Abstract
Previous studies have suggested that levels of transcripts for dsrA, a gene encoding a subunit of the dissimilatory sulfite reductase, are not directly related to the rates of sulfate reduction in sediments under all conditions. This phenomenon was further investigated with chemostat-grown Desulfovibrio vulgaris. Under sulfate-limiting conditions, dsrA transcript levels increased as the bulk rates of sulfate reduction in the chemostat increased, but transcript levels were similar at all sulfate reduction rates under electron donor-limiting conditions. When both electron donor- and electron acceptor-limiting conditions were considered, there was a direct correspondence between dsrA transcript levels and the rates of sulfate reduction per cell. These results suggest that dsrA transcript levels may provide important information on the metabolic state of sulfate reducers.
Quantifying levels of transcripts for key genes in anaerobic sedimentary environments can provide insights into the metabolic state of microorganisms carrying out important processes in those environments (4, 5, 16, 17, 20, 24). Information can be gained about the relative rates of microbial metabolism under different conditions or the factors limiting the rates of growth and metabolism. Such information is especially useful in the design of bioremediation strategies because it indicates the level to which the genes for the desired bioremediation enzymes are being expressed and can suggest modifications in amendments to overcome nutrient limitations or other environmental stresses (24).
Methods to evaluate the metabolic state of sulfate-reducing bacteria are of interest because of the important role that sulfate reducers play in the biogeochemistry of aquatic sediments (32) and corrosion (8, 14), as well as in the degradation of organic contaminants (9). For example, some sulfate reducers can degrade aromatic hydrocarbons (13), including prevalent contaminants such as benzene (23) and polycyclic aromatic hydrocarbons (2, 3, 6, 12, 25), and stimulating the activity of aromatic hydrocarbon-degrading sulfate reducers can be an effective bioremediation strategy (1, 29).
Dissimilatory sulfite reductase catalyzes the final step in sulfate reduction, and sequences for this enzyme are highly conserved among sulfate reducers (18, 19, 31). Pure culture studies provided a preliminary indication that transcript levels of dissimilatory sulfite reductase genes might be related to rates of sulfate reduction (15, 26). However, relating transcript levels to rates of sulfate reduction in marine sediments was more problematic (5). Transcripts for dsrA, which encodes the alpha subunit of dissimilatory sulfite reductase, were quantified. There was a general trend that sediments with no sulfate reduction had low dsrA transcript levels and that dsrA levels were higher with active sulfate reduction. In sediments with similar sulfate concentrations, there was a direct correlation between the rates of sulfate reduction and dsrA transcript levels. However, in laboratory sediment incubations in which sulfate was continually being depleted over time, dsrA transcripts increased over time even though sulfate reduction rates did not increase (5). These results suggested that dsrA levels can only be related to sulfate reduction rates in sediments under very similar conditions, limiting the predictive value of dsrA transcript measurements over a range of environmental conditions.
In order to better understand the factors controlling dsrA transcript levels in sulfate reducers, Desulfovibrio vulgaris Hildenborough (ATCC 29579) was grown under strict anaerobic conditions (N2:CO2, 80:20 [vol/vol]) at 30°C in a freshwater medium (2.5 g liter−1 NaHCO3, 0.25 g liter−1 NH4Cl, 0.6 g liter−1 or 0.06 g liter−1 [for batch or continuous cultures, respectively] NaH2PO4·H2O, 0.1 g liter−1 KCl, and trace mineral and vitamin solutions) (21) supplemented with a selenite-tungstate solution (400 mg liter−1 NaOH, 6 mg liter−1 Na2SO3·5H2O, 8 mg liter−1 Na2WO4·2H2O) and reduced with sodium sulfide (1 mM final concentration). Continuous culture conditions under electron donor (10 mM lactate, 10 mM sulfate) and electron acceptor (10 mM lactate, 2.5 mM sulfate) limitations were described previously (10). The methods for measuring substrates and products were as follows: for sulfate, ion chromatography (22); for fatty acids, high-pressure liquid chromatography (10); for sulfide, colorimetry (7); for protein, a bicinchoninic acid method (30); and for cell numbers, epifluorescence microscopy with acridine orange staining (27).
Total RNA was isolated with the Qiagen RNeasy mini kit (Qiagen, Inc., Valencia, CA) after digestion of the cell pellet with 50 mg ml−1 lysozyme. Primers DSR1F (forward 5′-AAG GAA CCC CGC ACC AAC-3′) and DSR1R (reverse 5′-TTA TCT CAG GTG TCT CTT GCG GT-3′) (position 1 to 102, dsrA gene) were evaluated and optimized for quantitative PCR as previously described (4). Reverse transcription was performed with an Enhanced Avian First Strand synthesis kit (Sigma-Aldrich, St. Louis, MO). A dilution series of purified PCR products (105 to 1011 molecules) was the calibration standard for the real-time PCR quantification as previously described (16). The cDNA obtained from the reverse transcription reaction was quantified in a GeneAmp 5700 sequence detection system with a reaction mixture of primers (150 nM each), 9.5 μl of cDNA (20 to 250 ng μl−1), 12.5 μl of Power Sybr green PCR master mix (Applied Biosystems, Foster City, CA) to a final volume of 25 μl. dsr transcript levels were normalized to the total RNA levels to help account for different biomass-specific sulfate reduction rates (28).
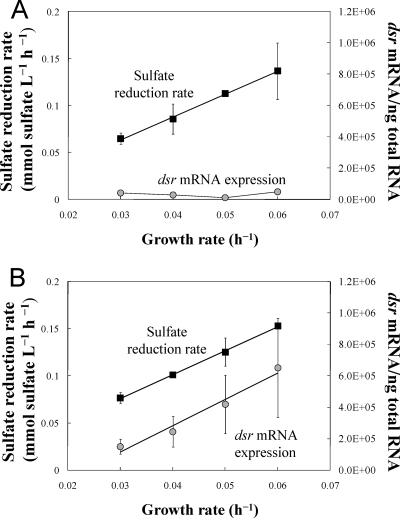
D. vulgaris was initially grown in chemostats in which electron-donor availability limited growth (Table 1). This was designed to simulate the conditions in superficial marine sediments in which sulfate is abundant. Under such conditions, the growth of sulfate reducers is primarily controlled by the rates at which complex organic matter is broken down into simpler substrates that sulfate reducers can utilize. In this system, bulk sulfate reduction rates (moles of sulfate reduced per liter per h) at steady state are analogous to the bulk sulfate reduction rates measured in sediments. The levels of dsrA transcripts were comparable over the range of sulfate reduction rates evaluated (Fig. 1A), suggesting that, as previously found in sediment studies (5), dsrA transcript levels are not always correlated with bulk rates of sulfate reduction. In order to investigate this further, a second set of chemostats (Table 1) were run in which the electron acceptor, sulfate, was the limiting nutrient (Fig. 1B). In contrast to the results from the electron donor-limited chemostats, in the electron acceptor-limited chemostats there was an increase in dsrA transcript levels that paralleled increasing sulfate reduction rates at increasing dilution rates (Fig. 1B).
TABLE 1.
Data at steady-state conditions during continuous growth of Desulfovibrio vulgaris
| Growth type and ratea | Acetate (mM) | Sulfate (mM) | Lactate (mM) | Biomassb |
|---|---|---|---|---|
| Electron donor (lactate) limited | ||||
| 0.03 | 6.8 ± 0.2 | 7.6 ± 0.2 | 0 ± 0 | 48.5 |
| 0.04 | 7.4 ± 0.1 | 7.9 ± 0.4 | 0 ± 0 | 56.7 |
| 0.05 | 6.9 ± 0.1 | 7.4 ± 0.03 | 0 ± 0 | 56.2 |
| 0.06 | 7.5 ± 0.5 | 7.7 ± 0.5 | 0 ± 0 | 59.3 |
| Electron acceptor (sulfate) limited | ||||
| 0.03 | 5.3 ± 0.2 | 0.4 ± 0.2 | 1.8 ± 0.9 | 50.1 |
| 0.04 | 6.4 ± 0.3 | 0 ± 0 | 1.3 ± 0.5 | 54.5 |
| 0.05 | 5 ± 0.4 | 0.4 ± 0.3 | 2.5 ± 0.7 | 58.0 |
| 0.06 | 6.5 ± 0.4 | 0 ± 0 | 2.5 ± 0.2 | 43.7 |
Specific growth rate (h−1) or dilution rate in the continuous culture. Steady-state concentrations of acetate, sulfate, and lactate (average of the results from triplicate continuous cultures at a certain growth rate × 3 sampling events and standard deviation). Initial conditions: electron donor-limited growth, 10 mM lactate and 10 mM sulfate; electron acceptor-limited growth, 10 mM lactate and 2.5 mM sulfate.
Biomass values are given in milligrams of dry weight per liter.
FIG. 1.
Sulfate reduction rate (mmol sulfate consumed at steady state per liter and h; black squares) and number of dsrA mRNA transcripts (gray circles) expressed by D. vulgaris cells grown under electron donor (A) or electron acceptor (B) limitation at different growth rates. Each point is the average of the results for three samples from triplicate continuous cultures at each growth rate. (A) Sulfate reduction rate, R2 = 0.997. (B) Sulfate reduction rate, R2 = 0.998; dsrA mRNA expression, R2 = 0.967.
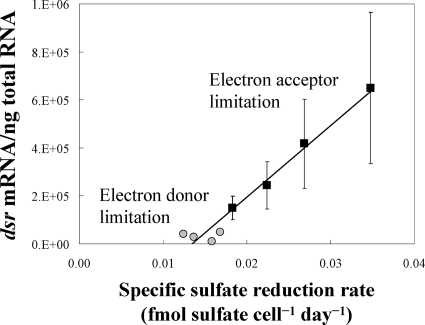
Differences in the results of the electron donor- versus electron acceptor-limited chemostats could be reconciled when the levels of dsrA transcripts were compared with the rate of sulfate reduction per cell (Fig. 2). There was a strong correlation between this cell-specific sulfate reduction rate and dsrA transcript levels.
FIG. 2.
dsr mRNA transcripts expressed by D. vulgaris cells grown under electron donor (gray circles) and acceptor (black squares) limitation at a specific sulfate reduction rate (femtomol sulfate consumed per cell and day). Each point is the average of the results for three samples from triplicate continuous cultures at each growth rate. R2 = 0.964.
High levels of dsrA transcripts in the sulfate-limited chemostats could not be attributed solely to low sulfate levels. For example, when D. vulgaris grown in sulfate-free medium either with pyruvate as a fermentative substrate or with lactate as the electron donor in coculture with Methanospirillum hungatei (DSM864), dsrA levels were low (1.06·104 ± 2,000 and 8.6·104 ± 8,000 transcripts per nanogram of RNA, respectively).
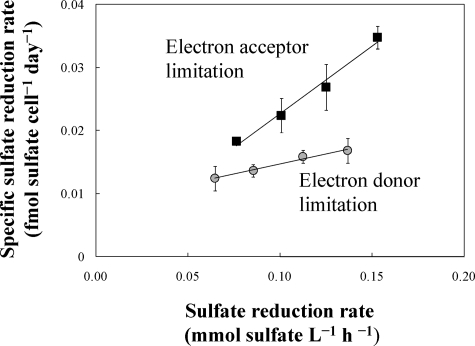
A comparison of specific sulfate reduction rates and bulk sulfate reduction rates in the electron donor-limited and electron acceptor-limited chemostats demonstrated that electron acceptor-limited cells consistently have a higher respiration rate per cell (Fig. 3). This is consistent with an apparently higher maintenance energy requirement (calculated as previously described by Esteve-Nuñez et al. [10]) under electron acceptor-limited conditions (6.3 mmol electrons per gram dry weight per h) than under electron donor-limited conditions (2.7 mmol electrons per gram per h). Higher expression of respiratory genes under electron acceptor-limiting versus electron donor-limiting conditions was previously noted in chemostat cultures of Geobacter sulfurreducens grown with either fumarate or Fe(III) as the electron acceptor (4).
FIG. 3.
Relationship between the specific sulfate reduction rate (femtomol sulfate consumed per cell and day) and the bulk sulfate reduction rate (mmol sulfate consumed per liter and h) in D. vulgaris continuous culture under electron donor (gray circles; R2 = 0.9894) and acceptor (black squares; R2 = 0.983) limitation. Each point is the average of the results for three samples from triplicate continuous cultures at each growth rate.
Implications.
These results suggest that the level of dsrA transcripts in an environmental sample cannot be expected to be related to the bulk rates of sulfate reduction because the relationship between dsrA transcript levels and the rates of sulfate reduction can be influenced by the metabolic state of the cell. This may explain why in a previous study (5) there was a good correlation between dsrA transcript levels and sulfate reduction rates at comparable nonlimiting sulfate concentrations, but dsrA levels continued to increase as sulfate concentrations declined even though sulfate reduction rates decreased. The inability of dsrA transcript levels to serve as a proxy for bulk sulfate reduction rates may not be a significant limitation for the study of sulfate reduction in most environments because several other more straightforward methods, such as monitoring the reduction of [35S]sulfate to [35S]sulfide (11), are available for estimating bulk sulfate reduction rates.
However, the results from this pure culture study suggest that dsrA transcript levels can provide insight into the metabolic state of sulfate reducers because dsrA levels are related to the rates of sulfate reduction per cell. This basic information on the metabolic state of sulfate reducers cannot readily be determined with any other method currently applicable to sedimentary environments. For studies with natural communities, the normalization of dsrA transcript levels to total RNA levels, as used here for pure cultures, might not be appropriate because many organisms will contribute to the RNA pool. One solution that has proven successful in diagnosing the metabolic status of subsurface Fe(III)-reducing communities is to normalize to the transcript levels of a housekeeping gene sequence specific for the organisms under study (16, 17). Thus, further studies are warranted to determine whether the relationship between dsrA expression and cell-specific rates of sulfate reduction observed here in pure culture holds for diverse communities typically found in sedimentary environments.
Acknowledgments
This research was supported by Office of Naval Research award no. N00014-03-1-0315 and Office of Science (BER), U.S. Department of Energy, cooperative agreement no. DE-FC02-02ER63446.
Footnotes
Published ahead of print on 25 July 2008.
REFERENCES
- 1.Anderson, R. T., and D. R. Lovley. 2000. Anaerobic bioremediation of benzene under sulfate-reducing conditions in a petroleum-contaminated aquifer. Environ. Sci. Technol. 34:2261-2266. [Google Scholar]
- 2.Annweiler, E., A. Materna, M. Safinowski, A. Kappler, H. H. Richnow, W. Michaelis, and R. U. Meckenstock. 2000. Anaerobic degradation of 2-methylnaphthalene by a sulfate-reducing enrichment culture. Appl. Environ. Microbiol. 66:5329-5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, B. V., L. C. Schiung, and S. Y. Yuan. 2002. Anaerobic biodegradation of polycyclic aromatic hydrocarbons in soil. Chemosphere 48:717-724. [DOI] [PubMed] [Google Scholar]
- 4.Chin, K. J., A. Esteve-Núñez, C. Leang, and D. R. Lovley. 2004. Direct correlation between rates of anaerobic respiration and levels of mRNA for key respiratory genes in Geobacter sulfurreducens. Appl. Environ. Microbiol. 70:5183-5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chin, K. J., M. L. Sharma, L. A. Russell, K. R. O'Neill, and D. R. Lovley. 2008. Quantifying expression of a dissimilatory (bi)sulfite reductase gene in petroleum-contaminated marine harbor sediments. Microb. Ecol. 55:489-499. [DOI] [PubMed] [Google Scholar]
- 6.Coates, J. D., J. C. Woodward, J. Allen, P. Philp, and D. R. Lovley. 1997. Anaerobic degradation of polycyclic aromatic hydrocarbons and alkanes in petroleum-contaminated marine harbor sediments. Appl. Environ. Microbiol. 63:3589-3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cord-Ruwisch, R. 1985. A quick method for determination of dissolved and precipitated sulfides in cultures of sulfate-reducing bacteria. J. Microbiol. Methods 4:33-36. [Google Scholar]
- 8.Dinh, H. T., J. Kuever, M. Mussmann, A. W. Hassel, M. Stratmann, and F. Widdel. 2004. Iron corrosion by novel anaerobic microorganisms. Nature 427:829-832. [DOI] [PubMed] [Google Scholar]
- 9.Ensley, B. D., and J. M. Suflita. 1995. Metabolism of environmental contaminants by mixed and pure cultures of sulfate-reducing bacteria, p. 293-332. In L. L. Barton (ed.), Sulfate-reducing bacteria. Biotechnology handbooks, vol. 8. Plenum Press, New York, NY. [Google Scholar]
- 10.Esteve-Núñez, A., M. M. Rothermich, M. Sharma, and D. R. Lovley. 2005. Growth of Geobacter sulfurreducens under nutrient-limiting conditions in continuous culture. Environ. Microbiol. 7:641-648. [DOI] [PubMed] [Google Scholar]
- 11.Fossing, H., and B. B. Jørgensen. 1989. Measurement of bacterial sulfate reduction in sediments: evaluation of a single-step chromium reduction method. Biogeochemistry 8:205-222. [Google Scholar]
- 12.Galushko, A., D. Minz, B. Schink, and F. Widdel. 1999. Anaerobic degradation of naphthalene by a pure culture of a novel type of marine sulphate-reducing bacterium. Environ. Microbiol. 1:415-420. [DOI] [PubMed] [Google Scholar]
- 13.Gibson, J., and C. S. Harwood. 2002. Metabolic diversity in aromatic compound utilization by anaerobic microorganisms. Annu. Rev. Microbiol. 56:345-369. [DOI] [PubMed] [Google Scholar]
- 14.Hamilton, W. A. 1985. Sulphate-reducing bacteria and anaerobic corrosion. Annu. Rev. Microbiol. 39:195-217. [DOI] [PubMed] [Google Scholar]
- 15.Hayes, L., and D. R. Lovley. 2002. Quantifying expression of dissimilatory sulfite reductase as an estimate of the rate of sulfate reduction, abstr. N-95. Abstr. 102nd Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC.
- 16.Holmes, D. E., K. P. Nevin, and D. R. Lovley. 2004. In situ expression of nifD in Geobacteraceae in subsurface sediments. Appl. Environ. Microbiol. 70:7251-7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holmes, D. E., K. P. Nevin, R. A. O'Neil, J. E. Ward, L. A. Adams, T. L. Woodard, H. A. Vrionis, and D. R. Lovley. 2005. Potential for quantifying expression of the Geobacteraceae citrate synthase gene to assess the activity of Geobacteraceae in the subsurface and on current-harvesting electrodes. Appl. Environ. Microbiol. 71:6870-6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karkhoff-Schweizer, R. R., D. P. Huber, and G. Voordouw. 1995. Conservation of the genes for dissimilatory sulfite reductase from Desulfovibrio vulgaris and Archaeoglobus fulgidus allows their detection by PCR. Appl. Environ. Microbiol. 61:290-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klein, M., M. Friedrich, A. J. Roger, P. Hugenholtz, S. Fishbain, H. Abicht, L. L. Blackall, D. A. Stahl, and M. Wagner. 2001. Multiple lateral transfers of dissimilatory sulfite reductase genes between major lineages of sulfate-reducing prokaryotes. J. Bacteriol. 183:6028-6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, P. K. H., D. R. Johnson, V. F. Holmes, J. He, and L. Alvarez-Cohen. 2006. Reductive dehalogenase gene expression as a biomarker for physiological activity of Dehalococcoides spp. Appl. Environ. Microbiol. 72:6161-6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lovley, D. R., R. C. Greening, and J. G. Ferry. 1984. Rapidly growing rumen methanogenic organism that synthesizes coenzyme M and has a high affinity for formate. Appl. Environ. Microbiol. 48:81-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lovley, D. R., and E. J. P. Phillips. 1994. Novel processes for anaerobic sulfate production from elemental sulfur by sulfate-reducing bacteria. Appl. Environ. Microbiol. 60:2394-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lovley, D. R., J. D. Coates, J. C. Woodward, and E. J. P. Phillips. 1995. Benzene oxidation coupled to sulfate reduction. Appl. Environ. Microbiol. 61:953-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lovley, D. R. 2003. Cleaning up with genomics: applying molecular biology to bioremediation. Nat. Rev. Microbiol. 1:35-44. [DOI] [PubMed] [Google Scholar]
- 25.Meckenstock, R. U., E. Annweiler, W. Michaelis, H. H. Richnow, and B. Schink. 2000. Anaerobic naphthalene degradation by a sulfate-reducing enrichment culture. Appl. Environ. Microbiol. 66:2743-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neretin, L. N., A. Schippers, A. Pernthaler, K. Hamann, R. Amann, and B. B. Jørgensen. 2003. Quantification of dissimilatory (bi)sulphite reductase gene expression in Desulfobacterium autotrophicum using real-time RT-PCR. Environ. Microbiol. 5:660-671. [DOI] [PubMed] [Google Scholar]
- 27.Rapposch, S., P. Zangerl, and W. Ginzinger. 2000. Influence of fluorescence of bacteria stained with acridine orange on the enumeration of microorganisms in raw milk. J. Dairy Sci. 83:2753-2758. [DOI] [PubMed] [Google Scholar]
- 28.Ravenschlag, K., K. Sahm, C. Knoblauch, B. B. Jørgensen, and R. Amann. 2000. Community structure, cellular rRNA content, and activity of sulfate-reducing bacteria in marine Arctic sediments. Appl. Environ. Microbiol. 66:3592-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rothermich, M. M., L. A. Hayes, and D. R. Lovley. 2002. Anaerobic, sulfate-dependent degradation of polycyclic aromatic hydrocarbons in petroleum-contaminated harbor sediment. Environ. Sci. Technol. 36:4811-4817. [DOI] [PubMed] [Google Scholar]
- 30.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 31.Wagner, M., A. J. Roger, J. L. Flax, G. A. Brusseau, and D. A. Stahl. 1998. Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J. Bacteriol. 180:2975-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Widdel, F., and T. A. Hansen. 1999. The dissimilatory sulfate- and sulfur-reducing bacteria. In M. Dworkin, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes: an evolving electronic database for the microbiological community, 3rd ed., release 3.0. Springer-Verlag, New York, NY.