Abstract
This study explored whether wildlife species serve as the reservoir for human Candida albicans strains in a given geographic area. C. albicans isolates were collected from nonmigratory wildlife admitted to the University of Illinois Wildlife Medical Clinic. A geographically and temporally matched set of C. albicans oral isolates was collected from healthy human volunteers. Multilocus sequence typing was used to assign strains to genetic clades. Clade 1 isolates, particularly diploid sequence type 69 (DST 69), were most common in humans. Clade 1 strains were less frequently recovered from wildlife, while clade 8 strains, particularly DST 90, were overrepresented in the wildlife collection. All instances where a wildlife and human isolate shared the same DST occurred within clade 1. Clade distributions between human and wildlife isolates were significantly different, demonstrating population isolation between the groups. These differences may indicate limited strain transfer between groups or differential selection of C. albicans isolates in humans and wildlife. Wildlife strains had an amphotericin B MIC significantly lower than that of human isolates; strains with increased susceptibility were from several clades. C. albicans isolates were collected from domestic animals to provide comparisons with human and wildlife data sets. C. albicans isolation from canine and feline oral and anal swabs was infrequent; companion animal isolates were closely related to clade 1 human isolates. Collectively, the data suggest a greater likelihood of C. albicans transfer from humans to animals than from animals to humans. The nontransient human population may maintain the connection between geography and the C. albicans genetic groups recovered from humans.
Candida species are common commensals of mucosal surfaces in the human body and are the leading cause of human fungal infections. Candida species are the fourth most common cause of hospital-acquired bloodstream infections in the United States, affecting as many as 10,000 to 40,000 patients annually (reviewed in reference 36). Candida bloodstream infections have a crude mortality rate of approximately 40%. Candida species are also a common cause of vulvovaginitis, which is experienced by approximately two-thirds of women, with nearly 50% of women suffering multiple episodes (reviewed in references 42 and 43). For each type of disease, Candida albicans is the most commonly isolated Candida species.
Although C. albicans is commonly regarded as a commensal of domestic animals and wildlife (11, 18, 41, 48), the prevalence of C. albicans in animal populations is far from being as well documented as it is in humans. Compared to the incidence of candidiasis in humans, it is clear that Candida-caused disease is encountered relatively infrequently in veterinary medicine. In fact, instances of systemic disease are noteworthy enough to be described in published case reports (9, 17, 25). Cutaneous disease, mastitis, stomatitis, and candidiasis of the mucous membranes of the digestive, genital, and urinary tracts have also been described for various species (23, 29, 38, 51). Many other species of Candida in addition to C. albicans have been isolated from clinical cases (12, 24, 26). However, the literature does not contain a report of a systematic attempt to determine the frequency of Candida carriage in animals.
Previous epidemiological studies using genetic fingerprinting with a complex probe (Ca3 [44]) and with the more recently developed multilocus sequence typing (MLST) method (8) have shown that C. albicans strains can be divided into distinct genetic clades (33, 44). While strains from different clades can be found side by side in the same locale, clade distribution varies between geographical locations (33, 44). This finding is perhaps unanticipated, considering that human travel has the potential to homogenize the worldwide distribution of C. albicans clades. These observations suggest that a local reservoir of C. albicans strains may maintain the association between certain clades and a specific geographic area (39, 40, 44). This work investigated the possibility that local nonmigratory wildlife species serve as a reservoir for human C. albicans isolates. We utilized MLST to construct a molecular phylogeny of C. albicans isolates collected from predominantly healthy wildlife species and from normally healthy adult humans who live in the same geographic area in central Illinois. Antifungal susceptibilities were also determined for these isolates. Because domestic animals may play a role in the transfer of C. albicans strains between humans and wildlife, the analysis was expanded to include isolates from domestic animals that reside in the same geographic area.
MATERIALS AND METHODS
Collection of wildlife C. albicans specimens.
Oral and anal/cloacal swabs were collected from live nonmigratory wildlife species immediately upon their arrival at the University of Illinois at Urbana-Champaign (UIUC) College of Veterinary Medicine (CVM) Wildlife Medical Clinic during the years 2003, 2004, and 2007. Information was collected regarding the health status of each animal, as well as the location at which it was found, its species, and its approximate age. Of the 210 animals sampled for this study, 60% presented as the result of trauma (animal attack, hit by car, etc.), 18% were healthy orphans, and 19% had signs of disease. The condition of the remaining animals was not specified. The study group was 43% avian (the most common species were crows and red-tailed hawks [11% each]), 52% mammalian (48% rabbits, 21% squirrels), and 5% reptilian (91% turtles, 9% snakes). Swabs were collected immediately upon presentation at the clinic to minimize the possibility that resulting C. albicans isolates were derived from human contact. The use of protective exam gloves to handle animals further reduced the possibility that C. albicans isolates were of human origin. Swabs were plated on CHROMagar Candida (34) and incubated at 37°C for 48 h. Green colonies were presumed to be C. albicans and streaked for purification. Verification that the isolate was C. albicans was accomplished using two PCR-based methods (3, 28). Isolates were frozen in 15% glycerol at −80°C. A total of 28 isolates from 22 animals are detailed in Table 1. Many other yeast species (not C. albicans) were recovered from the CHROMagar plates. C. albicans was not the most frequent yeast isolate collected from wildlife species; however, a detailed inventory of the other yeast species was not compiled.
TABLE 1.
C. albicans isolates from wildlife and domestic animalsa
| Species | Codeb | Swab sitec | Suscep. testd | Agee | Locationf | Healthg | ABC type | MAT locus | Cladeh | DSTi | Genotype designation for indicated genei
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AAT1a | ACC1 | ADP1 | PMI1b | SYA1 | VPS13 | ZWF1b | |||||||||||
| Dog | ZMK9 | O | Priv. owner | H | A | a/α | 1 | 66 | 2 | 3 | 5 | 2 | 2 | 6 | 5 | ||
| Cat | C69M | O | CCHS | H | A | a/α | 1 | 69 | 2 | 5 | 5 | 2 | 2 | 6 | 5 | ||
| Cat | C79M | O | CCHS | H | A | a/α | 1 | 69 | 2 | 5 | 5 | 2 | 2 | 6 | 5 | ||
| Squirrel | SqB056 | O | J | Urbana | H | A | a/α | 1 | 69 | 2 | 5 | 5 | 2 | 2 | 6 | 5 | |
| Rabbit | RbA025 | O | X | A | Champaign | T | A | a/α | 1 | 103 | 2 | 3 | 6 | 2 | 2 | 6 | 5 |
| Squirrel | SqA010 | O | X | A | Champaign | T | A | a/α | 1 | 127 | 2 | 3 | 6 | 2 | 2 | 20 | 5 |
| Dog | D19M | O | Priv. owner | H | A | a/α | 1 | 408 | 2 | 5 | 5 | 2 | 2 | 20 | 5 | ||
| Squirrel | SqB030 | A | X | J | Urbana | H | A | a/α | 1 | 485 | 2 | 5 | 5 | 2 | 2 | 6 | 20 |
| Raccoon | RcB036 | O | X | J | Oakland | T | A | a/α | 1 | 1089* | 2 | 3 | 6 | 9 | 2 | 6 | 5 |
| Starling | StA0524 | O | X | N | Champaign | H | A | a/α | 1 | 1102* | 4 | 67* | 5 | 2 | 126* | 148 | 5 |
| Rabbit | RbC078 | O | X | J | Rantoul | T | A | a/α | 1 | 1111* | 2 | 5 | 5 | 77* | 2 | 6 | 5 |
| Raccoon | RcA013* | O | X | J | Champaign | D | B | a/α | 3 | 1100* | 13 | 66* | 15 | 6 | 7 | 32 | 15 |
| Raccoon | RcC0556 | O | X | J | Champaign | H | B | a/α | 3 | 1104* | 13 | 10 | 15 | 4 | 7 | 32 | 12 |
| Squirrel | SqC075* | O | X | A | Urbana | T | B | a/α | 3 | 1110* | 13 | 10 | 15 | 6 | 7 | 32 | 15 |
| Rabbit | RbB043* | O | X | A | Urbana | T | B | a/α | 4 | 1101* | 8 | 14 | 6 | 4 | 7 | 10 | 8 |
| Cow | 7452 | O | UIUC dairy | H | A | a/α | 8 | 90 | 25 | 7 | 6 | 3 | 6 | 27 | 37 | ||
| Deer | DB053 | O | X | J | Villa Grove | T | C | a/α | 8 | 90 | 25 | 7 | 6 | 3 | 6 | 27 | 37 |
| Opossum | OpA052 | A | X | A | Bloomington | T | A | a/α | 8 | 90 | 25 | 7 | 6 | 3 | 6 | 27 | 37 |
| Squirrel | SqH001 | O | X | A | Champaign | T | C | a/α | 8 | 90 | 25 | 7 | 6 | 3 | 6 | 27 | 37 |
| Woodchuck | WcA017 | A | X | A | Urbana | D | C | a/α | 8 | 90 | 25 | 7 | 6 | 3 | 6 | 27 | 37 |
| Woodchuck | WcA019 | O | A | Urbana | D | C | a/α | 8 | 90 | 25 | 7 | 6 | 3 | 6 | 27 | 37 | |
| Squirrel | SqH099 | A | A | Champaign | T | C | a/a | 8 | 729 | 25 | 7 | 6 | 3 | 24 | 27 | 37 | |
| Deer | DA047 | O | X | J | Ludlow | T | A | a/α | 8 | 785 | 25 | 7 | 6 | 3 | 6 | 45 | 37 |
| Opossum | OpA059 | O | A | Bloomington | T | B | a/α | 8 | 1105* | 33 | 14 | 38 | 2 | 78 | 47 | 12 | |
| Opossum | OpA060 | O | A | Bloomington | T | A | a/α | 8 | 1106* | 33 | 7 | 6 | 3 | 6 | 27 | 37 | |
| Squirrel | SqD080 | O | X | A | Rantoul | T | B | a/α | 8 | 1112* | 33 | 3 | 38 | 2 | 78 | 122 | 15 |
| Red-tailed hawk | RhA029 | O | X | NS | Champaign | T | A | a/α | 9 | 918 | 62 | 3 | 3 | 3 | 3 | 39 | 95 |
| Squirrel | SqF087 | O | X | A | Champaign | NS | A | a/α | 9 | 1103* | 62 | 3 | 3 | 3 | 26 | 39 | 95 |
| Red-eared slider | RsA027* | O | X | A | Springfield | T | B | a/α | 10 | 304 | 4 | 7 | 16 | 7 | 13 | 19 | 14 |
| Squirrel | SqE086* | O | X | J | Urbana | T | C | a/α | 11 | 461 | 60 | 10 | 21 | 1 | 7 | 11 | 15 |
| Squirrel | SqE097* | O | J | Urbana | T | C | a/α | 11 | 1113* | 60 | 7 | 10 | 1 | 7 | 11 | 15 | |
| Crow | CrA038* | O | X | A | Champaign | D | A | a/α | Sing. | 732 | 5 | 32 | 21 | 34 | 7 | 55 | 5 |
| Squirrel | SqG098 | O | X | J | Urbana | T | B | a/α | Sing. | 1114* | 5 | 7 | 24 | 3 | 34 | 65 | 6 |
Entries corresponding to wildlife isolates are in lightface; boldface indicates isolates from domestic animals.
The code for each strain includes a species abbreviation and also strain number. This code is used to identify the isolate on the UPGMA tree in Fig. 2. Strains marked with an asterisk are missing one ALS5 allele as determined by a PCR-based assay (50). None of the animal isolates was missing both ALS5 alleles.
O, oral swab; A, anal swab.
C. albicans isolates used for antifungal susceptibility testing (suscept. test) are marked with an X. Only one isolate from each animal was included in the statistical analysis.
J, juvenile; A, adult; N, nestling; NS, not specified.
The approximate geographic location of the animal is indicated. City names are used to designate the approximate locations where wildlife were found. City names can be found on the map in Fig. 1. For domestic animals, animals belonging to private owners are indicated by “Priv. owner.” CCHS, Champaign County Humane Society. One cow isolate was obtained from the university's dairy farm (see Materials and Methods).
H, healthy; T, trauma; D, signs of disease; NS, not specified.
Sing., singleton; could not be assigned to a currently defined clade.
DSTs and genotype designations were determined from comparisons between experimental sequences and the MLST database at http://test1.mlst.net. Asterisks indicate new DSTs and genotypes that were identified in this study.
Collection of human C. albicans specimens.
Oral swabs were collected from normally healthy adult volunteers in 2007. All procedures were conducted under the guidance of the UIUC Institutional Review Board (IRB). Informed consent was obtained from each study volunteer. Volunteers were recruited from among those attending the annual UIUC CVM Open House. This event draws people from around the region, providing a reasonably unbiased sample of individuals from the same geographical region from which the wildlife species originated. Volunteers provided basic demographic information (gender, age, race/ethnicity) as well as information about the location of current and previous residences, the length of residence in each location, and recent travel history. Of the 246 study volunteers, 70% were female. Ages were represented nearly equally within the group, with 20% of volunteers in each of the following categories: 18 to 25 years of age, 26 to 35 years of age, and 36 to 45 years of age. The 46- to 55-year group had 24% of the study subjects, while the remaining subjects were in the 56- to 65-year group. Ninety-one percent of study subjects reported white as their racial group, while 4% reported African-American, 1% reported Asian, 2% chose the “more than one race” option, and 2% did not answer. Three percent of volunteers indicated Hispanic or Latino ethnicity. Volunteers collected an oral swab, which was then plated on CHROMagar Candida and incubated for 48 h at 37°C. A total of 72 plates (29%) were positive for C. albicans, and the identity of the isolates was verified using the PCR procedures described above. The final group of C. albicans-positive volunteers was 68% female and 96% white, with one Hispanic/Latino individual. Slightly more than half of the final volunteer pool was from the 26- to 35-year and 36- to 45-year age categories (30% and 25%, respectively). Forty-four of these specimens were from volunteers who lived in the geographic region defined by the wildlife isolates; details about these isolates are shown in Table 2. The remaining 28 isolates were frozen into the laboratory strain collection and not analyzed further.
TABLE 2.
Oral C. albicans isolates from healthy volunteers
| Codea | Residenceb | Curr. res. (mo)c | Geo. area (mo)c | Travel (wks/yr)d | ABC type | MAT locus | Cladee | DSTf | Genotype designation for indicated genef
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AAT1a | ACC1 | ADP1 | PMI1b | SYA1 | VPS13 | ZWF1b | |||||||||
| 1-21 | Savoy | 20 | 20 | 7.1 | A | a/α | 1 | 24 | 2 | 5 | 5 | 2 | 2 | 24 | 5 |
| 1-39 | Atwood | 156 | 516 | 0 | A | a/α | 1 | 24 | 2 | 5 | 5 | 2 | 2 | 24 | 5 |
| 1-5 | Mahomet | 48 | 84 | NA | A | a/α | 1 | 66 | 2 | 3 | 5 | 2 | 2 | 6 | 5 |
| 1-221 | Champaign | 24 | 84 | 1 | A | a/α | 1 | 66 | 2 | 3 | 5 | 2 | 2 | 6 | 5 |
| 2-117 | A | a/α | 1 | 66 | 2 | 3 | 5 | 2 | 2 | 6 | 5 | ||||
| 1-24 | Tolono | 108 | >108 | 3 | A | a/α | 1 | 69 | 2 | 5 | 5 | 2 | 2 | 6 | 5 |
| 1-25 | Tolono | 108 | >108 | 3 | A | a/α | 1 | 69 | 2 | 5 | 5 | 2 | 2 | 6 | 5 |
| 1-93 | Champaign | 156 | >156 | 1 | A | a/α | 1 | 69 | 2 | 5 | 5 | 2 | 2 | 6 | 5 |
| 1-151 | Champaign | 8 | 128 | 2 | A | a/α | 1 | 69 | 2 | 5 | 5 | 2 | 2 | 6 | 5 |
| 1-161 | Champaign | 36 | 96 | 2 | A | a/α | 1 | 69 | 2 | 5 | 5 | 2 | 2 | 6 | 5 |
| 1-216 | Urbana | 24 | 24 | 4 | A | a/α | 1 | 69 | 2 | 5 | 5 | 2 | 2 | 6 | 5 |
| 1-155 | Champaign | 12 | 12 | 2 | A | a/α | 1 | 79 | 2 | 5 | 5 | 9 | 2 | 6 | 5 |
| 1-177* | Champaign | 12 | 12 | 0 | A | a/α | 1 | 103 | 2 | 3 | 6 | 2 | 2 | 6 | 5 |
| 1-95 | Champaign | 12 | >12 | 1 | A | a/α | 1 | 184 | 3 | 5 | 5 | 2 | 2 | 6 | 5 |
| 1-107 | Tolono | 72 | >72 | 0.5 | A | a/α | 1 | 185 | 2 | 5 | 5 | 9 | 2 | 24 | 5 |
| 2-138 | A | a/α | 1 | 277 | 2 | 2 | 5 | 2 | 2 | 6 | 5 | ||||
| 1-86 | Springfield | 180 | 180 | 6 | A | a/α | 1 | 417 | 2 | 3 | 5 | 2 | 2 | 24 | 5 |
| 1-238 | Urbana | 180 | 180 | 1 | A | a/α | 1 | 426 | 2 | 5 | 5 | 2 | 2 | 106 | 5 |
| 1-204 | Champaign | 10 | 10 | 4 | A | a/α | 1 | 572 | 8 | 5 | 5 | 2 | 2 | 6 | 5 |
| 1-44 | Rantoul | 30 | 30 | 3 | A | a/α | 1 | 939 | 2 | 2 | 2 | 4 | 2 | 24 | 5 |
| 1-23* | Urbana | 60 | 180 | 4 | A | a/α | 1 | 1087* | 2 | 3 | 5 | 76* | 2 | 20 | 5 |
| 1-52 | Urbana | 31 | 31 | 12 | A | a/α | 1 | 1088* | 2 | 5 | 5 | 9 | 2 | 24 | 146* |
| 1-54 | Champaign | 240 | >240 | 2 | A | a/α | 1 | 1089* | 2 | 3 | 6 | 9 | 2 | 6 | 5 |
| 1-110 | Rantoul | 36 | >36 | 1 | A | a/α | 1 | 1091* | 2 | 3 | 6 | 2 | 2 | 6 | 49 |
| 1-139 | Normal | 132 | >132 | 1 | A | a/α | 1 | 1092* | 2 | 5 | 5 | 2 | 50 | 6 | 5 |
| 1-163 | Champaign | 36 | 36 | 2 | A | a/α | 1 | 1093* | 101* | 5 | 5 | 2 | 2 | 6 | 12 |
| 1-197 | Fisher | 360 | 432 | 1 | A | a/α | 1 | 1116* | 13 | 2 | 5 | 2 | 2 | 6 | 5 |
| 1-92 | Champaign | 180 | 204 | 0.5 | A | a/α | 1 | 1117* | 2 | 3 | 5 | 2 | 2 | 175* | 147* |
| 2-210 | A | a/α | 1 | 1131* | 2 | 69* | 5 | 2 | 2 | 6 | 5 | ||||
| 2-215 | A | a/α | 1 | 1133* | 2 | 5 | 5 | 2 | 2 | 5 | 5 | ||||
| 1-178 | Bloomington | 156 | 156 | 4 | A | α/α | 2 | 1094* | 35 | 2 | 14 | 4 | 4 | 24 | 4 |
| 1-203* | Savoy | 12 | 12 | 2 | A | a/α | 2 | 1096* | 35 | 4 | 14 | 4 | 4 | 4 | 4 |
| 1-246* | Sidney | 72 | 72 | 4 | A | a/α | 2 | 1096* | 35 | 4 | 14 | 4 | 4 | 4 | 4 |
| 1-234 | Champaign | 48 | 48 | 1 | A | a/α | 2 | 1098* | 35 | 4 | 4 | 25 | 4 | 35 | 4 |
| 2-237 | A | a/a | 2 | 1134* | 35 | 2 | 14 | 4 | 4 | 4 | 4 | ||||
| 1-32* | Champaign | 20 | 68 | 4 | B | a/α | 3 | 236 | 13 | 10 | 15 | 6 | 7 | 32 | 12 |
| 1-226* | Tuscola | 360 | >360 | 0 | B | a/α | 3 | 344 | 13 | 10 | 15 | 6 | 7 | 37 | 15 |
| 1-120* | Champaign | 72 | 72 | 6 | B | a/α | 3 | 476 | 13 | 10 | 15 | 4 | 7 | 37 | 15 |
| 1-132* | Champaign | 11 | 11 | 8.5 | B | a/α | 3 | 726 | 13 | 7 | 15 | 6 | 7 | 37 | 15 |
| 1-233* | Mahomet | 324 | 372 | NA | B | a/α | 3 | 957 | 13 | 7 | 15 | 6 | 7 | 32 | 15 |
| 1-19* | St. Joseph | 120 | 360 | 0 | B | a/α | 3 | 1085* | 13 | 10 | 15 | 6 | 58 | 37 | 15 |
| 1-55* | Champaign | 48 | 72 | 1 | B | a/α | 3 | 1090* | 13 | 7 | 15 | 6 | 7 | 55 | 5 |
| 1-171** | Champaign | 36 | 36 | 4 | B | a/α | 3 | 1115* | 13 | 7 | 15 | 6 | 127* | 140 | 15 |
| 2-175* | B | a/α | 3 | 1130* | 13 | 7 | 14 | 6 | 7 | 55 | 15 | ||||
| 2-231* | B | a/α | 4 | 124 | 8 | 14 | 8 | 4 | 7 | 10 | 8 | ||||
| 2-212* | B | a/α | 5 | 1132* | 4 | 22 | 6 | 18 | 64 | 53 | 15 | ||||
| 1-237 | Urbana | 36 | 36 | 0.5 | A | a/α | 7 | 1099* | 6 | 3 | 10 | 12 | 125* | 46 | 12 |
| 1-28 | Champaign | 20 | 20 | 1 | B | a/α | 8 | 666 | 33 | 14 | 38 | 2 | 78 | 122 | 15 |
| 2-74 | B | a/α | 8 | 666 | 33 | 14 | 38 | 2 | 78 | 122 | 15 | ||||
| 1-20 | Mahomet | 156 | 204 | 1 | B | a/α | 8 | 1086* | 33 | 3 | 38 | 2 | 78 | 47 | 15 |
| 1-190 | St. Joseph | 9 | 396 | 1 | B | a/α | 8 | 1095* | 33 | 14 | 38 | 2 | 78 | 122 | 22 |
| 2-127* | A | a/α | 11 | 461 | 60 | 10 | 21 | 1 | 7 | 11 | 15 | ||||
| 2-213* | B | a/α | 11 | 1056 | 60 | 10 | 21 | 5 | 7 | 11 | 15 | ||||
| 1-18** | St. Joseph | 120 | 360 | 0 | A | a/α | Sing. | 1084* | 13 | 2 | 41 | 12 | 34 | 76 | 27 |
| 1-225 | Bloomington | 12 | 36 | 2 | A | a/α | Sing. | 1097* | 13 | 26 | 5 | 3 | 93 | 53 | 12 |
Strains coded with a “1-” prefix were collected from volunteers recruited at the CVM Open House. Strains coded with a “2-” prefix were collected from pet owners in an attempt to collect matched sets of isolates from humans and their pets. The pet owners did not provide information about their residence or travel habits. Strains marked with one asterisk are missing one ALS5 allele; both ALS5 alleles are missing in strains marked with two asterisks.
Approximate residential locations are noted. City names match those on the map in Fig. 1. Pet owners (strain code 2) were not asked to provide information about their residence or travel habits.
Volunteers indicated the amount of time at their current residence (curr. res.) as well as the location of their previous residence and amount of time lived there. This information was used to determine the number of months the individual spent in the geographic area of study. In many instances, the amount of time at the previous residence was not specified. Individuals whose current and previous residence was in the geographic area of study, but who did not specify length of time at the previous residence are reported as more than the time spent at the current residence.
NA, not answered in the question-naire.
Sing., singleton; could not be assigned to a currently defined clade.
DSTs and genotype designations were determined from comparisons between experimental sequences and the MLST database at http://test1.mlst.net. Asterisks indicate new DSTs and genotypes that were identified in this study.
Collection of C. albicans specimens from domestic animals.
C. albicans isolates from domestic animals were sought in a variety of settings. All samples from companion animals were collected in 2007. Oral and anal swabs were collected from 45 veterinary student-owned dogs during a classroom exercise. The swabs were plated on CHROMagar Candida. One isolate (D19M) was recovered from the mouth of a dog. Swabbing of a graduate student-owned dog yielded another oral C. albicans isolate (ZMK9).
In another attempt, matched sets of C. albicans isolates were sought from humans and their pets. UIUC IRB guidelines were followed and informed consent was obtained prior to enrollment in the study. Veterinary students who had completed a clinical microbiology course were invited to participate in the study. Forty-three individuals volunteered. They owned an average of 2 pets each (range, 1 to 5), and a total of 89 pets were tested (45% dogs, 43% cats, 12% other). Oral swabs from the students and either oral or anal swabs from the pets were plated on Sabouraud agar with 50 μg ml−1 chloramphenicol (SabCml) and incubated for 48 h. Yeast colonies were streaked to CHROMagar Candida and further verified as C. albicans by use of the methods described above. This study yielded 11 C. albicans isolates from the pet owners (26%) but no isolates from the animals. The C. albicans-positive pet owners included one male volunteer and one person of Asian descent. Other volunteers were female, white, and non-Hispanic/Latino. SabCml agar was used in this study to determine whether the primary culture medium had an effect on the percentage of C. albicans isolates collected. The similar percentages of positive cultures between two studies involving normally healthy humans (29% versus 26%) suggest that the choice of culture medium did not affect our ability to culture C. albicans from clinical sources.
Oral and anal swabs were collected from dogs, cats, and rabbits at the Champaign County Humane Society. Thirty dogs (mean of 33 days at the shelter [range, 9 to 103 days]), 60 cats (mean days at the shelter, 83 [range, 4 to 304]), and 4 rabbits (mean days at the shelter, 99 [range, 28 to 156]) were tested. The swabs were plated on SabCml and incubated at 37°C for 48 h and yeast colonies processed as described above. Two feline oral C. albicans isolates (C69M, C79M) were recovered.
C. albicans isolates were also sought among yeast colonies recovered from the UIUC dairy in 2006. High somatic cell counts at the dairy prompted culture of the cows and revealed the presence of yeasts. Repeated culture of yeasts suggested a persistent problem in some of the cows. These animals, as well as the dairy environment and workers, were cultured for yeasts by use of SabCml agar. Thirteen of approximately 240 cows at the dairy were studied. Oral, nasal, rectal, and vaginal swabs were collected. The teat skin was also swabbed and milk was sampled from at least one quarter of the udder. A single isolate of C. albicans (from the oral cavity of cow 7452) was found among the specimens from this study.
Geographic information systems mapping.
To determine the approximate geographic origins of the C. albicans isolates, we used the address where each wildlife species was found and the self-reported home addresses from human volunteers at the CVM Open House. We used the ArcGIS address geocoding tools and Streetmap USA street files from ESRI (Redlands, CA) to determine the latitude and longitude of each address. Of 66 available addresses, 53 (80%) were geocoded successfully with the software. We determined the location of five more addresses manually using Google Maps (http://maps.google.com). The remaining eight addresses were incomplete and included a street without a number, only a zip code, or, in one case, only the city. These isolates were placed on the map in the approximate center of the street, zip code, or city as available.
MLST analysis.
Genomic DNA was extracted from each C. albicans isolate by use of a previously described method (20). MLST analysis was conducted as described by Bougnoux et al. (8) by PCR amplification of fragments from seven C. albicans genes (AAT1a, ACC1, ADP1, PMI1b, SYA1, VPS13, ZWF1b). PCR products were purified using the MultiScreen-PCR 96-well filtration system (Millipore). DNA sequences were determined at the W. M. Keck Center for Comparative and Functional Genomics (Roy J. Carver Biotechnology Center, University of Illinois, Urbana-Champaign, IL). DNA sequences were verified for both strands. Strain SC5314 was included in each experiment to test the reproducibility of the MLST method in our lab; in each case, the SC5314 sequences matched those reported in the C. albicans MLST database (http://test1.mlst.net). The genotype for each sequenced PCR product was identified by a query of the MLST database; the diploid sequence type (DST) for each isolate was identified by a database query using all genotype information for each strain. Frank Odds kindly assigned genotype designations and DSTs based on our original data. Nucleotide polymorphisms within the DNA fragments were coded according to the method described by Bougnoux et al. (7). This information was used to generate a phylogenetic tree from matrices of pairwise Nei's genetic distances by use of the unweighted-pair group method using average linkages (UPGMA) in the Phylogeny Inference Package, PHYLIP (14), available at http://evolution.genetics.washington.edu/phylip.html. C. albicans isolates were assigned to genetic groups by use of known reference strains recently analyzed by Odds et al. (33). Clade assignments were made by using the Nei's genetic distance threshold of 0.045, which clustered reference strains in the same manner as the P distance threshold of 0.04 used by Odds et al. (33). The nonrandomness of allelic combinations was calculated using Fisher's exact test (http://www.physics.csbsju.edu/stats/).
Additional strain-typing methods.
Additional characterization of C. albicans isolates was conducted by PCR. One method, now commonly referred to as ABC typing, detects the presence or absence of a transposable intron in 25S rRNA-encoding DNA (27). With this method, strains are designated as genotype A, B, or C depending on the size of PCR products generated with primers CA-INT-L and CA-INT-R (27). PCR was also used to determine the mating type (MAT) for each C. albicans isolate (47). PCR primers were used to detect the presence of the a or the α allele, giving three possible resulting genotypes: a/a, a/α, and α/α. Amplification of the central tandem repeat domain from the agglutinin-like sequence genes ALS3, ALS5, and ALS6 was used to distinguish between strains with similar DSTs. Primers and methods for these PCRs have been published previously (35, 50). PCR primers flank the tandem repeat domain and amplify the length of this region in each gene. The size of the band indicates the number of tandem copies of the 108-bp repeat unit present in each allele. In some isolates, alleles encode the same number of repeat copies and produce a single PCR product. In other isolates, alleles with different numbers of repeat copies are amplified and produce two PCR products. Previous work showed that some C. albicans isolates are missing one or both ALS5 alleles and that the deletion is due to direct repeats that flank the ALS5 locus (50). Each C. albicans isolate in this study was assessed for the presence of ALS5 using the previously published method (50).
Antifungal susceptibility testing.
Antifungal MICs were determined using the broth microdilution method recommended by the Clinical and Laboratory Standards Institute (CLSI document M27-A2) (30, 37). The incubation times and MIC endpoint criteria used were those recommended by the CLSI: for amphotericin B, a complete inhibition of visible growth, and for echinocandins and azoles, a prominent reduction in growth (>50%) compared with what was seen for the growth control well (partial inhibition). Data for human and wildlife isolates were compared using the Wilcoxon rank-sum test. MIC distributions of amphotericin B, caspofungin, and fluconazole were examined, since they represent commonly used agents within each of the three major antifungal drug classes. Statistical analyses were performed using SPSS version 13 (Chicago, IL). In instances where more than one isolate was available from an individual, MIC data for only one isolate were included in the analysis to avoid bias in the statistical comparisons. In nearly all cases, MICs were the same among the multiple isolates or differed by only one dilution. For the small number of cases where MICs among multiple isolates showed greater differences, the results of the analysis were the same regardless of the isolate that was included.
RESULTS
Common geographic origins of human and wildlife C. albicans isolates.
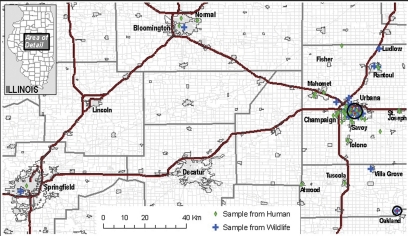
Fig. 1 shows the geographical origins of wildlife species from which C. albicans oral and/or anal isolates were obtained. The geographical origins of healthy human volunteers from whom C. albicans isolates were collected at the CVM Open House encompassed a much larger region, including the Chicago area and neighboring states. Only strains from humans who reside in the geographic region defined by the wildlife collection were included in the study. Figure 1 demonstrates the common geographic distribution of the two strain collections. Table 1 shows detailed information about the C. albicans wildlife isolates. Most were derived from healthy orphaned animals or those that were brought to the clinic with a traumatic injury. Table 2 displays detailed information about the oral C. albicans isolates collected from healthy human volunteers. Wildlife species included in the study were nonmigratory, which suggests that each spent its entire life in this geographic region. The human volunteers included in the study group reported a mean of 132 months (11 years; median, 72 months) of residence in the region.
FIG. 1.
Map of the geographic region of study. The inset (top left) shows the relationship between the area of study and the state of Illinois. Self-reported locations of the residence of C. albicans culture-positive humans and locations at which C. albicans-positive wildlife species were found are indicated. The geographic region defined for this study includes cities such as Champaign and Urbana (approximate total population, 186,000), Bloomington and Normal (161,000), Springfield (111,000), and Rantoul (13,000) as well as many smaller towns with populations that range from a few hundred to several thousand residents. Shaded areas on the map indicate cities and towns. Those from which human and/or wildlife specimens originated are labeled by name and correspond to the information provided in Tables 1 and 2. Much of the land in this geographic area is used for agricultural purposes; wooded areas are also common. Circled areas indicate the origins of humans and animals that shared a DST; these cases are highlighted in the text.
Genotypes, clade assignments, and analysis.
Genotypes and DSTs were assigned to MLST data by comparison to known sequences at http://test1.mlst.net. Our strain collection added 12 new genotypes and 36 new DSTs to the database (Tables 1 and 2). DST 69 was the most abundant in the current data set, consistent with previous reports that showed DST 69 as the most common strain of human origin (33, 46). DST 90 (clade 8) was frequently observed but only among the wildlife isolates. Two DST 90 isolates derived from humans are found at http://test1.mlst.net: one from a Malaysian urine specimen and another from a South African blood culture. In the current work, isolates of the same DST were distributed across the geographic area rather than being clustered in a smaller area (Fig. 1 and Tables 1 and 2).
Clade assignments for C. albicans isolates in this study were made (Table 3) by comparison to MLST data published by Odds et al. (33). In our work, a Nei's genetic distance threshold of 0.045 distributed known isolates into the same genetic clades that Odds et al. (33) designated using a P distance threshold of 0.04. The study of Odds et al. (33) incorporated nearly 1,400 C. albicans isolates, of which 201 were from North America (North American isolates in Table 3), and included 3 isolates from animal sources. Comparison between the two sets of strains showed that the clade distribution of our collection from healthy human volunteers (“human group 1” in Table 3) was similar to that of the North American isolates from the work of Odds et al. (33). In both collections, clade 1 is the most populous, followed by clade 3 and clade 2 (Table 3). Using Fisher's exact test, only the proportions of the clade 4 isolates differed significantly between the two collections (P = 0.024). The observed difference could indicate that clade 4 strains are not distributed homogeneously within North America. A precedent for this idea is set by the nonhomogenous distribution of clade 2 strains within the United States (40). Overall, these comparisons demonstrate that our human isolate collection is very similar to the one with broader distribution in North America.
TABLE 3.
Comparison between clade assignments, ABC genotypes, and mating types for isolates from the present study and those reported by Odds et al. (33)
| Group characteristic (reference or source) | % of isolates from the work of Odds et al. (33) | No. (%) of isolates
|
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Totala | Clade
|
Singleton | ||||||||||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | ||||
| North American isolates (33) | 201 (14.7) | 99 (49.3) | 20 (10.0) | 31 (15.4) | 18 (9.0) | 1 (0.5) | 4 (2.0) | 1 (0.5) | 4 (2.0) | 5 (2.5) | 0 (0.0) | 6 (3.0) | 1 (0.5) | 0 (0.0) | 1 (0.5) | 2 (1.0) | 0 (0.0) | 2 (1.0) | 6 (3.0) | |
| Human group 1 (this study) | 44 (100) | 26 (59.1) | 4 (9.1) | 8 (18.2) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (2.3) | 3 (6.8) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (4.5) | |
| All human isolates (this study) | 55 (100) | 30 (54.5) | 5 (9.1) | 9 (16.4) | 1 (1.8) | 1 (1.8) | 0 (0.0) | 1 (1.8) | 4 (7.3) | 0 (0.0) | 0 (0.0) | 2 (3.6) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (3.6) | |
| Wildlife isolates (this study) | 28 (100) | 7 (25.0) | 0 (0.0) | 3 (10.7) | 1 (3.6) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 10 (35.7) | 2 (7.1) | 1 (3.6) | 2 (7.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (7.1) | |
| Wildlife isolates, no replicates | 22 (100) | 6 (27.3) | 0 (0.0) | 3 (13.6) | 1 (4.5) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 6 (27.3) | 2 (9.1) | 1 (4.5) | 1 (4.5) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (9.1) | |
| Domestic animal isolates | 5 (100) | 4 (80.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (20.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| All isolates (this study) | 88 (100) | 41 (46.6) | 5 (5.7) | 12 (13.6) | 2 (2.3) | 1 (1.1) | 0 (0.0) | 1 (1.1) | 15 (17.0) | 2 (2.3) | 1 (1.1) | 4 (4.5) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 4 (4.5) | |
| ABC genotype | 83 (100) | |||||||||||||||||||
| A | 64.1 | 55 (66.3) | 40 (72.7) | 5 (9.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (1.8) | 3 (5.5) | 2 (3.6) | 0 (0.0) | 1 (1.8) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 3 (5.5) |
| B | 23.0 | 24 (28.9) | 0 (0.0) | 0 (0.0) | 12 (50.0) | 2 (8.3) | 1 (4.2) | 0 (0.0) | 0 (0.0) | 6 (25.0) | 0 (0.0) | 1 (4.2) | 1 (4.2) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (4.2) |
| C | 12.9 | 4 (4.8) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 3 (75.0) | 0 (0.0) | 0 (0.0) | 1 (25.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Mating type | 83 (100) | |||||||||||||||||||
| a/α | 91.5 | 80 (96.4) | 40 (50.0) | 3 (3.8) | 12 (15.0) | 2 (2.5) | 1 (1.3) | 0 (0.0) | 1 (1.3) | 11 (13.8) | 2 (2.5) | 1 (1.3) | 3 (3.8) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 4 (5.0) |
| a/a | 2.5 | 2 (2.4) | 0 (0.0) | 1 (50.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (50.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| α/α | 6.0 | 1 (1.2) | 0 (0.0) | 1 (100) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
Total numbers of isolates are different in terms of some variables. If two strains from the same animal were different with respect to the variable considered, both strains were included in the count. If the strains were the same for the given variable, only one strain was included in the count.
The main purpose of the present study was to determine whether the clade distributions of C. albicans isolates were similar between humans and wildlife when geographical location was removed as a variable. Comparisons between the human isolates collected from CVM Open House visitors (human group 1 in Table 3) and those from wildlife (wildlife isolates with no replicates in Table 3) showed significant differences in the proportions of clade 1 (P = 0.014) and clade 8 (P = 0.031) strains. No other significant difference was observed. This significant difference in clade composition results from a deficit of clade 1 wildlife isolates and an overrepresentation of clade 8 among wildlife isolates. This result suggests that although some overlap is evident, the clade distributions of strains differ between humans and wildlife in the geographic region studied. Similar results were obtained when clade proportions were compared between our wildlife isolate collection and that of the North American collection of Odds et al. (33) (deficit of clade 1 isolates in the wildlife sample [P = 0.039]; excess of clade 8 [P = 0.000075]). This result demonstrated that the two collections of isolates from human origin were more similar to each other than either was to the wildlife isolate collection. This enrichment in clade 8 is noteworthy. About 27% of wildlife isolates were of clade 8. To our knowledge, this is the highest proportion of clade 8 isolates reported to date. In the most comprehensive study of worldwide clade distribution yet available, Odds et al. (33) analyzed 1,371 isolates, mostly from humans, which were separated into 10 regions of origin, including Scotland, England/Wales, France, elsewhere in Europe, the Middle East, Africa, Asia, North America, South America, and Australia. We compared the proportion of clade 8 isolates for each of these regions to the proportion for our wildlife collection. Nine of the 10 comparisons indicated an excess of clade 8 isolates in the wildlife collection. In these regions, the percentages of clade 8 isolates ranged from 0% to 6.4%, with a mean percentage of 1.8% ± 2.2% (± standard deviation) and a median of 1.5%. South America (81 isolates, 16% of clade 8) was the only region that did not show a significantly different proportion of clade 8 isolates.
MLST sequence data comparisons between individual strains.
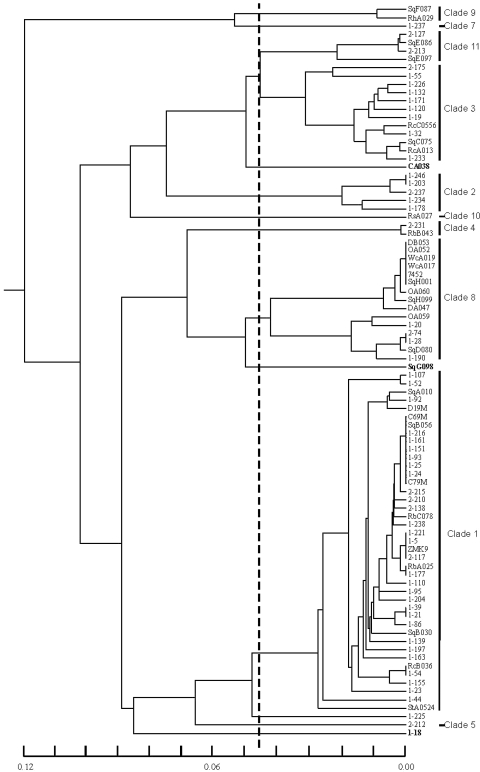
The phylogenetic tree generated from sequence data illustrates the relationships among various human C. albicans isolates, among wildlife isolates, and between human and wildlife isolates (Fig. 2). Indications of loss of heterozygosity (LOH) were common among multiple isolates from the same animal. For example, two isolates from squirrel B (heterozygous oral isolate SqB056 and homozygous anal isolate SqB030) coclustered among clade 1 strains but remained relatively distant from each other (Table 1; Fig. 2). Isolates from squirrel E (SqE086 and SqE097, both oral isolates) varied at three positions within the ACC1 locus and at eight within ADP1; all were heterozygous in isolate SqE086 and homozygous in SqE097 (Table 1). Because ACC1 and ADP1 are approximately 1,000 kb apart on chromosome R, this result suggests long-range LOH in strain SqE097, possibly due to the loss of one chromosome R homolog. Isolates from squirrel H (heterozygous oral isolate SqH001 and homozygous anal isolate SqH099) showed similar LOH patterns. SqH099 was also homozygous at the MAT locus (a/a), while SqH001 was heterozygous. Strain differences detected for multiple isolates from the same animal source were never sufficient to place an isolate into a different clade. Similar genetic variations and LOH between strains from the same or different body sites were documented previously with human patients (4, 5, 32, 46).
FIG. 2.
Dendrogram based on MLST data obtained for 88 C. albicans isolates. Sequence data obtained at seven loci were used to generate a UPGMA dendrogram from matrices of pairwise Nei's genetic distances. A threshold value of 0.045 was used to identify the clades described by Odds et al. (33). The scale bar indicates Nei's genetic distance values.
Our data set provided several examples of humans with identical C. albicans DSTs (Table 2). In all but one of these cases, the individuals who provided the isolates recorded residential addresses that were distant from each other within the context of the geographical region studied, and there was no evidence for contact between the individuals. The exception was two individuals with DST 69 (1-24 and 1-25), who reported an identical street address and were of different genders and a similar age group. This information suggested that the C. albicans strain may have been transferred between these individuals due to their close proximity and likely close contact. This trend was not universally true, however, since two other sets of volunteers who fit these criteria (same address, different genders, similar age), 1-92 and 1-93 along with 1-18 and 1-19, provided specimens that were distinct genetically.
Examples of the same DST being exhibited by wildlife were limited to DST 90. These isolates were from mammals located in distant parts of our geographic area (Table 1 and Fig. 1). The complete data set contained several examples of humans and animals that were colonized with identical DSTs (Tables 1 and 2). Two of the animals were located within the Champaign-Urbana city limits. The third was a raccoon from a rural area (Fig. 1 and Table 1). Raccoons, however, are known for seeking contact with the by-products of human life, such as garbage cans. All isolates with DSTs in common between a human and wildlife species were in clade 1. It is important to view the DSTs we observed as representative of those in central Illinois and not to imply that shared DSTs indicate contact between a specific human and animal.
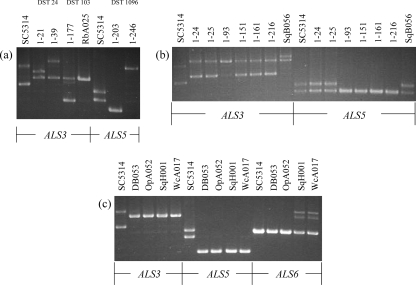
Use of ALS gene tandem repeat copy numbers as a genotyping tool.
Because many of the C. albicans isolates were indistinguishable by the methods used, we employed another method to distinguish between seemingly identical isolates. Previous studies demonstrated a significant association between the C. albicans genetic group and the number of copies of the 108-bp tandemly repeated sequence present in the central domain of several genes in the ALS family (35, 50). ALS genes encode large cell surface glycoproteins, some of which have an adhesive function (19). We used published methods to amplify the tandem repeat domains from ALS3, ALS5, and ALS6 (Fig. 3). Amplification of the ALS3 tandem repeat domain was sufficient to differentiate between strains 1-21 and 1-39 (DST 24) (Fig. 3a) and between DST 103 isolates 1-177 and RbA025. ALS5 tandem repeat domain length differences distinguished between strains 1-203 and 1-246 (DST 1096) (Fig. 3a). Amplification of the ALS3 and ALS5 tandem repeat domains differentiated between many of the strains with DST 69 (clade 1) (Fig. 3b) and distinguished the squirrel B strain with a unique ALS3 pattern. Strains 1-24 and 1-25, isolated from the individuals who shared a street address, were still indistinguishable, as were strains 1-151, 1-161, and 1-216 (Fig. 3b). DST 90 strains (clade 8) were also examined using this method (Fig. 3c). Although amplification of the ALS3 and ALS5 tandem repeat domains could not distinguish between the strains, amplification of the ALS6 tandem repeat domain produced an unexpected result (Fig. 3c). Primers that are positioned outside of the ALS tandem repeats are expected to yield no more than two fragments, one from each chromosome of the diploid organism. Instead, strains SqH001 and WcA017 produced three fragments (Fig. 3c). The presence of three fragments suggests the possibility of a second ALS6 locus in these strains. Because many C. albicans strains have lost at least one ALS5 allele (50), all isolates in the study were also screened for this property. Strains in clades 1, 2, 3, 4, 5, 10, and 11 as well as singleton strains lacked one ALS5 allele (Tables 1 and 2). Two human isolates (1-171, clade 3; 1-18, singleton) were missing both ALS5 alleles (Table 1).
FIG. 3.
Ethidium bromide-stained agarose gel of PCR products derived from amplification of genomic DNA with primer pairs that hybridize to sequences flanking the tandem repeat regions of ALS3, ALS5, and ALS6 (50). The tandem repeat region of each gene consists of head-to-tail copies of a conserved 108-bp sequence (19). ALS alleles in diploid C. albicans often contain different numbers of copies of the 108-bp unit in the central domain. Allelic differences in the tandem repeat copy number in these genes were explored for their utility in distinguishing between C. albicans isolates with the same DST. Control lanes using SC5314 genomic DNA are shown for each set of PCR products. For SC5314, one ALS3 allele has 9 tandem repeat copies, while the other has 12 copies (35). One ALS5 allele has four tandem repeat copies, while the other has five, and both ALS6 alleles have four tandem repeat copies (50). (a) Use of ALS3 or ALS5 tandem repeat copy numbers to distinguish between three different pairs of isolates, each sharing the same DST. (b) Use of ALS3 and ALS5 tandem repeat copy numbers to distinguish between DST 69 strains. (c) Use of ALS3, ALS5, and ALS6 tandem repeat copy numbers to distinguish between DST 90 strains.
Analysis of C. albicans isolates from domestic animals.
The clade distributions of human and wildlife isolates were significantly different, as described above. These results raised the question of what would be observed among isolates collected from domestic animals that, relative to wildlife species, were in closer contact with humans and possibly could serve as a route of transmission of C. albicans isolates from wildlife to humans. The first attempt to collect C. albicans isolates from domestic animals involved oral and/or anal swabs from veterinary student-owned dogs. One oral isolate, D19M, was recovered. Next, we attempted to collect matched sets of C. albicans isolates from humans and their pets. All pets tested were culture negative, but 11 more human oral isolates were recovered (designated with the prefix “2-” in Table 2 and included in the “all human isolates” category in Table 3). Another canine isolate was obtained from a graduate student-owned dog (ZK9M). Two oral feline isolates (C69M, C79M) were obtained from animals at the Champaign County Humane Society. A final C. albicans isolate (called 7452) was recovered from a cow at the UIUC dairy. MLST analysis of the domestic animal C. albicans isolates showed that the cats and dogs were colonized with common human isolates, while the cow isolate type was the most common type among local wildlife (DST 90) (Table 1). The pets tested were all indoor animals that were in close contact with humans. At the time the cow isolate was collected, the cow occupied a tie stall in a barn. Although the cow had contact with humans daily, the presence of cattle feed in the barn attracts a large and diverse population of wildlife species to the cow's environment, providing ample opportunity for direct interaction or for ingestion of wildlife feces from contaminated feed.
Antifungal susceptibility data.
The collection of human and wildlife C. albicans isolates was tested for antifungal susceptibility (Table 4). Antifungal MICs were determined using a broth dilution method. Azole resistance was not observed for any of the strains tested (Table 4). C. albicans MIC distributions for human and wildlife isolates were not statistically different except for those for amphotericin B. A significantly lower amphotericin B MIC was observed for the wildlife isolates compared to what was seen for the human isolates (P = 0.001), although the difference amounted to only approximately 1 dilution. A previous study showed that amphotericin B-resistant C. albicans isolates are found in several clades but that clade SA, which corresponds to clade 4 in the MLST phylogeny of Odds et al. (33), had significantly more isolates that were amphotericin B resistant than did other clades (2). Examination of data from the current study showed one clade 4 strain among the wildlife isolates (Table 1) and none in the human strain collection (Table 2). One human isolate (1-44; clade 1) had a MIC of 2 μg ml−1, while those of all others were 1 μg ml−1 (Table 4). Among the wildlife isolates (Table 1), strains with MICs of 0.5 μg ml−1 were clade 4 (RbB043), clade 8 (DA047, DB053), and a singleton (SqG098). The clade 9 strain RhA029 had a MIC of 0.25 μg ml−1. Testing larger numbers of isolates would be required to determine if strains with lower amphotericin B MICs are found more frequently in deer (as for DA047 and DB053) or avian species (as for RhA029).
TABLE 4.
Antifungal susceptibilities of human and wildlife C. albicans isolates
| Antifungal agent | Endpoint inhibition criterion | Incubation time (h) | Specimen origina | Cumulative % of isolates with MICs (μg/ml) ofb:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.007 | 0.015 | 0.03 | 0.06 | 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | ||||
| Amphotericin B | Complete | 48 | Human | — | — | — | — | — | — | — | 97.7 | 100 | — |
| Wildlife | — | — | — | — | 4.5 | 22.7 | 100 | — | — | ||||
| Caspofungin | Partial | 24 | Human | 9.1 | 72.7 | 97.7 | 100 | — | — | — | — | — | — |
| Wildlife | 22.7 | 81.8 | 100 | — | — | — | — | — | — | — | |||
| Anidulafungin | Partial | 24 | Human | 31.8 | 65.9 | 95.5 | 100 | — | — | — | — | — | — |
| Wildlife | 27.3 | 68.2 | 95.5 | 100 | — | — | — | — | — | — | |||
| Micafungin | Partial | 24 | Human | 59.1 | 100 | — | — | — | — | — | — | — | — |
| Wildlife | 54.5 | 95.5 | 100 | — | — | — | — | — | — | — | |||
| Itraconazole | Partial | 48 | Human | — | 25.0 | 38.6 | 56.8 | 93.2 | 100 | — | — | — | — |
| Wildlife | 4.5 | 9.1 | 9.1 | 31.2 | 100 | — | — | — | — | — | |||
| Posaconazole | Partial | 48 | Human | 6.8 | 27.3 | 52.3 | 90.9 | 100 | — | — | — | — | — |
| Wildlife | — | 9.1 | 18.2 | 95.5 | 100 | — | — | — | — | — | |||
| Voriconazole | Partial | 48 | Human | 63.6 | 97.7 | 100 | — | — | — | — | — | — | — |
| Wildlife | 90.9 | 100 | — | — | — | — | — | — | — | — | |||
| Fluconazole | Partial | 48 | Human | — | — | — | — | 29.5 | 95.5 | 100 | — | — | — |
| Wildlife | — | — | — | — | 54.5 | 100 | — | — | — | — | |||
For human isolates, the number was 44; for wildlife isolates, the number was 22.
—, no isolates.
DISCUSSION
This study stems from the observation that distinct C. albicans genetic clades are associated with specific geographical areas (33, 44). Analysis of a collection of C. albicans isolates from wildlife and humans residing in central Illinois revealed a significant difference in the clade distributions between the two populations. The wildlife collection showed a deficit in clade 1 strains, the clade most commonly found in humans, and an excess of clade 8 strains, a clade more rarely observed in humans. Testing of additional wildlife isolates on a worldwide basis is required to determine if clade 8 strains are more often associated with wildlife populations. In the current study, the statistically significant difference in the clade compositions of the human and wildlife isolates demonstrates population isolation between the two groups. This isolation may be caused by the limited transfer of strains between the groups or the differential selection of isolates in humans and wildlife. Other clade differences between human and wildlife species in this geographic region may exist but would require a larger sample size to be detected. Despite these global differences in clade distribution, we observed some overlap in clade assignments between human and wildlife isolates. This trend continued to the level of sequence data, where the DSTs of some human and wildlife isolates were the same. At this time, the extent of genomic differences between C. albicans isolates with the same DST is unknown. Assessment of ABC type, MAT locus alleles, and number of tandem repeat sequence copies in the central domain of ALS3, ALS5, and ALS6 provided an added level of strain discrimination that was sufficient to distinguish between identical DSTs for some isolates but not for others (Fig. 3). Although the tandem repeat copy numbers within ALS3, ALS5, and ALS6 are quite stable in vitro (50), the stability of these sequences has not been assessed in vivo.
Similarities between human and animal C. albicans isolates have been demonstrated in other studies. Bougnoux et al. (6) used MLST to type strains from invasive candidiasis in humans, from healthy humans, and from starlings in France. While the starling isolates tended to group together genetically, some of the human isolates also were found in the same cluster. In two cases, a bird and a human isolate shared the same DST. Edelmann et al. (13) used Ca3 fingerprinting to analyze C. albicans from humans with candidemia and from a diverse group of animals including wildlife, domestic animals, and zoo species. The authors found that isolates from animal species can be as closely related to human strains as human strains are to each other. Using MLST and a data set that accounted for the geographical origins of the isolates, Jacobsen et al. (22) showed that the clade distributions of C. albicans strains from humans and animals were significantly different. Taken together, these analyses support the conclusion that C. albicans isolates from humans and animals show some common characteristics but overall tend to be separate genetically. Whether these conclusions can be extrapolated to all societies, however, remains in question, especially for cultures in which animals and humans live in very close proximity to each other and where C. albicans is not the major Candida species in the human normal flora (49). It is likely important to consider wildlife and domestic animal species separately when addressing questions of C. albicans transfer between human and animal populations. In addition, it is also likely necessary to consider species individually within each category. For example, although the nature of our data set allows us to make a global conclusion regarding the clade distribution of C. albicans among human and wildlife sources, we cannot rule out the possibility that a particular wildlife species serves as a reservoir in a more limited fashion. Also, previous studies have raised the question of whether C. albicans isolates from animals are found as species-specific monophyletic groups (13). Our current data demonstrate polyphyletic groups from squirrels, raccoons, and rabbits but do not rule out the possibility that isolates are monophyletic for a specific animal species.
In order for strain transfer to occur between animals and humans, the recipient must come into contact with a C. albicans-colonized animal or animal-derived material. In most previously published work and in our initial hypothesis, the directionality of transfer was considered from a human perspective, i.e., whether wildlife species serve as a reservoir for humans (13). This perspective is understandable given the paucity of published information regarding the frequency of C. albicans isolation from animal species and the human-centered focus of the Candida research community. Data from the current study provide a clearer perspective on how strain transfers may occur. Direct encounter with wildlife is a relatively rare event in the lives of most humans, although encounter with animal-derived material (feces, for example) is more common, perhaps particularly for children. Direct encounter with wildlife is a more common event for dogs and cats, and transfer of a wildlife-derived C. albicans strain from pet to human is a believable scenario. However, our data argue against this transmission mechanism, because pet isolates were quite rare and genetically more similar to those from humans than to those from wildlife. We observed overlap between isolates from humans and wildlife, limited to clade 1 strains, which comprise the most common genetic group in humans. This observation suggests that the encounter between wildlife and the by-products of human life (such as filled garbage cans that contain dirty diapers and half-eaten food) is perhaps a more common scenario for strain transfer and that the directionality of transfer is more likely from humans to wildlife.
The extremely low frequency of isolation of C. albicans from domestic animals (particularly dogs and cats) and the presence of the most common human DSTs in animals from which an isolate was recovered are important data to note. Combined, these pieces of information support the conclusion that transfer is from human to animal and suggest that pet owners are at little risk of contracting C. albicans from an animal in their home. This conclusion stands in contrast to a statement in an earlier report which suggested that immunodeficient humans should consider animals as a potential source of Candida infections (13). Data presented in the current study suggest that maintaining the companionship of a family pet presents a relatively low risk with respect to contracting candidiasis and that a greater risk is likely presented by contact with other humans. These data also suggest the possibility that the nontransient human population serves as the reservoir that links C. albicans genetic groups recovered from humans to a specific geographic region. In our analysis, human volunteers reported a mean of 11 years of residence (median, 6 years) with a mean of 2.5 weeks (median, 2 weeks) of travel per year (Table 2).
Data from this study address the question of where C. albicans is found. Our observations suggest that C. albicans is more readily recovered from human sources. C. albicans was recovered from 29% of oral swabs from normally healthy human volunteers at the CVM Open House and from 26% of pet owners sampled in a subsequent effort. These results compare favorably to observations made in previous studies of similar experimental groups. Odds (31) compiled a summary of 16 studies of healthy individuals and showed a range of 1.9 to 41.4% (median, 17.6%) recovery of C. albicans from oral swabs. Our testing of animals produced fewer C. albicans isolates. However, although only 11% of wildlife cases provided a C. albicans isolate, 19% of the mammals were culture positive, suggesting that C. albicans is nearly as common among mammalian wildlife as among humans. Surveys of domestic animals provided far fewer isolates (2% of classroom dogs, 0% in the pet study, and 2% of animals at the Champaign County Humane Society). Numerous other yeast species were present in wildlife, but no other yeasts were recovered from the classroom dogs or from animals in the pet study. Approximately 20% of the plated oral swabs from dogs at the Champaign County Humane Society showed yeast colonies of the same morphology that clearly were not C. albicans.
The consensus drawn from various texts is that C. albicans is associated mainly with humans, other mammals, and avian species (1, 15, 21, 31, 45). Isolates from reptiles were not cited, making our red-eared slider strain unique. Some texts avoid discussing whether C. albicans can be found environmentally, while others claim that reports of environmental C. albicans isolates are rare. Some texts mention C. albicans isolation from water (primarily as a measure of pollution with human waste), soil, air, and plants (1, 16). Veterinary microbiology textbooks largely seem to take their cues from the human medical literature and present C. albicans as part of the normal mammalian and avian mucosal and/or skin microbiota and therefore as a common commensal (11, 18, 41, 48). This view of C. albicans is not consistent with the results of our strain isolation efforts, which provided rare positive C. albicans cultures from domestic animals. Perhaps a different sample collection method would provide a higher percentage of positive results. Alternatively, it may be that C. albicans only transiently colonizes the species we studied, making the detection of C. albicans-positive animals a greater challenge. Experimental C. albicans infection of a wild gull, by allowing the bird to eat contaminated fish, suggested that the bird serves as an asymptomatic C. albicans carrier that eventually clears the fungal cells, resulting in culture-negative fecal specimens (10). Serial sampling of C. albicans-positive animals from both the wildlife and domestic populations would resolve these questions and further contribute to our understanding of the association between C. albicans and animal species.
Acknowledgments
We thank the students at the University of Illinois College of Veterinary Medicine Wildlife Medical Clinic for collection of the C. albicans isolates. We thank students in the College of Veterinary Medicine for participating in our strain collection efforts. We thank the Champaign County Humane Society for providing access to their resident animal population. We thank Dawn Morin for the yeast isolate from the dairy cow and for information regarding the wildlife population at the UIUC dairy. We thank Bill Brown for his assistance with geocoding.
This work was supported by grant R01 AI39735 from the National Institutes of Health to D.R.S. and by the Developmental Studies Hybridoma Bank. L.W. was supported by a Merck-Merial summer fellowship. This investigation was conducted in a facility constructed with support from Research Facilities Improvement Program grant number C06 RR16515-01 from the National Center for Research Resources, National Institutes of Health. This work is dedicated to the memory of our friend and mentor Jim Corbin.
Footnotes
Published ahead of print on 11 July 2008.
REFERENCES
- 1.Ahearn, D. G., and R. L. Schlitzer. 1984. Key to yeasts pathogenic for man and animal, p. 998-1003. In N. J. W. Kregar-van Rij (ed.), The yeasts: a taxonomic study, 3rd ed. Elsevier Science Publishers, Amsterdam, The Netherlands.
- 2.Blignaut, E., J. Molepo, C. Pujol, D. R. Soll, and M. A. Pfaller. 2005. Clade-related amphotericin B resistant among South African Candida albicans isolates. Diagn. Microbiol. Infect. Dis. 5329-31. [DOI] [PubMed] [Google Scholar]
- 3.Boucher, H., S. Mercure, S. Montplaisir, and G. Lemay. 1996. A novel group I intron in Candida dubliniensis is homologous to a Candida albicans intron. Gene 180189-196. [DOI] [PubMed] [Google Scholar]
- 4.Bougnoux, M. E., D. Diogo, C. Pujol, D. R. Soll, and C. d'Enfert. 2007. Molecular epidemiology and population dynamics in Candida albicans, p. 51-70. In C. d'Enfert and B. Hube (ed.), Candida: comparative and functional genomics. Caister Academic Press, Wymondham, United Kingdom.
- 5.Bougnoux, M.-E., D. Diogo, N. Francois, B. Sendid, S. Veirmeire, J. F. Colombel, C. Bouchier, H. Van Kruiningen, C. d'Enfert, and D. Poulain. 2006. Multilocus sequence typing reveals intrafamilial transmission and microevolutions of Candida albicans isolates from the human digestive tract. J. Clin. Mirobiol. 441810-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bougnoux, M.-E., D. M. Aanensen, S. Morand, M. Theraud, B. G. Spratt, and C. d'Enfert. 2004. Multilocus sequence typing of Candida albicans: strategies, data exchange and applications. Infect. Genet. Evol. 4243-252. [DOI] [PubMed] [Google Scholar]
- 7.Bougnoux, M. E., C. Pujol, D. Diogo, C. Bouchier, D. R. Soll, and C. d'Enfert. 2008. Mating is rare within as well as between clades of the human pathogen Candida albicans. Fung. Genet. Biol. 45221-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bougnoux, M.-E., A. Tavanti, C. Bouchier, N. A. R. Gow, A. Magnier, A. D. Davidson, M. C. J. Maiden, C. d'Enfert, and F. C. Odds. 2003. Collaborative consensus for optimized multilocus sequence typing of Candida albicans. J. Clin. Microbiol. 415265-5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, M. R., C. A. Thompson, and F. M. Mohamed. 2005. Systemic candidiasis in an apparently immunocompetent dog. J. Vet. Diagn. Investig. 17272-276. [DOI] [PubMed] [Google Scholar]
- 10.Buck, J. D. 1986. A note on the experimental uptake and clearance of Candida albicans in a young captive gull (Larus sp.). Mycopathologia 9459-61. [DOI] [PubMed] [Google Scholar]
- 11.Carter, G. R., and D. Wise. 2003. Infections caused by yeasts and yeastlike fungi, p. 257-261. In Essentials of veterinary bacteriology and mycology, 6th ed. Blackwell Publishing, Oxford, United Kingdom.
- 12.Duarte, E. R., and J. S. Hamdan. 2006. Susceptibility of yeast isolates from cattle with otitis to aqueous solution of povidone iodine and to alcohol-ether solution. Med. Mycol. 44369-373. [DOI] [PubMed] [Google Scholar]
- 13.Edelmann, A., M. Kruger, and J. Schmid. 2005. Genetic relationship between human and animal isolates of Candida albicans. J. Clin. Microbiol. 436164-6166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felsenstein, J. 1993. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5164-166. [Google Scholar]
- 15.Gentles, J. C., and C. J. LaTouche. 1969. Yeasts as human and animal pathogens, p. 107-182. In A. H. Rose and J. S. Harrison (ed.), The yeasts, volume 1. Biology of yeasts. Academic Press, London, United Kingdom.
- 16.Hagler, A. N. 2006. Yeasts and indicators of environmental quality, p. 515-532. In C. A. Rosa and G. Peter (ed.), Biodiversity and ecophysiology of yeasts. Springer, Berlin, Germany.
- 17.Heseltine, J. C., D. L. Panciera, and G. K. Saunders. 2003. Systemic candidiasis in a dog. J. Am. Vet. Med. Assoc. 223821-824. [DOI] [PubMed] [Google Scholar]
- 18.Hirsh, D. C., and E. L. Biberstein. 2004. Yeast—Cryptococcus, Malassezia, and Candida, p. 265-272. In D. C. Hirsch, N. J. MacLachlan, and R. L. Walker (ed.), Veterinary microbiology, 2nd ed. Blackwell Publishing, Oxford, United Kingdom.
- 19.Hoyer, L. L., C. B. Green, S.-H. Oh, and X. Zhao. 2008. Discovering the secrets of the Candida albicans agglutinin-like sequence (ALS) gene family—a sticky pursuit. Med. Mycol. 461-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoyer, L. L., S. Scherer, A. R. Shatzman, and G. P. Livi. 1995. Candida albicans ALS1: domains related to a Saccharomyces cerevisiae sexual agglutinin separated by a repeating motif. Mol. Microbiol. 1539-54. [DOI] [PubMed] [Google Scholar]
- 21.Hurley, R., J. de Louvois, and A. Mulhall. 1987. Yeasts as human and animal pathogens, p. 207-281. In A. H. Rose and J. S. Harrison (ed.), The yeasts, 2nd ed. Academic Press, London, United Kingdom.
- 22.Jacobsen, M. D., M.-E. Bougnoux, C. d'Enfert, and F. C. Odds. 30 May 2008. Multilocus sequence typing of Candida albicans isolates from animals. Res. Microbiol. doi: 10.1016/j.resmic.2008.05.003. [DOI] [PubMed]
- 23.Jadhav, V. J., and M. Pal. 2006. Canine mycotic stomatitis due to Candida albicans. Rev. Iberoam. Micol. 23233-234. [DOI] [PubMed] [Google Scholar]
- 24.Krukowski, H., M. Tietze, T. Majewski, and P. Rózanski. 2001. Survey of yeast mastitis in dairy herds of small-type farms in the Lublin region, Poland. Micropathologia 1505-7. [DOI] [PubMed] [Google Scholar]
- 25.Kuwamura, M., M. Ide, J. Yamate, Y. Shiraishi, and T. Kotani. 2006. Systemic candidiasis in a dog, developing spondylitis. J. Vet. Med. Sci. 681117-1119. [DOI] [PubMed] [Google Scholar]
- 26.Ledbetter, E. C., V. H. Patten, J. M. Scarlett, and F. M. Vermeylen. 2007. In vitro susceptibility patterns of fungi associated with keratomycosis in horses of the northeastern United States: 68 cases (1987-2006). J. Am. Vet. Med. Assoc. 2311086-1091. [DOI] [PubMed] [Google Scholar]
- 27.McCullough, M. J., K. V. Clemons, and D. A. Stevens. 1999. Molecular and phenotypic characterization of genotypic Candida albicans subgroups and comparison with Candida dubliniensis and Candida stellatoidea. J. Clin. Microbiol. 37417-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mirhendi, H., K. Makimura, K. Zomorodian, N. Maeda, T. Ohshima, and H. Yamaguchi. 2005. Differentiation of Candida albicans and Candida dubliniensis using a single-enzyme PCR-RFLP method. Jpn. J. Infect. Dis. 58235-237. [PubMed] [Google Scholar]
- 29.Moretti, A., B. Posteraro, L. Boncio, L. Mechelli, E. De Gasperis, F. Agnetti, and M. Raspa. 2004. Diffuse cutaneous candidiasis in a dog. Diagnosis by PCR-REA. Rev. Iberoam. Micol. 21139-142. [PubMed] [Google Scholar]
- 30.National Committee for Clinical Laboratory Standards. 2002. Reference method for broth dilution antifungal susceptibility testing of yeast; approved standard—second edition. NCCLS document M27-A2. NCCLS, Wayne, PA.
- 31.Odds, F. C. 1988. Candida and candidosis. Bailliere Tindall, London, United Kingdom.
- 32.Odds, F. C., A. D. Davidson, M. D. Jacobsen, A. Tavanti, J. A. Whyte, C. C. Kibbler, D. H. Ellis, M. C. J. Maiden, D. J. Shaw, and N. A. R. Gow. 2006. Candida albicans strain maintenance, replacement, and microvariation demonstrated by multilocus sequence typing. J. Clin. Microbiol. 443647-3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Odds, F. C., M.-E. Bougnoux, D. J. Shaw, J. M. Bain, A. D. Davidson, D. Diogo, M. D. Jacobsen, M. Lecomte, S.-Y. Li, A. Tavanti, M. C. J. Maiden, N. A. R. Gow, and C. d'Enfert. 2007. Molecular phylogenetics of Candida albicans. Eukaryot. Cell 61041-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Odds, F. C., and R. Bernaerts. 1994. CHROMagar Candida, a new differential isolation medium for presumptive identification of clinically important Candida species. J. Clin. Microbiol. 321923-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oh, S.-H., G. Cheng, J. A. Nuessen, R. Jajko, K. M. Yeater, X. Zhao, C. Pujol, D. R. Soll, and L. L. Hoyer. 2005. Functional specificity of Candida albicans Als3p proteins and clade specificity of ALS3 alleles discriminated by the number of copies of the tandem repeat sequence in the central domain. Microbiology 151673-681. [DOI] [PubMed] [Google Scholar]
- 36.Pfaller, M. A., and D. J. Diekema. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20133-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfaller, M. A., S. A. Messer, L. Boyken, R. J. Hollis, C. Rice, S. Tendolkar, and D. J. Diekema. 2004. Further standardization of broth microdilution methodology for in vitro susceptibility testing of caspofungin against Candida species by use of an international collection of more than 3,000 clinical isolates. J. Clin. Microbiol. 423117-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pressler, B. M., S. L. Vaden, I. F. Lane, L. D. Cowgill, and J. A. Dye. 2003. Candida spp. urinary tract infections in 13 dogs and seven cats: predisposing factors, treatment, and outcome. J. Am. Anim. Hosp. Assoc. 39263-270. [DOI] [PubMed] [Google Scholar]
- 39.Pujol, C., A. R. Dodgson, and D. R. Soll. 2005. Population genetics of ascomycetes pathogenic to humans and animals, p. 149-188. In J. Xu (ed.), Evolutionary genetics of fungi. Horizon Scientific Press, Norfolk, United Kingdom.
- 40.Pujol, C., M. Pfaller, and D. R. Soll. 2002. Ca3 fingerprinting of Candida albicans bloodstream isolates from the United States, Canada, South America, and Europe reveals a European clade. J. Clin. Microbiol. 402729-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quinn, P. J., B. K. Markey, M. E. Carter, W. J. Donnelly, and F. C. Leonard. 2002. Yeasts and disease production, p. 233-239. In Veterinary microbiology and microbial disease. Blackwell Publishing, Oxford, United Kingdom.
- 42.Richter, S. S., R. P. Galask, S. A. Messer, R. J. Hollis, D. J. Diekema, and M. A. Pfaller. 2005. Antifungal susceptibilities of Candida species causing vulvovaginitis and epidemiology of recurrent cases. J. Clin. Microbiol. 432155-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sobel, J. D. 1998. Vulvovaginal candidiasis: epidemiologic, diagnostic, and therapeutic considerations. Am. J. Obstet. Gynecol. 178203-211. [DOI] [PubMed] [Google Scholar]
- 44.Soll, D. R., and C. Pujol. 2003. Candida albicans clades. FEMS Immunol. Med. Microbiol. 391-7. [DOI] [PubMed] [Google Scholar]
- 45.Spencer, J. F. T., and D. M. Spencer. 1997. Ecology: where yeasts live, p. 33-58. In J. F. T. Spencer and D. M. Spencer (ed.), Yeasts in natural and artificial habitats. Springer, Berlin, Germany.
- 46.Tavanti, A., A. D. Davidson, M. J. Fordyce, N. A. R. Gow, M. C. J. Maiden, and F. C. Odds. 2005. Population structure and properties of Candida albicans, as determined by multilocus sequence typing. J. Clin. Microbiol. 435601-5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tavanti, A., N. A. R. Gow, S. Senesi, M. C. J. Maiden, and F. C. Odds. 2003. Optimization and validation of multilocus sequence typing for Candida albicans. J. Clin. Microbiol. 413765-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Timoney, J. F., J. H. Gillespie, F. W. Scott, and J. E. Barlough. 1998. The opportunistic fungal infections, p. 407-424. In Hagen and Bruner's microbiology and infectious diseases of domestic animals, 8th ed. Comstock, Ithaca, NY.
- 49.Xu, J., and T. G. Mitchell. 2003. Geographical differences in human oral yeast flora. Clin. Infect. Dis. 36221-224. [DOI] [PubMed] [Google Scholar]
- 50.Zhao, X., S.-H. Oh, R. Jajko, D. J. Diekema, M. A. Pfaller, C. Pujol, D. R. Soll, and L. L. Hoyer. 2007. Analysis of ALS5 and ALS6 allelic variability in a geographically diverse collection of Candida albicans isolates. Fungal Genet. Biol. 441298-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zlotowski, P., D. B. Rozza, C. A. Pescador, D. E. Barcellos, L. Ferreiro, E. M. Sanches, and D. Driemeier. 2006. Muco-cutaneous candidiasis in two pigs with postweaning multisystemic wasting syndrome. Vet. J. 171566-569. [DOI] [PubMed] [Google Scholar]