Abstract
Members of the genus Acanthamoeba, amphizoic protozoan parasites, are causative agents of granulomatous amoebic encephalitis and amoebic keratitis. Proteinases play a role in various biologic actions in Acanthamoeba, including host tissue destruction, pathogenesis, and digestion of phagocytosed food. Interestingly, we found that encystation of Acanthamoeba was inhibited by the serine proteinase inhibitor phenylmethanesulfonyl fluoride. In this study, we characterize a serine proteinase that is involved in mediating the encystation of Acanthamoeba. This encystation-mediating serine proteinase (EMSP) is shown to be highly expressed during encystation by real-time PCR and Western blot analysis. Chemically synthesized small interfering RNA against EMSP inhibited the expression of EMSP mRNA and significantly reduced the encystation efficiency of Acanthamoeba. An EMSP-enhanced green fluorescent protein fusion protein localized to vesicle-like structures within the amoeba. Using LysoTracker analysis, these vesicular structures were confirmed to be lysosomes. After incubation of the transfected amoeba in encystment media, small fluorescent vesicle-like structures gathered and formed ball-like structures, which were identified as colocalizing with the autophagosome. Taken together, these results indicate that EMSP plays an important role in the differentiation of Acanthamoeba by promoting autolysis.
Acanthamoeba is an opportunistic pathogen responsible for several diseases in humans, such as granulomatous amoebic encephalitis and amoebic keratitis (16). Acanthamoeba exists in two stages, trophozoite and cyst forms. Trophozoites demonstrate movement, phagocytosis, metabolism, and cell division (11). This form becomes a cellulose-walled cyst by a process called encystation that occurs under unfavorable conditions such as desiccation, starvation, hyperosmolarity, or extreme temperature (5). Encystation is one of the survival strategies employed by the parasite to protect against the host immune response or chemical treatments (17). Because the cyst wall protects the parasite against harmful conditions, as well as against most antibiotics, it makes the treatment of this infection more difficult. Thus, disrupting the encystation process is essential to reducing the transmission of the disease and is a potential target for drug development.
Recently, encystation-mediating factors have been investigated intensively in an effort to discover the encystation mechanism of protozoan parasites. The cyst wall proteins 1, 2, and 3 of Giardia intestinalis (6, 26, 28), regulated in development A and B proteins (red A and red B) of Physarum polycephalum (1), and the lactate dehydrogenase and enolase of Toxoplasma gondii (7) have all been reported to mediate encystment. In the case of Acanthamoeba, only the cyst-specific protein 21 (4, 8) has been reported to function in encystation. Investigations of encystation-related gene profiles have identified xylose isomerase, Na P-type ATPase, and subtilisin-like serine proteinase genes as having increased expression during encystation of Acanthamoeba, compared to the trophozoite forms (19).
Proteases from various protozoan parasites have been characterized at the molecular and cellular levels, and the roles these enzymes play are now coming into focus (13). Central roles have been proposed for proteases in diverse processes such as host cell invasion and egress, encystation, excystation, catabolism of host proteins, differentiation, cell cycle progression, cytoadherence, and both stimulation and evasion of the host immune responses (13). Until now, the proteinases of Acanthamoeba have been shown to be involved primarily in pathogenesis or phagocytosis (9, 10, 12, 18, 24). However, little is known about the role of proteinases during the encystation of Acanthamoeba.
In this study, we demonstrate a role for proteinases during encystation of Acanthamoeba. We identified and characterized a serine proteinase as an encystation-mediated proteinase. Studies of this proteinase may serve to broaden our understanding of the encystation mechanism of Acanthamoeba.
MATERIALS AND METHODS
Amoeba cultivation and induction of encystation.
Two strains of Acanthamoeba obtained from the American Type Culture Collection (ATCC), A. castellanii strain Castellani (ATCC 30011; genotype T4), from contaminated eukaryotic cell culture, and A. healyi OC-3A (ATCC 30866; genotype T12), from the brain of a granulomatous amoebic encephalitis patient, were used in this study. They were cultured axenically in PYG (peptone-yeast-glucose) medium at 25°C in an incubator (Sanyo, San Diego, CA). Encystment was induced by the procedure of Bowers and Korn (3).
Zymogram analysis.
Protease activity in the trophozoites and cysts was determined using gelatin substrate gel electrophoresis. Culture media and cell lysates of A. castellanii were subjected to 11% sodium dodecyl sulfate-polyacrylamide gel electrophoresis with 0.1% gelatin at 4°C. After electrophoresis, gels were washed with agitation for 60 min at room temperature with 2.5% Triton X-100 and incubated overnight at 37°C in 0.1 M Tris-HCl buffer (pH 7.6). The gel was then stained with Coomassie blue. To check the inhibition of the proteolytic activity of A. castellanii, 10 μM trans-epoxysuccinyl-l-leucylamido(4-guanidin)butane (E64) or 1 mM phenylmethanesulfonyl fluoride (PMSF) was added (Sigma, St. Louis, MO).
Real-time PCR.
Total RNA was purified using Trizol reagent (Gibco BRL, Rockville, MD), and cDNA synthesis was conducted using a RevertAid first-strand cDNA synthesis kit (Fermentas, Hanover, IN). Real-time PCR (RT-PCR) was performed with the GeneAmp 5700 sequence detection system (Biosystems, Barcelona, Spain), using the default thermocycler program for all genes: 10 min of preincubation at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. Individual reactions were carried out in 20-μl volumes in a 96-well plate containing 1× buffer, 3.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, different concentrations of sense and antisense primers (Table 1), 0.025 U/μl enzyme, and Sybr green. All reaction mixtures were made using Sybr Premix Ex Taq (Takara, Otsu, Shiga, Japan). 18S ribosomal DNA was used for the reference gene.
TABLE 1.
Primer sequences for RT-PCR of proteinases
| Organism | Proteinase used for identificationa | Primer sequence
|
|
|---|---|---|---|
| Sense | Antisense | ||
| A. castellanii | Cysteine proteinase | 5′-CCGAGCAGAACCTCATCGATT | 5′-CCATGCCCTTGTTGTTGATGAT |
| Serine-type endopeptidase | 5′-GAAGGCGCTCACCGAATACAT | 5′-GGTTCGTCATCTGCTGATAGCC | |
| Peptidase M28 | 5′-TCGGTATCGTGACGGACTACGT | 5′-TAGCCGCACTTGGTGTTCTTG | |
| Aspartyl aminopeptidase-like protein | 5′-ACCATCGACATCGGTTTGC | 5′-TGAAGTCGGAGTAGAAGGCCTT | |
| Subtilisin-like serine proteinase | 5′-CAACAAGATGCTCTACACGCC | 5′-CGTAATCGGAGCCATCCAACA | |
| 18S rDNA | 5′-TCCAAT TTTCTGCCACCGAA | 5′-ATCATTACCCTAGTCCTCGCGC | |
| A. healyi | Subtilisin-like serine proteinase | 5′-TCAAGGTGCTCGGATGCAAT | 5′-ATGTTAGCCACAGACTGCGTC |
rDNA, ribosomal DNA.
Western blotting.
For production of the antibody against the encystation-mediating serine proteinase (EMSP), the GST gene fusion system was utilized (Amersham Biosciences, Buckinghamshire, England). The PCR product of the full-length EMSP of A. castellanii was ligated into the pGEX 4T-2 expression vector. Purified recombinant serine proteinase (>100 μg) was subcutaneously injected into 6-week-old SD rats (Bionics, Seoul, South Korea). Blood samples were collected from the rats before administering boosters, to monitor antibody production. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting were performed as described previously (15).
Transient transfection.
To investigate the intracellular localization of EMSP, the gene was cloned into the pUb vector with an enhanced green fluorescent protein (EGFP) as a marker (14). This plasmid was then transfected into live cells of A. healyi and A. castellanii. These strains were grown to mid-log phase, washed with phosphate-buffered saline, and then resuspended in PYG culture medium. Approximately 4 × 105 cells per well were seeded into a six-well culture plate in 3 ml of PYG medium and incubated overnight at 25°C. Transient transfection was performed by SuperFect transfection reagent (Qiagen, Hilden, Germany) as previously described (14).
Gene silencing methodology.
The small interfering RNA (siRNA) targeting the EMSP gene of A. healyi was synthesized by Sigma-Proligo (Boulder, CO), based on the cDNA sequence. The siRNA duplex with sense (5′-AGGAUCAGAACGCCUGUAAdTdT) and antisense (5′-UUACAGGCGUUCUGAUCCUdTdT) sequences was used. The siRNA (4 μg) was added to A. healyi trophozoites at a density of 4 × 105 cells. As a control, siRNA with a scrambled sequence absent in Acanthamoeba was used.
Microscopy.
Amoebae expressing EGFP were selected and allowed to adhere to a cell culture dish (BD Falcon). The cells were observed using an Olympus IX70 fluorescent microscope with a cooled charge-coupled-device camera (Roper Scientific, Tucson, AZ). EGFP fluorescence was achieved with a 500- to 530-nm band-pass filter. Images and time lapse photographs were acquired and analyzed through the Metamorph imaging system (Universal Imaging Corp., Downingtown, PA).
Transmission electron microscopy.
The cell suspensions were centrifuged for 10 min at 2,000 rpm and the sediments washed three times in cold phosphate-buffered saline. The sediments were prefixed with 4% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.2 to 7.4) for 2 h. After rinsing with 0.1 M cacodylate buffer, the sediments were postfixed with 1% osmium tetraoxide for 3 h, rinsed twice with 0.1 M maleate buffer (pH 5.2), dehydrated with ethyl alcohol, treated with propylene oxide-resin (1:1 dilution) overnight with continuous shaking, and then embedded in resin and incubated overnight at 60°C. Ultrathin sections cut on a Reichert-Jung ultramicrotome were stained with uranyl acetate and lead citrate. The sections were observed under a transmission electron microscope (H-7000; Hitachi, Tokyo, Japan).
RESULTS
Proteolytic activity during Acanthamoeba encystation.
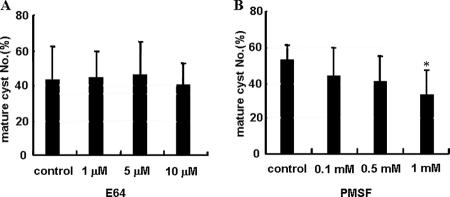
To compare the patterns of total proteolytic activity during encystation, gelatin zymograms were conducted with the cultured media of trophozoites and the cell lysate of trophozoite or cyst stages. Many new proteolytic bands appeared during encystation compared to the trophozoite stage (Fig. 1A). Almost all proteolytic activity induced by encystation was completely inhibited by the serine protease inhibitor PMSF (Fig. 1B). To examine the effects of proteinase activity on cyst formation, cysteine proteinase inhibitor (E64) or serine proteinase inhibitor (PMSF) was added to trophozoites in encystment media. While E64 did not demonstrate any effect on the formation of mature cysts (Fig. 2A), PMSF reduced the number of mature cysts in a dose-dependent manner (Fig. 2B).
FIG. 1.
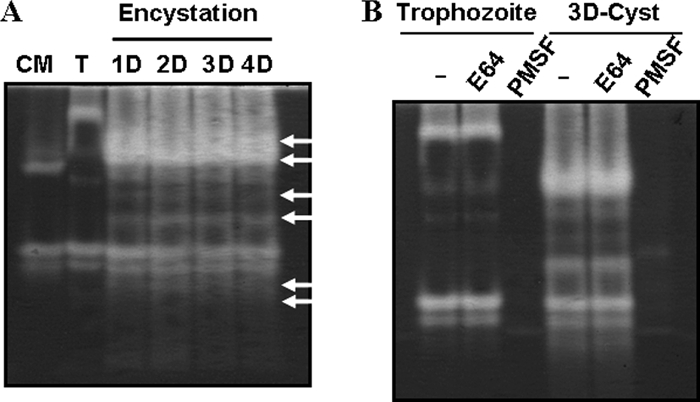
Gelatin zymogram analysis comparing the proteinase activities during encystation. The patterns of total proteolytic activity within culture media (CM), trophozoite (T), and cyst cell lysates (Day 1 [1D], 2D, 3D, and 4D) were compared by gelatin zymogram analysis (A). Arrows indicate new proteolytic bands that appear during encystation. Almost all proteolytic activity, induced by encystations, was inhibited by the serine protease inhibitor PMSF (B).
FIG. 2.
The effect of proteinase activity on encystation. E64 (cysteine proteinase inhibitor) did not affect the formation of mature cyst (A). PMSF (serine proteinase inhibitor) reduced the number of mature cysts in a dose-dependent manner (B). The experiments were repeated three times, and the average values are presented with error bars representing standard deviations. *, means are significantly different at a P value of <0.05 by Student's t test.
EMSP expression pattern during encystation.
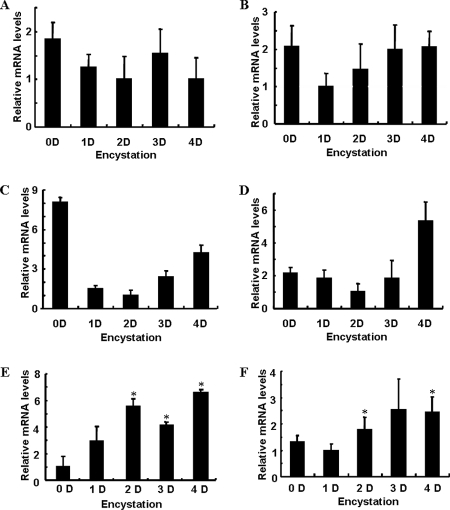
To identify the serine proteinase associated with encystation of Acanthamoeba, we screened expressed sequence tag (EST) data of encysted Acanthamoeba (20). Five kinds of proteinases were identified in cyst cDNA library ESTs. These included a cysteine proteinase, a serine-type endopeptidase, peptidase M28, an aspartyl aminopeptidase-like protein, and a subtilisin-like serine proteinase. To compare the mRNA expression level of each of these proteinases during encystation, we conducted RT-PCR (Fig. 3) during encystations. The expression levels of the cysteine proteinase, the serine-type endopeptidase, peptidase M28, and the aspartyl aminopeptidase-like protein were similar or lower than those measured at the trophozoite stage (Fig. 3A to D). However, the expression of the serine proteinase was highly induced during encystation (Fig. 3E). We named this gene EMSP. The EMSP of A. healyi also showed an increased expression during encystation compared to during the trophozoite stage (Fig. 3F).
FIG. 3.
Proteinase mRNA expression levels as measured by RT-PCR. The cysteine proteinase, serine-type endopeptidase, peptidase M28, and aspartyl aminopeptidase-like protein all showed similar or lowered mRNA expression levels during encystation (A to D). The serine proteinases of A. castellanii and A. healyi were highly expressed during encystation (E and F). *, means are significantly different at a P value of <0.05 by Student's t test.
EMSP genes of A. castellanii and A. healyi.
We identified the full-length open reading frame of this serine proteinase in A. castellanii (GenBank accession no. EU365402) and A. healyi (GenBank accession no. EU365404) by rapid amplification of cDNA ends PCR. Based on homology searches, the deduced amino acid sequences of the EMSP cDNAs of A. castellanii and A. healyi showed 77% and 78% similarities, respectively, with subtilisin-like serine proteinase. The phylogenetic tree with other subtilases, including prokaryotic, extracellular, intracellular fungal, and eukaryotic subtilases, indicated that EMSP belonged to the serine protease family subtilisin group (data not shown). Western blot analysis using antibodies against recombinant EMSP of A. healyi showed higher expression levels of the proteinase in encysting cells than to trophozoites (Fig. 4). The mature form of EMSP was detected as a band of about 33 kDa.
FIG. 4.
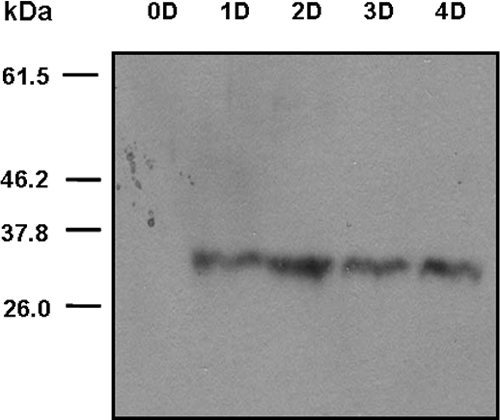
Western blot analysis of the polyclonal antibody prepared against a recombinant PMSF protein. The mature form of EMSP was detected as a 33-kDa band during encystation (Day 1 [1D], 2D, 3D, and 4D).
Effect of the EMSP gene on the encystation.
To confirm the role of EMSP during encystation of Acanthamoeba, gene silencing via siRNA was performed. As indicated by fluorescence-activated cell sorter analysis, 99% of the amoebae were determined to have been transfected. In siRNA-transfected cells, EMSP was not expressed at a higher level than that measured in trophozoites by RT-PCR (Fig. 5). Furthermore, the formation of mature cysts was almost completely inhibited in EMSP siRNA-transfected cells (Fig. 6).
FIG. 5.
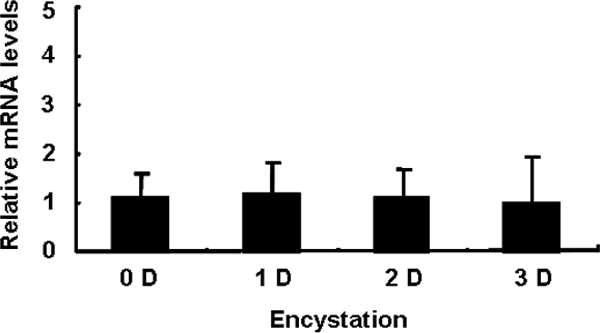
RT-PCR after transfection with EMSP siRNA. After transfection, cells were transferred to encystment media and gathered during encystation from 0 to 3 days. RT-PCR analysis demonstrates that EMSP levels of the trophozoite and cyst forms were similar. The means are not significantly different at a P value of <0.05 by Student's t test.
FIG. 6.
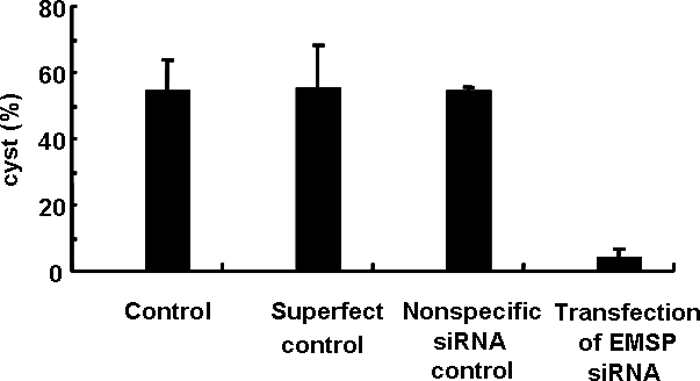
Inhibition of encystation by EMSP siRNA. Cells transfected with SuperFect transfection reagent, nonspecific negative siRNA, or EMSP siRNA were incubated in encystment media for 4 days, and the numbers of mature cysts were scored. The formation of mature cysts was almost completely inhibited in EMSP siRNA-transfected cells.
Intracellular localization of EMSP during encystation.
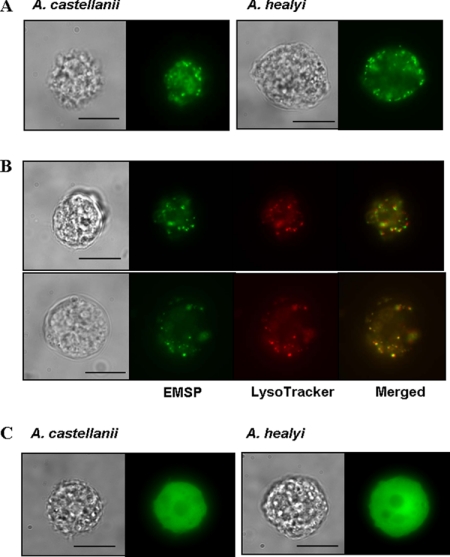
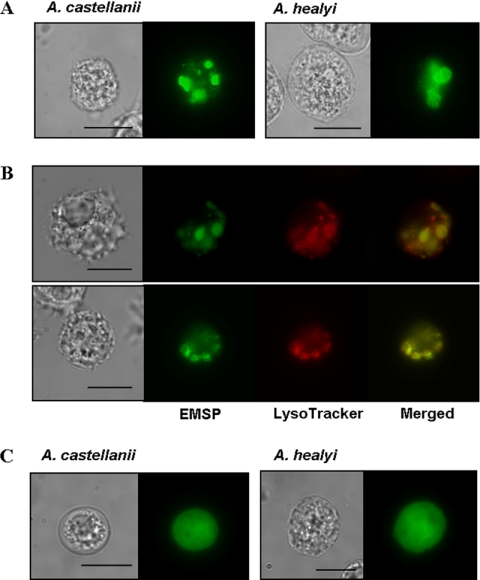
The amoebae expressing an EMSP-EGFP fusion protein showed fluorescent vesicle-like structures (Fig. 7A) distributed in the cytoplasm of trophozoites and that colocalized with the lysosome stain LysoTracker red DND-99 (Fig. 7B). In contrast, the fluorescence of EGFP alone showed dispersed distribution in the cytoplasm (Fig. 7C). When transfected amoebae were transferred to encystment media, the small fluorescent vesicles gathered together and formed ball-like structures within 24 h (Fig. 8A). These ball-like structures (green) were colocalized with the autophagosome marker, LysoTracker (red) (Fig. 8B) (2, 22). We propose that these structures are likely the autophagosome. The fluorescence of EGFP alone showed dispersed distribution in the cytoplasm during encystation (Fig. 8C). The 65-h encystment cells were observed with electron microscopy (Fig. 9). In the control cells, there were large vacuoles that include digested cytoplasmic contents (Fig. 9A). However, EMSP siRNA-transfected cells showed large vacuoles containing undigested mitochondria and cytoplasmic materials within the autophagosome (Fig. 9B).
FIG. 7.
Transfection of pUbEMSPg for the intracellular localization of EMSP. The localization of the EMSP-EGFP fusion protein showed fluorescent vesicle-like structures distributed within the cytoplasm (A). Expressed EMSP-EGFP fusion proteins (green signal) completely overlapped with lysosomes stained with LysoTracker red DND-99 (red signal) in A. castellanii (panel B, top) and A. healyi (panel B, bottom). Amoebae with the fluorescence of EGFP alone showed dispersed distribution in the cytoplasm (C). Bar = 10 μm.
FIG. 8.
Localization of EMSP during encystation. After transfection of plasmid encoding the EMSP-EGFP fusion protein, cells were transferred to encystment media and incubated for 24 to 48 h. Small fluorescent vesicle-like structures gathered and formed ball-like structures (A). These ball-like structures (green signal) colocalized with LysoTracker red DND-99 staining in A. castellanii (panel B, top) and A. healyi (panel B, bottom). Amoebae with the fluorescence of EGFP alone showed dispersed distribution in the cytoplasm during encystation (C). Bar = 10 μm.
FIG. 9.
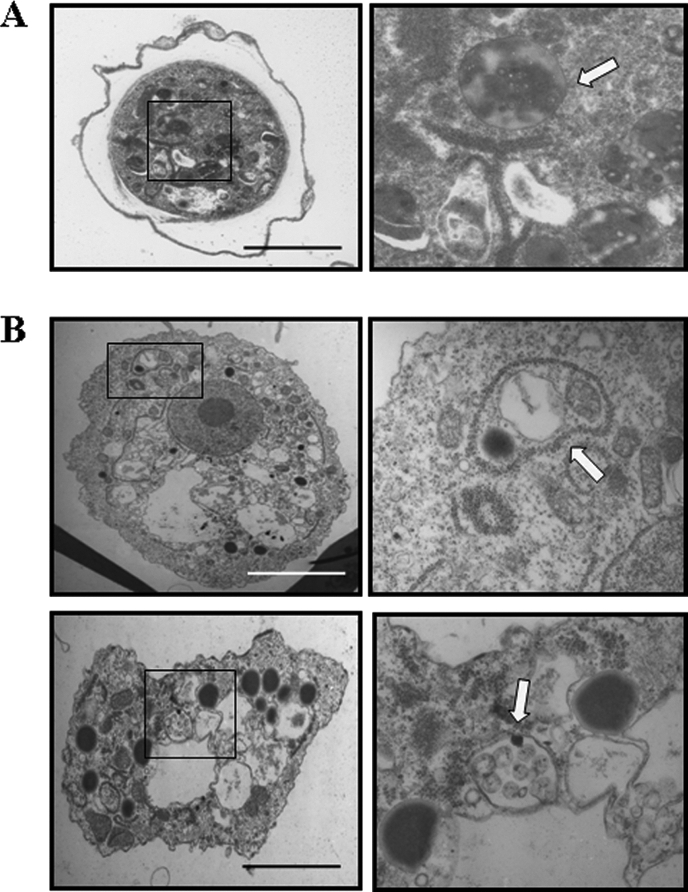
The morphology of the autophagosome after transfection of EMSP specific siRNA. Control cells and cells transfected with EMSP siRNA were fixed after 65 h of incubation in encystment media. As shown in panel A, autophagosome in mature cyst digested almost all of the cytoplasmic contents. siRNA-transfected cells showed undigested cytoplasmic materials and organelles within the autophagosome (B). Arrows, autophagosome; bar, 10 μm.
DISCUSSION
In our previous report, EST data of the encysting Acanthamoeba identified three copies of subtilisin-like serine proteinases (20). Using differentially expressed gene (DEG) screening, comparing trophozoite and cyst stages, we also identified DEG14, which showed a 77% similarity to subtilisin-like serine proteinases (19). DEG14 was found to be identical to EMSP by sequence analysis. It is possible that the common expression of EMSP during encystation indicates an important role of this proteinase during encystation of Acanthamoeba.
Most research on the role of proteolytic enzymes in parasitic protozoa, including Acanthamoeba, was focused on pathogenesis and phagocytosis. Recently, the roles of proteases during differentiation, including encystation and excystation, were confirmed. The encystation-specific cysteine proteinase of Giardia lamblia was shown to be required for cyst wall formation (28). In Entamoeba invadens, specific cysteine proteinase inhibitors significantly reduced the efficiency of encystation (25). In filamentous fungi, the subtilisin-like serine protease of Podospora anserina (PSPA) was shown to be induced during the vegetative incompatibility reaction (23).
Most subtilases have been reported to be secretory enzymes with roles during pathogenesis (10, 21), with the exception of PSPA, and protease B of S. cerevisiae has been associated with the autophagosome (23, 27). The result of phylogenetic analysis of 11 subtilisin sequences also supported the intracellular localization of PSPA. Protease B is also a vacuolar protease involved in autophagy (27). The phylogenetic tree (data not shown) indicated that EMSP formed a clade with a serine proteinase of A. healyi, a reported secretory enzyme and possible virulence factor (10). Based on the intracellular localization and trafficking of EMSP during encystation, EMSP may be associated with autolysis functions during the process of molecular recycling.
Electron microscopic results supplemented the evidence for EMSP being an autophagosomal enzyme. Transmission electron microscopy analysis of EMSP siRNA-treated amoebae revealed the defects in substance degradation and autophagy maturation during encystation. However, further study will be necessary to confirm this premise.
Furthering our understanding of the molecular mechanisms required during encystation may provide important targets for drug development that prevents cyst formation in pathogenic protozoa. As demonstrated in this study, many new proteolytic bands appear during encystation. A detailed investigation on these proteolytic bands may identify other factors mediating encystation of Acanthamoeba. Based on our results, it is possible that inhibition of EMSP may improve the therapeutic efficacy of amoebicidal drugs by interrupting encystation of Acanthamoeba.
Acknowledgments
This work was supported by the Korea Research Foundation grant funded by the Korean government (MOEHRD, Basic Research Promotion Fund; KRF-2005-204-E00027) and the Brain Korea 21 Project in 2007.
Footnotes
Published ahead of print on 1 August 2008.
REFERENCES
- 1.Bailey, J., L. J. Cook, R. Kilmer-Barber, E. Swanston, L. Solnica-Krezel, K. Lohman, W. F. Dove, J. Dee, and R. W. Anderson. 1999. Identification of three genes expressed primarily during development in Physarum polycephalum. Arch. Microbiol. 172364-376. [DOI] [PubMed] [Google Scholar]
- 2.Bampton, E. T., C. G. Goemans, D. Niranjan, N. Mizushima, and A. M. Tolkovsky. 2005. The dynamics of autophagy visualized in live cells: from autophagosome formation to fusion with endo/lysosomes. Autophagy 123-36. [DOI] [PubMed] [Google Scholar]
- 3.Bowers, B., and E. D. Korn. 1969. The fine structure of Acanthamoeba castellanii (Neff strain). II. Encystment. J. Cell Biol. 41786-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, L., T. Orfeo, G. Gilmartin, and E. Bateman. 2004. Mechanism of cyst specific protein 21 mRNA induction during Acanthamoeba differentiation. Biochim. Biophys. Acta 169123-31. [DOI] [PubMed] [Google Scholar]
- 5.Cordingley, J. S., R. A. Wills, and C. L. Villemez. 1996. Osmolarity is an independent trigger of Acanthamoeba castellanii differentiation. J. Cell Biochem. 61167-171. [DOI] [PubMed] [Google Scholar]
- 6.Davis-Hayman, S. R., J. R. Hayman, and T. E. Nash. 2003. Encystation-specific regulation of the cyst wall protein 2 gene in Giardia lamblia by multiple cis-acting elements. Int. J. Parasitol. 331005-1012. [DOI] [PubMed] [Google Scholar]
- 7.Dzierszinski, F., M. Mortuaire, N. Dendouga, O. Popescu, and S. Tomavo. 2001. Differential expression of two plant-like enolases with distinct enzymatic and antigenic properties during stage conversion of the protozoan parasite Toxoplasma gondii. J. Mol. Biol. 3091017-1027. [DOI] [PubMed] [Google Scholar]
- 8.Hirukawa, Y., H. Nakato, S. Izumi, T. Tsuruhara, and S. Tomino. 1998. Structure and expression of a cyst specific protein of Acanthamoeba castellanii. Biochim. Biophys. Acta 139847-56. [DOI] [PubMed] [Google Scholar]
- 9.Hong, Y. C., M. Y. Hwang, H. C. Yun, H. S. Yu, H. H. Kong, T. S. Yong, and D. I. Chung. 2002. Isolation and characterization of a cDNA encoding a mammalian cathepsin L-like cysteine proteinase from Acanthamoeba healyi. Korean J. Parasitol. 4017-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hong, Y. C., H. H. Kong, M. S. Ock, I. S. Kim, and D. I. Chung. 2000. Isolation and characterization of a cDNA encoding a subtilisin-like serine proteinase (ahSUB) from Acanthamoeba healyi. Mol. Biochem. Parasitol. 111441-446. [DOI] [PubMed] [Google Scholar]
- 11.Khan, N. A. 2007. Acanthamoeba invasion of the central nervous system. Int. J. Parasitol. 37131-138. [DOI] [PubMed] [Google Scholar]
- 12.Kim, W. T., H. H. Kong, Y. R. Ha, Y. C. Hong, H. J. Jeong, H. S. Yu, and D. I. Chung. 2006. Comparison of specific activity and cytopathic effects of purified 33 kDa serine proteinase from Acanthamoeba strains with different degree of virulence. Korean J. Parasitol. 44321-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klemba, M., and D. E. Goldberg. 2002. Biological roles of proteases in parasitic protozoa. Annu. Rev. Biochem. 71275-305. [DOI] [PubMed] [Google Scholar]
- 14.Kong, H. H., and T. D. Pollard. 2002. Intracellular localization and dynamics of myosin-II and myosin-IC in live Acanthamoeba by transient transfection of EGFP fusion proteins. J. Cell Sci. 154993-5002. [DOI] [PubMed] [Google Scholar]
- 15.Legesse-Miller, A., Y. Sagiv, R. Glozman, and Z. Elazar. 2000. Aut7p, a soluble autophagic factor, participates in multiple membrane trafficking processes. J. Biol. Chem. 2032966-32973. [DOI] [PubMed] [Google Scholar]
- 16.Marciano-Cabral, F., and G. Cabral. 2003. Acanthamoeba spp. as agents of disease in humans. Clin. Microbiol. Rev. 16273-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McClellan, K., K. Howard, E. Mayhew, J. Niederkorn, and H. Alizadeh. 2002. Adaptive immune responses to Acanthamoeba cysts. Exp. Eye Res. 75285-293. [PubMed] [Google Scholar]
- 18.Mitro, K., A. Bhagavathiammai, O. M. Zhou, G. Bobbett, J. H. McKerrow, R. Chokshi, B. Chokshi, and E. R. James. 1994. Partial characterization of the proteolytic secretions of Acanthamoeba polyphaga. Exp. Parasitol. 78377-385. [DOI] [PubMed] [Google Scholar]
- 19.Moon, E. K., D. I. Chung, Y. C. Hong, and H. H. Kong. 2007. Differentially expressed genes of Acanthamoeba castellanii during encystation. Korean J. Parasitol. 45283-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moon, E. K., D. I. Chung, Y. C. Hong, T. I. Ahn, and H. H. Kong. 2008. Acanthamoeba castellanii: gene profile of encystations by ESTs analysis and KOG assignment. Exp. Parasitol. 119111-116. [DOI] [PubMed] [Google Scholar]
- 21.Moon, E. K., S. T. Lee, D. I. Chung, and H. H. Kong. 2006. Intracellular localization and trafficking of serine proteinase AhSub and cysteine proteinase AhCP of Acanthamoeba healyi. Eukaryot. Cell 5125-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munafó, D. B., and M. I. Colombo. 2001. A novel assay to study autophagy: regulation of autophagosome vacuole size by amino acid deprivation. J. Cell. Sci. 1143619-3629. [DOI] [PubMed] [Google Scholar]
- 23.Paoletti, M., M. Castroviejo, J. Begueret, and C. Clave. 2001. Identification and characterization of a gene encoding a subtilisin-like serine protease induced during the vegetative incompatibility reaction in Podospora anserina. Curr. Genet. 39244-252. [DOI] [PubMed] [Google Scholar]
- 24.Serrano-Luna, J. de, J., I. Cervantes-Sandoval, J. Calderon, F. Navarro-Garcia, V. Tsutsumi, and M. Shibayama. 2006. Protease activities of Acanthamoeba polyphaga and Acanthamoeba castellanii. Can. J. Microbiol. 5216-23. [DOI] [PubMed] [Google Scholar]
- 25.Sharma, M., K. Hirata, S. Herdman, and S. Reed. 1996. Entamoeba invadens: characterization of cysteine proteinases. Exp. Parasitol. 8484-91. [DOI] [PubMed] [Google Scholar]
- 26.Sun, C. H., J. M. McCaffery, D. S. Reiner, and F. D. Gillin. 2003. Mining the Giardia lamblia genome for new cyst wall proteins. J. Biol. Chem. 27821701-21708. [DOI] [PubMed] [Google Scholar]
- 27.Takeshige, K., M. Baba, S. Tsuboi, T. Noda, and Y. Ohsumi. 1992. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 119301-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Touz, M. C., M. J. Nores, I. Slavin, C. Carmona, J. T. Conrad, M. R. Mowatt, T. E. Nash, C. E. Coronel, and H. D. Lujan. 2002. The activity of a developmentally regulated cysteine proteinase is required for cyst wall formation in the primitive eukaryote Giardia lamblia. J. Biol. Chem. 2778474-8481. [DOI] [PubMed] [Google Scholar]