | 848 | 86 | Exidiopsis sp. strain RJB11821 (AY509549) |
The designations correspond to the band designations shown in Fig. 2B and C.
DDBJ accession number.
Phylogenetic analysis for uncultured clones of basidiomycetes.
A phylogenetic analysis was conducted to clarify the assignments of the sequences of clones F1 and F4 in the division Basidiomycota. As a result, these sequences were shown to be closely related to the Auriculariaceae but formed a cluster separated from known species in this family (Fig. 3). Interestingly, these sequences also formed a distinct cluster with an Exidiopsis-like sequence (accession number AB213415) that was isolated from the rhizosphere of chrysanthemum (10).
FIG. 3.
Phylogenetic analysis of basidiomycete sequences isolated from roots of nonnodulated soybean mutants. The internal transcribed spacer sequences of ribosomal intergenic regions were used to construct the tree. The numbers at nodes are bootstrap values, and values of <500 are not shown. Scale bar = 0.1 substitution per site. Clones F1 and F4 are the sequences corresponding to bands F1 (Fig. 2B) and F4 (Fig. 2C), respectively. AB213415 is a root-associated fungal sequence isolated from the rhizosphere of chrysanthemum. Uncultured basidiomycetes TP1 and TPC2 (accession numbers AY382817 and AY382818) were reported to be orchid mycorrhizae by McCormick et al. (16).
PCA for stem- and root-associated microbial communities.
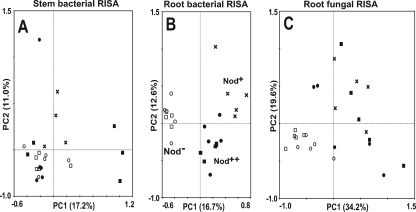
PCA was performed to compare the complex ARISA profiles of the microbial community structures for the three soybean nodulation phenotypes. For the stem bacterial ARISA, the majority of the samples were clustered around the center of the ordination diagram, and no clear cluster was formed based on nodulation phenotypes (Fig. 4A). PCA was not performed for the stem fungal communities, which did not vary among the plant genotypes.
FIG. 4.
Principal-component plots generated from ARISA profiles for soybean-associated microbial communities. (A) Bacterial ARISA with primers ITSF and ITSReub for stems. (B) Bacterial ARISA with primers ITSF and ITSReub for roots. (C) Fungal ARISA with primers ITS1F and ITS4 for roots. ARISA profiles were obtained with an ABI genetic analyzer (data not shown). Symbols: ○, Nod− cultivar En1282; □, Nod− cultivar En1314; ×, Nod+ cultivar Enrei; •, Nod++ cultivar En6500; ▪, Nod++ cultivar Sakukei 4.
The PCA for bacterial ARISA of root samples indicated that the community structures could be classified into three groups according to the nodulation phenotype (Fig. 4B). In addition, the bacterial community in roots of Nod− soybeans was more similar to that in roots of Nod++ soybeans than to that in roots of Nod+ soybeans. For fungi, as expected from RISA profiles, the results of PCA showed that the community structures could be divided into two groups corresponding to the nonnodulated (Nod−) and nodulated (Nod+ and Nod++) soybeans (Fig. 4C). For both bacterial and fungal communities in roots, the samples for Nod− soybeans were more tightly clustered than those for Nod+ and Nod++ soybeans (Fig. 4B and C).
DISCUSSION
Nodulation phenotype-specific microbes were surveyed by using RISA of the stems and roots of soybeans, including nonnodulating and hypernodulating mutants. To eliminate the effects of a genotype-specific mutation in the analyses, two independent lines derived from the same parental line were used for each mutant phenotype. The overall profiles of the bacterial and fungal RISA for both stems and roots showed considerable variation in the gels from sample to sample. This could be attributed to the long-term field cultivation (3 months), and the use of a composite sampling strategy may improve the quality of profiling in RISA. In the stem, there was no DNA band specific for a nodulation phenotype for either bacteria or fungi. In fact, the stem profiles were poor profiles, presumably because of the excess amount of plant DNA and the low microbial biomass in stems, a problem noted in other stem-associated community analyses (33). Therefore, technical improvements are required for appropriate analysis of the stem-associated microbial community.
Both bacterial and fungal RISA of root samples detected some polymorphic DNA bands among the nodulation phenotypes. Although we expected that the nodulation phenotype would affect the microbial community structure in roots, the overall structures of the bacterial communities were very similar for all the nodulation phenotypes (Fig. 2A). The nodulating bacterium B. japonicum was strictly associated with only nodulation phenotypes. Thus, the presence of nodulating bacteria or nodules may influence fungal colonization of roots to a large extent.
Meanwhile, RISA revealed a significant impact of nodulation phenotype on fungal communities in roots, as expected. Band F1 was consistently present in Nod− soybeans and less abundant in Nod+ and Nod++ soybeans, whereas the results for band F2 were just the opposite. One possible explanation for this phenomenon is that the fungi may be antagonistic to each other or may compete for common resources, where one fungus thrives in the roots more successfully in the absence of a nodulation system and the other thrives in the presence of such a system. Furthermore, the results of the fungal RISA with the ITS4-B primer suggested that the basidiomycetes preferentially occurred in Nod− soybean roots. In the fungal RISA, the profiles of Nod+ soybeans showed the simplest pattern (Fig. 2B and C), suggesting that the fungal community in the roots of Nod+ soybeans is more tightly controlled than the fungal communities in the roots of Nod− and Nod++ soybeans.
The sequence analyses of clone F1 revealed a high level of identity with F. solani, which can be considered a plant pathogen (9, 13, 28), while nonpathogenic strains do exist as common soil saprophytes (24). The results of the phylogenetic analysis of clones F1 and F4 indicated that these fungi closely resembled members of the Auriculariales but formed a cluster separate from known species of this taxon. Interestingly, a fungal sequence (accession number AB213415) isolated from the rhizosphere of chrysanthemum was included in the cluster (10). Ikeda et al. (10) also reported that Exidia/Exidiopsis-like sequences, including the accession number AB213415 sequence, were strongly associated with the roots of wild-type plants and that their population levels were reduced in the roots of Bacillus thuringiensis transgenic plants. If this is so, nodulation may have an ecological impact equivalent to B. thuringiensis toxin production. Thus, the results of the present study also suggest that this group of basidiomycetes could be recognized as a fungal group sensitive to a plant genotype or phenotype.
To date, members of the Auriculariales have generally been considered saprophytic fungi. In a terrestrial orchid, McCormick et al. (15a) reported the presence of mycorrhizae (accession numbers AY382817 and AY382818) that are closely related to members of the Auriculariales. However, as shown in Fig. 3, the sequences of clones F1 and F4 are more closely related to known species of Auricularia and Exidia/Exidiopsis. Thus, the cluster containing clones F1 and F4 in Fig. 3 may represent a new root-associated group in the Auriculariales. In light of these and previous findings, interactions of Auriculariales within the rhizosphere may be more common than previously thought.
The effect of nodulation phenotype on microbial community structure was also examined by PCA of ARISA profiles. Unexpectedly, the results of PCA for bacterial RISA revealed that the nonnodulated soybean roots had a microflora that was more similar to that of hypernodulated roots than to that of wild-type nodulated roots. This phenomenon was also observed with the root-associated fungi (Fig. 2C). These results indicate that the presence of nodules or symbiotic nitrogen fixation is not entirely responsible for the shifts in microbial community structure (Fig. 4B). One possible explanation for this phenomenon may be the lack of activation of an autoregulation system in both phenotypes. Thus, Nod− plants may not have enough activation of autoregulation systems in the field, since the rate of mycorrhizal infection in these plants was shown to be very low (less than 10%) (data not shown). The field used in the present study has been cultivated as a paddy field for rice every 2 years, and this rotation may be responsible for the low rate of mycorrhizal infection. Meanwhile, Nod++ plants have lost the function of autoregulation systems completely. Some of the genetic elements for nodulation and autoregulation systems in legumes may be involved in the accommodation of root-associated microbes other than rhizobia.
Acknowledgments
We thank M. Kokubun (Tohoku University), K. Sakamoto (Chiba University), and M. Hayashi (National Institute of Agrobiological Sciences) for providing soybean seeds and unpublished genetic information.
This work was supported in part by Special Coordination Funds for Promoting Science and Technology awarded to K.M. by the Ministry of Education, Culture, Sports, Science, and Technology of Japan and in part by a Grant-in-Aid for Scientific Research to K.M. from the Japan Society for the Promotion of Science (grant 17658034). L.E.E.R. was supported by a Japanese government scholarship (MEXT).
Footnotes
Published ahead of print on 25 July 2008.
REFERENCES
- 1.Akao, S., and H. Kouchi. 1989. A supernodulating mutant isolated from soybean cultivar Enrei. Soil Sci. Plant Nutr. 38:183-187. [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Cardinale, M., L. Brusetti, P. Quatrini, S. Borin, A. M. Puglia, A. Rizzi, E. Zanardini, C. Sorlini, C. Corselli, and D. Daffonchio. 2004. Comparison of different primer sets for use in automated ribosomal intergenic spacer analysis of complex bacterial communities. Appl. Environ. Microbiol. 70:6147-6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carroll, B. J., D. L. McNeil, and P. M. Gresshoff. 1985. Isolation and properties of soybean [Glycine max (L.) Merr.] mutants that nodulate in the presence of high nitrate concentrations. Proc. Natl. Acad. Sci. USA 82:4162-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 6.Francisco, P. B., and S. Akao. 1993. Autoregulation and nitrate inhibition of nodule formation in soybean cv. Enrei and its nodulation mutants. J. Exp. Bot. 44:547-553. [Google Scholar]
- 7.Gardes, M., and T. D. Bruns. 1993. ITS primers with enhanced specificity for basidiomycetes: application to the identification of mycorrhizae and rusts. Mol. Ecol. 2:113-118. [DOI] [PubMed] [Google Scholar]
- 8.Hung, P. Q., S. M. Kumar, V. Govindsamy, and K. Annapurna. 2007. Isolation and characterization of endophytic bacteria from wild and cultivated soybean varieties. Biol. Fertil. Soils 44:155-162. [Google Scholar]
- 9.Ikeda, S., S. Fujji, T. Sato, H. Furuya, H. Naito, N. Ytow, H. Ezura, K. Minamisawa, and T. Fujimura. 2007. Microbial diversity in milled rice as revealed by ribosomal intergenic spacer analysis. Microbes Environ. 22:165-174. [Google Scholar]
- 10.Ikeda, S., H. Shinoyama, Y. Yamanaka, N. Ytow, H. Ezura, and T. Fujimura. 2005. Application of ribosomal intergenic spacer analysis to the characterization of microbial communities in rhizospheric soils of the Bt transgenic chrysanthemum. Microbes Environ. 20:258-263. [Google Scholar]
- 11.Ikeda, S., K. N. Watanabe, K. Minamisawa, and N. Ytow. 2004. Evaluation of soil DNA from arable land in Japan using a modified direct-extraction method. Microbes Environ. 19:301-309. [Google Scholar]
- 12.Kerkhof, L., M. Santoro, and J. Garland. 2000. Response of soybean rhizosphere communities to human hygiene water addition as determined by community level physiological profiling (CLPP) and terminal restriction fragment length polymorphism (TRFLP) analysis. FEMS Microbiol. Lett. 184:95-101. [DOI] [PubMed] [Google Scholar]
- 13.Killebrew, J. F., K. W. Roy, G. W. Lawrence, K. S. McLean, and H. H. Hodges. 1988. Greenhouse and field evaluation of Fusarium solani pathogenicity to soybean seedlings. Plant Dis. 72:1067-1070. [Google Scholar]
- 14.Kuklinsky-Sobral, J., W. L. Araujo, R. Mendes, I. O. Geraldi, A. A. Pizzirani-Kleiner, and J. L. Azevedo. 2004. Isolation and characterization of soybean-associated bacteria and their potential for plant growth promotion. Environ. Microbiol. 6:1244-1251. [DOI] [PubMed] [Google Scholar]
- 15.Matsunami, T., A. Kaihatsu, T. Maekawa, M. Takahashi, and M. Kokubun. 2004. Characterization of vegetative growth of a supernodulating soybean genotype, Sakukei 4. Plant Prod. Sci. 7:165-171. [Google Scholar]
- 15a.McCormick, M. K., D. F. Whigham, and J. O'Neill. 2004. Mycorrhizal diversity in photosynthetic terrestrial orchids. New Phytol. 163:425-438. [DOI] [PubMed] [Google Scholar]
- 16.Meixner, C., G. Vegvari, J. Ludwig-Müller, H. Gagnon, S. Steinkellner, C. Staehelin, P. Gresshoff, and H. Vierheilig. 2007. Two defined alleles of the LRR receptor kinase GmNARK in supernodulating soybean govern differing autoregulation of mycorrhization. Physiol. Plant. 130:261-270. [Google Scholar]
- 17.Newman, T., F. J. de Bruijn, P. Green, K. Keegstra, H. Kende, L. McIntosh, J. Ohlrogge, N. Raikhel, S. Somerville, M. Thomashow, et al. 1994. Genes galore: a summary of methods for accessing results from large-scale partial sequencing of anonymous Arabidopsis cDNA clones. Plant Physiol. 106:1241-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishimura, R., M. Hayashi, G. J. Wu, H. Kouchi, H. Imaizumi-Anraku, Y. Murakami, S. Kawasaki, S. Akao, M. Ohmori, M. Nagasawa, K. Harada, and M. Kawaguchi. 2002. HAR1 mediates systemic regulation of symbiotic organ development. Nature 420:426-429. [DOI] [PubMed] [Google Scholar]
- 19.Offre, P., B. Pivato, S. Siblot, E. Gamalero, T. Corberand, P. Lemanceau, and C. Mougel. 2007. Identification of bacterial groups preferentially associated with mycorrhizal roots of Medicago truncatula. Appl. Environ. Microbiol. 73:913-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oka-Kira, E., and M. Kawaguchi. 2006. Long-distance signaling to control root nodule number. Curr. Opin. Plant Biol. 9:496-502. [DOI] [PubMed] [Google Scholar]
- 21.Oldroyd, G. E. D., and J. A. Downie. 2004. Calcium, kinases and nodule signalling in legumes. Nat. Rev. 5:566-576. [DOI] [PubMed] [Google Scholar]
- 22.Page, R. D. M. 1996. TreeView: an application to display phylogenetic trees on personal computers. Bioinformatics 12:357-358. [DOI] [PubMed] [Google Scholar]
- 23.Parniske, M. 2000. Intracellular accommodation of microbes by plants: a common developmental program for symbiosis and disease? Curr. Opin. Plant Biol. 3:320-328. [DOI] [PubMed] [Google Scholar]
- 24.Peters, R. D. 2002. Inoculation with nonpathogenic Fusarium solani increases severity of pea root rot caused by Aphanomyces euteiches. Plant Dis. 86:411-414. [DOI] [PubMed] [Google Scholar]
- 25.Pimentel, I. C., C. Glienke-Blanco, J. Gabardo, R. M. Stuart, and J. L. Azevedo. 2006. Identification and colonization of endophytic fungi from soybean (Glycine max (L.) Merril) under different environmental conditions. Braz. Arch. Biol. Technol. 49:705-711. [Google Scholar]
- 26.Radutoiu, S., L. H. Madsen, E. B. Madsen, H. H. Felle, Y. Umehara, M. Gronlund, S. Sato, Y. Nakamura, S. Tabata, N. Sandai, and J. Stougaard. 2003. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425:585-592. [DOI] [PubMed] [Google Scholar]
- 27.Rosenblueth, M., and E. Martínez-Romero. 2006. Bacterial endophytes and their interactions with hosts. Mol. Plant-Microbe Interact. 19:827-837. [DOI] [PubMed] [Google Scholar]
- 28.Roy, K. W., G. W. Lawrence, H. H. Hodges, K. S. McLean, and J. F. Killebrew. 1989. Sudden death syndrome of soybean: Fusarium solani as incitant and relation of Heterodera glycines to disease severity. Phytopathology 79:191-197. [Google Scholar]
- 29.Saito, A., M. Kawahara, S. Ikeda, M. Ishimine, S. Akao, and K. Minamisawa. 2008. Broad distribution and phylogeny of anaerobic endophytes of cluster XIVa clostridia in plant species including crops. Microbes Environ. 23:73-80. [DOI] [PubMed] [Google Scholar]
- 30.Saito, A., S. Ikeda, H. Ezura, and K. Minamisawa. 2007. Microbial community analysis of the phytosphere using culture-independent methodologies. Microbes Environ. 22:93-105. [Google Scholar]
- 31.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 32.Scallan, U., A. Liliensiek, N. Clipson, and J. Connolly. 2008. RIBOSORT: a program for automated data preparation and exploratory analysis of microbial community fingerprints. Mol. Ecol. Res. 8:95-98. [DOI] [PubMed] [Google Scholar]
- 33.Sessitsch, A., B. Reiter, U. Pfeifer, and E. Wilhelm. 2002. Cultivation-independent population analysis of bacterial endophytes in three potato varieties based on eubacterial and Actinomycetes specific PCR of 16S rRNA genes. FEMS Microbiol. Ecol. 39:23-32. [DOI] [PubMed] [Google Scholar]
- 34.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]