Abstract
The wood decay fungus Phanerochaete chrysosporium has served as a model system for the study of lignocellulose conversions, but aspects of its cellulolytic system remain uncertain. Here, we report identifying the gene that encodes the glycoside hydrolase (GH) family 45 endoglucanase (EG) from the fungus, cloning the cDNA, determining its heterologous expression in the methylotrophic yeast Pichia pastoris, and characterizing the recombinant protein. The cDNA consisted of 718 bp, including an open reading frame encoding a 19-amino-acid signal peptide, a 7-amino-acid presequence at the N-terminal region, and a 180-amino-acid mature protein, which has no cellulose binding domain. Analysis of the amino acid sequence revealed that the protein has a low similarity (<22%) to known fungal EGs belonging to the GH family 45 (EGVs). No conserved domain of this family was found by a BLAST search, suggesting that the protein should be classified into a new subdivision of this GH family. The recombinant protein has hydrolytic activity toward amorphous cellulose, carboxylmethyl cellulose, lichenan, barley β-glucan, and glucomannan but not xylan. Moreover, a synergistic effect was observed with the recombinant GH family 6 cellobiohydrolase from the same fungus toward amorphous cellulose as a substrate, indicating that the enzyme may act in concert with other cellulolytic enzymes to hydrolyze cellulosic biomass in nature.
Cellulose, a linear polymer of β-1,4-linked anhydrous glucose residues, is the major component of the plant cell wall and the most abundant biopolymer on earth. Therefore, its degradation process has a great impact on the natural carbon cycle. In nature, cellulose is degraded mainly by microorganisms, which have a series of cellulose-hydrolyzing enzymes such as cellobiohydrolases (CBHs; EC 3.2.1.91), endoglucanases (EGs; EC 3.2.1.4), and β-glucosidases (EC 3.2.1.21) (7). These enzymes are classified into glycoside hydrolase (GH) families, based on amino acid sequence similarities (16, 17). Details are available at the Carbohydrate-Active enZymes (CAZy) Web server (http://www.cazy.org/).
The filamentous ascomycete Hypocrea jecorina (anamorph, Trichoderma reesei) is the best-studied microorganism from the viewpoint of cellulose degradation, and a filtrate of its cellulose-degrading culture has been extensively marketed as a commercial enzyme mixture (3, 35). This fungus produces two CBHs belonging to the GH families 6 and 7 (Cel6A and Cel7A, or CBHI and CBHII, respectively) and at least five EGs (Cel7B, Cel5A, Cel12A, Cel61A, and Cel45A or EGI, EGII, EGIII, EGIV, and EGV) when cellulosic biomass is available as a substrate (39, 47), and these enzymes act cooperatively or synergistically to hydrolyze cellulose (18, 44). Compared to the genomes of other cellulolytic fungi, the recently sequenced T. reesei genome revealed relatively few cellulases and hemicellulases (32).
In contrast to our knowledge of ascomycetes, knowledge about cellulose degradation by basidiomycetes is limited. However, the wood-rotting fungus Phanerochaete chrysosporium is able to efficiently degrade lignocellulose, and it secretes an array of enzymes that hydrolyze cellulose and hemicellulose (1, 5, 6, 12, 13, 41). The culture filtrate of P. chrysosporium grown in the presence of cellulose contains CBHs belonging to GH families 6 and 7 (43) and four EGs in GH families 5 and 12 (15, 42), suggesting that the basidiomycete has a cellulolytic system that is similar to that of ascomycetes. In the total genome sequence of P. chrysosporium (33), moreover, many genes which encode homologues of H. jecorina EGIV (GH family 61) sequences have been found by using a BLAST search, and the cDNA of one of these genes has been cloned from a cellulolytic culture of this fungus (45). Thus, homologues of all H. jecorina cellulases have been cloned from P. chrysosporium, except for the GH family 45 EG.
The GH family 45 (formerly known as the cellulase family K) EG is distantly related to plant expansins or fungal swollenins, which have a dispersing effect on solid celluloses without hydrolyzing them. This family is composed mainly of β-1,4- and β-1,3/1,4-glucanases with a net inverting hydrolytic mechanism (24, 37). The enzymes classified into this family share a catalytic domain with a rather small molecular size (∼20 kDa) and broad substrate specificities for β-1,3/1,4-glucans, such as lichenan and barley β-glucan, as well as cellulose and its derivatives. As far as we know, only one basidiomycetes gene (egl1) encoding a putative GH family 45 protein has been cloned from the pathogenic fungus Ustilago maydis, and this protein has an important function for the filamentous growth of the fungus (38). However, there is no information about GH family 45 proteins in other basidiomycetes. Moreover, no cDNA encoding a member of this family has been cloned from P. chrysosporium so far.
In the present study, we report the identification, cloning, and heterologous expression of cDNA encoding the GH family 45 protein from P. chrysosporium. We also characterize the recombinant protein as an EG and demonstrate its synergism with CBH.
MATERIALS AND METHODS
Materials.
P. chrysosporium strain K-3 (22) was used as the source for target genes. Escherichia coli strain JM109 (Takara Bio, Shiga, Japan) and Pichia pastoris strain KM71H (Invitrogen, Carlsbad, CA) were used as the strains for subcloning the host and for the heterologous production of recombinant protein, respectively. Microcrystalline cellulose (MCC; Funacel II, Funakoshi, Tokyo, Japan), carboxylmethyl cellulose (CMC 7LFD; Hercules, Wilmington, DE), and glucomannan from konjac tuber (Wako Pure Chemical Industries, Osaka, Japan) were used for the enzymatic assay. Xylan from beech wood, β-d-glucan from barley, and lichenan were purchased from Sigma-Aldrich (St. Louis, MO). Phosphoric acid-swollen cellulose (PASC) was prepared from MCC as described previously (48).
Cultivation of and search for the gene encoding the 18-kDa protein.
P. chrysosporium was grown on Kremer and Wood medium (30) containing 2% cellulose (CF11; Whatman, Fairfield, NJ) as the sole carbon source, based on the method described in a previous report (14). After 3 days of cultivation, culture filtrate and mycelia (containing unused cellulose particles) were separated using a glass filter membrane (Advantec GA-100; Toyo Roshi Kaisha, Tokyo, Japan), and the mycelia were frozen with liquid nitrogen to extract RNA as described below. Culture filtrate (500 μl) was concentrated in an Ultrafree-0.5 model centrifugal filter device (Millipore, Billerica, MA) and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in 12% polyacrylamide gels (Mini-Protean II; Bio-Rad, Hercules, CA). After the 18-kDa proteins were transferred to a polyvinylidene difluoride membrane (Millipore), using a Trans-Blot SD cell (Bio-Rad), the N-terminal amino acid sequence was determined with a protein sequencer (model 491 cLC; Applied Biosystems, Foster City, CA) as described previously (29). The amino acid sequence thus obtained was subjected to a BLAST alignment search in the P. chrysosporium genome database, version 2.0 (http://genome.jgi-psf.org/Phchr1/Phchr1.home.html), using the TBLASTN alignment program with default settings, except for the expected value for 1e-1 and the scoring matrix of PAM30.
Cloning cDNA encoding the GH family 45 EG PcCel45A.
Total RNA was extracted from approximately 200 mg of frozen mycelial powder, using Isogen (Nippon Gene, Tokyo, Japan) according to the manufacturer's instructions. First-strand cDNA was synthesized from mRNA, which was purified from 1 μg of total RNA extracted from the above with Oligotex-dT30 Super (Takara Bio), using ReverTraAce (Toyobo, Osaka, Japan) and a 3′ rapid amplification of cDNA ends (RACE) adaptor primer (Invitrogen), according to the manufacturers' instructions. The oligonucleotide primers for the amplification of the cDNA fragment encoding the GH family 45 EG (cel45A) were designed based on the total genomic sequences Pccel45A-F1 (5′-ATGGCGAAGCTGTCGATGTTCTTGGG-3′) and Pccel45A-F2 (5′-CTGACCGTCTCCGAGAAGCGTG-3′), and PCR was performed using KOD-Plus (version 2; Toyobo) software according to the manufacturer's instructions. Since abridged universal amplification primer (Invitrogen) was used as the reverse primer, the coding region and the 3′ untranslated region were amplified at the same time. The nucleotide sequence of the 5′ untranslated region was amplified by using a GeneRacer kit with SuperScript III reverse transcriptase (RT; Invitrogen) with the gene-specific primers Pccel45A-5′-R1 (5′-CAGCCTTGCCGCAAGCAGGAGAGCCGC-3′) and Pccel45A-5′-R2 (5′-CGCAAGCAGGAGAGCCGCAGCCCGAAT-3′). All PCR products were cloned by using a Zero Blunt TOPO PCR cloning kit and E. coli JM109 and were sequenced by using a Thermo Sequenase primer cycle sequencing kit (GE Healthcare, Buckinghamshire, United Kingdom) with DNA sequencer model SQ5500E (Hitachi High Technologies, Tokyo, Japan) as described previously (23, 50).
Sequence analysis.
The NCBI protein database (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi) was searched for the amino acid sequence of PcCel45A, using the BLASTP algorithm with default settings. For the phylogenetic analysis, multiple alignment was performed with the aid of MAFFT version 6 software (http://align.bmr.kyushu-u.ac.jp/mafft/online/server/) with the E-INS-i algorithm (25, 26). This analysis included amino acid sequences of PcCel45A, the NCBI sequences producing significant alignments with the BLASTP search, the fungal enzymes in the GH family 45 on the CAZy server (http://www.cazy.org/), and fungal swollenins. Alignments were visualized with BOXSHADE software (http://www.ch.embnet.org/software/BOX_form.html). The phylogenetic tree file was then made from the multiple alignment results of the MAFFT server with the minimum linkage method, and the tree was drawn with FigTree version 1.1.2 (http://tree.bio.ed.ac.uk/software/figtree/). A search for the presence of signal peptides and N glycosylation and O glycosylation sites was performed with SignalP version 3.0 (http://www.cbs.dtu.dk/services/SignalP/) (2, 34), NetNGlyc version 1.0 (http://www.cbs.dtu.dk/services/NetNGlyc/), and NetOGlyc version 3.1 (http://www.cbs.dtu.dk/services/NetOGlyc/) server software at the Center for Biological Sequence Analysis.
Heterologous expression of cel45A and purification of PcCel45A.
Two different oligonucleotide primers were designed, based on the nucleotide sequence of the mature protein, for ligation into the XhoI and NotI sites of the Pichia expression vector pPICZα (Invitrogen), as follows: Pccel45A-XhoI-F, 5′-TTTCTCGAGAAAAGACTGACCGTCTCCGAGAAGCGTG-3′; and Pccel45A-NotI-R, 5′-TTTTGCGGCCGCTCACGAAGGGGCAGTCCCCTTGTT-3′. Approximately 5 μg of the DNA construct in pPICZα was linearized with Bpu1102I (Takara Bio) prior to the transformation of P. pastoris. The electroporation and selection of transformants were carried out according to the instruction manual of the EasySelect Pichia expression kit (version G; Invitrogen) as described previously (20, 21, 28, 49). The recombinant protein was produced using a mini-jar fermentor (TSC-M5L; Takasugi Seisakusho, Tokyo, Japan) equipped with a dissolved-oxygen controller (DJ-1033; Able Corp., Tokyo, Japan) according to the Pichia fermentation process guidelines (Invitrogen). After 4 days in a methanol-fed batch culture, the medium was collected by centrifugation (30 min at 5,000 × g), and ammonium sulfate and sodium acetate buffer (pH 5.0) were added to the cell-free culture medium to final concentrations of 1 M and 20 mM, respectively. The solution was fractionated on a phenyl-Toyopearl 650S column (26 mm by 120 mm) equilibrated with 20 mM sodium acetate buffer containing 1 M ammonium sulfate (pH 5.0). The protein was eluted with a 300-ml reverse gradient to 20 mM sodium acetate buffer (pH 5.0). The fractions containing the recombinant protein were collected, equilibrated against 20 mM potassium phosphate buffer (pH 7.0), and applied to a SuperQ-Toyopearl 650S column (9 mm by 120 mm) equilibrated with the same buffer. The protein was eluted from the column with a linear gradient from 0 to 0.5 M NaCl in 100 ml. The purity was confirmed by SDS-PAGE analysis on 12% polyacrylamide gels, and the N-terminal amino acid sequence was confirmed as described above.
Enzyme assay.
PcCel45A (1.0 μM) was incubated with 0.5% concentrations of various substrates (MCC, PASC, CMC, β-glucan, lichenan, glucomannan, and xylan) for up to 120 h in 250 μl of 50 mM sodium acetate (pH 5.0). The reaction was stopped by boiling the mixture for 5 min, for thin-layer chromatography (TLC) analysis. Each reaction mixture was centrifuged (15,000 × g), and the supernatant thus obtained was applied to a precoated silica gel 60 TLC plate (Merck, Darmstadt, Germany). After plates were developed with EtOAc-CH3COOH-H2O (3:2:1 [by volume]), reducing sugars were detected with orcinol reagent as described previously (27). To measure the amount of reducing sugar produced by PcCel45A, the reaction was stopped by the addition of an equal volume of 1.0 M NaOH after incubation for 1 h, and reducing sugar was measured with the p-hydroxybenzoic acid hydrazide method as described previously (31), using glucose (Wako) as a standard.
The soluble products from PASC were analyzed by high-performance liquid chromatography (HPLC; LC-2000 series; Jasco, Tokyo, Japan), using a corona charged aerosol detector (ESA Biosciences, Chelmsford, MA). PcCel45A (1.0 μM) was incubated with 0.5% PASC for up to 240 min in 250 μl of 50 mM sodium acetate (pH 5.0), and the reaction mixture was boiled for 5 min and then centrifuged (15,000 × g). The supernatant was separated on Shodex Asahipak NH2P-50 (Showa Denko K. K., Kanagawa, Japan) with a linear gradient of acetonitrile-H2O (60:40 to 50:50 [vol/vol]). The amount of each product was quantified by using cellooligosaccharides with degree of polymerization (DP) values of 2 to 7 (Seikagaku Corporation, Tokyo, Japan) as standards.
The hydrolysis of PASC and highly crystalline cellulose from Cladophora sp. (19) by 1 μM PcCel45A was also tested with 1 μM of recombinant P. chrysosporium Cel6A expressed in P. pastoris (K. Igarashi, T. Ishida, and M. Samejima, unpublished data). PcCel45A and PcCel6A were incubated for up to 240 min at molar ratios of 100:0, 75:25, 50:50, 25:75, and 0:100 in 50 mM sodium acetate buffer (pH 5.0), and the soluble products were separated by HPLC after centrifugation as described above.
Nucleotide sequence accession number.
The nucleotide sequence has been submitted to the DDBJ/EMBL/GenBank databases under accession number AB378504.
RESULTS
P. chrysosporium was cultivated using cotton cellulose as the sole carbon source, and the secreted protein in the 3-day culture solution was concentrated and separated by SDS-PAGE. In addition to the expected Cel7 isozymes of 50 to 75 kDa, a protein with a molecular mass of 18 kDa was observed (Fig. 1), and its N-terminal amino acid sequence was determined as ATGGYVQQAT. The peptide sequence was subjected to a BLAST search (TBLASTN mode) in the genome database of P. chrysosporium, and one match was found (scaffold 6, 1798240-1798263).
FIG. 1.
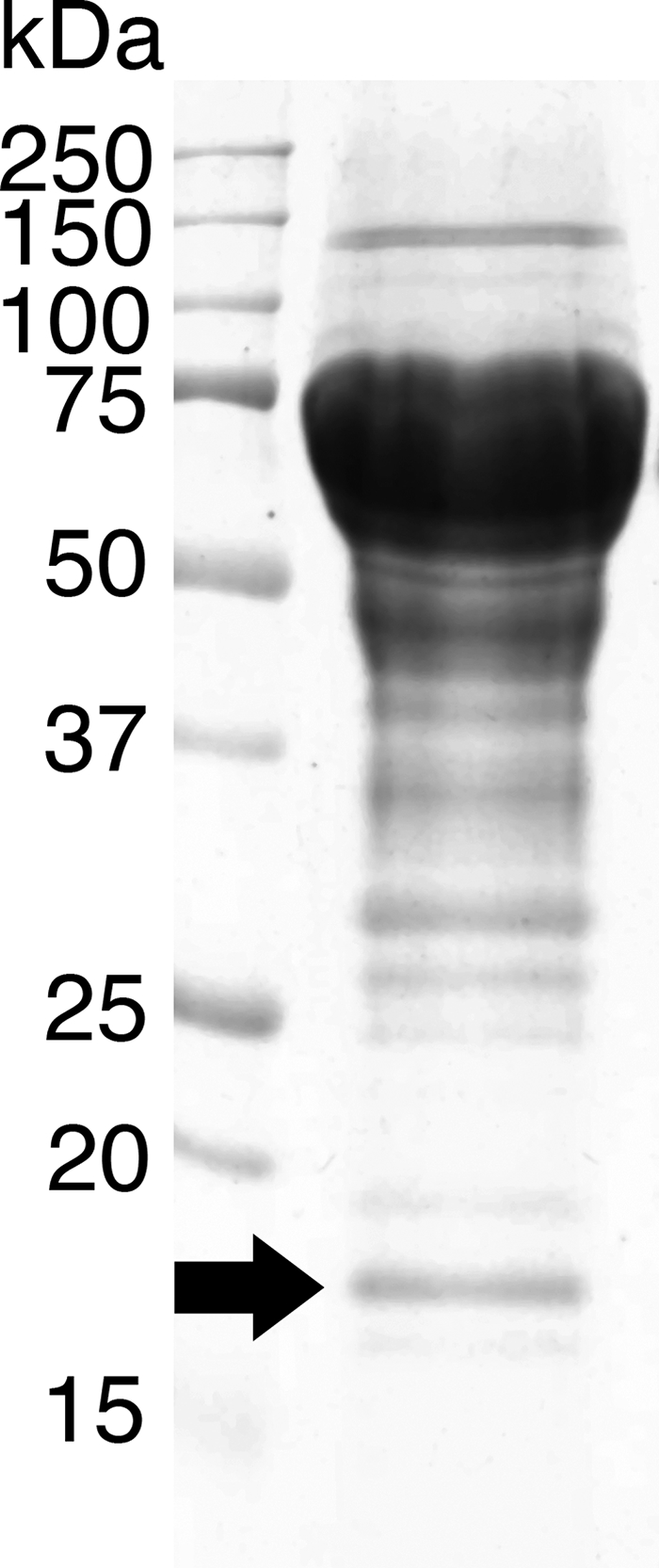
SDS-PAGE of culture filtrate from the cellulolytic culture of P. chrysosporium. The arrow indicates the protein subjected to N-terminal sequencing. Approximately 100 μg of crude protein from a 3-day culture was separated on a 12% polyacrylamide gel as described in Materials and Methods.
RT-PCR was carried out using oligonucleotide primers designed from the nucleotide sequence around the region and a 3′ RACE primer, and a fragment encoding the 18-kDa protein was cloned. After the 5′ RACE, cDNA encoding the whole protein was obtained (see Fig. S1A in the supplemental material; accession number AB378504). The cDNA consisted of 718 bp, including an open reading frame with 206 amino acids, which was divided into three exons by two introns in the genome of P. chrysosporium (see Fig. S1B in the supplemental material). Analysis of the deduced amino acid sequence, using SignalP version 3.0 server software, indicated that the first 19 amino acids may have been a signal peptide, since it was different from the native N-terminal sequence. The deduced amino acids included the peptide sequence, which has been found in the previous secretome work reported by Vanden Wymelenberg et al. (46). This peptide was initially assigned to an incomplete v1 gene model, pc.55.40.1, and an automated annotation of a later version of the genome (v2) failed to predict a model at this location. One possible N glycosylation site and four possible O glycosylation sites were predicted by the NetNGlyc and NetOGlyc servers, respectively. No putative cellulose-binding domain was found in the sequence.
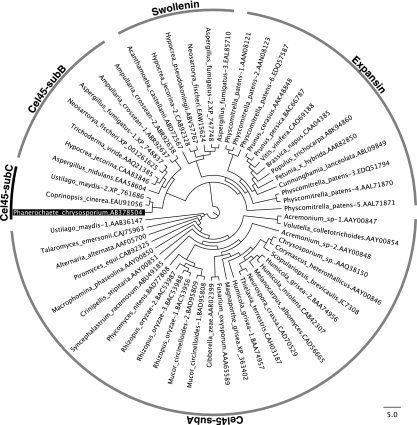
Analysis of the amino acid sequence in the NCBI protein database revealed that the protein has a low similarity to fungal EGV, though no putative conserved domain was found in the sequence. Except for hypothetical proteins, the highest homology was obtained for H. jecorina (T. reesei) EGV (22%). Five amino acid sequences with homology to the protein were analyzed by MAFFT multiple alignment (see Fig. S2 in the supplemental material). Asp140 may be one of the catalytic residues, based on the comparison with H. jecorina EGV. However, another possible catalytic residue (Asp27 in H. jecorina EGV) is missing, again indicating that this protein is different from the known enzymes. Phylogenetic analysis was performed for the sequences showing significant alignments with PcCel45A on the BLASTP search, including the fungal GH family 45 EGs (subfamily A), plant expansins, fungal enzymes of the GH family 45 on the CAZy server (subfamily B), and fungal swollenins (Fig. 2). In terms of amino acid sequence, PcCel45A was clearly distinct from that of the GH family 45 subfamily A but showed some similarity to that of subfamily B, as expected from the result of the BLAST search. This clade includes only hypothetical proteins from Coprinopsis cinerea (EAU91056) and U. maydis (XP_761686) among basidiomycetes, in addition to PcCel45A.
FIG. 2.
Phylogenetic tree of the sequences showing significant alignments with PcCel45A from a BLASTP search of the fungal GH family 45 EGs (subfamily A), plant expansins, fungal enzymes in the GH family 45 on the CAZy server (subfamily B), and fungal swollenins. The sequences were aligned using MAFFT, and the tree was drawn by FigTree.
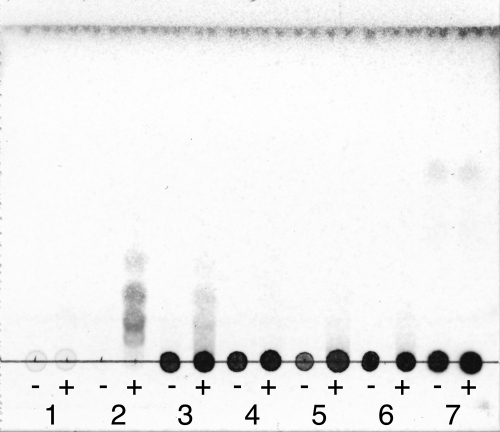
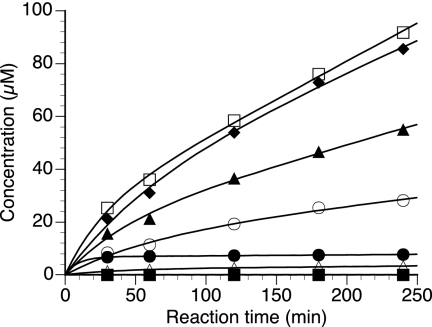
The cDNA encoding PcCel45A was heterologously expressed in the methylotrophic yeast P. pastoris, and the hydrolytic activity of the recombinant protein toward various glycans was examined (Table 1). The soluble products were analyzed by TLC, as shown in Fig. 3. The recombinant PcCel45A had apparent hydrolytic activity toward PASC, CMC, lichenan, β-glucan from barley, and glucomannan, whereas no sugar was detectable when MCC or xylan was used as a substrate. Although the initial velocity for β-1,3/1,4-glucans (lichenan and barley β-glucan) was higher than that for β-1,4-glucans (PASC and CMC) as shown in Table 1, oligosaccharide production was detected only in the case of β-1,4-glucans (Fig. 3). The time course of hydrolytic product formation from PASC was then quantified by HPLC (Fig. 4). Cellooligosaccharides with a DP value of 3 to 5 were similarly formed, whereas the formation of cellobiose (DP, 2) or glucose (DP, 1) was only slight. The production of cellooligosaccharides with DP values of 6 to 7 was also limited, possibly because of their poor solubility.
TABLE 1.
Hydrolytic activity of recombinant P. chrysosporium PcCel45A toward various glycan substratesa
| Substrate | Activity (min−1) |
|---|---|
| MCC | ND |
| PASC | 5.4 |
| CMC | 6.4 |
| Lichenan | 19 |
| β-Glucan from barley | 13 |
| Glucomannan | 1.3 |
| Xylan | ND |
PcCel45 (1.0 μM) was incubated with 0.5% substrate in 50 mM sodium acetate buffer (pH 5.0) at 30°C for 1 h. The formation of reducing sugar was quantified by the p-hydroxybenzoic acid hydrazide method as described in Materials and Methods. ND, not detected after 120 h of incubation.
FIG. 3.
TLC analysis of soluble products in the reaction mixtures of PcCel45A with various glycans. PcCel45 (1.0 μM) was incubated with 0.5% substrate in 50 mM sodium acetate buffer (pH 5.0) at 30°C for 1 h. The products were applied, developed, and detected as described in Materials and Methods. Lanes 1, MCC; 2, PASC; 3, CMC; 4, lichenan; 5, barley β-glucan; 6, glucomannan; 7, xylan. In each lane, reaction mixtures with (+) and without (−) enzyme were applied.
FIG. 4.
Time course of cellooligosaccharide production from PASC by PcCel45A. PcCel45A (1.0 μM) was incubated with 0.5% PASC in 50 mM sodium acetate buffer (pH 5.0) at 30°C, and the soluble products were quantified by HPLC. Filled square, glucose; filled circle, cellobiose; filled triangle, cellotriose; filled diamond, cellotetraose; open square, cellopentaose; open circle, cellohexaose; open triangle, cellopentaose.
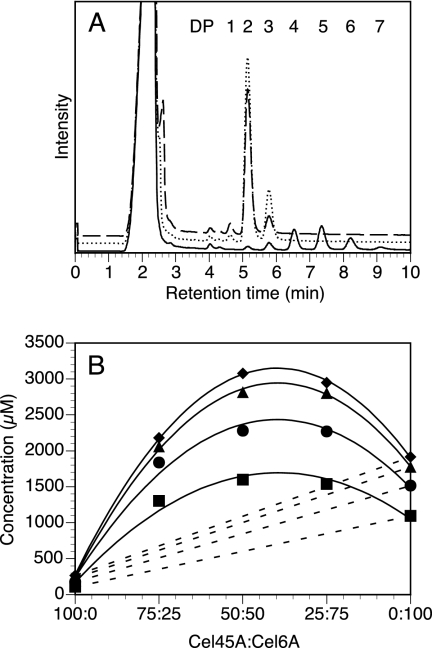
Recombinant PcCel45A, PcCel6A, and a mixture of these enzymes were each incubated with PASC, and the amounts of soluble products were analyzed by HPLC to examine whether or not these enzymes showed a synergistic effect. As shown in Fig. 5A, the main product generated by PcCel6A from PASC was cellobiose, and the pattern was the same in the presence and absence of PcCel45A. However, an apparent increase in the formation of soluble products was observed with a synergy plot (Fig. 5B). After a 120-min incubation, the amounts of products were 3.6- and 2.7-fold higher than the calculated sums (Fig. 5B, dotted line) for ratios of PcCel45A:PcCel6A of 75:25 and 50:50, respectively. However, when highly crystalline cellulose from green algae (Cladophora sp.) was used as a substrate, only a 1.3-fold enhancement was observed under the same reaction conditions (data not shown). This result may indicate that PcCel45A enhances the activity of PcCel6A by increasing the number of chain ends of hydrated cellulose compared with those of tightly packed crystalline cellulose.
FIG. 5.
(A) Production of cellooligosaccharides by PcCel45A (solid line), PcCel6A (dashed line), and a mixture of the two enzymes (ratio of PcCel45A:PcCel6A of 50:50 [dotted line]) monitored by HPLC after 120 min of incubation. (B) Synergy plot of cellooligosaccharides production by the two enzymes after incubation for 60 min (squares), 120 min (circles), 180 min (triangles), and 240 min (diamonds). Dashed lines indicates the calculated sum of products formed by the two enzymes separately.
DISCUSSION
Many filamentous fungi produce various kinds of cellulolytic enzymes, though cellulose itself is a simple β-1,4-linked anhydrous glucose polymer. The reason for the multiplicity of the enzymes may be that cellulose does not exist alone in plant cell walls but forms complexes with other cell wall components, such as hemicellulose and lignin. Cellulolytic fungi may recognize each complex as one substrate and, therefore, produce many types of enzymes with various functions. In the present study, we report cloning and characterizing the GH family 45 EG from the basidiomycete P. chrysosporium, and we show that this fungus has a cellulolytic system similar to those of ascomycetes.
PcCel45A has a molecular mass of 18 kDa and is one of the smallest EGs. A BLAST search of the amino acid sequence of PcCel45A showed that the protein may be classified into the GH family 45, which includes many small EGs (8, 9, 11, 36), and this conclusion is supported by the result of hydrophobic cluster analysis (performed by B. Henrissat, personal communication). Since only a low sequence similarity was found with other known GH family 45 EGs, the protein represents a new subdivision of this family. The EGs belonging to the GH family 45 have already been divided into two subfamilies named A and B. Humicola insolens Cel45A and H. jecorina Cel45A are representatives of these subfamilies. PcCel45A and other hypothetical proteins from basidiomycetes are rather similar to H. jecorina Cel45A, though one of the putative catalytic amino acids (Asp27) is missing in the N-terminal region, (36). Since the family consists of inverting EGs, it might be difficult to identify the catalytically important amino acid residues without mutational or structural studies.
Although we constructed a heterologous expression vector for PcCel45A according to the analytical results obtained with SignalP, the N-terminal sequence of the recombinant PcCel45A was also processed so that the starting position is the same as that of the native enzyme (ATGGYVQQAT). This is because there is a Kex2 protease site (Lys-Arg) just before the native N-terminal sequence (4). It is not clear whether the signal peptide predicted by the algorithm is incorrect or whether the protein is produced initially as a preprotein with the LTVSEKRATG N-terminal sequence and this is followed by processing of the preprotein by a Kex2-type protease. Moreover, when we performed a BLAST search of the genome database of P. chrysosporium, another isozyme was found in the same scaffold (scaffold 6, 1801470-1802160), and this isozyme seems to have no signal peptide at the N-terminal region. Attempts to RT-PCR amplify the corresponding cDNA from the same cellulose-derived mRNA were unsuccessful (data not shown), suggesting that the gene is expressed differently or is inactive.
The recombinant protein had apparent hydrolytic activity toward various glycan substrates. Specific activities for lichenan and barley β-glucan, which consist mainly of β-1,3/1,4-glucan, were higher than those for β-1,4-glucans (PASC and CMC), as is the case for the known GH family 45 EGs. However, product analysis by TLC showed no significant accumulation of shorter oligosaccharide (DP, ∼7) in the case of β-1,3/1,4-glucans, whereas cellooligosaccharides with DP values of 3 to 5 were produced from PASC. It is possible that PcCel45A could not recognize the repeating unit of lichenan and barley β-glucan, cellotriose connected by β-1,3 linkages (40), so that only partial hydrolysis occurred for these substrates. Moreover, PcCel45A did not hydrolyze xylan but did degrade glucomannan, indicating the importance of the C-6 methylol group of hexosans for effective hydrolysis. In the crystal structure of H. insolens Cel45A complexed with ligands (10), six glucose residues are held at the active site, and all C-6 methylol groups are recognized by amino acids. This suggests that EGs of the GH family 45 are generally designed for hydrolysis of hexosans.
When PcCel45A and PcCel6A were incubated together with PASC, an apparent synergy of more than 3-fold was observed for the production of cellooligosaccharides compared with the calculated sum of production by the two enzymes separately. However, when highly crystalline cellulose was used as a substrate, the synergy was less than 1.3-fold, suggesting that the synergy involved mainly the amorphous region but not the crystalline cellulose. In the study of swollenin from H. jecorina, Saloheimo and coworkers observed that the protein disrupts the structure of algal (Valonia sp.) cell walls without producing any detectable amount of reducing sugar (37). In the case of PcCel45A, we also observed dispersion of various cellulose suspensions (data not shown), even though this protein has apparent hydrolytic activity for cellulose. Since this phenomenon is often found for other GH family 45 EGs as well as expansins or swollenins, these proteins seem to have a general function in assisting the hydrolysis of glycosyl bonds by other cellulases.
In conclusion, we have reported the cloning and heterologous expression of a cDNA encoding a GH family 45 protein from P. chrysosporium. We characterized the recombinant protein as an EG that has a synergistic relationship with CBH, suggesting that this fungal enzyme acts in concert with other cellulolytic enzymes to hydrolyze cellulosic biomass in nature.
Supplementary Material
Acknowledgments
We thank Bernard Henrissat at CNRS and Universities of Aix-Marseille I and II in Marseille for sequence analysis of PcCel45A and Koichi Harazono in Nagase & Co., Ltd., for help in producing recombinant PcCel45A in a jar fermentor.
This research was supported by Grants-in-Aid for Scientific Research to K.I. (no. 19688016) and to M.S. (no. 17380102) from the Japan Society for the Promotion of Science (JSPS) and by a grant for development of technology for high-efficiency biomass energy conversion from the New Energy and Industrial Technology Development Organization (NEDO).
Footnotes
Published ahead of print on 1 August 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Almin, K. E., K. E. Eriksson, and B. Pettersson. 1975. Extracellular enzyme system utilized by the fungus Sporotrichum pulverulentum (Chrysosporium lignorum) for the breakdown of cellulose. 2. Activities of the five endo-1,4-beta-glucanases towards carboxymethylcellulose. Eur. J. Biochem. 51:207-211. [DOI] [PubMed] [Google Scholar]
- 2.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 3.Bhikhabhai, R., G. Johansson, and G. Pettersson. 1984. Isolation of cellulolytic enzymes from Trichoderma reesei QM 9414. J. Appl. Biochem. 6:336-345. [PubMed] [Google Scholar]
- 4.Brake, A. J., J. P. Merryweather, D. G. Coit, U. A. Heberlein, F. R. Masiarz, G. T. Mullenbach, M. S. Urdea, P. Valenzuela, and P. J. Barr. 1984. Alpha-factor-directed synthesis and secretion of mature foreign proteins in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 81:4642-4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broda, P., P. Birch, P. Brooks, J. L. Copa-Patino, M. L. Sinnott, C. Tempelaars, Q. Wang, A. Wyatt, and P. Sims. 1994. Phanerochaete chrysosporium and its natural substrate. FEMS Microbiol. Rev. 13:189-195. [DOI] [PubMed] [Google Scholar]
- 6.Broda, P., P. R. Birch, P. R. Brooks, and P. F. Sims. 1996. Lignocellulose degradation by Phanerochaete chrysosporium: gene families and gene expression for a complex process. Mol. Microbiol. 19:923-932. [DOI] [PubMed] [Google Scholar]
- 7.Clarke, A. J. 1997. Biodegradation of cellulose. Technomic Publishing Company, Inc., Lancaster, PA.
- 8.Davies, G. J., G. Dodson, M. H. Moore, S. P. Tolley, Z. Dauter, K. S. Wilson, G. Rasmussen, and M. Schulein. 1996. Structure determination and refinement of the Humicola insolens endoglucanase V at 1.5 Å resolution. Acta Crystallogr. Sect. D 52:7-17. [DOI] [PubMed] [Google Scholar]
- 9.Davies, G. J., G. G. Dodson, R. E. Hubbard, S. P. Tolley, Z. Dauter, K. S. Wilson, C. Hjort, J. M. Mikkelsen, G. Rasmussen, and M. Schulein. 1993. Structure and function of endoglucanase V. Nature 365:362-364. [DOI] [PubMed] [Google Scholar]
- 10.Davies, G. J., S. P. Tolley, B. Henrissat, C. Hjort, and M. Schulein. 1995. Structures of oligosaccharide-bound forms of the endoglucanase V from Humicola insolens at 1.9 Å resolution. Biochemistry 34:16210-16220. [DOI] [PubMed] [Google Scholar]
- 11.Eberhardt, R. Y., H. J. Gilbert, and G. P. Hazlewood. 2000. Primary sequence and enzymic properties of two modular endoglucanases, Cel5A and Cel45A, from the anaerobic fungus Piromyces equi. Microbiology 146:1999-2008. [DOI] [PubMed] [Google Scholar]
- 12.Eriksson, K. E., and B. Pettersson. 1975. Extracellular enzyme system utilized by the fungus Sporotrichum pulverulentum (Chrysosporium lignorum) for the breakdown of cellulose. 1. Separation, purification and physico-chemical characterization of five endo-1,4-beta-glucanases. Eur. J. Biochem. 51:193-206. [DOI] [PubMed] [Google Scholar]
- 13.Eriksson, K. E., and B. Pettersson. 1975. Extracellular enzyme system utilized by the fungus Sporotrichum pulverulentum (Chrysosporium lignorum) for the breakdown of cellulose. 3. Purification and physico-chemical characterization of an exo-1,4-beta-glucanase. Eur. J. Biochem. 51:213-218. [DOI] [PubMed] [Google Scholar]
- 14.Habu, N., K. Igarashi, M. Samejima, B. Pettersson, and K. E. Eriksson. 1997. Enhanced production of cellobiose dehydrogenase in cultures of Phanerochaete chrysosporium supplemented with bovine calf serum. Biotechnol. Appl. Biochem. 26:97-102. [PubMed] [Google Scholar]
- 15.Henriksson, G., A. Nutt, H. Henriksson, B. Pettersson, J. Stahlberg, G. Johansson, and G. Pettersson. 1999. Endoglucanase 28 (Cel12A), a new Phanerochaete chrysosporium cellulase. Eur. J. Biochem. 259:88-95. [DOI] [PubMed] [Google Scholar]
- 16.Henrissat, B. 1991. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280:309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henrissat, B., and A. Bairoch. 1993. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 293:781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henrissat, B., H. Driguez, C. Viet, and M. Schulein. 1985. Synergism of cellulases from Trichoderma reesei in the degradation of cellulose. BioTechnology 3:722-726. [Google Scholar]
- 19.Igarashi, K., M. Wada, R. Hori, and M. Samejima. 2006. Surface density of cellobiohydrolase on crystalline celluloses. A critical parameter to evaluate enzymatic kinetics at a solid-liquid interface. FEBS J. 273:2869-2878. [DOI] [PubMed] [Google Scholar]
- 20.Igarashi, K., M. Yoshida, H. Matsumura, N. Nakamura, H. Ohno, M. Samejima, and T. Nishino. 2005. Electron transfer chain reaction of the extracellular flavocytochrome cellobiose dehydrogenase from the basidiomycete Phanerochaete chrysosporium. FEBS J. 272:2869-2877. [DOI] [PubMed] [Google Scholar]
- 21.Ishida, T., K. Yaoi, A. Hiyoshi, K. Igarashi, and M. Samejima. 2007. Substrate recognition by glycoside hydrolase family 74 xyloglucanase from the basidiomycete Phanerochaete chrysosporium. FEBS J. 274:5727-5736. [DOI] [PubMed] [Google Scholar]
- 22.Johnsrud, S. C., and K. E. Eriksson. 1985. Cross breeding of selected and mutated homokaryotic strains of Phanerochaete chrysosporium K-3: new cellulase deficient strains with increased ability to degrade lignin. Appl. Microbiol. Biotechnol. 21:320-327. [Google Scholar]
- 23.Kajisa, T., M. Yoshida, K. Igarashi, A. Katayama, T. Nishino, and M. Samejima. 2004. Characterization and molecular cloning of cellobiose dehydrogenase from the brown-rot fungus Coniophora puteana. J. Biosci. Bioeng. 98:57-63. [DOI] [PubMed] [Google Scholar]
- 24.Karlsson, J., M. Siika-aho, M. Tenkanen, and F. Tjerneld. 2002. Enzymatic properties of the low molecular mass endoglucanases Cel12A (EG III) and Cel45A (EG V) of Trichoderma reesei. J. Biotechnol. 99:63-78. [DOI] [PubMed] [Google Scholar]
- 25.Katoh, K., K. Kuma, H. Toh, and T. Miyata. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33:511-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katoh, K., K. Misawa, K. Kuma, and T. Miyata. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawai, R., K. Igarashi, M. Kitaoka, T. Ishii, and M. Samejima. 2004. Kinetics of substrate transglycosylation by glycoside hydrolase family 3 glucan (1->3)-beta-glucosidase from the white-rot fungus Phanerochaete chrysosporium. Carbohydr. Res. 339:2851-2857. [DOI] [PubMed] [Google Scholar]
- 28.Kawai, R., K. Igarashi, and M. Samejima. 2006. Gene cloning and heterologous expression of glycoside hydrolase family 55 beta-1,3-glucanase from the basidiomycete Phanerochaete chrysosporium. Biotechnol. Lett. 28:365-371. [DOI] [PubMed] [Google Scholar]
- 29.Kawai, R., K. Igarashi, M. Yoshida, M. Kitaoka, and M. Samejima. 2006. Hydrolysis of beta-1,3/1,6-glucan by glycoside hydrolase family 16 endo-1,3(4)-beta-glucanase from the basidiomycete Phanerochaete chrysosporium. Appl. Microbiol. Biotechnol. 71:898-906. [DOI] [PubMed] [Google Scholar]
- 30.Kremer, S. M., and P. M. Wood. 1992. Evidence that cellobiose oxidase from Phanerochaete chrysosporium is primarily an Fe(III) reductase. Kinetic comparison with neutrophil NADPH oxidase and yeast flavocytochrome b2. Eur. J. Biochem. 205:133-138. [DOI] [PubMed] [Google Scholar]
- 31.Lever, M. 1972. A new reaction for colorimetric determination of carbohydrates. Anal. Biochem. 47:273-279. [DOI] [PubMed] [Google Scholar]
- 32.Martinez, D., R. M. Berka, B. Henrissat, M. Saloheimo, M. Arvas, S. E. Baker, J. Chapman, O. Chertkov, P. M. Coutinho, D. Cullen, E. G. Danchin, I. V. Grigoriev, P. Harris, M. Jackson, C. P. Kubicek, C. S. Han, I. Ho, L. F. Larrondo, A. L. de Leon, J. K. Magnuson, S. Merino, M. Misra, B. Nelson, N. Putnam, B. Robbertse, A. A. Salamov, M. Schmoll, A. Terry, N. Thayer, A. Westerholm-Parvinen, C. L. Schoch, J. Yao, R. Barbote, M. A. Nelson, C. Detter, D. Bruce, C. R. Kuske, G. Xie, P. Richardson, D. S. Rokhsar, S. M. Lucas, E. M. Rubin, N. Dunn-Coleman, M. Ward, and T. S. Brettin. 2008. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina). Nat. Biotechnol. 26:553-560. [DOI] [PubMed] [Google Scholar]
- 33.Martinez, D., L. F. Larrondo, N. Putnam, M. D. Gelpke, K. Huang, J. Chapman, K. G. Helfenbein, P. Ramaiya, J. C. Detter, F. Larimer, P. M. Coutinho, B. Henrissat, R. Berka, D. Cullen, and D. Rokhsar. 2004. Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat. Biotechnol. 22:695-700. [DOI] [PubMed] [Google Scholar]
- 34.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int. J. Neural Syst. 8:581-599. [DOI] [PubMed] [Google Scholar]
- 35.Rabinovich, M. L., M. S. Melnik, and A. V. Bolobova. 2002. Microbial cellulases. Appl. Biochem. Microbiol. 38:355-373. [PubMed] [Google Scholar]
- 36.Saloheimo, A., B. Henrissat, A. M. Hoffren, O. Teleman, and M. Penttila. 1994. A novel, small endoglucanase gene, egl5, from Trichoderma reesei isolated by expression in yeast. Mol. Microbiol. 13:219-228. [DOI] [PubMed] [Google Scholar]
- 37.Saloheimo, M., M. Paloheimo, S. Hakola, J. Pere, B. Swanson, E. Nyyssonen, A. Bhatia, M. Ward, and M. Penttila. 2002. Swollenin, a Trichoderma reesei protein with sequence similarity to the plant expansins, exhibits disruption activity on cellulosic materials. Eur. J. Biochem. 269:4202-4211. [DOI] [PubMed] [Google Scholar]
- 38.Schauwecker, F., G. Wanner, and R. Kahmann. 1995. Filament-specific expression of a cellulase gene in the dimorphic fungus Ustilago maydis. Biol. Chem. Hoppe-Seyler 376:617-625. [DOI] [PubMed] [Google Scholar]
- 39.Shoemaker, S., K. Watt, G. Tsitovsky, and R. Cox. 1983. Characterization and properties of cellulases purified from Trichoderma reesei strain L27. Biotechnology 1:687-690. [Google Scholar]
- 40.Stone, B. A., and A. E. Clarke. 1992. Chemistry and biology of (1->3)-beta-glucans. La Trobe University Press, Victoria, Australia.
- 41.Streamer, M., K. E. Eriksson, and B. Pettersson. 1975. Extracellular enzyme system utilized by the fungus Sporotrichum pulverulentum (Chrysosporium lignorum) for the breakdown of cullulose. Functional characterization of five endo-1,4-beta-glucanases and one exo-1,4-beta-glucanase. Eur. J. Biochem. 59:607-613. [DOI] [PubMed] [Google Scholar]
- 42.Uzcategui, E., G. Johansson, B. Ek, and G. Pettersson. 1991. The 1,4-beta-D-glucan glucanohydrolases from Phanerochaete chrysosporium. Re-assessment of their significance in cellulose degradation mechanisms. J. Biotechnol. 21:143-159. [DOI] [PubMed] [Google Scholar]
- 43.Uzcategui, E., A. Ruiz, R. Montesino, G. Johansson, and G. Pettersson. 1991. The 1,4-beta-D-glucan cellobiohydrolases from Phanerochaete chrysosporium. I. A system of synergistically acting enzymes homologous to Trichoderma reesei. J. Biotechnol. 19:271-285. [DOI] [PubMed] [Google Scholar]
- 44.Valjamae, P., V. Sild, A. Nutt, G. Pettersson, and G. Johansson. 1999. Acid hydrolysis of bacterial cellulose reveals different modes of synergistic action between cellobiohydrolase I and endoglucanase I. Eur. J. Biochem. 266:327-334. [DOI] [PubMed] [Google Scholar]
- 45.Vanden Wymelenberg, A., S. Denman, D. Dietrich, J. Bassett, X. Yu, R. Atalla, P. Predki, U. Rudsander, T. T. Teeri, and D. Cullen. 2002. Transcript analysis of genes encoding a family 61 endoglucanase and a putative membrane-anchored family 9 glycosyl hydrolase from Phanerochaete chrysosporium. Appl. Environ. Microbiol. 68:5765-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vanden Wymelenberg, A., G. Sabat, D. Martinez, A. S. Rajangam, T. T. Teeri, J. Gaskell, P. J. Kersten, and D. Cullen. 2005. The Phanerochaete chrysosporium secretome: database predictions and initial mass spectrometry peptide identifications in cellulose-grown medium. J. Biotechnol. 118:17-34. [DOI] [PubMed] [Google Scholar]
- 47.Vinzant, T. B., W. S. Adney, S. R. Decker, J. O. Baker, M. T. Kinter, N. E. Sherman, J. W. Fox, and M. E. Himmel. 2001. Fingerprinting Trichoderma reesei hydrolases in a commercial cellulase preparation. Appl. Biochem. Biotechnol. 93:99-107. [DOI] [PubMed] [Google Scholar]
- 48.Wood, T. M. 1988. Preparation of crystalline, amorphous, and dyed cellulase substrates. Methods Enzymol. 160:19-25. [Google Scholar]
- 49.Yoshida, M., K. Igarashi, M. Wada, S. Kaneko, N. Suzuki, H. Matsumura, N. Nakamura, H. Ohno, and M. Samejima. 2005. Characterization of carbohydrate-binding cytochrome b562 from the white-rot fungus Phanerochaete chrysosporium. Appl. Environ. Microbiol. 71:4548-4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoshida, M., T. Ohira, K. Igarashi, H. Nagasawa, and M. Samejima. 2002. Molecular cloning and characterization of a cDNA encoding cellobiose dehydrogenase from the wood-rotting fungus Grifola frondosa. FEMS Microbiol. Lett. 217:225-230. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.