Abstract
Appropriate responses of organisms to heat stress are essential for their survival. In eukaryotes, adaptation to high temperatures is mediated by heat shock transcription factors (HSFs). HSFs regulate the expression of heat shock proteins, which function as molecular chaperones assisting in protein folding and stability. In many model organisms a great deal is known about the products of hsf genes. An important exception is the filamentous fungus and model eukaryote Neurospora crassa. Here we show that two Neurospora crassa genes whose protein products share similarity to known HSFs play different biological roles. We report that heat shock factor 1 (hsf1) is an essential gene and that hsf2 is required for asexual development. Conidiation may be blocked in the hsf2 knockout (hsf2KO) strain because HSF2 is an integral element of the conidiation pathway or because it affects the availability of protein chaperones. We report that genes expressed during conidiation, for example fluffy, conidiation-10, and repressor of conidiation-1 show wild-type levels of expression in a hsf2KO strain. However, consistent with the lack of macroconidium development, levels of eas are much reduced. Cultures of the hsf2KO strain along with two other aconidial strains, the fluffy and aconidial-2 strains, took longer than the wild type to recover from heat shock. Altered expression profiles of hsp90 and a putative hsp90-associated protein in the hsf2KO strain after exposure to heat shock may in part account for its reduced ability to cope with heat stress.
The heat shock response is a protective mechanism against sudden temperature change and other forms of physical and chemical stress. The response is brought into effect by heat shock proteins (HSPs), molecular chaperones that assist protein folding and stability (35). Several highly conserved families of HSPs, named according to their molecular masses, are expressed in response to heat shock and include HSP110, HSP100, HSP90, HSP70, HSP60, and a number of small HSPs. Some of the family members are expressed in non-heat shock conditions, but the expression of others is induced only upon exposure to stress due to the action of heat shock transcription factors (HSFs). HSFs are a functionally conserved group of molecules. They were first identified as regulators of the heat shock response (reviewed in reference 28), but it is now evident that certain HSFs also regulate developmental pathways (for a review, see reference 36).
Drosophila melanogaster and Saccharomyces cerevisiae each have one essential HSF (7, 48). In S. cerevisiae, while the induction of genes encoding chaperones and associated proteins is under the control of its HSF, a subset of genes induced by a variety of environmental stresses and encoding metabolic enzymes, components of the ubiquitin-proteosome degradation pathway, and antioxidant defense proteins are under the control of the transcription factors Msn2, Msn4, Yap1, and Skn7 (5, 17, 6, 25). In Drosophila, temperature-dependent alternative splicing of dHSF gives rise to four forms of dHSF mRNA which encode HSF isoforms with different transcriptional activities (16). Although the functional significance of the HSF isoforms is not yet understood clearly, one possibility is that they activate different subsets of HSP or stress-responsive genes. Moreover, in species with multiple hsf genes, a functional specialization of certain HSFs toward developmental roles has occurred. For example, of the four vertebrate HSFs, HSF1 mediates heat shock gene expression, whereas HSF2 is reported to be unresponsive to temperature, while its activity changes throughout differentiation. In addition, alternative splicing of hsf1, hsf2, and hsf4 transcripts generates a variety of HSF isoforms (13, 18, 45). Since these splice forms show tissue-specific expression, it is likely that they also have distinct roles.
In contrast, no Neurospora HSFs have yet been isolated, although HSPs are expressed in this organism during heat shock and cell differentiation (14, 19) suggesting that, as seen for other organisms, the Neurospora HSF(s) is required for development. Neurospora crassa undergoes both sexual and asexual developmental cycles. When nitrogen is limiting, female sexual development ensues, and when nuclei of opposite mating types fuse numerous asci are produced, each containing eight haploid spores (42). In addition, two asexual developmental pathways are described: macroconidiation and microconidiation. Macroconidial development in Neurospora can be induced by desiccation, nutrient deprivation (47), light (23), oxidative stress (29), and signals relayed from the organism's circadian clock (37). Once induced, aerial hyphae are formed which branch and eventually form chains of macroconidia, each containing between one and five nuclei (44). In a distinctly different developmental process, microconidia containing one or rarely two nuclei are formed from smaller branched microconidiophores (30).
Several phases in the development of macroconidia have been recorded (31, 44). The first step is the production of branching aerial hyphae. Minor constrictions then develop, followed by the production of proconidia via budding from the terminal cells of each branch. Finally, proconidia are separated by major constrictions and mature into conidia, which are easily separated and dispersed by wind or water. A number of mutants affecting this developmental pathway are known. The fluffyoid (fld) and aconidial-2 (acon-2) mutants are blocked before and the fluffy (fl) mutant is blocked after the formation of minor constrictions (31, 44). The acon-3 mutant is blocked after the maturation of minor constrictions. conidial separation-1 (csp-1) and csp-2 are required for conidial separation (41). Genetic and molecular studies place acon-2 upstream of fl, with acon-3, csp-1, and csp-2 being downstream (9, 1).
We report here that Neurospora hsf1 is an essential gene and that a deletion strain can be maintained only as a heterokaryon. In strains with hsf2 deleted, no dramatic alteration of the heat shock response is detected. However, macroconidiation in hsf2 is defective; aerial hyphae emerge but no macroconidia are produced. Consistent with this, differences are seen between the wild-type and hsf2 knockout (hsf2KO) strains in the expression of two conidiation-associated transcripts, fl and eas.
MATERIALS AND METHODS
Bioinformatic analysis.
Genes were identified by searching the Broad Institute Neurospora crassa database. Protein prediction was performed using NCBI BLAST at http://www.ncbi.nlm.nih.gov/BLAST/ and COILS at http://www.ch.embnet.org/software/COILS_form.html. Sequence alignments were performed using ClustalW at http://www.ebi.ac.uk/clustalw/.
Strains and growth conditions.
mus-51a (FGSC9718), hsf2 a (FGSC 11671), acon-2 a (FGSC3263), fl a (FGSC818), and the hsp90a (NCU01792), hsp30 (NCU09364), hsp78 (NCU02630), hsp101 (NCU00104), hsp70 (NCU08683), and hsp90 (NCU04142) strains were obtained from the Fungal Genetics Stock Center (http://www.fgsc.net/) and crossed into either 30-7 bd A or 87-3 bd a to introduce the bd mutation. 87-3 bd a was obtained from Christian Heintzen (University of Manchester, United Kingdom). fl,bd was a kind gift from D. Bell-Pedersen (Texas A&M University). The hsf1KO strain was made by amplifying 1-kbp regions 5′ and 3′ of the hsf1 open reading frame (ORF) and fusing these to the hygromycin phosphotransferase (hph) gene. mus-51a was transformed with this fragment and transformants were selected on hygromycin-containing agar plates (8). Strains were maintained on minimal medium (1× Vogel's salts [10], 2% sucrose, 500 ng/ml biotin, 1.5% agar). Crossing medium contained 1× Westergaard's salts, 2% sucrose, 500 ng/ml biotin, and 1.5% agar (10). Spores were picked onto slants containing Vogel's minimal medium, heat shocked at 60°C for 1 h to induce germination, and then incubated at 30°C. Progeny were screened for hygromycin resistance on minimal medium containing 200 μg/ml of hygromycin B and/or for rhythmic conidiation on race tubes.
Race tube medium contained 1× Vogel's salts, 0.1% glucose, 0.17% arginine, 500 ng/ml biotin, and 1.5% agar. After inoculation with 5- to 10-day-old macroconidia, race tubes were placed in constant light (LL) at 25°C for 48 h and then transferred to constant darkness (DD). Growth fronts were marked daily. Heat shocks were given by immersing race tubes in a 50°C water bath for 1 h after 24 h in the dark at 25°C. The race tube cultures were then returned to 25°C to recover and examined daily for regrowth.
Microconidiation was induced according to the protocol of Ebbole and Sachs (11). Conidia were harvested by vortexing in 2 ml of sterile distilled water and filtered through 5-μm filters (Millipore) into sterile Eppendorf tubes. Microconidia were plated onto minimal (1× Vogel's salts, 1% sorbose, 0.005% fructose, 0.005% glucose, 1 M sorbitol) or minimal hygromycin-containing plates (200 μg/ml). Plates were incubated at 30°C until single colonies were detected (3 or 4 days later). Colonies were picked onto minimal agar slants, and this was followed by the transfer of conidia onto hygromycin-containing (200 μg/ml) slants.
Liquid culture medium for the growth of mycelial mats and shaking cultures contained 1× Vogel's salts, 2% glucose, and 500 ng/ml biotin. One hundred milliliters of liquid culture medium was inoculated with 2.3-cm-diameter discs cut from mycelial mats. These cultures were put into LL at 25°C with shaking at 150 rpm. After 24 h, cultures were placed into a shaking water bath at 45°C. Flasks were removed at regular intervals and their contents were immediately harvested by vacuum filtration onto filter paper and frozen in liquid nitrogen.
The experiment examining nitrogen starvation-induced conidial development was first described in the work of Bailey-Shrode and Ebbole (1). Strains cultured on Vogel's minimal medium slants (2% sucrose) were used to inoculate culture dishes of modified Vogel's minimal medium (1.5% sucrose) with 50 mM NH4Cl in place of NH4NO3. After 3 days at 30°C, a cork borer was used to punch discs from the resulting mycelial mats. These discs were then transferred to 100-ml flasks containing 50 ml of modified Vogel's minimal medium and incubated for 22 h at 34°C in LL plus shaking at 150 rpm. The cultures were then washed in sterile distilled water and transferred to flasks containing Vogel's minimal medium lacking any nitrogen source. Tissue was harvested before and 1 h, 6 h, 12 h, and 24 h after transfer.
Induction of conidiation by exposure to air was carried out essentially as described in the work of Lee and Ebbole (26), with the following modifications. Disks were cut from mycelial mats and placed into Vogel's minimal medium with 2% glucose. After 24 h in DD at 25°C, disks were blotted dry and placed on agar blocks (Vogel's minimal medium, 2% glucose, 2% agar) in open petri dishes containing 15 ml of fresh culture medium.
RT-PCR.
The RevertAid H− first-strand cDNA synthesis kit (Fermentas) was used for all reverse transcription-PCR (RT-PCR). Primers are listed in Table 1. For semiquantitative RT-PCR, 1 μg of total RNA was reverse transcribed using the poly(T) primer supplied with the kit, following the manufacturer's instructions. A limiting number of cycles were used to stop the subsequent PCRs during the exponential phase of amplification. After agarose gel electrophoresis, quantification was performed using Bio-Rad QuantityOne software and data were normalized against actin transcript levels.
TABLE 1.
Primers used
| Gene | Usea | Primerb
|
|
|---|---|---|---|
| Forward | Reverse | ||
| hsf1 | RT/cloning/cDNA probe | CGGGATCCCGCATGTCTTCTCCTAACC | CGGGATCCCGCGACAGAAGCAACAGAT |
| RNA probe/sqRT-PCR (1; 35) | CGGGATCCCGCATGTCTTCTCCTAACC | TAATACGACTCACTATAGGGACGTGAGGGCCTGCTGGAATAAGGTCG | |
| hsf2L | RT/cloning/cDNA probe | TAGGATCCTACAGTGCAGATGGCTACC | TAGGATCCACATAACGTCTCAAGATGC |
| RNA probe/sqRT-PCR (2; 30) | TAGGATCCTACAGTGCAGATGGCTACC | TAATACGACTCACTATAGGGATCCTCCATCTGGCGCTGAACCTGC | |
| hsf2S | RT/cloning | TAGGATCCTACAGTGCAGATGGCTACC | TAGGATCCCAAGCTAACCAGGTCTCCG |
| hsf2 5′ flank | Southern blot DNA probe | GTAGCCTCTCCATACCTACC | GTAGAGTCCATTCTACCTCG |
| hsp70 | DNA probe | GTCTCGACAAGAAGGTCG | TCTCGGAGGTGGACTTGG |
| hsp90 | DNA probe | GGATCCTTGTTGCCGACAGAGTTA | GGATCCCATCAATGGGGTCGACAAGG |
| NCU01792 | DNA probe | AGCGCTCTTCTGCTACCG | CATGGATCTTGGGTATGC |
| fluffy | RNA probe/sqRT-PCR (1; 30) | TGTGATGGCCAGATGCCATGC | TAATACGACTCACTATAGGGACGAGTTATCGCTGCCCTCACG |
| actin | sqRT-PCR (0.5; 20) | GGCTGGCCGTGATCTTACCGACTA | GAGAGCGAGGCGAGAATGGAACC |
| con-10 | sqRT-PCR (1; 26) | CGTCAACATGGCTGGCACTGGTAACG | TTGTAATACGACTCACTATAGGGCAAATTGTCAAAGCATTCAGTTGC |
| rco-1 | sqRT-PCR (1; 35) | GTACGAGGATGAGATTGCGCT | TCTGATGAGCTTGTCCTCGGCTC |
For semiquantitative RT-PCR (sqRT-PCR), the numbers in parentheses indicate the amounts of cDNA in μl followed by the cycle numbers.
Modifications are in lightface, and annealing portions are in boldface.
Construction of the hsf1 knockout strain.
A hsf1 deletion cassette was made containing the hph gene (confers resistance to hygromycin) flanked by approximately 1.6 kbp 5′ and 2 kbp 3′ of the NCU08512 ORF. One hundred microliters of a mus-51 conidial cell suspension (2 × 109 conidia/ml) in a 2-mm gap cuvette (Bio-Rad) was electroporated with 0.5 μg of the knockout construct. A Bio-Rad gene pulser was used with the following settings: 12.5 kV/cm, 400 Ω, 25 μF. Transformants were selected on hygromycin plates (200 μg/ml) incubated at 30°C for 4 days. Transformants were crossed with 87-3 bd a to obtain homokaryons.
RNA extraction and Northern blotting.
RNA was extracted using Trizol (Invitrogen) following the manufacturer's protocol. Equal amounts of RNA were run through a 1% agarose formaldehyde gel and blotted onto Hybond N+ nitrocellulose membrane (GE Healthcare). Blots were probed with randomly primed 32P-labeled (Roche labeling kit; GE Healthcare radionucleotide) cDNA or 32P-labeled RNA (MAXIscript kit; Ambion) and detected using phosphorimager screens (Fuji). Blots were prehybridized (NorthernMax; Ambion) and hybridized at 42°C for DNA probes and at 68°C for RNA probes. Blots were washed twice for 10 min in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) plus 0.1% sodium dodecyl sulfate (SDS), once for 15 min in 1× SSC plus 0.1% SDS at room temperature, and once for 15 min in 0.1× SSC plus 0.1% SDS at either 42 or 68°C.
DNA extraction and Southern blotting.
Genomic DNA was extracted from homogenized tissue by use of a cetyltrimethylammonium bromide protocol. DNA was digested with NdeI (hsf2) or NdeI and BamHI (hsf1), separated through 0.8% agarose gels, and transferred onto Hybond N+ nitrocellulose membrane. The DNA was cross-linked to membranes with UV light. Blots were hybridized to digoxigenin (DIG)-labeled (Roche) probes against the 5′-flanking region of the knockout cassettes (http://www.dartmouth.edu/∼neurosporagenome/knockouts_completed.html). Primers for the amplification of probe DNA are listed in Table 1. Each blot was incubated with prehybridization solution (NorthernMAX; Ambion) at 42°C for 1 h before the addition of 300 ng of denatured probe in 4 to 5 ml of fresh hybridization solution and incubation at 42°C overnight. After hybridization, the blots were washed according to the manufacturer's protocol, with the final washes carried out at 42°C. Detection of bound DIG-labeled probe was also carried out according to the manufacturer's protocol, and the blots were exposed to X-ray film for 5 to 25 min.
RESULTS
Identification of putative HSFs.
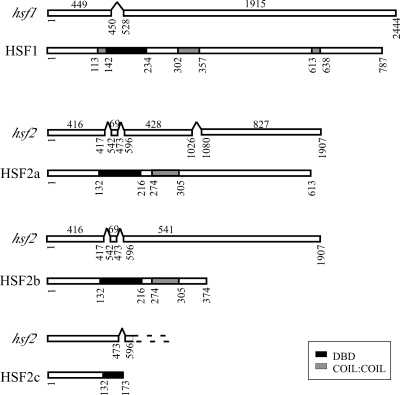
Two putative Neurospora HSFs encoded by hsf1 (NCU08512) and hsf2 (NCU08480) were identified in a BLAST search of the Neurospora genome. We cloned RNAs expressed from these genes and analyzed their sequences to determine the details of any introns. hsf1 contains one intron and encodes a 787-amino-acid (aa) protein. Alternative splicing of hsf2 RNA gives rise to at least three different transcripts encoding polypeptides of 613 aa, 374 aa, and 173 aa or of 579 aa, 340 aa, and 139 aa depending at which of the three possible in-frame ATGs translation is initiated (Fig. 1). HSFs from different species, though binding to similar DNA consensus sequences, are not highly conserved. However, all contain a DNA-binding domain at the amino terminus and an adjacent cluster of hydrophobic amino acids organized into heptad repeats required for oligomerization, and they often also contain a heptad repeat close to the carboxy terminus (49). In order to become functionally active, HSF protein monomers form homotrimers through the interaction of coiled-coil domains found toward the N terminus. HSF1 and the large HSF2 isoforms contain putative DNA-binding and trimerization domains. In addition, a carboxy heptad repeat is present in HSF1. In contrast, the 173-aa isoform of HSF2 contains only a partial DNA-binding domain (Fig. 1). Although the presence of these alternatively spliced transcripts in the five cDNAs we cloned and sequenced suggests that they are present at a high level relative to what is seen for the fully spliced transcript, further investigation revealed that in >99% of the transcripts, introns 1 to 3 have been removed (data not shown).
FIG. 1.
RNA splice forms and protein domain structures of HSF1 and HSF2 isoforms. For each pair of diagrams the top schematic represents RNA and the bottom schematic represents protein. Open rectangles in mRNAs represent exons. Nucleotide numbers below transcripts start from the ATGs of the respective ORFs. Amino acid numbers are indicated below proteins. DBD, DNA-binding domain.
hsf1 is an essential gene; hsf2 is required for asexual development.
To knock out hsf1, at least 1.5 kb of DNA 5′ and 3′ of the ORF was ligated to the hygromycin phosphotransferase gene (Fig. 2A). This construct was electroporated into conidia of a mus-51::bar strain which lacks an enzyme required for nonhomologous DNA joining. Due to the absence of mus-51, targeted transformation is almost 100% efficient (33). DNA from 20 transformants was extracted and recombination events at the hsf1 gene were checked by Southern blot analysis (Fig. 2B). All transformants were heterokaryons harboring both nuclei with hsf1 intact and nuclei with hsf1 deleted. To obtain homokaryons, microconidiation was induced in two of the heterokaryons (11). Only five colonies derived from microconidia were hygromycin resistant, and once again these were heterokaryons (data not shown). In parallel, two of the transformants were backcrossed to the bd strain. Sixty-five spores derived from these crosses were selected and all 39 that germinated were inoculated onto hygromycin slants. None of these strains were resistant to hygromycin, supporting our conclusion that hsf1 is an essential gene.
FIG. 2.
Analysis of mus-51::bar, ▵hsf1 transformants. (A) Schematic diagram showing the location of SacI restriction enzyme sites in the region of hsf1. (Left) Wild-type hsf1 locus; (right) locus with the hsf1 gene deleted and replaced with hph. (B) Southern blot of mus-51::bar, ▵hsf1 strains. Genomic DNA digested with SacI and probed with DIG-labeled DNA homologous to a region 5′ of the hsf1 ORF. Lanes: 1, 7, and 12, DNA ladder; 8, control parental mus-51 genomic DNA; 2 to 6, 9 to 11, 13, and 14, DNA from different transformants.
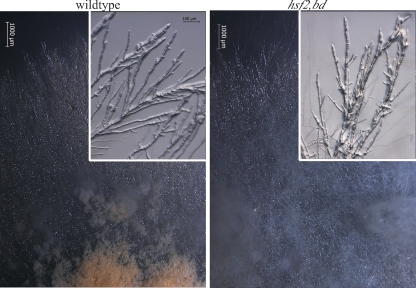
A hsf2 deletion strain (8) was obtained from the Fungal Genetics Stock Center (University of Missouri, Kansas City) and checked by Southern analysis (data not shown). The vegetative hyphal growth of the hsf2 mutant appears normal, as does the growth rate, but the deletion strain has a striking phenotype; it produces very few macroconidia (Fig. 3). Only after several weeks are a few proconidial chains present, perhaps induced by starvation or desiccation.
FIG. 3.
The hsf2KO strain is aconidial. Light microscopy showing growth of bd (left) and hsf2KO,bd (right) strains on minimal agar plates (low magnification) and at higher magnification (inset; magnification, ×80) at the growth front. Macroconidia (asexual spores) are bright orange.
The heat shock response in the hsf2,bd strain.
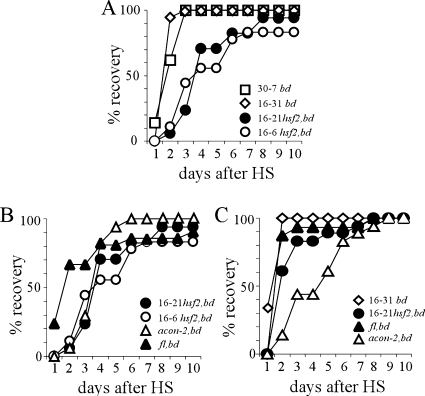
We first assayed the phenotypic response of the hsf2 mutant to heat shock by monitoring its growth along race tubes after exposure to high temperature. In order to assess the effect of heat shock on conidiation as well as on growth a hsf2,bd strain was initially used in these experiments. The bd mutation allows conidiation to occur in the presence of the high carbon dioxide levels which accumulate in the enclosed race tube. In response to a 1-h, 50°C heat shock during the third day in DD at 25°C, growth of both bd and hsf2,bd strains stopped. On the second day after the heat shock 100% of the bd cultures had resumed growth, while only 4 days after the heat shock did 50% of the hsf2,bd cultures resume growth (Fig. 4A). We wondered if the delayed recovery was correlated with the aconidial state of the hsf2,bd strain, and we therefore repeated the experiment, this time including the acon-2,bd and fl,bd strains. Of note is that the acon-2,bd strain is a temperature-sensitive strain and no spores develop at 34°C or above. Therefore, all of the strains were cultured at either 25°C or 34°C before and after a 50°C heat shock. In both sets of conditions, the aconidial strains took longer to recover from heat shock than did the bd strain (Fig. 4B and C). These data suggest either that the germination of conidia that fall to the agar surface after heat shock accounts for the faster regrowth of the bd strain and/or that all the aconidial strains are inherently temperature sensitive.
FIG. 4.
Response of wild-type and aconidial strains to heat shock. Strains inoculated onto race tubes were germinated and grown in LL at 25°C (A and B) or 34°C (C) for 48 h before transfer to DD at 25°C (A and B) or 34°C (C). After 24 h in DD, cultures were exposed to a 1-h 50°C heat shock and then transferred back to DD at 25°C (A and B) or 34°C (C). Growth was monitored twice daily over the following 9 days. The graph shows the percentages of recovery for the following strains against time (h) after heat shock. (A) 30-7 bd (n = 21), 16-31 bd (n = 18), 16-21 hsf2KO,bd (n = 17), and 16-6 hsf2KO,bd (n = 18). Numbers preceding the strain names refer to the cross and progeny identifiers, e.g., 16-31 indicates cross number 16 and progeny number 31. (B) 16-21 hsf2KO,bd (n = 17), 16-6hsf2KO,bd (n = 18), acon-2,bd (n = 17), and fl,bd (n = 21). (C) 16-31 bd (n = 17), 16-21hsf2KO,bd (n = 18), fl,bd (n = 15), and acon-2,bd (n = 18). The average results of three independent experiments are plotted. HS, heat shock.
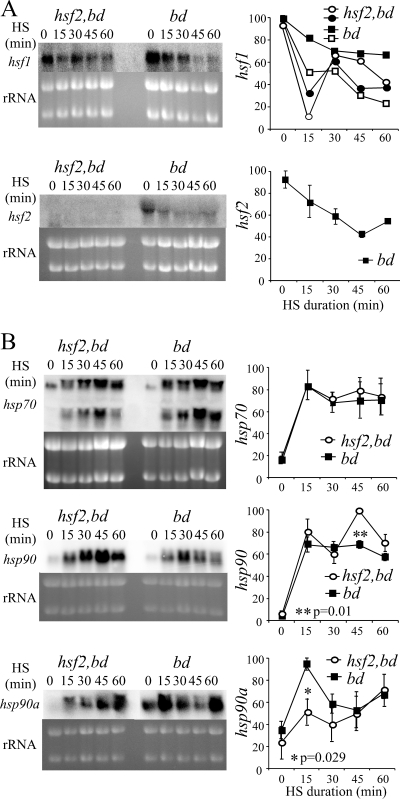
If the latter of these possibilities is true, then these results implicate HSF2 in the heat shock response. Therefore, we followed levels of both hsf1 and hsf2 and of three HSP transcripts in control and heat-shocked cultures. In a bd strain, the levels of hsfI and hsf2 mRNA declined during the first 45 min of a prolonged heat shock. In the hsf2,bd strain, hsf1 transcription also decreased and, as expected, hsf2 RNA was not expressed (Fig. 5A). In spite of this, the three HSP transcripts we assayed, namely, those for hsp70 (NCU09602) and hsp90 (NCU04142) and a transcript that arises from NCU01792 which we have named heat shock protein 90-associated (hsp90a), were upregulated in response to heat shock in both strains (Fig. 5B). Interestingly, two hsp70 transcripts can be distinguished after transfer to high temperatures in both the wild-type and hsf2,bd strains. The lower-molecular-weight transcript is most likely another member of the hsp70 family, ssb1 (NCU02075), which shares 87% identity over 239 nucleotides to our 901-nucleotide hsp70 probe. Of note is that hsp90 transcripts were significantly more abundant 15 min after the onset of heat shock in the hsf2,bd strain than in the bd strain. One other significant difference was detected after 45 min of exposure to heat shock between levels of hsp90a RNA in the bd and hsf2,bd strains, with the levels being higher in the bd strain.
FIG. 5.
Gene expression in wild-type and hsf2,bd knockout strains. Northern blots of bd and hsf2KO,bd RNA probed for hsf1 and hsf2 RNA (A) and hsp70, hsp90, and hsp90a RNA (B). Cultures transferred from LL at 25°C to LL at 45°C were harvested for RNA extraction after 0, 15, 30, 45, and 60 min. There are two sets of data for the hsf1 transcript and three for all other transcripts. The y axes represent the relative amounts of each transcript (percentages of the maximum). HS, heat shock.
RNA levels of genes associated with conidiation are affected in the hsf2 strain.
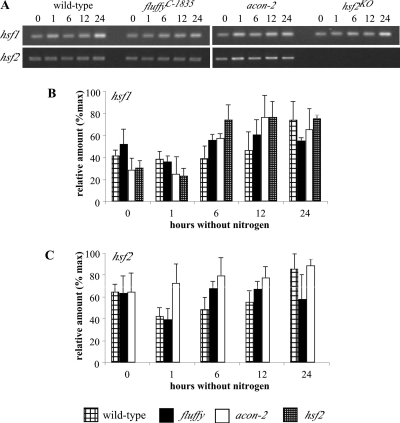
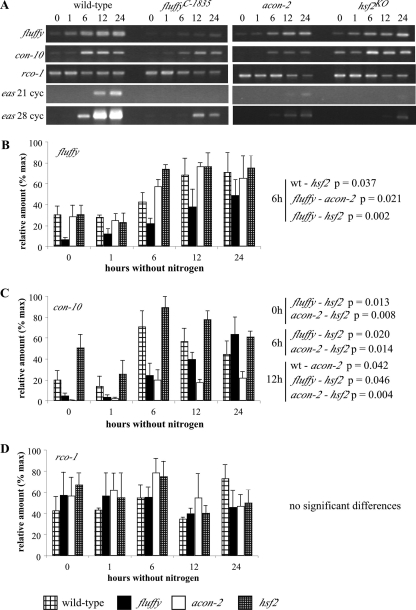
To test where HSF2 acts in the known conidiation pathway, we investigated the expression of hsf1, hsf2 (Fig. 6), fl, conidiation-10 (con-10), repressor of conidiation-1 (rco-1), and eas (Fig. 7) in the wild-type, fl, acon-2, and hsf2 strains. We assayed gene expression upon the transfer of cultures to medium lacking nitrogen, conditions known to induce the conidiation pathway (1). Although there was a general increase in hsf1 mRNA as development progressed, no significant difference in the levels of either hsf1 or hsf2 RNA was detected among the four strains. As previously reported, levels of fl and con-10 RNA in the wild type increase from 6 hours after the transfer of cultures to medium lacking nitrogen (1). Among the aconidial strains, fl transcript levels are higher in the acon-2 and hsf2 strains at 6 h, whereas con-10 transcripts show significantly increased levels of expression in the hsf2 strain (similar to con-10 levels in the wild-type) compared to those in the acon-2 and fl strains at 0, 6, and 12 h. To our surprise, no differences in the transcript levels of rco-1, a repressor of con-10, were detected between any of the strains at the time points sampled. In our study, by far the most striking difference between the aconidial strains and the wild type is the greatly reduced levels of eas in the fl, acon-2, and hsf2 strains.
FIG. 6.
Semiquantitative RT-PCR of hsf and hsf2 RNA in wild-type, flC-1835, acon-2, and hsf2KO mutants. Cultures were harvested before and at various times after transfer to medium lacking nitrogen, conditions that initiate conidiation in the wild-type strain. (A) Representative gels showing RT-PCR products of each transcript. (B and C) Relative amounts of each transcript graphed after normalization against actin. % max, percentages of the maximum.
FIG. 7.
Comparison of fl, con-10, rco-1, and eas expression in wild-type and conidiation-defective strains of Neurospora. Semiquantitative RT-PCR of fl, con-10, rco-1, and eas RNA in the wild type and the flC-1835, acon-2, and hsf2KO mutants. Cultures were harvested before and at various times after transfer to medium lacking nitrogen, conditions that initiate conidiation in the wild-type strain. (A) Representative gels showing RT-PCR products of each transcript. (B, C, and D) Relative amounts of each transcript graphed after normalization against actin. % max, percentages of the maximum; wt, wild type.
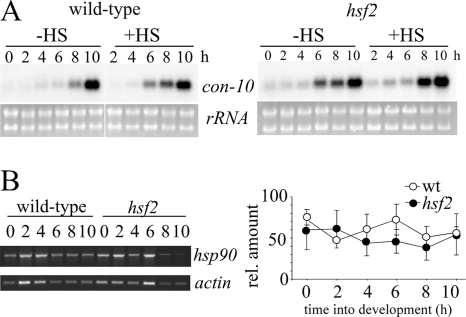
Conidiation can be induced by a number of environmental cues, for example, carbon and nitrogen deprivation and light (47). Synchronous conidiation can also be induced by the exposure of mycelia to air (44). Interestingly, although developmentally induced genes are expressed at similar times in cultures induced to conidiate by either nitrogen starvation or exposure to air, evidence suggests that their sensitivity to other stresses, such as heat shock, may change. For example, it is reported that in air-exposed cultures, con-10 is induced by heat shock, whereas in liquid cultures it is not (26). However, in our hands, no significant difference in the developmental induction of con-10 expression between the wild-type and hsf2 strains during air-induced development was revealed, and in neither strain was con-10 induced by heat shock (Fig. 8). con-10 has been shown to be regulated by numerous factors e.g., carbon and nitrogen deprivation and light, and it is probable that slight differences in experimental conditions account for the discrepancy between our results and those of Lee and Ebbole (26).
FIG. 8.
con-10 and hsp90 expression during development in air-exposed cultures. (A) Northern blots of wild-type and hsf2 total RNA probed for con-10. Mycelial mats were taken out of liquid cultures and exposed to air at time zero. Control mats (no heat shock [−HS]) were harvested at 2-h intervals. Heat-shocked samples (+HS) were exposed to 50°C for 1 hour before being harvested. (B) (Left) Representative gel showing semiquantitative RT-PCR products of hsp90 and actin transcripts; (right) quantitation of hsp90 RNA from the same samples (n = 3). rel., relative; wt, wild type.
In an effort to determine whether development in the hsf2 strain is indirectly affected through the misregulation of HSPs, we also assayed expression levels of the hsp90 transcript. Again, no significant differences in expression were detected (Fig. 7B). Furthermore, strains with hsp90a (NCU01792), hsp30 (NCU09364), hsp78 (NCU02630), and hsp101 (NCU00104) deleted showed no overt defect in conidiation (data not shown). While we can conclude that these HSPs are not essential for normal asexual development, hsp70 (NCU08683) and hsp90 (NCU04142) are essential genes, and thus, as for hsf1, their role in development could not be assessed by simple phenotypic observation of the respective gene knockout strains.
DISCUSSION
The following three genes encoding proteins with domains sharing similarity to known HSFs are present in Neurospora (4): hsf1 and hsf2, which are discussed in this paper, and a third gene not included in our study, NCU02413, which encodes response regulator-2 (rrg-2). rrg-2 contains a truncated HSF DNA-binding domain and is involved in Neurospora's response to oxidative stress (2). HSF1, HSF2a, and HSF2b all contain conserved HSF binding domains and coiled-coil regions that in other HSFs are required for trimerization (43). HSF binding domains recognize and bind to heat shock elements (HSE), consensus sequences found in the promoters of hsp genes (28). The basic HSE shows similarity across a wide range of organisms and is based around GAA repeats, the spacing and orientation of which vary and may influence which sites are recognized by different HSFs or differently phosphorylated HSFs (20). For Neurospora, HSE are not yet well defined, but nGAAn.. nTTCn motifs (where “n” can be any nucleotide) are present in the promoters of hsp30 (38) and hsp70 (21). Additionally, a factor in Neurospora protein extracts binds specifically to labeled oligonucleotides containing the classic yeast HSF binding sequence nTTCnnGAAnnTTCn (32). Band shift assays with purified Neurospora HSF(s) should aid in the identification of the discriminating consensus binding sites for HSF1 and HSF2.
Our finding that various splice forms of hsf2 exist is consistent with recent publications from other laboratories. For example, alternative splicing of hsf RNA is developmental stage specific in Schistosoma mansoni (22), and in Drosophila different splice forms are made depending on the stress to which the organism is exposed (16). Moreover, in both Drosophila and humans, alternative splicing of hsf RNA produces either an activator or a repressor of transcription (45). The repressors retain the DNA-binding and trimerization domains but lack the C-terminal activation domain, similar to Neurospora HSF2b and HSF2c. Nonetheless, further work is required to elucidate the conditions under which the three splice variants of Neurospora hsf2 are expressed and their purpose. Intriguingly, a possible function for HSF2b and HSF2c arises from a study reporting that in response to heat shock, proteins bind to putative HSE and/or stress elements in the glycogen synthase gene of Neurospora and repress transcription (15). Although we found only low levels of alternatively spliced hsf transcripts and observed that these did not correlate with reduced levels of glycogen synthase mRNA (data not shown), this does not rule out the possibility that forms of HSF2 protein change with time and in response to heat shock and/or development.
Whereas hsf2KO strains are viable but have an aconidial phenotype, we report that hsf1 is an essential gene. No homokaryon hsf1KO transformants were obtained, and no viable hsf1KO progeny were obtained from crosses of hsf1KO and hsf1+ heterokaryons with the wild type. The single S. cerevisiae and Drosophila HSF genes are also essential. In yeast, evidence suggests that HSF is required for the expression of both the constitutive and heat-inducible forms of Hsp90 (Hsc82 and Hsp82), and in the S. cerevisiae hsf-82 mutant, in which Hsp82 is no longer induced upon heat shock, spindle pole body formation is affected and completion of the cell cycle at 37°C is blocked (51). Moreover, indirect effects of Hsp90 misexpression on cell wall integrity, for example, occurs via a reduction in Slt2 kinase activity and a consequent lack of transcriptional activation of a subset of cell wall genes (46). The essential nature of Neurospora hsf1 would be easily explained if future experiments to elucidate its targets reveal a subset of genes that encode constitutive as well as heat shock-induced protein chaperones.
Our results suggest that HSF2 may have a role in regulating the level of HSP transcripts. We looked at the levels of hsp70, hsp90, and hsp90a specifically, because in other organisms the interaction of chaperones hsp70 and hsp90 with hormone receptors, kinases, and other signaling molecules is well known (reviewed in reference 34) and the inhibition or overexpression of these chaperones is associated with defects in development in Drosophila and in yeast (40, 12, 51, 46). The putative protein encoded by NCU01792 contains a CS domain, a domain that binds to Hsp90 and which is found in p23, a Hsp90 cochaperone (27). In our experiments, there were no significant differences between hsp70 transcript levels in the bd and hsf2,bd strains. However, immediately following heat shock, hsp90a RNA is significantly lower in the hsf2,bd strain compared to levels in the wild type, and hsp90 transcript levels are higher than those seen for the wild type 45 min after exposure to heat shock. Both of these departures from wild-type levels of gene expression could be directly due to the lack of HSF2.
Deleting hsf2 had a dramatic effect on asexual development; aerial hyphae developed but no conidial chains formed. The following six mutants blocked at different stages of conidial development have been placed in a conidiation pathway: the fld (linkage group IV [LGIV]), fl (LGII), acon-2 (LGIII), and acon-3 (LGIV) mutants and two conidial separation mutants, the csp-1 and csp-2 mutants. In addition, several recently generated knockout strains have been recorded as aconidial, including strains with mutations in vegetative growth and asexual development (the vad-2 [LGI] and vad-4 [LGIII] strains). hsf1 and hsf2 are both located on LGII, separated by approximately 100 kbp, and do not map to any of the previously described aconidial mutants. Bailey-Shrode and Ebbole place acon-2 upstream of fl and acon-3 in the pathway to conidiation (1). In this scheme, fl regulates acon-3 and eas, a gene encoding a hydrophobin found on the surfaces of macroconidia (3, 24), while acon-3 regulates con-6 and con-10. The rco-1 gene product represses the expression of eas, con-6, and con-10 (50, 26, 1). We were interested to find out if HSF2 is an integral element of the proposed pathway or whether it acts as a facilitator ensuring that the necessary chaperones are present to ensure the correct conformation of pathway components.
After initiation of development induced by the transfer of cultures to medium lacking nitrogen we found, in agreement with previous reports, that fl transcript levels in the wild type increase after 6 hours (1). fl is a transcriptional activator and a major regulator of conidiation in Neurospora (1, 39), whose expression has previously been reported to be low in the acon-2 strain (1). However, we found that fl was increased in all three aconidial strains, data which suggest that ACON2 regulates some posttranscriptional aspect of fl gene expression. In keeping with the lack of spore formation in the acon-2 and fluffy mutants, levels of con-10 are low in these strains. With the exception of eas, and the higher levels of fl at 6 h, all of the transcripts assayed throughout development showed similar levels of expression in the hsf2 and wild-type strains.
One scenario that could account for the aconidial phenotype of the hsf2KO strain is that the induction of HSPs that interact with components of the conidiation pathway are no longer sufficiently abundant for conidiation to proceed. Alternatively, HSF2 or a HSF2-induced protein is an integral component of the developmental pathway. We were unable in this study to distinguish conclusively between these possibilities. On the one hand, asexual development is normal in strains with different hsp genes deleted, and no significant differences in hsp90 expression over the course of development were detected. Moreover, levels of many of the transcripts we assayed show wild-type expression patterns in the hsf2 strain, which would be not be expected if protein folding is affected. On the other hand, some hsp genes are essential for survival, and their effect on asexual development could not be easily tested. However, if HSF2 or a HSF2-induced protein is indeed an integral part of the conidiation program, the gene expression data we have place HSF2 in a pathway parallel to that of fluffy. In support of this conclusion, wild-type fl and con-10 (repressed in a fluffy mutant) transcript levels are seen throughout development in the hsf2 strain. Hence, while acon-2 and fluffy can regulate the level of both con-10 and eas, the hsf2 strain must regulate levels of eas and other as yet unidentified genes essential for normal development via an independent pathway.
Acknowledgments
We thank Christian Heintzen for helpful discussions, Christian Heintzen and Suzanne Hunt for critical reading of the manuscript, and the reviewers for helpful suggestions.
This work was supported by a grant from The Leverhulme Trust, F/00120Z.
Footnotes
Published ahead of print on 27 June 2008.
REFERENCES
- 1.Bailey-Shrode, L., and D. J. Ebbole. 2004. The fluffy gene of Neurospora crassa is necessary and sufficient to induce conidiophore development. Genetics 1661741-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banno, S., R. Noguchi, K. Yamashita, F. Fukumori, M. Kimura, I. Yamaguchi, and M. Fujimura. 2007. Roles of putative His-to-Asp signaling modules HPT-1 and RRG-2, on viability and sensitivity to osmotic stress and oxidative stresses in Neurospora crassa. Curr. Genet. 51197-208. [DOI] [PubMed] [Google Scholar]
- 3.Bell-Pedersen, D., J. C. Dunlap, and J. J. Loros. 1992. The Neurospora circadian clock-controlled gene, ccg-2, is allelic to eas and encodes a fungal hydrophobin required for formation of the conidial rodlet layer. Genes Dev. 62382-2394. [DOI] [PubMed] [Google Scholar]
- 4.Borkovich, K. A., L. A. Alex, O. Yarden, M. Freitag, G. E. Turner, N. D. Read, S. Seiler, D. Bell-Pedersen, J. Paietta, N. Plesofsky, M. Plamann, M. Goodrich-Tanrikulu, U. Schulte, G. Mannhaupt, F. E. Nargang, A. Radford, C. Selitrennikoff, J. E. Galagan, J. C. Dunlap, J. J. Loros, D. Catcheside, H. Inoue, R. Aramayo, M. Polymenis, E. U. Selker, M. S. Sachs, G. A. Marzluf, I. Paulsen, R. Davis, D. J. Ebbole, A. Zelter, E. R. Kalkman, R. O'Rourke, F. Bowring, J. Yeadon, C. Ishii, K. Suzuki, W. Sakai, and R. Pratt. 2004. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 681-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boy-Marcotte, E., G. Lagniel, M. Perrot, F. Bussereau, A. Boudsocq, M. Jacquet, and J. Labarre. 1999. The heat shock response in yeast: differential regulations and contributions of the Msn2p/Msn4p and Hsf1p regulons. Mol. Microbiol. 33274-283. [DOI] [PubMed] [Google Scholar]
- 6.Causton, H. C., B. Ren, S. S. Koh, C. T. Harbison, E. Kanin, E. G. Jennings, T. I. Lee, H. L. True, E. S. Lander, and R. A. Young. 2001. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12323-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clos, J., J. T. Westwood, P. B. Becker, S. Wilson, K. Lambert, and C. Wu. 1990. Molecular cloning and expression of a hexameric Drosophila heat shock factor subject to negative regulation. Cell 631085-1097. [DOI] [PubMed] [Google Scholar]
- 8.Colot, H. V., G. Park, G. E. Turner, C. Ringelberg, C. M. Crew, L. Litvinkova, R. L. Weiss, K. A. Borkovich, and J. C. Dunlap. 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 10310352-10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Correa, A., and D. Bell-Pedersen. 2002. Distinct signaling pathways from the circadian clock participate in regulation of rhythmic conidiospore development in Neurospora crassa. Eukaryot. Cell 1273-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis, R. H., and F. J. deSerres. 1970. Genetic and microbiological research techniques for Neurospora. Methods Enzymol. 17A79-143. [Google Scholar]
- 11.Ebbole, D., and M. S. Sachs. 1990. A rapid and simple method of isolation of Neurospora crassa homokaryons using microconidia. Fungal Genet. Newsl. 3717-18. [Google Scholar]
- 12.Feder, J. H., J. M. Rossi, J. Solomon, N. Solomon, and S. Lindquist. 1992. The consequences of expressing hsp70 in Drosophila cells at normal temperatures. Genes Dev. 61402-1413. [DOI] [PubMed] [Google Scholar]
- 13.Fiorenza, M. T., T. Farkas, M. Dissing, D. Kolding, and V. Zimarino. 1995. Complex expression of murine heat shock transcription factors. Nucleic Acids Res. 23467-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fracella, F., C. Scholle, A. Kallies, T. Häfker, T. Schröder, and L. Rensing. 1997. Differential HSC70 expression during asexual development of Neurospora crassa. Microbiology 1433615-3624. [DOI] [PubMed] [Google Scholar]
- 15.Freitas, F. Z., and M. C. Bertolini. 2004. Genomic organization of the Neurospora crassa gsn gene: possible involvement of the STRE and HSE elements in the modulation of transcription during heat shock. Mol. Genet. Genomics 272550-561. [DOI] [PubMed] [Google Scholar]
- 16.Fujikake, N., Y. Nagai, H. A. Popiel, H. Kano, M. Yamaguchi, and T. Toda. 2005. Alternative splicing regulates the transcriptional activity of Drosophila heat shock transcription factor in response to heat/cold stress. FEBS Lett. 5793842-3848. [DOI] [PubMed] [Google Scholar]
- 17.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 112241-2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodson, M. L., and K. D. Sarge. 1995. Regulated expression of heat shock factor 1 isoforms with distinct leucine zipper arrays via tissue-dependent alternative splicing. Biochem. Biophys. Res. Commun. 211943-949. [DOI] [PubMed] [Google Scholar]
- 19.Hafker, T., D. Techel, G. Steier, and L. Rensing. 1998. Differential expression of glucose-regulated (grp78) and heat-shock-inducible (hsp70) genes during asexual development of Neurospora crassa. Microbiology 14437-43. [DOI] [PubMed] [Google Scholar]
- 20.Hashikawa, N., N. Yamamoto, and H. Sakurai. 2007. Different mechanisms are involved in the transcriptional activation by yeast heat shock transcription factor through two different types of heat shock elements. J. Biol. Chem. 28210333-10340. [DOI] [PubMed] [Google Scholar]
- 21.Kapoor, M., C. A. Curle, and C. Runham. 1995. The hsp70 gene family of Neurospora crassa: cloning, sequence analysis, expression, and genetic mapping of the major stress-inducible member. J. Bacteriol. 177212-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lantner, F., E. Ziv, D. Ram, and I. Schechter. 1998. Different forms of the mRNA encoding the heat-shock transcription factor are expressed during the life cycle of the parasitic helminth Schistosoma mansoni. Eur. J. Biochem. 253390-398. [DOI] [PubMed] [Google Scholar]
- 23.Lauter, F., and V. E. Russo. 1991. Blue light induction of conidiation-specific genes in Neurospora crassa. Nucleic Acids Res. 196883-6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lauter, F. R., V. E. Russo, and C. Yanofsky. 1992. Developmental and light regulation of eas, the structural gene for the rodlet protein of Neurospora. Genes Dev. 12A2373-2381. [DOI] [PubMed] [Google Scholar]
- 25.Lee, J., C. Godon, G. Lagniel, D. Spector, J. Garin, J. Labarre, and M. B. Toledano. 1999. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 27416040-16046. [DOI] [PubMed] [Google Scholar]
- 26.Lee, K., and D. J. Ebbole. 1998. Tissue-specific repression of starvation and stress responses of the Neurospora crassa con-10 gene is mediated by RCO1. Fungal Genet. Biol. 23269-278. [DOI] [PubMed] [Google Scholar]
- 27.Lee, Y.-T., J. Jacob, W. Michowski, M. Nowotny, J. Kuznicki, and W. J. Chazin. 2004. Human Sgt1 binds HSP90 through the CHORD-Sgt1 domain and not the tetratricopeptide repeat domain. J. Biol. Chem. 27916511-16517. [DOI] [PubMed] [Google Scholar]
- 28.Lindquist, S. 1986. The heat shock response. Annu. Rev. Biochem. 551151-1191. [DOI] [PubMed] [Google Scholar]
- 29.Lledías, F., P. Rangel, and W. Hansberg. 1999. Singlet oxygen is part of a hyperoxidant state generated during spore germination. Free Radic. Biol. Med. 261396-1404. [DOI] [PubMed] [Google Scholar]
- 30.Lowry, R., T. L. Durkee, and A. S. Springer. 1967. Ultrastructural studies of microconidium formation in Neurospora crassa. J. Bacteriol. 941757-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuyama, S. S., R. E. Nelson, and R. W. Siegel. 1974. Mutations specifically blocking differentiation of macroconidia Neurospora crassa. Dev. Biol. 41278-287. [DOI] [PubMed] [Google Scholar]
- 32.Meyer, U., C. Monnerjahn, D. Techel, and L. Rensing. 2000. Interaction of the Neurospora crassa heat shock factor with the heat shock element during heat shock and different developmental stages. FEMS Microbiol. Lett. 185255-261. [DOI] [PubMed] [Google Scholar]
- 33.Ninomiya, Y., K. Suzuki, C. Ishii, and H. Inoue. 2004. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc. Natl. Acad. Sci. USA 10112248-12253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nollen, E. A., and R. L. Morimoto. 2002. Chaperoning signaling pathways: molecular chaperones as stress-sensing ‘heat shock’ proteins. J. Cell Sci. 1152809-2816. [DOI] [PubMed] [Google Scholar]
- 35.Parsell, D., and S. Lindquist. 1993. The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu. Rev. Genet. 27437-496. [DOI] [PubMed] [Google Scholar]
- 36.Pirkkala, L., P. Nykanen, and L. Sistonen. 2001. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 151118-1131. [DOI] [PubMed] [Google Scholar]
- 37.Pittendrigh, C., V. Bruce, N. Rosensweig, and M. Rubin. 1959. Growth patterns in Neurospora. Nature 184169-170. [Google Scholar]
- 38.Plesofsky-Vig, N., and R. Brambl. 1987. Two developmental stages of Neurospora crassa utilize similar mechanisms for responding to heat shock but contrasting mechanisms for recovery. Mol. Cell. Biol. 73041-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rerngsamran, P., M. B. Murphy, S. A. Doyle, and D. J. Ebbole. 2005. Fluffy, the major regulator of conidiation in Neurospora crassa, directly activates a developmentally regulated hydrophobin gene. Mol. Microbiol. 56282-297. [DOI] [PubMed] [Google Scholar]
- 40.Rutherford, S., and S. Lindquist. 1998. Hsp90 as a capacitor for morphological evolution. Nature 396336-342. [DOI] [PubMed] [Google Scholar]
- 41.Selitrennikoff, C. P., R. E. Nelson, and R. W. Siegel. 1974. Phase-specific genes for macroconidiation in Neurospora crassa. Genetics 78679-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shear, C. L., and B. O. Dodge. 1927. Life histories and heterothallism of the red bread mold fungi of the Monlia sitipholoia group. J. Agric. Res. 341019-1042. [Google Scholar]
- 43.Sorger, P. K., and H. C. M. Nelson. 1989. Trimerization of a yeast transcriptional activator via a coil-coil motif. Cell 59807-813. [DOI] [PubMed] [Google Scholar]
- 44.Springer, M., and C. Yanofsky. 1989. A morphological and genetic analysis of conidiophore development in Neurospora crassa. Genes Dev. 3559-571. [DOI] [PubMed] [Google Scholar]
- 45.Tanabe, M., N. Sasai, K. Nagata, X. D. Liu, P. C. Liu, D. J. Thiele, and A. Nakai. 1999. The mammalian HSF4 gene generates both an activator and a repressor of heat shock genes by alternative splicing. J. Biol. Chem. 27427845-27856. [DOI] [PubMed] [Google Scholar]
- 46.Truman, A. W., S. H. Millson, J. M. Nuttall, M. Mollapour, C. Prodromou, and P. W. Piper. 2007. In the yeast heat shock response, Hsf1-directed induction of Hsp90 facilitates the activation of the Slt2 (Mpk1) mitogen-activated protein kinase required for cell integrity. Eukaryot. Cell 6744-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turian, G., and D. E. Bianchi. 1972. Conidiation in Neurospora. Bot. Rev. 38119-153. [Google Scholar]
- 48.Wiederrecht, G., D. Seto, and C. S. Parker. 1988. Isolation of the gene encoding the S. cerevisiae heat shock transcription factor. Cell 54841-853. [DOI] [PubMed] [Google Scholar]
- 49.Wu, C. 1995. Heat shock transcription factors: structure and regulation. Annu. Rev. Cell Dev. Biol. 11441-469. [DOI] [PubMed] [Google Scholar]
- 50.Yamashiro, C., D. Ebbole, B.-U. Lee, R. E. Brown, C. Bourland, L. Madi, and C. Yanofsky. 1996. Characterization of rco-1 of Neurospora crassa, a pleiotropic gene affecting growth and development that encodes a homolog of Tup1 of Saccharomyces cerevisiae. Mol. Cell. Biol. 166218-6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zarzov, P., H. Boucherie, and C. Mann. 1997. A yeast heat shock transcription factor (Hsf1) mutant is defective in both Hsc82/Hsp82 synthesis and spindle pole body duplication. J. Cell Sci. 1101879-1891. [DOI] [PubMed] [Google Scholar]