Abstract
The symbiosis between plant roots and arbuscular mycorrhizal (AM) fungi has been shown to affect both the diversity and productivity of agricultural communities. In this study, we characterized the AM fungal communities of Solanum tuberosum L. (potato) roots and of the bulk soil in two nearby areas of northern Italy, in order to verify if land use practices had selected any particular AM fungus with specificity to potato plants. The AM fungal large-subunit (LSU) rRNA genes were subjected to nested PCR, cloning, sequencing, and phylogenetic analyses. One hundred eighty-three LSU rRNA sequences were analyzed, and eight monophyletic ribotypes, belonging to Glomus groups A and B, were identified. AM fungal communities differed between bulk soil and potato roots, as one AM fungal ribotype, corresponding to Glomus intraradices, was much more frequent in potato roots than in soils (accounting for more than 90% of sequences from potato samples and less than 10% of sequences from soil samples). A semiquantitative heminested PCR with specific primers was used to confirm and quantify the AM fungal abundance observed by cloning. Overall results concerning the biodiversity of AM fungal communities in roots and in bulk soils from the two studied areas suggested that potato roots were preferentially colonized by one AM fungal species, G. intraradices.
Arbuscular mycorrhizal fungi (AMF) are obligate biotrophs, forming mutualistic relationships with a broad range of host plant species (42). Arbuscular mycorrhizal (AM) associations are generally considered to be nonspecific, as the same AMF species is able to colonize the roots of different host plants, which in turn can be colonized by various AMF species (13, 38, 52). However, results from some studies show that preferential associations between plants and AMF may exist (4, 7, 13, 32, 56), and reciprocal interactions between AMF and the plant community, based on reciprocal feedback, have been proposed (2, 3, 4). In particular, differential responses by plant species to individual isolates and species of AMF (43), leading to variations in the plant community composition and productivity and depending on the diversity and identity of the AMF (23, 51), have been observed previously.
Although most plants used in agriculture and horticulture form arbuscular mycorrhizae, agricultural management practices may affect the composition and diversity of AMF communities, as well as spore and mycelium densities, in both temperate and tropical agroecosystems (24). In general, agricultural practices have a negative impact on the AM association (8, 14); AMF abundance is reduced by phosphorus (P) fertilization (14, 33) and by crop cultivation, due mainly to either mechanical disturbance by tillage or a change of host plants in crop rotation systems. Moreover, seasonal variations in the abundance of AMF in agricultural soil have been observed (22).
Few data are available concerning potato plants and AMF. The influence of AMF species on the potato response to P nutrition (25, 26) and the effect on tuber yield and size (9) or on productivity (27) have been analyzed previously. Also, potato plants were used as trap cultures in order to study plants that, in crop rotation, could be used to enhance potato AM fungal inocula (5). However, to our knowledge, no data are available about the host specificities of AM fungal species with regard to potato plants.
In our study, we analyzed an agricultural site renowned for potato and onion production, where potatoes have been grown for about 150 years and a 5-year crop rotation (wheat, potatoes, maize or onion, wheat, and sugar beets) has been applied for at least 40 years. The indigenous AM fungal communities in the soil and in the roots of potatoes were detected by molecular methods and compared, in order to verify if these typical land use practices have selected particular AMF ecotypes with host specificity to potato plants.
MATERIALS AND METHODS
Study site and soil and potato sampling.
The study was performed in an agricultural area around Castelnuovo Scrivia, Italy (geographical position: latitude, 44°58′53"N; longitude, 08°52′56"E; altitude, 85 m above sea level). Considering the geographical features (position relative to rivers and streams and orientation), chemical features (pH, P content, and organic matter content), and pedological features (percentages of silt, clay, and sand) of the territory, two homogenous areas of 40 km2, called Castelnuovo Scrivia North (CSN; pH 7.98; total Pi content, 731 mg/kg; available Pi, 18 mg/kg; organic matter content, 1.56%; silt content, 48.44%; clay content, 23.12%; sand content, 28.44%; U.S. Department of Agriculture classification, loam) and Castelnuovo Scrivia South (CSS; pH 8.00; total Pi content, 758 mg/kg; available Pi, 17 mg/kg; organic matter content, 2.04%; silt content, 56.00%; clay content, 32.85%; sand content, 11.15%; U.S. Department of Agriculture classification, silt-clay-loam), were recognized, and in each of them four fields were selected. In each of these fields, potatoes (Solanum tuberosum L.) were planted in April 2005 at a depth of 15 cm and at a distance of 28 cm from one another in furrows separated by 75 cm. Potato plants and soils were sampled in June 2005. Four potato plants in each field were randomly selected, and their roots were pooled together to obtain a mixed root sample for each area. In each field, four core samples of soil (500 g each) were also collected from the first 20-cm layer. The soil samples were sieved through 4-mm pores to eliminate large particles and homogenized to achieve a pooled sample for every field.
Assessment of root colonization.
Mycorrhizal frequency, the level of mycorrhizal colonization, arbuscule abundance, and arbuscule abundance in the colonized area were evaluated microscopically according to the method of Trouvelot et al. (47). Briefly, 30 randomly chosen 1-cm-long pieces of potato roots were cut, cleared for 20 min at 60°C in 10% KOH, stained with 1% methyl blue in lactic acid, and mounted onto slides. Results were statistically analyzed by analysis of variance followed by Fisher's protected least-significant-difference test, with the cutoff for significance at a P value of 0.05.
DNA extraction from potato roots.
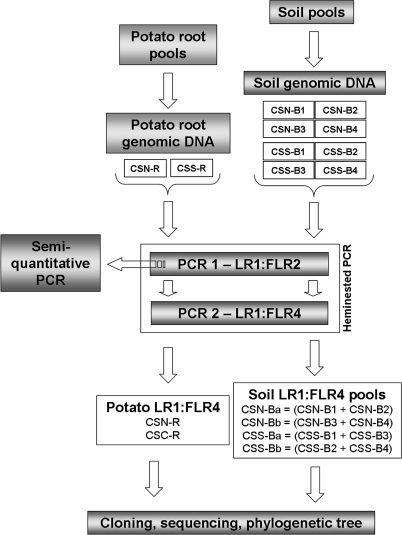
All the procedures concerning DNA extraction and analyses are summarized in Fig. 1. For each area, one root genomic DNA extraction was performed (yielding samples CSN-R and CSS-R) (Fig. 1). The previously pooled root samples were rinsed in water and dried at 50°C overnight before being processed as described by Farmer et al. (11). A 1-g sample of roots was assembled by randomly taking pieces from different parts of the pooled root samples obtained from each field and homogenizing the mixture in liquid nitrogen and then in 8 ml of extraction buffer (0.1 M Tris-HCl [pH 8.0], 0.1 M NaCl, 0.1 M EDTA [pH 8.0], 2% sodium dodecyl sulfate, 1% polyvinyl polypyrrolidone). After 15 min of incubation at 70°C and 10 min of centrifugation at 14,000 × g at 4°C, the supernatant was recovered and mixed with 1/10 (vol/vol) 5 M sodium acetate, pH 5.5. After a second centrifugation (5 min at 14,000 × g and 4°C), DNA was precipitated at −20°C by adding 2 volumes of isopropanol. After centrifugation, the pellet was rinsed with 500 μl of 75% ethanol, dried, resuspended in 50 μl of sterile distilled water, and purified using the Geneclean Turbo kit (Qbiogene).
FIG. 1.
Diagram of work flow to prepare the PCR fragment pools used in this study. To prepare PCR fragments from soil, one aliquot of soil for each of the four selected fields per area was used for genomic DNA extraction. Four genomic DNA samples per area (CSN-B1, CSN-B2, CSN-B3, and CSN-B4 and CSS-B1, CSS-B2, CSS-B3, and CSS-B4) were obtained and used in a heminested PCR in which the primer pairs LR1 and FLR2 for the first amplification step and LR1 and FLR4 for the second step were applied. The four LR1/FLR4 heminested PCR products (CSN-B1, CSN-B2, CSN-B3, and CSN-B4 and CSS-B1, CSS-B2, CSS-B3, and CSS-B4) were pooled two by two in equal molar quantities as follows: CSN-B1 and CSN-B2 were pooled to yield CSN-Ba, CSN-B3 and CSN-B4 were pooled to yield CSN-Bb, CSS-B1 and CSS-B3 were pooled to yield CSS-Ba, and CSS-B2 and CSS-B4 were pooled to yield CSS-Bb. The PCR products were then cloned into the pCR4-TOPO vector. To prepare PCR fragments from potato roots, a pooled root sample from each area was obtained by mixing potato roots randomly selected from each field. One root genomic DNA sample (CSN-R or CSS-R) per area was assessed, and one LR1/FLR4 heminested PCR product (CSN-R or CSS-R) per area was cloned into the pCR4-TOPO vector. Finally, inserts from randomly selected clones were sequenced and analyzed by using bioinformatics tools.
The quality and quantity of DNA in the samples were verified spectrophotometrically at 260 and 280 nm, and the DNA was visualized after being stained with ethidium bromide on a 0.8% agarose gel in TAE buffer (40 mM Tris, pH 7.8, 20 mM acetic acid, 2 mM EDTA) under UV light (37).
DNA extraction from soil.
For each of the four fields per area, the soil samples were pooled and an aliquot (250 mg) of soil was used for genomic DNA extraction with the Power soil DNA isolation kit according to the recommendations of the manufacturer (MO BIO). The quality and quantity of DNA in the four soil genomic DNA samples per area (CSN-B1, CSN-B2, CSN-B3, and CSN-B4 and CSS-B1, CSS-B2, CSS-B3, and CSS-B4) were checked as described above (Fig. 1).
PCR amplification of a partial LSU rRNA gene region.
Five-microliter samples of genomic DNA at different dilutions (1:1, 1:5, 1:10, 1:100, 1:500, and 1:1,000) were used for the amplification of a partial large-subunit (LSU) rRNA gene region. In order to enhance the efficiency of the amplification and increase the amount of DNA available for cloning, a heminested PCR was carried out using the primer pairs LR1 (52) and FLR2 (48) for the first amplification step and LR1 and FLR4 (13) for the second one. The reactions were performed in a final volume of 20 μl containing 2 μl of 10× PCR buffer (Qbiogene), 125 μM (each) deoxynucleoside triphosphates (dNTPs), 500 nM (each) primers, and 0.4 U of Taq polymerase (Qbiogene). In the first PCR, 1 μl of T4 bacteriophage 32 (Qbiogene) was also added and 5 μl of diluted or undiluted root or soil genomic DNA extract was added to 15 μl of the PCR mix. Each reaction mix was overlaid with mineral oil, and the amplifications were performed in a thermal cycler (T3000 thermocycler; Biometra). The PCR program was as follows: 93°C for 1 min, 58°C for 1 min, and 72°C for 1 min (30 cycles), followed by 5 min at 72°C. For the second amplification, 5-μl aliquots of the products of the first PCR were diluted 1:500 and 1:1,000 and used as templates under the same PCR conditions. PCR products were separated by gel electrophoresis on a 1.4% agarose gel in TAE buffer, and DNA was visualized under UV light after being stained with ethidium bromide (37).
Cloning of PCR products and sequencing.
The LR1/FLR4 heminested PCR products were cloned into the pCR4-TOPO vector by using the TOPO TA cloning kit for sequencing according to the recommendations of the manufacturer (Invitrogen), and chemically competent Escherichia coli TOP 10 cells (Invitrogen) were transformed with the vector.
For the soil from each area, the four LR1/FLR4 products (CSN-B1, CSN-B2, CSN-B3, and CSN-B4 or CSS-B1, CSS-B2, CSS-B3, and CSS-B4) generated by four reactions were pooled two by two, in equimolar quantities, in the following way: for CSN soil, CSN-B1 and CSN-B2 were pooled to yield CSN-Ba, and CSN-B3 and CSN-B4 were pooled to yield CSN-Bb, whereas for CSS soil, CSS-B1 and CSS-B3 were pooled to yield CSS-Ba, and CSS-B2 and CSS-B4 were pooled to yield CSS-Bb. In this way, two cloning reactions per area (CSN and CSS) were performed. In contrast, for the potato roots from each area, one LR1/FLR4 product (CSN-R or CSS-R) was obtained and cloned. The steps involved in preparing the PCR fragment pools are schematically represented in Fig. 1.
The presence of cloned inserts was checked by PCR using the LR1 and FLR4 primers directly on the bacterial colonies diluted in water and lysed at 100°C for 5 min. The reactions were performed in a final volume of 25 μl containing 2.5 μl of 10× PCR buffer (Qbiogene), 125 μM (each) dNTPs, 500 nM (each) primers, and 0.5 U of Taq polymerase (Qbiogene). An aliquot (2 μl) of diluted bacterial colonies was added to a 23-μl PCR mix. Amplifications were performed as described before, except for the number of cycles (27 cycles). One strand of inserts obtained from randomly selected clones in each LSU rRNA gene library was sequenced using the LR1 primer, in order to analyze about 50 sequences from each soil or potato root sample. In particular, 24, 24, 22, 22, 48, and 43 PCR fragments obtained from CSN-Ba, CSN-Bb, CSS-Ba, and CSS-Bb soil libraries and from CSN and CSS potato root libraries, respectively, were sequenced. Sequencing was performed using Genome Express (Meylan, France) according to the BigDye Terminator protocol. Results were compared to known sequences using the BLASTN algorithm (1).
Sequence analyses and reconstruction of phylogenetic trees.
An alignment of the sequences over 710 bp was performed using ClustalW 1.8.1, and the alignment was optimized manually using the Se-Al version 2.0 software (University of Oxford). Phylogenetic analyses were performed using the neighbor-joining (NJ) algorithm and G. versiforme as the out-group. The reliability of the internal branches of the NJ tree was assessed using the bootstrap method with 1,000 replicates. Trees were drawn using NJplot (http://biom3.univ-lyon1.fr).
Diversity analyses.
Rarefaction analyses (16, 34) were applied in order to determine if the number of tested clones sufficiently represented Glomeromycota diversity in the soils and in the roots. The rarefaction curves were produced by plotting the number of ribotypes observed against the number of sequences obtained using the freely available Analytic Rarefaction version 1.3 software (http://www.uga.edu/∼strata/software/anRareReadme.html).
To compare the degrees of richness and evenness of AMF in soils and in potato roots, the Shannon and Simpson diversity indices (H and D, respectively) and the Shannon equitability index (EH) were applied using the following formulas:
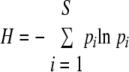 |
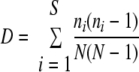 |
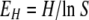 |
For Shannon's diversity index, pi is the proportion of sequences belonging to each ribotype relative to the total number of sequences. For Simpson's diversity index, ni is the number of ribotypes and N is the total number of sequences. In both formulas, i represents each species and, as for Shannon's equitability index, S is the total number of ribotypes (31).
Semiquantitative PCR analyses.
PCR products obtained from the first reaction using LR1 and FLR2 primers were used as templates in a new reaction using taxon-specific primers in combination with LR1 or FLR4. The pairs of primers for each taxon were as follows: FLR4 and 5.25 for G. mosseae (53), LR1 and 8.24 for G. intraradices (11), LR1 and Getunsp2 for G. etunicatum and G. claroideum (these two species, belonging to Glomus group B, are closely related and cannot be distinguished with the primers used [11, 36]), and LR1 and 53.12 for G. geosporum (19). Five microliters of the first PCR amplification product diluted 1:500 was added to a 15-μl PCR mix. The reactions were performed in a final volume of 20 μl containing 2 μl of 10× PCR buffer (Qbiogene), 125 μM (each) dNTPs, 500 nM (each) primers, and 0.4 U of Taq polymerase (Qbiogene). PCR conditions were identical to those described above, but the number of cycles was 15, 17, 19, 21, 23, or 25 in order to characterize the exponential phase of the PCR. Five-microliter samples of the PCR products were analyzed by gel electrophoresis on a 1.4% agarose gel in TAE buffer, and the products were stained with ethidium bromide and quantified by densitometry using Quantity One version 4.4.1 software (Bio-Rad Laboratories). In particular, the PCR products were quantified at different step points of the amplification process to ensure that the quantification of the most abundant product was performed during the exponential phase of the reaction. After it was established that the most abundant PCR product was quantified in the exponential phase of the PCR, the other PCR fragments were quantified at the same step point. The band intensity was expressed as the relative optical absorbance.
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study have been deposited in the EMBL database under the accession numbers AM947863 to AM947934.
RESULTS
Soil composition.
The soils in the two areas had similar pHs and amounts of total and available Pi. They differed in terms of pedological structure, with soil from the CSN area being more sandy than that from the CSS area; the Pi concentrations were high in all fields.
Root colonization by AMF.
The levels of colonization of the root systems in areas CSN and CSS were 5.5 and 2.1%, respectively (Table 1). In contrast, the proportions of root fragments which were in contact with an AMF (the mycorrhizal frequencies) (Table 1) were 30.7 and 16.7%, respectively, revealing the presence of active fungi in the soil. Although relatively small fractions of the root systems were colonized, the levels of arbuscule abundance in the colonized areas were high.
TABLE 1.
Mycorrhizal frequency, mycorrhizal colonization, arbuscule abundance, and arbuscule abundance in the colonized area at the time of harvesta
| Sample source | F (%) | M (%) | A (%) | a (%) |
|---|---|---|---|---|
| CSN | 30.7 ± 6.5 | 5.5 ± 2.1 | 4.9 ± 2.2 | 80.9 ± 9.2 |
| CSS | 16.7 ± 1.9 | 2.1 ± 0.7 | 1.8 ± 0.6 | 83.1 ± 5.5 |
F, mycorrhizal frequency (percentage of root fragments in contact with AMF); M, level of mycorrhizal colonization (percentage of root system colonized); A, arbuscule abundance (percentage of root system with arbuscules); a, arbuscule abundance in the colonized area (percentage of colonized root system with arbuscules). Values are means ± standard deviations. Differences between values for the two areas were statistically significant (P < 0.05) in all cases.
Amplification of AM fungal LSU rRNA gene sequences from soil and root tissues and diversity analyses.
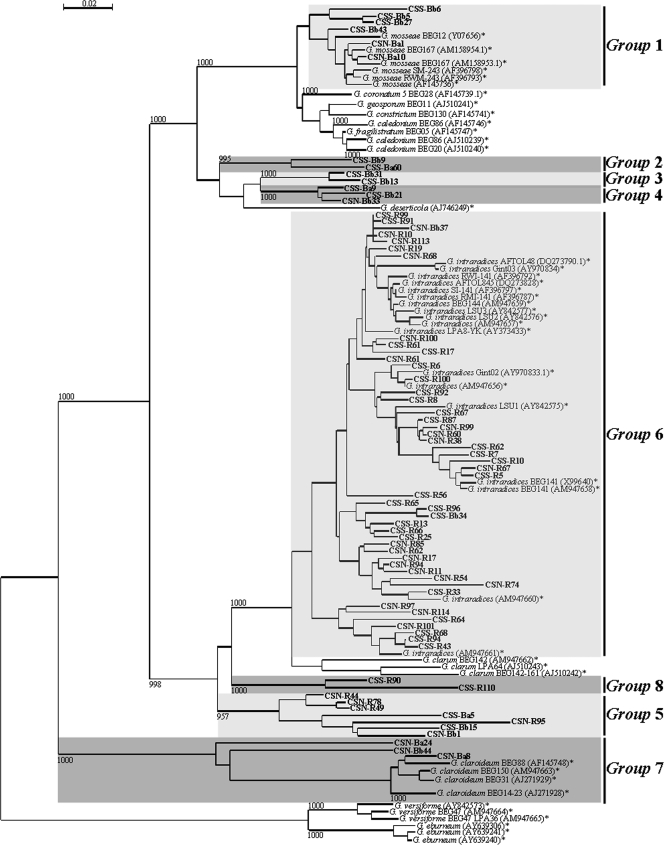
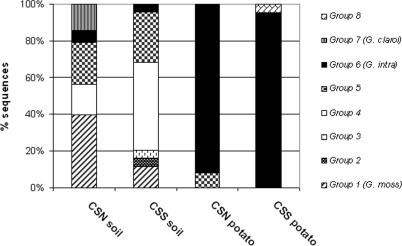
LR1/FLR4 PCR products obtained from the DNA in potato root samples or soil extracts were used to construct LSU rRNA gene libraries, from each of which randomly selected clones were sequenced. No chimeric clones were detected by BLAST analyses, and the sequences obtained had high degrees of similarity to Glomeromycota LSU rRNA gene sequences. The size of the amplified LR1/FLR4 fragment was around 720 bp for all sequences analyzed, corresponding to that expected for Glomus species; no sequences corresponding to Gigasporaceae or Acaulosporaceae were detected. After phylogenetic analyses of the 183 sequences, eight monophyletic ribotypes could be distinguished on the basis of bootstrap values of more than 980‰ (Fig. 2). Three groups, G. mosseae (group 1), G. intraradices (group 6), and G. claroideum (group 7) sequences, clustered with previously identified AMF sequences. Five other groups did not cluster with any known Glomus sequence (Fig. 2). Most of the sequences obtained from the LSU rRNA gene originating from potato root samples clustered with the G. intraradices group (92% for CSN roots and 95% for CSS roots), whereas among the sequences obtained from the soils, only 3 (6.25%) of 48 and 2 (4.5%) of 44 sequences from CSN and CSS, respectively, clustered with the G. intraradices group (Fig. 3).
FIG. 2.
NJ tree representing AM fungal sequences isolated from CSN and CSS soils and potato roots in this study (designations in bold) in comparison to known sequences (*). Bootstrap values were estimated from 1,000 replicates. Ribotypes, defined as sequence groups showing bootstrap values of ≥980‰, are indicated.
FIG. 3.
Bar plot showing the relative proportions of monophyletic groups originating from soils and from potato roots, as identified by the NJ algorithm and the bootstrap method with 1,000 replicates. G. claroi, G. claroideum; G. intra, G. intraradices; G. moss, G. mosseae.
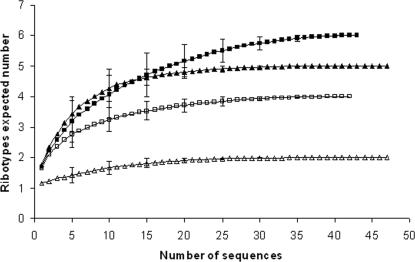
To determine if the number of clones sequenced was sufficient to represent Glomeromycota diversity in the soil and in the potato roots, rarefaction curves were constructed (Fig. 4). The data indicated that the number of sequences analyzed per sample provided coverage of AM fungal diversity. In fact, all the rarefaction curves almost reached a plateau, and the range of the standard deviation decreased as the sample size increased.
FIG. 4.
Rarefaction curves for LSU rRNA gene libraries from CSN soil (closed triangles), CSS soil (closed squares), CSN potato roots (open triangles), and CSS potato roots (open squares) determined using the Analytical Rarefaction program version 1.3 (http://www.uga.edu/∼strata/software/anRareReadme.html).
The Shannon and Simpson indices of diversity and evenness were applied in order to compare the degrees of richness and evenness of AMF in soils and in potato roots (Table 2). The numbers of expected ribotypes in soils (five ribotypes for CSN and six ribotypes for CSS) were higher than those in potato roots (two ribotypes for both CSN and CSS); D and H values for soils corresponded to a higher level of sequence diversity (richness) than those for potato roots, and EH values for soils showed a higher degree of evenness than those for potato roots.
TABLE 2.
Shannon and Simpson diversity indices and Shannon equitability values to compare degrees of soil and root richness and evenness
| Sample type | H | EH | D |
|---|---|---|---|
| CSN soil | 1.176 | 0.731 | 0.308 |
| CSS soil | 1.376 | 0.768 | 0.227 |
| CSN potato | 0.287 | 0.414 | 0.777 |
| CSS potato | 0.188 | 0.271 | 0.728 |
Monitoring of AMF in field trials using specific PCR primers.
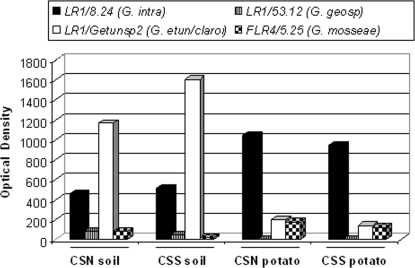
Previously developed specific PCR primers in combination with LR1 or FLR4 were used in a semiquantitative PCR to detect G. mosseae, G. intraradices, G. geosporum, and G. etunicatum/G. claroideum. Soil and potato PCR products obtained from the first reaction with LR1 and FLR2 primers were used as templates for a semiquantitative heminested PCR with the taxon-specific primers, the PCR products obtained at different cycles of amplification were quantified, and quantification was confirmed to be performed during the exponential phase. Figure 5 illustrates the signal intensities of soil and potato PCR fragments obtained during an exponential step point of a semiquantitative heminested PCR. G. intraradices was more abundant in potato roots than in the corresponding soil, whereas G. claroideum/G. etunicatum was more abundant in the soil than in the potato roots. The absolute values obtained for the four fungal species or groups using different primers cannot be compared directly, as the amplification efficiency of each primer pair was not estimated. Nevertheless, the ratios of the species in soil versus root samples can be compared. The abundance of G. etunicatum/G. claroideum in potato roots was approximately equal to that of G. mosseae, whereas in the soil, G. etunicatum/claroideum was much more abundant than G. mosseae. In potato roots, G. intraradices was five to six times more abundant than G. etunicatum/claroideum, whereas in the soil, G. etunicatum/G. claroideum was 2.5- to 3-fold more abundant than G. intraradices. G. geosporum, detected in both soils at a level similar to that of G. mosseae, could not, contrary to G. mosseae, be detected in the roots of potato plants grown in either field.
FIG. 5.
Optical density measurements obtained for the PCR fragments during the exponential phase of semiquantitative PCR to detect G. intraradices (G. intra; primer pair LR1/8.24), G. geosporum (G. geosp; primer pair LR1/53.12), G. etunicatum/G. claroideum (G. etun/claroi; primer pair LR1/Getunsp2), and G. mosseae (primer pair FLR4/5.25) in the soil and potato root samples. The data shown are representative of results for at least three replicas.
DISCUSSION
In this study, the diversity of the AM fungal community was examined by assessing the diversity of the LSU rRNA gene (52) in two soils with different features in areas where crop rotation is applied.
To optimize the cloning reaction and reduce the possible cloning bias, a strategy similar to that described by Renker et al. (35) was used. Several DNA extractions from samples from the same area were performed, and the extracted DNA was combined before cloning. From the phylogenetic analyses of 183 AM fungal sequences obtained from soils and potato roots, eight monophyletic ribotypes defined on the basis of the bootstrap level (980‰) were identified. All sequences belonged to the genus Glomus (groups A and B) (41), which usually dominates the AMF communities in European agricultural soils (24). We obtained a smaller number of groups than other studies on AM fungal diversity, such as those of Pivato et al. (32) and Gollotte et al. (13), who identified 12 and 10 groups, respectively, using our molecular approach. However, the number of sequences analyzed appeared to be sufficient to describe AM fungal diversity, as shown by the rarefaction curves, indicating that the level of diversity detected in this study fell in the same range as those found in previous diversity studies (13, 32, 50).
The genetic diversity of AMF associated with CSN and CSS soils was evaluated by analyzing 48 and 44 sequences, respectively. Five monophyletic ribotypes were identified in CSN samples, three of them belonging to known AMF. Similarly, three of six monophyletic ribotypes detected in CSS soil were already known. The low level of AMF biodiversity in soil was likely due to the typical agricultural management practices of the Castelnuovo Scrivia area, as an inverse relationship between management intensity and AMF diversity has been observed previously (8, 14, 28, 29, 33). It is also known that, in arable lands, rapidly sporulating AMF species, such as G. mosseae, G. geosporum, and G. etunicatum, are among the dominating species (28).
The overall levels of root colonization in plants from CSN and CSS were rather low, although the percentages of root fragments in contact with an AMF (the mycorrhizal frequencies) revealed the presence of active fungi in the soils. The high level of arbuscule abundance in the colonized area suggested active AM symbiosis in the mycorrhizal fraction of the root system. Agricultural management practices and the high Pi concentrations of both areas may explain the relatively low level of root mycorrhization (25), even though the time interval (April to June) was sufficient for the establishment of extensive colonization. Physiological conditions can also affect colonization, as reported by McArthur and Knowles (26), who observed that the percentage of potato roots colonized by G. fasciculatum declined during the period of rapid tuber growth corresponding to 54 to 60 days after planting.
The genetic diversity of AMF associated with CSN and CSS potato roots was assessed through the analysis of, respectively, 48 and 43 sequences. In both root samples, two monophyletic ribotypes were identified, one of which belonged to a known AMF species, G. intraradices. Specifically, 92 and 95% of the sequences identified from CSN and CSS potato roots, respectively, corresponded to G. intraradices. Even if this fungus was not the most abundant in the soils, it largely dominated in potato roots. Studies conducted on both natural and agricultural ecosystems have revealed AM fungal communities dominated by taxa belonging to Glomus group A (17, 28, 32, 39, 40, 49), to which G. intraradices belongs. Similar data, but not as evident as those in our study, were obtained for root nodules (40), but in that case only 63% of the obtained sequences corresponded to G. intraradices. In the latter study, G. intraradices was more frequent in legumes, which had higher nitrogen (N) concentrations than nonlegumes, and among the legumes, it was more frequent in nodules, where atmospheric N fixation takes place, than in roots. The reasons for this association are not known, but previous observations showed that changes in AMF community composition occur after N fertilization (10) and that G. intraradices is related to N-enriched soils (20, 21). G. mosseae, belonging to group A, was also present in both CSN and CSS soils, where it was more abundant than G. intraradices; in potato roots it was not detected (Fig. 3).
In our root or soil samples, only Glomus species were detected, despite some pedological differences between the two soils. According to previous experiments on four different plant species, inoculated with a single fungal isolate, the proportion of intraradical and soil hyphae is specific to each AMF family (15). Nevertheless, within each family, different isolates colonize roots to a different extent; variations are much more limited if the hyphal length in soil or the fungal biomass (measured as micrograms of ergosterol per gram [dry weight] of soil or roots) is considered. Differences observed previously in the concomitant presence of several fungi, as in the present study, could be ascribed to various factors, including competition among the various species/strains, specificity in the interaction with the host, or some form of selective pressure in the environment.
The assessment of AM fungal diversity in potato roots and in the two soils, based on Shannon and Simpson indices, revealed a higher level of evenness in the soils than in the roots, suggesting the dominance of some AM taxa among the fungi colonizing the roots. In addition, Bharadwaj et al. (5) showed, using a potato trap culture, that G. intraradices and G. mosseae are the most common fungi in potato roots (but also in the soil), while McArthur and Knowles (26) demonstrated that inoculation with G. intraradices results in a stronger growth response by the potato than inoculation with G. dimorphicum or G. mosseae. Taken together, these data suggest that there may be a preferential interaction between S. tuberosum and G. intraradices, in agreement with the fact that host specificity has been observed previously in many systems, including agricultural fields (8).
In order to confirm the cloning results, a more sensitive method was applied. In particular, the abundance of four Glomus species or groups, G. intraradices, G. etunicatum/G. claroideum (which cannot be differentiated on the basis of a single primer [11, 36]), G. geosporum, and G. mosseae, was assessed by a semiquantitative heminested PCR (6) using taxon-specific primers (11, 19, 53) with PCR products originating from soil and potato root samples. Three and two Glomus species, all of them belonging to Glomus group A, were detected by semiquantitative PCR in the two soils and in the potato roots, respectively (Fig. 5). The levels of abundance of each species in soils and roots were different, thus confirming our cloning data. G. intraradices was the most abundant fungus in roots, and G. etunicatum/G. claroideum was most abundant in soils. The greater sensitivity of this approach was confirmed by the detection, in both soils, of G. geosporum, a fungus belonging to Glomus group A, not detected by the cloning method.
Several factors may affect the development of AMF communities in roots and soil, for example, seasonal changes (46), as shown previously for G. intraradices, possibly in relation to fluctuations in N mineralization (39). Photosynthate allocation, which in the potato is affected by temperature, light intensity, N availability, and the plant developmental stage (12, 44, 45), may also play a role by affecting C availability for the mycobionts. Finally, early colonization by one AM isolate may prevent further colonization by other fungi, a phenomenon known as autoregulation (18, 30, 54, 55).
In conclusion, we propose that the typical land use practices in Castelnuovo Scrivia have led to a very specific interaction between G. intraradices and potato plants. This finding suggests that the plant species and the agricultural practices used in this area have exerted selective pressure and that some form of specificity between AM fungal and plant genotypes exists. The reasons for this specific association between G. intraradices and S. tuberosum require further investigation.
Acknowledgments
This work was funded partly by Progetto Alfieri “I prodotti tipici agroalimentari dell'Alessandrino e la loro comunicazione a livello nazionale e internazionale” (Fondazione Cassa di Risparmio di Torino).
We thank Elisa Gamalero and Stefania Biondi for critical reading of the manuscript.
Footnotes
Published ahead of print on 1 August 2008.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bever, J. D. 2002. Host-specificity of AM fungal population growth rates can generate feedback on plant growth. Plant Soil 244:281-290. [Google Scholar]
- 3.Bever, J. D. 2002. Negative feedback within a mutualism: host-specific growth of mycorrhizal fungi reduces plant benefit. Proc. R. Soc. B 269:2595-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bever, J. D., A. Pringle, and P. A. Schultz. 2002. Dynamics within the plant-arbuscular mycorrhizal fungal mutualism: testing the nature of community feedback, p. 267-292. In M. G. A. van der Heijden and I. E. Sanders (ed.), Mycorrhizal ecology. Springer-Verlag, Berlin, Germany.
- 5.Bharadwaj, D. P., P. Lundquist, and S. Alströma. 2007. Impact of plant species grown as monocultures on sporulation and root colonization by native arbuscular mycorrhizal fungi in potato. Appl. Soil Ecol. 35:213-225. [Google Scholar]
- 6.Brechenmacher, L., S. Weidmann, D. van Tuinen, O. Chatagnier, S. Gianinazzi, P. Franken, and V. Gianinazzi-Pearson. 2004. Expression profiling of up-regulated plant and fungal genes in early and late stages of Medicago truncatula-Glomus mosseae interactions. Mycorrhiza 14:253-262. [DOI] [PubMed] [Google Scholar]
- 7.Copetta, A., G. Lingua, and G. Berta. 2006. Effects of three AM fungi on growth, distribution of glandular hairs, and essential oil production in Ocimum basilicum L. var. Genovese. Mycorrhiza 16:485-494. [DOI] [PubMed] [Google Scholar]
- 8.Douds, D. D., and P. D. Millner. 1999. Biodiversity of arbuscular mycorrhizal fungi in agroecosystems. Agric. Ecosyst. Environ. 74:77-93. [Google Scholar]
- 9.Duffy, E. M., and A. C. Cassells. 2000. The effect of inoculation of potato (Solanum tuberosum L.) microplants with arbuscular mycorrhizal fungi on tuber yield and tuber size distribution. Appl. Soil Ecol. 15:137-144. [Google Scholar]
- 10.Eom, A. H., D. C. Hartnett, G. W. T. Wilson, and D. A. H. Figge. 1999. The effect of fire, mowing and fertilizer amendment on arbuscular mycorrhizas in tallgrass prairie. Am. Midl. Nat. 142:55-70. [Google Scholar]
- 11.Farmer, M. J., X. Li, G. Feng, B. Zhao, O. Chatagnier, S. Gianinazzi, V. Gianinazzi-Pearson, and D. van Tuinen. 2007. Molecular monitoring of field-inoculated AMF to evaluate persistence in sweet potato crops in China. Appl. Soil. Ecol. 35:599-609. [Google Scholar]
- 12.Frommer, W. B., and U. Sonnewald. 1995. Molecular analysis of carbon partitioning in solanaceous species. J. Exp. Bot. 46:587-607. [Google Scholar]
- 13.Gollotte, A., D. van Tuinen, and D. Atkinson. 2004. Diversity of arbuscular mycorrhizal fungi colonising roots of the grass species Agrostis capillaris and Lolium perenne in a field experiment. Mycorrhiza 14:111-117. [DOI] [PubMed] [Google Scholar]
- 14.Gosling, P., A. Hodge, G. Goodlass, and G. D. Bending. 2006. Arbuscular mycorrhizal fungi and organic farming. Agric. Ecosyst. Environ. 113:17-35. [Google Scholar]
- 15.Hart, M. M., and R. J. Reader. 2002. Taxonomic basis for variation in the colonization strategy of arbuscular mycorrhizal fungi. New Phytol. 153:335-344. [Google Scholar]
- 16.Heck, K. L., Jr., G. Van Belle, and D. Simberloff. 1975. Explicit calculation of the rarefaction diversity measurement and the determination of sufficient sample size. Ecology 56:1459-1461. [Google Scholar]
- 17.Helgason, T., A. H. Fitter, and J. P. W. Young. 1999. Molecular diversity of arbuscular mycorrhizal fungi colonising Hyacinthoides non-scripta (bluebell) in a seminatural woodland. Mol. Ecol. 8:659-666. [Google Scholar]
- 18.Hepper, C. M., C. Azcón-Aguilar, S. Rosendahl, and R. Sen. 1988. Competition between three species of Glomus used as spatially separated introduced and indigenous mycorrhizal inocula for leek (Allium porrum L.). New Phytol. 110:207-215. [Google Scholar]
- 19.Jacquot-Plumey, E., D. van Tuinen, O. Chatagnier, S. Gianinazzi, and V. Gianinazzi-Pearson. 2001. 25S rDNA-based molecular monitoring of glomalean fungi in sewage sludge-treated field plots. Environ. Microbiol. 3:525-531. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, N. C. 1993. Can fertilization of soil select less mutualistic mycorrhizae? Ecol. Appl. 3:749-757. [DOI] [PubMed] [Google Scholar]
- 21.Jumpponen, A., J. Trowbridge, K. Mandyam, and L. Johnson. 2005. Nitrogen enrichment causes minimal changes in arbuscular mycorrhizal colonization but shifts community composition: evidence from rDNA data. Biol. Fertil. Soils 41:217-224. [Google Scholar]
- 22.Kabir, Z., I. P. O'Halloran, J. W. Fyles, and C. Hamel. 1997. Seasonal changes of arbuscular mycorrhizal fungi as affected by tillage practices and fertilization: hyphal density and mycorrhizal root colonization. Plant Soil 192:285-293. [Google Scholar]
- 23.Klironomos, J. N., J. McCune, M. Hart, and J. Neville. 2000. The influence of arbuscular mycorrhizae on the relationship between plant diversity and productivity. Ecol. Lett. 3:137-141. [Google Scholar]
- 24.Mathimaran, N., R. Ruh, P. Vullioud, E. Frossard, and J. Jansa. 2005. Glomus intraradices dominates arbuscular mycorrhizal communities in a heavy textured agricultural soil. Mycorrhiza 16:61-66. [DOI] [PubMed] [Google Scholar]
- 25.McArthur, D. A. J., and N. R. Knowles. 1992. Resistance responses of potato to vesicular-arbuscular mycorrhizal fungi under varying abiotic phosphorus levels. Plant Physiol. 100:341-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McArthur, D. A. J., and N. R. Knowles. 1993. Influence of vesicular-arbuscular mycorrhizal fungi on the response of potato to phosphorus deficiency. Plant Physiol. 101:147-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niemira, B. A., G. R. Safir, R. Hammerschmidt, and G. W. Bird. 1995. Production of prenuclear minitubers of potato with peat-based arbuscular mycorrhizal fungal inoculum. Agron. J. 87:942-946. [Google Scholar]
- 28.Oehl, F., E. Sieverding, K. Ineichen, P. Mäder, T. Boller, and A. Wiemken. 2003. Impact of land use intensity on the species diversity of arbuscular mycorrhizal fungi in agroecosystems of central Europe. Appl. Environ. Microbiol. 69:2816-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oehl, F., E. Sieverding, P. Mäder, D. Dubois, K. Ineichen, T. Boller, and A. Wiemken. 2004. Impact of long-term conventional and organic farming on the diversity of arbuscular mycorrhizal fungi. Oecologia 138:574-583. [DOI] [PubMed] [Google Scholar]
- 30.Pearson, J. N., L. K. Abbott, and D. A. Jasper. 1993. Mediation of competition between two colonizing VA mycorrhizal fungi by host plants. New Phytol. 123:93-98. [DOI] [PubMed] [Google Scholar]
- 31.Pielou, E. C. 1966. Shannon's formula as a measure of specific diversity: its use and misuse. Am. Nat. 102:243-282. [Google Scholar]
- 32.Pivato, B., S. Mazurier, P. Lemanceau, S. Siblot, G. Berta, C. Mougel, and D. van Tuinen. 2007. Medicago species affect the community composition of arbuscular mycorrhizal fungi associated with roots. New Phytol. 176:197-210. [DOI] [PubMed] [Google Scholar]
- 33.Preger, A. C., M. C. Rillig, A. R. Johns, C. C. Du Preez, I. Lobe, and W. Amelung. 2007. Losses of glomalin-related soil protein under prolonged arable cropping: a chronosequence study in sandy soils of the South African Highveld. Soil Biol. Biochem. 39:445-453. [Google Scholar]
- 34.Raup, D. M. 1975. Taxonomic diversity estimation using rarefaction. Paleobiology 1:333-342. [Google Scholar]
- 35.Renker, C., K. Weiβhuhn, H. Kellner, and F. Buscot. 2006. Rationalizing molecular analysis of field-collected roots for assessing diversity of arbuscular mycorrhizal fungi: to pool, or not to pool, that is the question. Mycorrhiza 16:525-553. [DOI] [PubMed] [Google Scholar]
- 36.Rodriguez, A., J. P. Clapp, L. Robinson, and J. C. Dodd. 2004. Studies on the diversity of the distinct phylogenetic lineage encompassing Glomus claroideum and Glomus etunicatum. Mycorrhiza 92:986-989. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 38.Sanders, I. R. 2002. Specificity in the arbuscular mycorrhizal symbiosis, p. 415-437. In M. G. A. van der Heijden and I. E. Sanders (ed.), Mycorrhizal ecology. Springer-Verlag, Berlin, Germany.
- 39.Santos-González, J. C., D. R. Finlay, and A. Tehler. 2007. Seasonal dynamics of arbuscular mycorrhizal fungal communities in roots in a seminatural grassland. Appl. Environ. Microbiol. 73:5613-5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scheublin, T. R., K. P. Ridgway, J. P. W. Young, and M. G. A. van der Heijden. 2004. Nonlegumes, legumes, and root nodules harbor different arbuscular mycorrhizal fungal communities. Appl. Environ. Microbiol. 70:6240-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwarzott, D., C. Walker, and A. Schuessler. 2001. Glomus, the largest genus of the arbuscular mycorrhizal fungi (Glomales), is nonmonophyletic. Mol. Phylogenet. Evol. 21:190-197. [DOI] [PubMed] [Google Scholar]
- 42.Smith, S. E., and D. J. Read. 1997. Mycorrhizal symbiosis, 2nd ed. Academic Press, London, United Kingdom.
- 43.Streitwolf-Engel, R., T. Boller, A. Wiemken, and I. R. Sanders. 1997. Clonal growth traits of two Prunella species are determined by co-occurring arbuscular mycorrhizal fungi from a calcareous grassland. J. Ecol. 85:181-191. [Google Scholar]
- 44.Sweetlove, L. J., J. Kossmann, J. W. Riessmeier, R. N. Trethewey, and S. A. Hill. 1998. The control of source to sink carbon flux during tuber development in potato. Plant J. 15:697-706. [DOI] [PubMed] [Google Scholar]
- 45.Sweetlove, L. J., and S. A. Hill. 2000. Source metabolism dominates the control of source to sink carbon flux in tuberizing potato plants throughout the diurnal cycle and under a range of environmental conditions. Plant Cell Environ. 23:523-529. [Google Scholar]
- 46.Sýkorová, Z., K. Ineichen, A. Wiemken, and D. Redecker. 2007. The cultivation bias: different communities of arbuscular mycorrhizal fungi detected in roots from the field, from bait plant transplanted to the field, and from a greenhouse trap experiment. Mycorrhiza 18:1-14. [DOI] [PubMed] [Google Scholar]
- 47.Trouvelot, A., J. L. Kough, and V. Gianinazzi-Pearson. 1986. Mesure du taux de mycorhization VA d'un système radiculaire. Recherche de methods d'estimation ayant une signification fonctionnelle, p. 217-221. In V. Gianinazzi-Pearson and S. Gianinazzi (ed.), Physiological and genetical aspects of mycorrhizae. INRA Presse, Paris, France.
- 48.Trouvelot, S., D. van Tuinen, M. Hijri, and V. Gianinazzi-Pearson. 1999. Visualization of ribosomal DNA loci in spore interphasic nuclei of glomalean fungi by fluorescence in situ hybridization. Mycorrhiza 8:203-206. [Google Scholar]
- 49.Vandenkoornhuyse, P., S. Baldauf, C. Leyval, J. Straczek, and J. P. W. Young. 2002. Extensive fungal diversity in plant roots. Science 295:2051. [DOI] [PubMed] [Google Scholar]
- 50.Vandenkoornhuyse, P., R. Husband, T. J. Daniell, I. J. Watson, J. M. Duck, A. H. Fitter, and J. P. W. Young. 2002. Arbuscular mycorrhizal community composition associated with two plant species in a grassland ecosystem. Mol. Ecol. 11:1555-1564. [DOI] [PubMed] [Google Scholar]
- 51.van der Heijden, M. G. A., J. N. Klironomos, M. Ursic, P. Moutoglis, R. Streitwolf-Engel, T. Boller, A. Wiemken, and I. R. Sanders. 1998. Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature 396:69-72. [Google Scholar]
- 52.van Tuinen, D., E. Jacquot, B. Zhao, A. Gollotte, and V. Gianinazzi-Pearson. 1998. Characterization of root colonization profiles by a microcosm community of arbuscular mycorrhizal fungi using 25S rDNA-targeted nested PCR. Mol. Ecol. 7:879-887. [DOI] [PubMed] [Google Scholar]
- 53.van Tuinen, D., B. Zhao, and V. Gianinazzi-Pearson. 1998. PCR in studies of AM fungi: from primers to application, p. 387-400. In A. K. Varma (ed.), Mycorrhiza manual. Springer-Verlag, Heidelberg, Germany.
- 54.Vierheilig, H., J. M. Garcia-Garrido, U. Wyss, and Y. Piché. 2000. Systemic suppression of mycorrhizal colonization of barley roots already colonized by AM fungi. Soil Biol. Biochem. 32:589-595. [Google Scholar]
- 55.Vierheilig, H. 2004. Further root colonization by arbuscular mycorrhizal fungi in already mycorrhizal plants is suppressed after a critical level of root colonization. J. Plant Physiol. 161:339-341. [DOI] [PubMed] [Google Scholar]
- 56.Zhu, Y. G., A. S. Laidlaw, P. Christie, and M. E. R. Hammond. 2000. The specificity of arbuscular mycorrhizal fungi in perennial ryegrass-white clover pasture. Agric. Ecosyst. Environ. 77:211-218. [Google Scholar]