Abstract
There is considerable heterogeneity among the Shiga toxin type 2 (Stx2) toxins elaborated by Shiga toxin-producing Escherichia coli (STEC). One such Stx2 variant, the Stx2d mucus-activatable toxin (Stx2dact), is rendered more toxic by the action of elastase present in intestinal mucus, which cleaves the last two amino acids of the A2 portion of the toxin A subunit. We screened 153 STEC isolates from food, animals, and humans for the gene encoding Stx2dact by using a novel one-step PCR procedure. This method targeted the region of stx2dact that encodes the elastase recognition site. The presence of stx2dact was confirmed by DNA sequencing of the complete toxin genes. Seven STEC isolates from cows (four isolates), meat (two isolates), and a human (one isolate) that carried the putative stx2dact gene were identified; all were eae negative, and none was the O157:H7 serotype. Three of the isolates (CVM9322, CVM9557, and CVM9584) also carried stx1, two (P1332 and P1334) carried stx1 and stx2c, and one (CL-15) carried stx2c. One isolate, P1130, harbored only stx2dact. The Vero cell cytotoxicities of supernatants from P1130 and stx1 deletion mutants of CVM9322, CVM9557, and CVM9584 were increased 13- to 30-fold after treatment with porcine elastase. Thus, Stx2dact-producing strains, as detected by our one-step PCR method, can be isolated not only from humans, as previously documented, but also from food and animals. The latter finding has important public health implications based on a recent report from Europe of a link between disease severity and infection with STEC isolates that produce Stx2dact.
Shiga toxin-producing Escherichia coli (STEC) strains have been considered significant food-borne pathogens since 1982, when E. coli O157:H7 was first associated with outbreaks of hemorrhagic colitis (24). These pathogens can cause severe clinical manifestations ranging from mild to bloody diarrhea and in rare cases (5%) postinfection hemolytic-uremic syndrome. A variety of foods have been implicated as vehicles in the transmission of STEC, including ground beef, sprouts, dairy products, unpasteurized apple juice, and sausage (1, 3, 8, 18, 21). Studies have shown that Shiga toxins 1 (Stx1) and 2 (Stx2) play an important role in bloody diarrhea and hemolytic-uremic syndrome (7). Stx1 is highly conserved, although two sequence variants have been reported (6, 32). In contrast, the Stx2 group is quite heterogeneous and is comprised of a growing list of variants. The Stx2 variants reported to date include Stx2c (Stx2v-a), Stx2d (Stx2d-OX3a and Stx2d-Ount), activatable Stx2d (Stx2dact) (Stx2vh-a and Stx2vh-b), Stx2e, Stx2f, and Stx2g (14, 19, 25, 26). The variant toxins exhibit strong homology to Stx2 but differ in biological activities, such as relative cytotoxicities for Vero and HeLa cells. Stx2d activatable toxin was originally detected in STEC strain B2F1, an O91:H21 strain that has two toxin alleles, stx2d1 and stx2d2 (formerly designated vh-a and vh-b [28]). This toxin can be rendered more active by treatment with intestinal mucus. Elastase found in mucus cleaves two amino acids from the C terminus of the Stx2d A2 peptide. This modification results in “activation” or an increase in the cytotoxicity of Stx2dact of up to 1,000-fold (16, 19). The enhanced toxicity is directly linked to the fact that the StxA subunit of the activatable toxin has two amino acid substitutions at Ser291 and Glu297 compared to the prototypical sequence of Stx2; Ser291 and Glu297 may serve as recognition motifs for elastase. Bielaszewska and colleagues proposed that Stx2dact may be more pathogenic to humans than other Shiga toxins due to elastase activation in vivo (5).
The true incidence of STEC strains that produce Stx2dact is unclear because Stx2dact is not distinguishable from Stx2 or Stx2c immunologically, and some strategies to detect the Stx2d activatable genotype do not distinguish between Stx2dact and Stx2c (29). PCR and restriction fragment length polymorphism (RFLP) analysis have been used to discriminate among the Stx2 variant genotypes (2, 10, 15). Some confusion resulted from the identification by Piérard et al. (23) of a divergent Stx2-like toxin produced by E. coli strain EH250 that was designated sequence variant VT-2d (verocytotoxin 2d). This toxin (also known as Stx2d) is not mucus activated (unpublished observations). Consequently, the low percentage of human-pathogenic isolates reported to produce Stx2d (23) likely does not represent the incidence and pathogenic niche of strains that produce the Stx2d activatable toxin variant. Indeed, Gobius et al. (13) subsequently demonstrated by using toxin gene sequencing that the stx2dact genotype is relatively common in isolates from sheep and cattle in Australia, and later Vu-Khac and Cornick found that this genotype occurs in Vietnam (30).
In the present study, a novel one-step PCR method specific for stx2dact detection was developed to screen for the Stx2dact gene in a collection of 153 STEC isolates derived from food, cattle, and human patients. These isolates were previously described as Stx2 producers. Isolates containing stx2dact were then further characterized to determine their Vero cytotoxicity and the activation phenotype.
MATERIALS AND METHODS
Bacterial strains.
A total of 153 STEC isolates that were recovered from animals, humans, and foods and were stored at the University of Maryland and the Center for Veterinary Medicine of the U.S. Food and Drug Administration were selected for this study based on prior indications that they possessed stx2 genes. This collection included 135 serotype O157 isolates, 13 O111 isolates, and one isolate each belonging to serotypes O22:H8, O46:H38, O91:H21, O103, and O113:K75:H21. Virulence determinants of these isolates, including the stx1, stx2, and eae genes, were identified by PCR assays as previously described (20).
DNA sequencing of Stx2 operons.
The Stx2 genes of the 153 STEC isolates were amplified and sequenced. Briefly, chromosomal DNA was prepared using a Wizard genomic DNA purification kit (Promega, Madison, WI) according to the manufacturer's instructions. The entire stx2 operon was amplified by PCR with two pairs of primers, primers Stx2F-21328 and Stx2R-22087 and primers Stx2F-22017 and Stx2R-22711 (Table 1). Both strands of each amplicon were sequenced at the Biotechnology Center of the University of Maryland. The stx2 sequences were analyzed with Sequencher 4.0 software (Gene Codes Corporation, Ann Arbor, MI). Nucleotide sequence alignment was performed using the BLAST program (http://www.ncbi.nlm.nih.gov/BLAST/). Predicted amino acid alignments were prepared with the CLUSTALW program (version 1.8; http://www.ebi.ac.uk/clustalw/) and the BOXSHADE program (version 3.21; http://www.ch.embnet.org/software/BOX_form.html).
TABLE 1.
Primers used in PCR assays to identify stx1, stx2, stx2d, and eae in STEC
| Target | Primer | Sequence (5′-3′) | Annealing temp (°C) | Product size (bp) |
|---|---|---|---|---|
| stx1 | VT1-fa | TGTAACTGGAAAGGTGGAGTATACA | 60 | 210 |
| VT1-ra | GCTATTCTGAGTCAACGAAAAATAAC | |||
| stx2 | VT2-fa | GTTTTTCTTCGGTATCCTATTCC | 60 | 484 |
| VT2-ra | GATGCATCTCTGGTCATTGTATTAC | |||
| stx2 | Stx2F-21328 | TTCTGAGCAATCGGTCACTG | 60 | 779 |
| Stx2R-22087 | CGGCGTCATCGTATACACAG | |||
| stx2 | Stx2F-22017 | GTCACAGCAGAAGCCTTACG | 60 | 714 |
| Stx2R-22711 | ACCCACATACCACGAATCAG | |||
| stx2d | Stx2d-activatable | CTTTATATACAACGGGTG | 54 | 359 |
| CKS2 | CTGAATTGTGACACAGATTAC | |||
| stx2/2c | Stx2/2c | TTTTATATACAACGGGTA | 54 | 359 |
| eae | EAE-fa | GACTGTCGATGCATCAGGCAAAG | 54 | 366 |
| EAE-ra | TTGGAGTATTAACATTAACCCCAGG | |||
| stx1 inactivation | stx1-KP1 | ACAGCTGAAGCTTTACGTTTTCGGCAAATACAGAGGGTGTAGGCTGGAGCTGCTTCG | 55 | 1,600 |
| stx1-KP2 | GTAATTTGCGCACTGAGAAGAAGAGACTGAAGATTCCATCTCCATATGAATATCCTCCTTAG | |||
| stx1-F | TAATCCCACGGACTCTTCC | 54 | ||
| stx1-R | GCGATGTTACGGTTTGTTACTG | |||
| V-stx1-F | TAATGTGGTTGCGAAGGAAT | 54 | 1,071 | |
| k1 | CAGTCATAGCCGAATAGCCT | |||
| k2 | CGGTGCCCTGAATGAACTGC | 54 | 928 | |
| V-stx1-R | CGCACTGAGAAGAAGAGACTG | |||
| u49-xbaI | TCATCTAGAGTGGTTGCGAAGGAATTTACC | 56 | 1,168 | |
| L1188-xbaI | TAATCTAGATGAATCCCCCTCCATTATGA | |||
| rpu2 | CTGGATGATCTCAGTGGGCGTTCTTATGT | 58 | 4,159 | |
| rpl4132 | AGACTGCGTCAGTGAGGTTCCACTATGC | |||
| pDS132-F | CCTGTTTGAAGATGGCAAG | 50 | 1,708 | |
| pDS132-R | GGATGTAACGCACTGAGAAG |
Data from reference 20.
PCR screening for the stx2dact variant.
An stx2dact-specific forward 21-mer primer homologous to the sequence that encodes the region of the A2 peptide recognized by elastase (Table 1) was used with CKS2 (28), a primer homologous to the sequence encoding a conserved Stx2 motif 3′of the B-subunit-encoding region. The presence of stx2 or stx2c was determined by PCR using a similar strategy, in which the forward primer was homologous to the sequence in the Stx2 and Stx2c toxin genes that differs from the elastase recognition-encoding sequence in stx2dact (Stx2/2c; 5′-TTTTATATACAACGGGTA-3′). The PCR amplification conditions were as follows. Briefly, template DNA (1 μl) was added to 24 μl of a reaction mixture that contained 0.1 mmol of each deoxyribonucleotide, 10 mmol of Tris-HCl (pH 8.3), 50 mmol of KCl, 10 mmol of Na2EDTA, 2 mmol of MgCl2, and 1.0 U of AmpliTaq Gold DNA polymerase (Applied Biosystems, Branchburg, NJ). The PCR was performed using a GeneAmp PCR system 9700 thermal cycler (Perkin-Elmer, Foster City, CA). The program consisted of an initial template denaturation step of 94°C for 10 min, followed by 30 cycles of denaturation at 94°C for 1 min, primer annealing at 54°C for 1 min, and primer extension at 72°C for 1 min. The final extension step consisted of 72°C for 10 min. Amplified products were electrophoresed in 2% agarose gels that contained 0.5 μg ml ethidium bromide. The controls included stx2-positive E. coli O157:H7 strain UMD8 recovered from a ground beef sample from an outbreak in 1993 and stx2dact-positive strain CL-15. This method was further evaluated by testing 153 STEC isolates.
Southern hybridization for detection of stx2dact.
A Southern hybridization analysis was performed to determine the number of stx2-like sequences in strains containing stx2dact. Genomic DNA was extracted from stx2dact-containing bacterial strains by the sodium dodecyl sulfate-cetyltrimethylammonium bromide-proteinase K method (31), digested with PstI and EcoRI, separated by electrophoresis on a 0.7% (wt/vol) agarose gel, and transferred to a positively charged Hybond nylon membrane (GE Healthcare, Buckinghamshire, United Kingdom) by capillary blotting. A 359-bp fragment from stx2dact was amplified by PCR with primers Stx2d-activatable and CKS2 and then used as a hybridization probe. The probe was labeled with alkaline phosphatase according to the instructions of the AlkPhos direct labeling and detection system with CDP-Star (Amersham Pharmacia Biotech, Buckinghamshire, England). The membrane was hybridized with the probe and washed according to the manufacturer's instructions. The hybridization signals were detected with an AFP mini medical imaging analyzer (AFP Imaging, New York, NY). For the strains for which two Stx2 types were detected by Southern hybridization, further identification was performed using PCR-restriction fragment length polymorphism analysis with PstI digestion.
Construction of Stx1 null mutants.
The Stx1 operon in CVM9322 was inactivated by lambda red recombinase-mediated allelic exchange as described by Datsenko and Wanner (9) to create a Δstx1 deletion mutant of CVM9322. Briefly, the kanamycin resistance (Km) cassette of plasmid pKD4 was PCR amplified using oligonucleotides stx1-KP1 and stx1-KP2 (Table 1). These oligonucleotides were 62-mers in which the 5′ 40 bases were complementary to regions inside stx1, followed 3′ by 22 bases that flanked the Km cassette open reading frame. The resulting 1.6-kb amplicon was used to transform CVM9332 (Table 1) via electroporation. Replacement of the native Stx1 operon with the Km cassette amplicon was facilitated by transformation of CVM9332 with the low-copy-number plasmid pKD46, which expresses the lambda red recombinase (and contains an ampicillin resistance marker). Kanamycin-resistant clones were then cured of the pKD46 plasmid by repeated growth at 37°C in the absence of ampicillin. Replacement of the Stx1 gene with the Km cassette was verified by PCR using three pairs of primers, stx1-F and stx1-R, V-stx1-F and k1, and k2 and V-stx1-R (Table 1). One positive clone for transformation was selected for further study.
Δstx1 deletion mutants of CVM9557 and CVM9584 were constructed using a suicide plasmid-mediated approach described previously, with modifications (22). Briefly, stx1 was amplified using primers u49-xbaI and L1188-xbaI and ligated into the pGEM-T Easy vector (Promega). A mutant stx1 allele with an 84-bp deletion (stx1Δ84) was obtained by reverse PCR using primers rpu2 and rpl4132. The stx1Δ84 allele was treated with XbaI and cloned into the corresponding sites in pDS132 to obtain pDS132/stx1. This plasmid was used to transform E. coli DH5α λpir via electroporation. The insertion was confirmed by PCR using primers pDS132-F and pDS132-R. The pDS132/stx1 construct was then introduced into CVM9557 and CVM9584 via electroporation. Chloramphenicol-resistant transformants were selected and grown in the absence of chloramphenicol to allow for loss of the pDS132 plasmid. The transformants that had undergone the second recombination event were identified as chloramphenicol-sensitive derivatives capable of growth on LB agar with 5% sucrose and without NaCl. Chromosomal mutations were confirmed by PCR using primers u49 and L1188.
Vero cell cytotoxicity assay.
Seven stx2dact-positive STEC isolates and nine representative E. coli O157:H7 isolates were selected and tested for cytotoxicity. E. coli K-12, a nontoxic laboratory strain, was used as the negative control. Preparations were obtained by using Vero cells and the method of Gentry and Dalrymple (12). Briefly, Vero cells suspended in tissue culture medium (Dulbecco modified Eagle medium containing 10% fetal bovine serum) were seeded into wells (approximately104 cells/well) of a 96-well microtiter plate, leaving the exterior rows empty, and incubated for 24 h at 37°C in the presence of 5% CO2. The tissue culture medium was removed by aspiration and replaced with 100 μl fresh medium. On the other hand, a single colony of each bacterial strain was removed, inoculated into 5 ml LB broth, and shaken overnight at 37°C. After the cell concentration was adjusted to 1 × 109 CFU/ml, bacterial cultures were centrifuged and filtered with a 0.45-μm-pore-size filter. Filter-sterilized bacterial culture supernatants (100 μl) were then added to the first row of wells. Serial dilutions (1:5) of each supernatant were prepared in 96-well microtiter plates using tissue culture medium. The last row was not inoculated and served as a control for unintoxicated cell background. After incubation for 48 h, the cells were fixed in buffered formalin and stained with crystal violet. The intensity of the color of the fixed and stained cells was measured with an Elx800 microplate reader (Biotek, Winooski, VT) at 600 nm (A600). The staining intensity was proportional to the number of viable, attached tissue culture cells present before they were fixed to the well. The 50% cytotoxic dose (CD50) was the amount of toxin required to kill 50% of the cells in a well. The CD50 for each bacterial strain was determined by plotting the optical density at 600 nm of each dilution well after subtraction of the optical density at 600 nm for the blank against the log-transformed toxin dilution (12) using Originpro7.5 software (OriginLab, Northampton, MA). The data shown below are the averages of three cytotoxicity assays.
Activation assay.
STEC isolates possessing only stx2dact were also tested for toxin activation by elastase (16). Bacterial culture supernatants were incubated with commercial porcine pancreatic elastase with a specific activity of 45 U/mg protein (Calbiochem, San Diego, CA) at a final concentration of 1 mg/ml for 1 to 2 h at 37°C in 50 mM potassium phosphate buffer. The CD50 of the treated toxin samples were determined using Vero cells as described above. As a control, a separate sample of each preparation was treated with the buffer and incubated in the same manner. The level of activation of cytotoxicity by various samples was determined by dividing the cytotoxicity of the elastase-treated sample by the cytotoxicity of the buffer-treated sample. The data shown below are the averages of at least three activation assays.
Nucleotide sequence accession numbers.
The sequences of stx2dact subunit A and subunit B from strains P1332, P1334, P1330, CL-15, CVM9322, CVM9557, and CVM9584 have been deposited in the GenBank database under accession numbers EU816436 to EU816449.
RESULTS
DNA sequencing of stx2.
To determine the prevalence of Stx2 variant toxins in a library of previously identified STEC isolates, PCR products derived with primers known to amplify the entire stx2, stx2c, and stx2dact operons were sequenced. Of the153 STEC isolates tested, 137 contained sequences homologous to the prototypical sequence of stx2 and 16 isolates contained variant stx2 operons. The predicted amino acid sequences of the StxA2 subunit showed that seven of the latter isolates had carboxy-terminal modifications typical of Stx2act, whereas the remaining nine isolates had amino acid sequences characteristic of Stx2c. Comparison of the predicted StxA2 subunit amino acid sequences from these STEC isolates with Stx2dact sequences from O91:H21 strain B2F1 (28) showed that seven isolates encoded an StxA2 subunit with Phe313Ser and Lys319Glu amino acid substitutions, which is characteristic of activatable Stx2d (Fig. 1). Comparison of the predicted amino acid sequences of the B subunits with Stx2d and Stx2c sequences from O91:H21 strain B2F1 and O157:H- strain E32511 (26), respectively, showed that all 16 isolates had Asp35Asn and Asp43Ala substitutions characteristic of both the StxB2dact and StxB2c subunits. The nucleotide sequences of the stx2 genes of P1334, P1330, CL-15, CVM9322, CVM9557, and CVM9584 were identical to that of stx2d2. The nucleotide sequence of the stx2 variant from P1332 differed by one nucleotide from the sequence of stx2d2. Nine of the STEC isolates with Stx2 variant genotypes were O157:H7 isolates that contained the stx2c variant. None of the stx2dact variants were found in O157 isolates (Fig. 1).
FIG. 1.
Alignment of predicted amino acid sequences of the A and B subunits of Stx2 variants with the published sequences of Stx2dact of STEC strain B2F1 (Stx2d2) (GenBank accession no. AF479829) and Stx2 (GenBank accession no. AB035143). Dots indicate residues identical to those in Stx2dact. Residues Ser313 and Glu319 in the A subunit and residues Asn35 and Asp43 in the B subunit, which distinguish Stx2d from Stx2, are indicated by bold type and underlining.
stx2dact detection by PCR.
An stx2dact-specific PCR detection method was also used to screen the STEC library for stxA2 genotypes consistent with the toxin activation phenotype. In this analysis, subsequent restriction enzyme digestion and RFLP comparison were not necessary to distinguish stx2dact from stx2 or stx2c; however, the method did not discriminate between the stx2 and stx2c genotypes. Three stx2dact-positive variants identified by PCR screening of the STEC isolates were confirmed by sequencing to have the activatable genotype. No false positives were identified by PCR. The PCR method was further evaluated by testing 14 other non-O157 STEC isolates suspected of containing stx2, and four stx2dact variants were identified in four of these additional isolates. The amino acid substitutions in these four isolates were further confirmed by stx2 sequencing (Fig. 1). P1332 possessed a new stx2dact sequence that has not been reported previously.
Additional characteristics of the 16 STEC isolates carrying Stx2 variants.
The seven stx2dact-positive isolates belonged to six different serotypes (Table 2). The remaining nine STEC isolates were all serotype O157:H7, and most of them carried both the stx1 and stx2c genes; the one exception was isolate CVM3961, which was stx1 negative. It is also notable that the stx2dact-positive isolates were all eae negative. In addition, three of the E. coli O157:H7 isolates lacked the intimin gene.
TABLE 2.
Characteristics of 16 STEC isolates possessing stx2 variants
| Isolate | Serotype | Genotype
|
Source | |||
|---|---|---|---|---|---|---|
| stx2dact | stx2c | stx1 | eae | |||
| P1332 | O46:H38 | + | + | + | − | Beef |
| P1334 | O91:H21 | + | + | + | − | Cow |
| P1330 | O22:H8 | + | − | − | − | Ground beef |
| CL-15 | O113:K75:H21 | + | + | − | − | Human |
| CVM9322 | O103 | + | − | + | − | Cow |
| CVM9557 | O111 | + | − | + | − | Cow |
| CVM9584 | O111 | + | − | + | − | Cow |
| CVM3605 | O157:H7 | − | + | + | − | Food |
| CVM3676 | O157:H7 | − | + | + | − | Cow |
| CVM3880 | O157:H7 | − | + | + | − | Cow |
| CVM3961 | O157:H7 | − | + | − | + | Cow |
| CVM3939 | O157:H7 | − | + | + | + | Cow |
| CVM3541 | O157:H7 | − | + | + | + | Cow |
| CVM3639 | O157:H7 | − | + | + | + | Cow |
| CVM3756 | O157:H7 | − | + | + | + | Human |
| CVM3962 | O157:H7 | − | + | + | + | Cow |
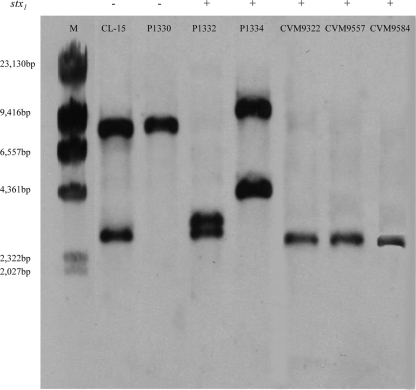
A Southern blot hybridization analysis was done to determine the number of copies of stx2 or its variant genes in the isolates (Fig. 2). Chromosomal DNA from the seven isolates that produced Stx2d activatable toxin were digested with EcoRI and PstI and probed with an stx2dact gene fragment. The probe was a 359-bp PCR-derived DNA product amplified with primers Stx2d-activatable and CKS2. It spanned the region from the 3′ end of the A subunit-encoding region through the B subunit open reading frame, a sequence that is 97% homologous to the corresponding region in stx2; therefore, hybridization should have occurred with stx2, stx2c, or stx2dact. The DNA probe hybridized with two bands in CL-15, P1332, and P1334. Amplification of the complete stx2-like toxin genes from these isolates followed by PstI digestion of the product and RFLP analysis showed that in addition to stx2dact Stx2c was also present in these isolates (data not shown). The remaining isolates produced only one band, a finding that indicated that they contained only one allele encoding the Stx2d activatable toxin.
FIG. 2.
Hybridization of EcoRI- and PstI-digested genomic DNA from Stx2d activatable toxin-producing isolates with the stx2dact probe. Lane M contained a molecular weight marker (HindIII-digested λ DNA ladder; New England Biolabs). The remaining lanes contained isolates CL-15 (serotype O113:K75:H21; stx2d+ stx1), P1330 (serotype O22:H8; stx2d+ stx1), P1332 (serotype O46:H38; stx2d+ stx1+), P1334 (serotype O91:H21; stx2d+ stx1+), CVM9322 (serotype O103; stx2d+ stx1+), CVM9557 (serotype O111; stx2d+ stx1+), and CVM9584 (serotype O111; stx2d+ stx1+) (the presence of the stx1 and stx2d genes was determined by PCR).
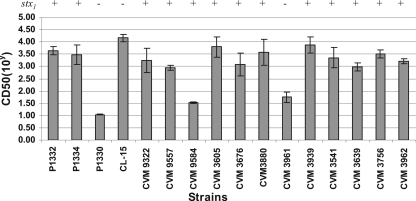
Cytotoxicity.
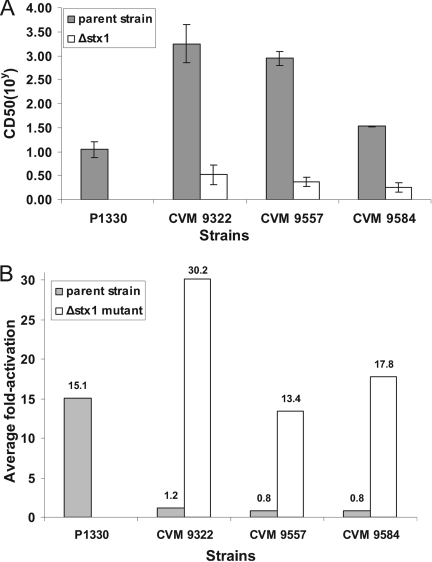
The Vero cell cytotoxicity of selected isolates was analyzed using a microtiter plate cytotoxicity assay. The supernatants of all isolates were cytotoxic to Vero cells, as expected; however, the CD50 of the isolates varied from 101 to 104 cells/ml (Fig. 3). Most of the isolates (13/16) had fairly high CD50, as predicted by the presence of stx1, stx2, or stx2c in their virulence profiles (Fig. 3). It has been shown previously that Stx2dact is less toxic to Vero cells than Stx1 or Stx2 and Stx2c despite the lethality of the isolates in mice in vivo (17). Consistent with these data and our Southern blot data, P1330 had the lowest CD50 (101 cells/ml) and was the only isolate that produced only Stx2dact (Fig. 3 and Fig. 4A). Following deletion of the Stx1 genes in CVM9322, CVM9557, and CVM9584, the cytotoxicities of the strains for Vero cells were reduced 20- to 500-fold, and the residual cytotoxicity was attributable to expression of the remaining single copy of stx2dact in each strain (Fig. 4A).
FIG. 3.
Results of the Vero cell cytotoxicity assay for 16 Stx2-positive E. coli isolates. The bars indicate means of three individual experiments. E. coli K-12, a nontoxic laboratory strain, was used as a negative control and did not induce a measurable response in Vero cells. +, isolates containing stx2c in addition to stx2dact.
FIG. 4.
(A) Vero cell cytotoxicity of STEC isolate P1330, which contained only stx2dact, three STEC parent strains containing a combination of stx1 and stx2dact, and stx1 knockout mutants of these strains before the treatment with elastase. (B) Average activation of cytotoxicity of the strains before and after treatment with elastase. Culture supernatants were incubated with porcine pancreatic elastase for 1 to 2 h at 37°C and transferred to Vero cells to determine the CD50. The level of activation was calculated by dividing the CD50 for each elastase-treated sample by the CD50 for the buffer-treated control. The bars indicate means of three individual experiments.
Activation capacity of stx2d-positive isolates.
Four STEC isolates containing only one copy stx2dact, P1330, CVM9322, CVM9557, and CVM9584, were used to demonstrate that the genes identified by PCR and sequencing as genes encoding the elastase recognition motifs produced functionally activatable toxins. Culture supernatants of the Stx2dact toxin producers were treated with porcine pancreatic elastase. The cytotoxicity to Vero cells of elastase-treated supernatants was compared with that of buffer-treated supernatants. Activation was determined by measuring the increase in cytotoxicity after elastase treatment compared with the cytotoxicity after mock treatment. There were two distinct categories of results (Fig. 4B). For P1330, the average activation detected after porcine elastase treatment was 15.1-fold. In contrast, for the rest of the isolates (CVM9322, CVM9557, and CVM9584), there was no difference with the porcine elastase treatment (Fig. 4B). In contrast to P1330, these isolates also produced type 1 Shiga toxin. We postulated that their high levels of cytotoxicity (Fig. 4A) masked most of the influence of activation of Stx2dact on total cytotoxicity. To test this hypothesis, we inactivated stx1 so that we were able to assess the activation phenotype in the isolates (Fig. 4B). Elastase treatment increased the cytotoxicities of the Δstx1 mutants of CVM9322, CVM9557, and CVM9584 by an average of 13.3- to 30.2-fold.
DISCUSSION
In this study, we analyzed a collection of 153 STEC isolates recovered from food, cattle, and human patients to further characterize the stx2 that they contained. Two of the six main stx2 subtypes were detected (stx2c in nine isolates and stx2dact in seven isolates). Of particular concern to us was identification of the mucus-activatable toxin type because of its potential to become more toxic in the gut and because of evidence from Europe indicating that infection with STEC producing such a toxin results in more severe disease in humans. The seven stx2dact-containing isolates belonged to various serotypes, including O46, O91, O22, O113, O103, and O111, and no stx2dact was found in the O157 serotype isolates. In addition, none of the isolates that harbored stx2dact was shown to also encode intimin. These findings are in agreement with the findings of other investigators, who have observed stx2dact only in non-O157 STEC (4, 11, 13). We speculate that the distribution of stx2dact among non-STEC isolates is indicative of the clonal nature of the emergence of STEC in general. However, because the stx2dact of at least one strain, O91 strain B2F1, is bacteriophage borne, it is possible that this potentially more toxic Stx2 variant could be transduced into an STEC strain that contains eae and has the related locus of enterocyte effacement traits.
The prevalence of stx2dact in STEC serotypes other than O157:H7 is not known and is difficult to assess because current diagnostic procedures may not detect non-O157 (sorbitol-fermenting) isolates. Likewise, toxin detection methods do not distinguish between Stx2 and Stx2 variants. Nonetheless, our study showed that humans may be at risk of acquiring Stx2act-producing isolates from livestock or as food contaminants. By screening with the novel one-step PCR method described in this study, we were able to identify activatable stx2d variants in STEC isolates previously recorded as Stx2 producers. Nucleotide sequencing of the stx2 operons from the seven PCR-identified stx2d isolates further confirmed the presence of the activatable stx2d genotype. Because this method does not require subsequent RFLP analysis, it is a rapid and specific way to detect the Stx2d activatable toxin variant. A drawback of the method is that it does not simultaneously amplify other toxin variant genes. Detection of genes encoding other toxin types would require additional PCRs with alternative primers or restriction enzyme digestion.
Although all of the Stx2 variant-expressing isolates that we characterized were cytotoxic to Vero cells, isolates possessing stx1 or stx2 tended to be more cytotoxic than isolates that contained stx2dact alone (17, 27). We demonstrated that supernatants from four representative isolates that carried stx2dact were functionally activatable with porcine elastase and also demonstrated that coexpression of more highly cytotoxic Stx types, in this case Stx1, masked detection of the activation phenotype in vitro. In conclusion, our studies revealed some additional sequence variability among the Stx2 variant genes. We demonstrated that the activatable toxin variant Stx2dact was present in approximately 5% of the STEC strains, and we describe here a simple and rapid PCR assay to detect the presence of the activatable stx2d genotype, which we believe increases the risk to the health of humans who become infected with such STEC strains.
Acknowledgments
This study was made possible by grants from the Joint Institute for Food Safety and Applied Nutrition (JIFSAN) of the University of Maryland and the U.S. Food and Drug Administration. We also acknowledge funding of the O'Brien laboratory at USUHS through National Institutes of Health National Institute for Allergy and Infectious Diseases grant AI20148-25.
Footnotes
Published ahead of print on 25 July 2008.
REFERENCES
- 1.Ammon, A., L. R. Petersen, and H. Karch. 1999. A large outbreak of hemolytic uremic syndrome caused by an unusual sorbitol-fermenting strain of Escherichia coli O157:H. J. Infect. Dis. 179:1274-1277. [DOI] [PubMed] [Google Scholar]
- 2.Bertin, Y., K. Boukhors, N. Pradel, V. Livrelli, and C. Martin. 2001. Stx2 subtyping of Shiga toxin-producing Escherichia coli isolated from cattle in France: detection of a new Stx2 subtype and correlation with additional virulence factors. J. Clin. Microbiol. 39:3060-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Besser, R. E., S. M. Lett, J. T. Weber, M. P. Doyle, T. J. Barrett, J. G. Wells, and P. M. Griffin. 1993. An outbreak of diarrhea and hemolytic uremic syndrome from Escherichia coli O157:H7 in fresh-pressed apple cider. JAMA 269:2217-2220. [PubMed] [Google Scholar]
- 4.Beutin, L., A. Miko, G. Krause, K. Pries, S. Haby, K. Steege, and N. Albrecht. 2007. Identification of human-pathogenic strains of Shiga toxin-producing Escherichia coli from food by a combination of serotyping and molecular typing of Shiga toxin genes. Appl. Environ. Microbiol. 73:4769-4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bielaszewska, M., A. W. Friedrich, T. Aldick, R. Schurk-Bulgrin, and H. Karch. 2006. Shiga toxin activatable by intestinal mucus in Escherichia coli isolated from humans: predictor for a severe clinical outcome. Clin. Infect. Dis. 43:1160-1167. [DOI] [PubMed] [Google Scholar]
- 6.Burk, C., R. Dietrich, G. Acar, M. Moravek, M. Bulte, and E. Martlbauer. 2003. Identification and characterization of a new variant of Shiga toxin 1 in Escherichia coli ONT:H19 of bovine origin. J. Clin. Microbiol. 41:2106-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butler, T., M. R. Islam, M. A. Azad, and P. K. Jones. 1987. Risk factors for development of hemolytic uremic syndrome during shigellosis. J. Pediatr. 110:894-897. [DOI] [PubMed] [Google Scholar]
- 8.Centers for Disease Control. 1994. Addressing emerging infectious disease threats: a prevention strategy for the United States. Executive summary. MMWR Recommend. Rep. 43:1-18. [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Baets, L., I. Van der Taelen, M. De Filette, D. Pierard, L. Allison, H. De Greve, J. P. Hernalsteens, and H. Imberechts. 2004. Genetic typing of Shiga toxin 2 variants of Escherichia coli by PCR-restriction fragment length polymorphism analysis. Appl. Environ. Microbiol. 70:6309-6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia-Aljaro, C., M. Muniesa, J. E. Blanco, M. Blanco, J. Blanco, J. Jofre, and A. R. Blanch. 2005. Characterization of Shiga toxin-producing Escherichia coli isolated from aquatic environments. FEMS Microbiol. Lett. 246:55-65. [DOI] [PubMed] [Google Scholar]
- 12.Gentry, M. K., and J. M. Dalrymple. 1980. Quantitative microtiter cytotoxicity assay for Shigella toxin. J. Clin. Microbiol. 12:361-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gobius, K. S., G. M. Higgs, and P. M. Desmarchelier. 2003. Presence of activatable Shiga toxin genotype (stx2d) in Shiga-toxigenic Escherichia coli from livestock sources. J. Clin. Microbiol. 41:3777-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito, H., A. Terai, H. Kurazono, Y. Takeda, and M. Nishibuchi. 1990. Cloning and nucleotide sequencing of Vero toxin 2 variant genes from Escherichia coli O91:H21 isolated from a patient with the hemolytic uremic syndrome. Microb. Pathog. 8:47-60. [DOI] [PubMed] [Google Scholar]
- 15.Jelacic, J. K., T. Damrow, G. S. Chen, S. Jelacic, M. Bielaszewska, M. Ciol, H. M. Carvalho, A. R. Melton-Celsa, A. D. O'Brien, and P. I. Tarr. 2003. Shiga toxin-producing Escherichia coli in Montana: bacterial genotypes and clinical profiles. J. Infect. Dis. 188:719-729. [DOI] [PubMed] [Google Scholar]
- 16.Kokai-Kun, J. F., A. R. Melton-Celsa, and A. D. O'Brien. 2000. Elastase in intestinal mucus enhances the cytotoxicity of Shiga toxin type 2d. J. Biol. Chem. 275:3713-3721. [DOI] [PubMed] [Google Scholar]
- 17.Lindgren, S. W., J. E. Samuel, C. K. Schmitt, and A. D. O'Brien. 1994. The specific activities of Shiga-like toxin type II (SLT-II) and SLT-II-related toxins of enterohemorrhagic Escherichia coli differ when measured by Vero cell cytotoxicity but not by mouse lethality. Infect. Immun. 62:623-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacDonald, K. L., M. J. O'Leary, M. L. Cohen, P. Norris, J. G. Wells, E. Noll, J. M. Kobayashi, and P. A. Blake. 1988. Escherichia coli O157:H7, an emerging gastrointestinal pathogen. Results of a one-year, prospective, population-based study. JAMA 259:3567-3570. [PubMed] [Google Scholar]
- 19.Melton-Celsa, A. R., J. F. Kokai-Kun, and A. D. O'Brien. 2002. Activation of Shiga toxin type 2d (Stx2d) by elastase involves cleavage of the C-terminal two amino acids of the A2 peptide in the context of the appropriate B pentamer. Mol. Microbiol. 43:207-215. [DOI] [PubMed] [Google Scholar]
- 20.Meng, J., S. Zhao, and M. P. Doyle. 1998. Virulence genes of Shiga toxin-producing Escherichia coli isolated from food, animals and humans. Int. J. Food Microbiol. 45:229-235. [DOI] [PubMed] [Google Scholar]
- 21.Michino, H., K. Araki, S. Minami, S. Takaya, N. Sakai, M. Miyazaki, A. Ono, and H. Yanagawa. 1999. Massive outbreak of Escherichia coli O157:H7 infection in school children in Sakai City, Japan, associated with consumption of white radish sprouts. Am. J. Epidemiol. 150:787-796. [DOI] [PubMed] [Google Scholar]
- 22.Philippe, N., J. P. Alcaraz, E. Coursange, J. Geiselmann, and D. Schneider. 2004. Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid 51:246-255. [DOI] [PubMed] [Google Scholar]
- 23.Piérard, D., G. Muyldermans, L. Moriau, D. Stevens, and S. Lauwers. 1998. Identification of new verocytotoxin type 2 variant B-subunit genes in human and animal Escherichia coli isolates. J. Clin. Microbiol. 36:3317-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riley, L. W., R. S. Remis, S. D. Helgerson, H. B. McGee, J. G. Wells, B. R. Davis, R. J. Hebert, E. S. Olcott, L. M. Johnson, N. T. Hargrett, P. A. Blake, and M. L. Cohen. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 308:681-685. [DOI] [PubMed] [Google Scholar]
- 25.Schmidt, H., J. Scheef, S. Morabito, A. Caprioli, L. H. Wieler, and H. Karch. 2000. A new Shiga toxin 2 variant (Stx2f) from Escherichia coli isolated from pigeons. Appl. Environ. Microbiol. 66:1205-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmitt, C. K., M. L. McKee, and A. D. O'Brien. 1991. Two copies of Shiga-like toxin II-related genes common in enterohemorrhagic Escherichia coli strains are responsible for the antigenic heterogeneity of the O157:H- strain E32511. Infect. Immun. 59:1065-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strockbine, N. A., L. R. Marques, J. W. Newland, H. W. Smith, R. K. Holmes, and A. D. O'Brien. 1986. Two toxin-converting phages from Escherichia coli O157:H7 strain 933 encode antigenically distinct toxins with similar biologic activities. Infect. Immun. 53:135-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teel, L. D., A. R. Melton-Celsa, C. K. Schmitt, and A. D. O'Brien. 2002. One of two copies of the gene for the activatable Shiga toxin type 2d in Escherichia coli O91:H21 strain B2F1 is associated with an inducible bacteriophage. Infect. Immun. 70:4282-4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tyler, S. D., W. M. Johnson, H. Lior, G. Wang, and K. R. Rozee. 1991. Identification of verotoxin type 2 variant B subunit genes in Escherichia coli by the polymerase chain reaction and restriction fragment length polymorphism analysis. J. Clin. Microbiol. 29:1339-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vu-Khac, H., and N. A. Cornick. 2008. Prevalence and genetic profiles of Shiga toxin-producing Escherichia coli strains isolated from buffaloes, cattle, and goats in central Vietnam. Vet. Microbiol. 126:356-363. [DOI] [PubMed] [Google Scholar]
- 31.Wilson, K. 1990. Preparation of genomic DNA from bacteria, p. 241-245. In F. M. Ausubel and R. Brent (ed.), Current protocols in molecular biology. Greene Publishing Associates and Wiley Interscience, New York, NY. [DOI] [PubMed]
- 32.Zhang, W., M. Bielaszewska, T. Kuczius, and H. Karch. 2002. Identification, characterization, and distribution of a Shiga toxin 1 gene variant (stx1c) in Escherichia coli strains isolated from humans. J. Clin. Microbiol. 40:1441-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]