Abstract
We describe regulation of the subcellular localization of cyclic AMP (cAMP)-dependent protein kinase (PKA) regulatory (Cgs1p) and catalytic (Pka1p) subunits in the fission yeast Schizosaccharomyces pombe in response to physiological stresses and during sexual differentiation as determined by fluorescence microscopy of the Cgs1-green fluorescent protein (GFP) and Pka1-GFP fusion proteins, respectively. In wild-type S. pombe cells cultured to log phase under normal growth conditions, Cgs1p and Pka1p are concentrated in the nucleus and more diffusely present in the cytoplasm. Nuclear localization of both proteins is dependent on cAMP, since in cells lacking adenylate cyclase they are detectable only in the cytoplasm. In cells lacking Cgs1p or both Cgs1p and adenylate cyclase, Pka1p is concentrated in the nucleus, demonstrating a role for Cgs1p in the nuclear exclusion of Pka1p. Nuclear-cytoplasmic redistribution of Cgs1p and Pka1p is triggered by growth in glucose-limited or hyperosmotic media and in response to stationary-phase growth. In addition, both proteins are excluded from the nucleus in mating cells undergoing karyogamy and subsequently concentrated in postmeiotic spores. Cgs1p is required for subcellular redistribution of Pka1p induced by growth in glucose-limited and hyperosmotic media and during karyogamy but is not required for Pka1p redistribution triggered by stationary-phase growth or for the enrichment of Pka1p in spores. Our results demonstrate that PKA localization is regulated by cAMP and regulatory subunit-dependent and -independent mechanisms in S. pombe.
Cyclic AMP (cAMP)-dependent protein kinase, also referred to as protein kinase A (PKA), is a ubiquitous serine/threonine kinase that functions in a variety of signal transduction networks involved in the regulation of a broad diversity of biological processes in eukaryotic organisms, including cell growth and differentiation, nutrient sensing, physiological stress responses, and phospholipid metabolism (32, 33, 37). In its inactive state, PKA exists as a tetrameric holoenzyme complex comprised of two catalytic subunits bound with high affinity to two regulatory subunits (37). PKA catalytic activity is induced by the binding of two molecules of cAMP to each regulatory subunit, resulting in their dissociation as a dimer from the catalytic subunits, which are released as catalytically active monomers. Given its diverse molecular and cellular functions, it is perhaps not surprising that in addition to regulation by its cAMP-binding regulatory subunits, PKA has been shown in some organisms to be regulated via posttranslational modifications (13, 15, 36), as well protein-protein interactions with factors that modulate its functions (3, 38). Two types of PKA-interacting proteins, A kinase anchoring proteins (AKAPs) (3) and A kinase interacting protein (AKIP) (30, 37, 38), have been shown to play roles in targeting PKA to specific subcellular sites and/or to specific protein complexes in mammalian cells. AKAPs bind to PKA regulatory subunits, whereas AKIP, also known as BCA3p (breast cancer associated gene 3 protein) (22), binds to the amino terminus of the PKA catalytic subunit. Several members of the AKAP family have been identified that bind not only to PKA but also to adenylate cyclases, phosphodiesterases that hydrolyze cAMP, and other signaling factors, thus implicating these proteins as potential scaffolds that spatially organize and fine-tune PKA-mediated cAMP signaling. AKAPs have been shown to target PKA to the plasma membrane, perinuclear space, and centrosomes, while AKIP plays a role localizing PKA to the nucleus. A third type of PKA regulatory protein, termed protein kinase inhibitor peptide, has been shown to inhibit PKA activity in neuronal cells by directly binding to the PKA catalytic subunit (9).
Interestingly, the genomes of the budding yeast Saccharomyces cerevisiae and its evolutionarily distant relative, the fission yeast Schizosaccharomyces pombe, do not contain open reading frames encoding proteins sharing sequence homology with mammalian AKAPs, AKIP, or protein kinase inhibitor peptides (unpublished BLAST search results) despite the fact that in both yeasts PKA plays a central role in the regulation of a variety of cellular processes (12, 18, 20, 28, 29, 34, 42). The S. cerevisiae genome contains three genes, TPK1, TPK2, and TPK3, encoding PKA catalytic subunits (40), and a single gene, BCY1, encoding a PKA regulatory subunit (39). Although S. cerevisiae strains carrying null mutations in any two TPK genes are viable, the deletion of all three TPK genes is lethal (40), demonstrating that PKA, like adenylate cyclase (25), is essential for cell viability in this organism. PKA has been shown to contribute to the regulation of a variety of cellular processes in S. cerevisiae, including nutrient sensing, extracellular stress response, cell integrity, sporulation, cell cycle regulation, and autophagy (4, 5, 12, 20, 29, 31).
Griffioen et al. (14) have provided evidence that, in log-phase S. cerevisiae cells cultured under normal growth conditions, the catalytically inactive Bcy1p-Tpk1p PKA holoenzyme complex is localized in the nucleus. These investigators showed that upon stimulation by cAMP, the PKA catalytic subunit Tpk1p is exported to the cytoplasm, whereas the regulatory subunit, Bcy1p, remains nuclear. In S. cerevisiae cells carrying a mutant allele of BCY1 encoding an N-terminally-truncated Bcy1p protein that is more evenly distributed between the cytoplasm and nucleus, Tpk1p was likewise found to be more evenly distributed within the cell, providing evidence that Bcy1p plays a central role in the nuclear targeting of Tpk1p. Griffioen et al. also showed that nuclear-cytoplasmic redistribution of Bcy1p and Tpk1p is induced by growth of S. cerevisiae cells in nonfermentable carbon sources and in response to stationary-phase growth (14). In a separate study, it was shown that normal nuclear-cytoplasmic redistribution of Bcy1p induced by growth of S. cerevisiae cells under glucose-derepressed conditions is dependent on direct protein-protein interaction with Zds1p, as well as phosphorylation by the protein kinase Yak1p (15).
The fission yeast S. pombe has a single gene, cgs1, encoding a PKA regulatory subunit, as well as single gene, pka1, encoding a PKA catalytic subunit (11, 24). Unlike S. cerevisiae, PKA is not essential for the viability of S. pombe cells under normal growth conditions, nor is its constitutive activation, as rendered by deletion of the cgs1 gene, detrimental to cell growth (11, 24). As in S. cerevisiae, PKA has been shown to play roles in a number of varied cellular processes in S. pombe, including sexual differentiation, spore germination, glucose sensing, stationary-phase adaptation, cell cycle regulation, and osmotic stress response (18, 24, 28, 41, 42). In addition, it has been shown that overexpression of pka1 leads to the arrest of S. pombe cells in G2 of the cell cycle (35), although the biological significance of this phenotype has not yet been established. The multifunctional but nonessential characteristics of PKA in S. pombe make this genetically tractable eukaryote an ideal model organism for investigating mechanisms of PKA regulation and function.
Since the subcellular localization of S. pombe PKA has not previously been described, we set out to investigate the localization of its regulatory and catalytic subunits under normal growth conditions, in response to physiological stresses, and during the major phases of sexual differentiation in this model eukaryote. Our results demonstrate that the subcellular localization of S. pombe PKA is regulated by cAMP and regulatory subunit-dependent and -independent mechanisms and that there are both similarities and striking differences in the regulation of PKA localization in this organism and its distant relative, S. cerevisiae.
MATERIALS AND METHODS
Yeast strains, media, and genetic methods.
The following S. pombe strains were used in the present study: SP870 (h90 ade6-210 leu1-32 ura4-D18) (from D. Beach), YMSM101 [h90 ade6-210 leu1-32 ura4-D18 pka1-GFP(S65T)-kanMX6] (the present study), YMSM102 [h90 ade6-210 leu1-32 ura4-D18 cyr1::ura4 pka1-GFP(S65T)-kanMX6] (the present study), YMSM103 [h90 leu1-32 ura4-D18 cgs1::ura4 pka1-GFP(S65T)-kanMX6] (the present study), YMSM104 [h90 leu1-32 ura4-D18 cyr1::LEU2 cgs1::ura4 pka1-GFP(S65T)-kanMX6] (the present study), YMSM105 [h90 ade6-210 leu1-32 ura4-D18 cgs1-GFP(S65T)-kanMX6] (the present study), YMSM106 [h90 ade6-210 leu1-32 ura4-D18 cyr1::ura4 cgs1-GFP(S65T)-kanMX6] (the present study), YMSM107 [h90 ade6-210 leu1-32 ura4-D18 pka1::ura4 cgs1-GFP(S65T)-kanMX6] (the present study), SPCGS1U (h90 ade6-210 leu1-32 ura4-D18 cgs1::ura4) (the present study), SPPKA1U (h90 ade6-210 leu1-32 ura4-D18 pka1::ura4) (the present study), DY114 (h90 ade6-210 leu1-32 ura4-D18 cyr1::ura4) (21), MK7 (h90 ade6-210 leu1-32 ura4-D18 cyr1::LEU2) (21), RWP38 (h− leu1-32 ura4::fbp1-lacZ cgs1::ura4) (from C. Hoffman), and CHP453 (h− leu1-32 ura4-D18 his7-366 pka1::ura4) (from C. Hoffman). Standard yeast genetic methods were used for strain constructions (1, 27). S. pombe cultures were grown in either YEAU (0.5% yeast extract, 3% dextrose, adenine [250 mg/liter], uracil [250 mg/liter]), YEAU-glycerol (0.5% yeast extract, 3% glycerol, 0.1% dextrose, adenine [250 mg/liter], uracil [250 mg/liter]), or synthetic minimal medium (EMM) (1) containing required auxotrophic supplements at concentrations of 250 mg/liter each (1). Where indicated, mating and sporulation were assayed on malt extract (ME) medium (1).
Construction of cgs1-GFP and pka1-GFP S. pombe strains.
The recombinant PCR method described by Krawchuk and Wahls (23) was used to generate cgs1-GFP(S65T)-kanMX6 (cgs1-GFP) and pka1-GFP(S65T)-kanMX6 (pka1-GFP) cassettes in which the green fluorescent protein (GFP) coding sequence is fused to the 3′ end of the cgs1 and pka1 protein coding sequences, respectively. The PCRs used plasmid pFA6a-GFP(S65T)-kanMX6 (2) as the template DNA together with the oligonucleotide primer sets cgs1-F1 (5′-GGTTCTGAACGCTATGACTG-3′)/cgs1-R1 (5′-GGGGATCCGTCGACCTGCAGCGTACGATGCTTTAGTTGATGGAGGTG-3′) and cgs1-F2 (5′-GTTTAAACGAGCTCGAATTCATCGATTTGTTTACCATTATCCCTGG-3′)/cgs1-R2 (5′-TAATTTGCACCCCTTTACTC-3′) for construction of the cgs1-GFP cassette and the primer pairs pka1-F1 (5′-GGACTTTGGTTTTGCCAAAC-3′)/pka1-R1 (5′-GGGGATCCGTCGACCTGCAGCGTACGAAAAGTCCTTAAAGATAGAAG-3′) and pka1-F2 (5′-GTTTAAACGAGCTCGAATTCATCGATAAGTGACGTTTGTAGCACTG-3′)/pka1-R2 (5′-TAATGATCCTTCGTCGTTGC-3′) for construction of the pka1-GFP cassette. S. pombe strain SP870 was transformed with cgs1-GFP and pka1-GFP cassettes, and transformants were isolated on YEAU containing G418. Transformants in which the endogenous cgs1 and pka1 genes were replaced by cgs1-GFP (YMSM105) and pka1-GFP (YMSM101) fusion genes, respectively, were identified by colony PCR (26). S. pombe cgs1-GFP and pka1-GFP strains harboring cgs1Δ, cyr1Δ, and/or pka1Δ mutations were constructed by genetic crosses using standard yeast genetic techniques (1, 27).
Phenotypic validation of cgs1-GFP and pka1-GFP S. pombe strains.
The functionality of Cgs1-GFP fusion protein was determined by assaying the cgs1-GFP strain YMSM105 for mating on ME medium. Since Pka1p is a repressor of sexual differentiation in S. pombe, cgs1Δ mutants exhibit a semisterile phenotype relative to wild-type S. pombe cells. We determined that cgs1-GFP (YMSM105) cells exhibited mating and sporulation frequencies similar to wild-type S. pombe cells on ME medium, whereas a cgs1Δ mutant exhibited very low levels of mating and sporulation under the same conditions (see Fig. S1A in the supplemental material). We determined further that cyr1Δ cgs1-GFP cells (YMSM106), similar to cyr1Δ cells (34, 42), were arrested for growth on YEAU medium containing 1.2 M KCl, whereas wild-type S. pombe cells grew under the same conditions (see Fig. S1B in the supplemental material). The functionality of the Pka1-GFP fusion protein was similarly assessed by examining the growth of pka1-GFP strain YMSM101 on YEAU medium containing 1.2 M KCl (YEAU+KCl), which is inhibitory to the growth of pka1-null (pka1Δ) mutants. The pka1-GFP strain grew similarly to wild-type S. pombe cells in YEAU+KCl, whereas pka1Δ cells did not grow under the same conditions (see Fig. S1C in the supplemental material). We also determined that pka1-GFP cells, like wild-type S. pombe cells, do not mate on nutrient rich YEAU growth medium (see Fig. S1D in the supplemental material). In contrast, the pka1Δ mutant, which is derepressed for sexual differentiation in comparison to wild-type cells, exhibited a high frequency of mating on the same medium (see Fig. S1D in the supplemental material). Based on these results, we conclude that the cgs1-GFP and pka1-GFP strains are phenotypically similar to wild-type S. pombe cells, indicating that the respective Cgs1-GFP and Pka1-GFP fusion proteins they encode retain functions of their wild-type counterparts.
Epifluorescence microscopy of GFP fusion proteins.
Epifluorescence microscopy was carried out using a Nikon 90i automated epifluorescence microscope system equipped with a 100× wide numerical aperture (1.45) TIRF objective lens and operated using Nikon NIS-Elements software. Images were captured by using a CoolSNAP HQ2 monochrome charge-coupled device camera (Photometrics). Because the fluorescence of both Cgs1-GFP and Pka1-GFP was weak, effective digital image capturing of cells expressing these proteins required that the UV light source be reduced with an ND4 neutral density filter during sample focusing. The filter was then disengaged just prior image acquisition by the charge-coupled device camera. RAW images were level-adjusted by using the Adobe PhotoShop CS3 software application.
Preparation of cell lysates and detection of GFP fusion proteins by immunoblotting.
S. pombe cell lysates were prepared as described previously (8). GFP fusion proteins were detected in S. pombe cell extracts by resolving proteins in sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis gels and subsequent immunoblotting with anti-GFP monoclonal antibody (catalog no. 11814460; Roche) and the SuperSignal West Dura chemiluminescence detection system (Pierce).
RESULTS
The subcellular distribution of S. pombe PKA subunits during log-phase growth varies in glucose-repressed and -derepressed conditions.
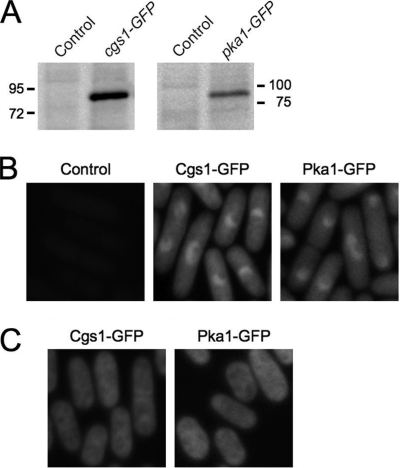
To investigate the subcellular localization of PKA regulatory and catalytic subunits in S. pombe, we constructed the strains YMSM105 and YMSM101 in which the protein coding sequences of the endogenous cgs1 and pka1 genes, respectively, are fused at the 3′ ends of their respective protein coding sequences to the protein coding sequence of the S65T mutant allele of GFP (17; see also Materials and Methods). For convenience, these strains shall henceforth be referred to as cgs1-GFP and pka1-GFP, respectively. The cgs1-GFP and pka1-GFP strains were judged to be phenotypically indistinguishable from wild-type S. pombe cells with respect to sexual differentiation and growth in hyperosmotic medium (see Materials and Methods and Fig. S1 in the supplemental material). In addition, anti-GFP immunoblotting of extracts prepared from cultures of cgs1-GFP and pka1-GFP cells showed that each expressed an immunoreactive protein with a molecular size within a range consistent with that for Cgs1-GFP (predicted size of 75 kDa) and Pka1-GFP (predicted size of 85 kDa) fusion proteins, respectively (Fig. 1A). Taken together, these results suggest that the cgs1-GFP and pka1-GFP strains express functional Cgs1-GFP and Pka1-GFP fusion proteins, respectively.
FIG. 1.
Subcellular localization of S. pombe PKA regulatory and catalytic subunits. (A) Immunoblot analysis of GFP fusion protein expression in cell lysates prepared from SP870 (Control), YMSM105 (cgs1-GFP), and YMSM101 (pka1-GFP) strains cultured overnight to log phase in YEAU medium. (B) Fluorescence photomicrographs of SP870 (Control), cgs1-GFP (Cgs1-GFP), and pka1-GFP (Pka1-GFP) S. pombe strains cultured in YEAU medium to mid-log phase. (C) Fluorescence photomicrographs of cgs1-GFP (Cgs1-GFP) and pka1-GFP (Pka1-GFP) cells were cultured in glucose-limited YEAU medium (YEAU-Glycerol) to mid-log phase.
To determine the localization of PKA subunits in S. pombe cells grown under normal conditions in medium containing glucose as a carbon source, the cgs1-GFP and pka1-GFP strains were cultured to mid-log phase in YEAU medium and analyzed by fluorescence microscopy. We observed that in both strains GFP fluorescence was concentrated in the nucleus and more diffusely present in the cytoplasm in both interphase and mitotic cells (Fig. 1B and see Fig. S2 in the supplemental material). Although the overall GFP fluorescence detected in both strains was relatively weak, it appeared somewhat brighter in cgs1-GFP cells relative to pka1-GFP cells, an observation consistent with the relative abundance of the GFP fusion proteins detected by immunoblotting of extracts prepared from the respective strains (Fig. 1A).
Since PKA is a key mediator of glucose sensing in S. pombe (18), we investigated whether the subcellular localization of its regulatory and catalytic subunits is altered when cells are cultured in glucose-derepressed conditions. To do this, the cgs1-GFP and pka1-GFP strains were cultured in YEAU medium containing 3% glycerol as the primary carbon source and only 0.1% glucose. Interestingly, we found that in this glucose-limited medium, Cgs1-GFP and Pka1-GFP were evenly distributed throughout the cell (Fig. 1C). Taken together, these results demonstrate that the nuclear-cytoplasmic distribution of PKA regulatory and catalytic subunits is regulated by a glucose-responsive mechanism in S. pombe.
cAMP is required for nuclear localization of PKA in wild-type S. pombe cells.
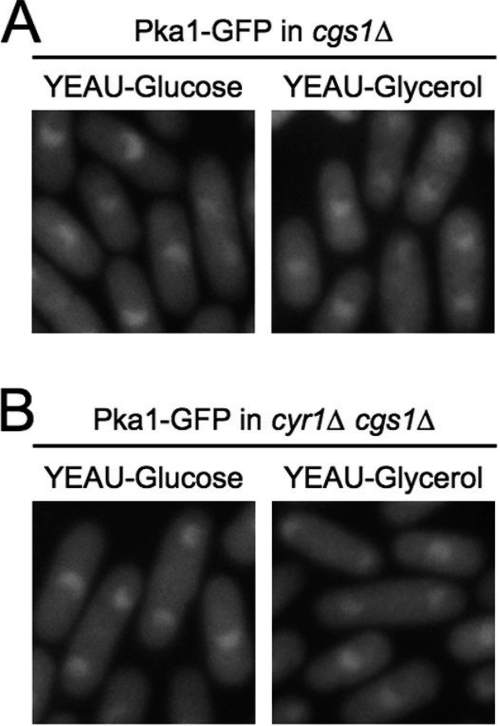
To determine whether subcellular distribution of the PKA regulatory and catalytic subunits is regulated in a cAMP-dependent fashion, we constructed S. pombe cgs1-GFP (YMSM106) and pka1-GFP (YMSM102) strains carrying a deletion of the single adenylate cyclase encoding gene in this organism, cyr1 (also known as git2) (19, 43). We observed that in YEAU medium containing either glucose or glycerol as the primary carbon source, Cgs1-GFP and Pka1-GFP were both localized primarily in the cytoplasm in cyr1Δ cells, with no nuclear localization detectable for either protein (Fig. 2A). To determine whether the altered localization detected for Cgs1-GFP and Pka1-GFP in cyr1Δ cells was due solely to the absence of cAMP, we cultured the cyr1Δ cgs1-GFP (YMSM106) and cyr1Δ pka1-GFP (YMSM102) strains in YEAU medium supplemented with cAMP. As shown in Fig. 2B, the pattern of Cgs1-GFP and Pka1-GFP in cyr1Δ cells cultured in medium supplemented with cAMP was similar to that observed in wild-type S. pombe cells, with both proteins being concentrated in the nucleus and diffusely present in the cytoplasm. These results demonstrate that cAMP is essential for nuclear localization of the PKA regulatory and catalytic subunits in wild-type S. pombe cells. It is worth noting that we found that restoration of Cgs1-GFP nuclear localization in cyr1Δ cgs1-GFP cells required incubation with a significantly higher concentration of cAMP for a longer duration of time (100 mM cAMP, 1.5 h) than was required for inducing nuclear localization of Pka1-GFP in cyr1Δ pka1-GFP cells (5 mM cAMP, 0.5 h).
FIG. 2.
cAMP is required for nuclear localization of Cgs1-GFP and Pka1-GFP. (A) Fluorescence photomicrographs of cyr1Δ cgs1-GFP (left panels) and cyr1Δ pka1-GFP (right panels) cells cultured in YEAU (YEAU-glucose) or YEAU-glycerol to mid-log phase. (B) Fluorescence photomicrographs of cyr1Δ cgs1-GFP and cyr1Δ pka1-GFP cells cultured in YEAU with 100 mM cAMP for 1.5 h and YEAU with 5 mM cAMP for 0.5 h, respectively. (C) Fluorescence photomicrographs of pka1Δ cgs1-GFP cells cultured in YEAU to mid-log phase.
Our finding that the induction of nuclear localization of Cgs1p in a cyr1Δ strain requires a higher concentration of cAMP for a longer period of time than that required for nuclear localization of Pka1p suggests that different proteins might regulate nuclear localization of one or both S. pombe PKA subunits. To determine whether the PKA catalytic subunit, Pka1p, is required for nuclear localization its regulatory subunit, Cgs1p, we constructed the pka1Δ cgs1-GFP strain YMSM107 and analyzed the localization of Cgs1-GFP in cells cultured in YEAU medium. As shown in Fig. 2C, the localization of Cgs1-GFP in pka1Δ cells was indistinguishable from that detected in wild-type S. pombe cells cultured under the same conditions, being concentrated in the nucleus and more diffusely present in the cytoplasm. This result demonstrates that cAMP-induced nuclear localization of the S. pombe PKA regulatory subunit is not dependent on the PKA catalytic subunit.
Role of the S. pombe PKA regulatory subunit in regulating nuclear-cytoplasmic distribution of the PKA catalytic subunit.
To determine whether the S. pombe PKA regulatory subunit, Cgs1p, contributes to regulation of subcellular localization of its catalytic subunit, Pka1p, we constructed a pka1-GFP strain carrying a deletion of the cgs1 gene (YMSM103). The cgs1Δ pka1-GFP strain was cultured in YEAU media containing either glucose or glycerol as a primary carbon source and observed by fluorescence microscopy. Interestingly, unlike wild-type S. pombe cells, we observed that Pka1-GFP was concentrated in the nucleus and diffusely present in the cytoplasm in cgs1Δ (YMSM103) cells cultured in YEAU medium containing either glucose or glycerol as the primary carbon source (Fig. 3A). These results indicate that Cgs1p is required for nuclear-cytoplasmic redistribution of Pka1p induced by growth of S. pombe cells in glucose-limited medium.
FIG. 3.
Cgs1p contributes to regulation of subcellular localization of Pka1p. Fluorescence photomicrographs of cgs1Δ pka1-GFP (A) and cyr1Δ cgs1Δ pka1-GFP (B) strains cultured in YEAU (YEAU-Glucose) (left panels) or YEAU-Glycerol (right panels) to mid-log phase are shown.
To address whether Cgs1p plays a role in nuclear exclusion of Pka1p in cells that do not produce cAMP, we constructed the S. pombe pka1-GFP strain YMSM104 carrying deletions of both the cgs1 and cyr1 genes. The cgs1Δ cyr1Δ pka1-GFP strain was cultured to log phase in YEAU medium containing either glucose or glycerol as a primary carbon source and observed by fluorescence microscopy. As shown in Fig. 3B, Pka1-GFP was concentrated in the nucleus in cgs1Δ cyr1Δ (YMSM104) cells cultured in either medium, thus demonstrating that Cgs1p is required for nuclear exclusion of Pka1p in cells lacking cAMP. This result also reveals that the S. pombe PKA catalytic subunit, Pka1p, can be imported into the nucleus by a mechanism that is independent of its regulatory subunit and cAMP.
S. pombe PKA regulatory and catalytic subunits are subcellularly redistributed in response to high osmolarity.
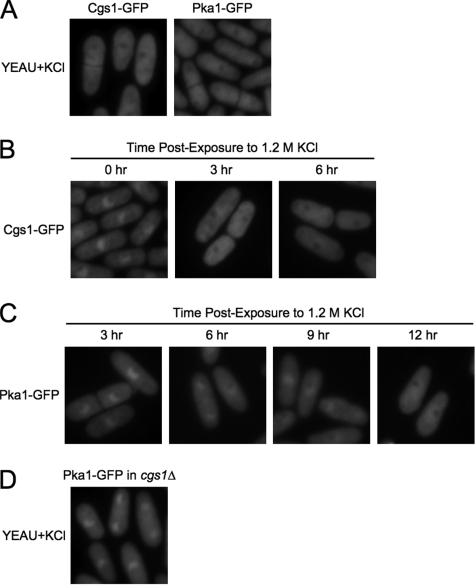
Since the pka1 gene has been shown to be essential for the growth of S. pombe cells in media containing a high concentration of inorganic osmolyte (42) and to be upregulated in response to organic osmolyte-induced hypertonic stress (7), we investigated whether the localization of PKA regulatory and catalytic subunits is altered when cells are cultured in high-osmolarity media. To do this, S. pombe cgs1-GFP (YMSM105) and pka1-GFP (YMSM101) strains were cultured overnight to mid-log phase in YEAU containing 1.2 M KCl and then analyzed by fluorescence microscopy. We found that Cgs1-GFP (Fig. 4A, left panel) and Pka1-GFP (Fig. 4A, right panel) were evenly distributed between the cytoplasm and nucleus under these conditions, although both proteins were reduced in concentration in the presumptive nucleolar space, as judged by DAPI staining of the respective cultures (see Fig. S3 in the supplemental material). To determine the time course for KCl-induced redistribution of S. pombe PKA subunits, cgs1-GFP (YMSM105) and pka1-GFP (YMSM101) strains were cultured in YEAU to mid-log phase and then diluted with YEAU containing 3 M KCl to a final concentration of 1.2 M KCl. The cultures were then examined by fluorescence microscopy at regular intervals. Nuclear-cytoplasmic redistribution of Cgs1-GFP was first detectable 3 h after KCl addition to the cgs1-GFP culture (Fig. 4B). In contrast, Pka1-GFP retained a predominantly nuclear localization until approximately 12 h after addition of KCl to the pka1-GFP culture, although the ratio of nuclear to cytoplasmic Pka1-GFP was discernibly reduced within 6 h after KCl exposure (Fig. 4C).
FIG. 4.
KCl-induced nuclear-cytoplasmic redistribution of Cgs1-GFP and Pka1-GFP. (A) Fluorescence photomicrographs of cgs1-GFP (Cgs1-GFP) and pka1-GFP (Pka1-GFP) cells cultured to mid-log phase in YEAU containing 1.2 M KCl (YEAU+KCl). cgs1-GFP (B) and pka1-GFP (C) strains were cultured in YEAU to 2 × 106 cells/ml and then diluted with YEAU containing 3 M KCl for a final concentration of 1.2 M KCl. The cultures were then examined by fluorescence microscopy at the indicated time points. (D) Fluorescence photomicrograph of cgs1Δ pka1-GFP cells cultured to mid-log phase in YEAU containing 1.2 M KCl.
To determine whether Cgs1p is required for KCl-induced redistribution of Pka1p, the cgs1Δ pka1-GFP strain YMSM103 was cultured in YEAU+KCl to mid-log phase and examined by fluorescence microscopy. Interestingly, we observed that the localization of Pka1-GFP in cgs1Δ pka1-GFP cells cultured in YEAU+KCl was indistinguishable from that detected in wild-type cells cultured in YEAU (Fig. 4D). This result demonstrates that KCl-induced redistribution of the S. pombe PKA catalytic subunit is dependent on the PKA regulatory subunit.
To determine whether changes in S. pombe PKA subunit localization are also triggered by growth in medium containing a high concentration of organic osmolyte, cgs1-GFP (YMSM105) and pka1-GFP (YMSM101) strains were cultured to mid-log phase in YEAU containing 2 M sorbitol (YEAU+Sorb). Under these conditions, Cgs1-GFP was found to be evenly distributed throughout the cell (Fig. 5A, left panel). Surprisingly, we found that 2 M sorbitol had no obvious effect on the distribution of Pka1-GFP, which as in cells cultured in YEAU, was concentrated in the nucleus and more diffusely present in the cytoplasm (Fig. 5A, right panel). Additional experiments demonstrated that nuclear-cytoplasmic redistribution of Cgs1-GFP is detectable within 2 h of exposure of cgs1-GFP cells to 2 M sorbitol (Fig. 5B). Taken together, these results indicate that high concentrations of organic and inorganic osmolytes have different effects on the nuclear-cytoplasmic distribution of the PKA regulatory and catalytic subunits.
FIG. 5.
Sorbitol-induced nuclear-cytoplasmic redistribution of Cgs1p. (A) Fluorescence photomicrographs of cgs1-GFP (Cgs1-GFP) and pka1-GFP (Pka1-GFP) cells cultured to mid-log phase in YEAU containing 2 M sorbitol. (B) The cgs1-GFP strain was cultured in YEAU to 2 × 106 cells/ml and then diluted with an equal volume of YEAU containing 4 M sorbitol for a final concentration of 2 M sorbitol. The culture was then examined by fluorescence microscopy at the indicated time points.
Nuclear-cytoplasmic redistribution of S. pombe PKA is induced during stationary phase.
Experiments were next carried out to determine whether changes in the distribution of S. pombe PKA regulatory and catalytic subunits are triggered in response to stationary-phase growth. To do this, the cgs1-GFP (YMSM105) and pka1-GFP (YMSM101) strains were cultured in YEAU medium and examined by fluorescence microscopy in late log phase (6 × 107 cells/ml) and at 3-h intervals upon reaching stationary-phase density (about 108 cells/ml). Since S. pombe cells are known to autofluoresce after prolonged incubation at stationary-phase densities, the wild-type S. pombe strain SP870 was monitored under the same conditions. We observed that by late log phase, Cgs1-GFP was evenly distributed throughout the cell (Fig. 6A). By 6 h after reaching stationary-phase density, partial nuclear exclusion of Cgs1-GFP was detected, and by 9 h after stationary-phase entry it was predominantly, if not exclusively cytoplasmic in localization (Fig. 6A). In contrast, a change in Pka1-GFP localization was not detected until 6 h into stationary phase, at which time the difference between nuclear and cytoplasmic GFP signals was less than that detected in log-phase cultures (Fig. 6B). By 9 h into stationary-phase density, Pka1-GFP appeared evenly distributed between the cytoplasm and nucleus (Fig. 6B). Meaningful results were not obtained for later time points, since we detected only marginal differences between the total cytoplasmic fluorescence of pka1-GFP cells and the autofluorescence of control cells cultured under the same conditions (data not shown). This was not the case for Cgs1-GFP, which exhibited cytoplasmic fluorescence considerably brighter than the autofluorescence exhibited by the corresponding control culture even after prolonged incubation at stationary-phase density (data not shown). These observations are in keeping with our aforementioned finding that Cgs1-GFP protein levels in cgs1-GFP cells are significantly higher than those of Pka1-GFP in pka1-GFP cells, as judged by anti-GFP immunoblot analysis of cell extracts prepared from the respective strains (see Fig. 1A).
FIG. 6.
Nuclear-cytoplasmic redistribution of Cgs1 and Pka1 is induced during stationary phase. SP870 wild-type (A to C), cgs1-GFP (A), pka1-GFP (B), and cgs1Δ pka1-GFP (C) S. pombe strains were cultured in YEAU and examined by fluorescence microscopy at late log phase (6 × 107 cells/ml) and at 3-h intervals upon reaching stationary-phase density (about 108 cells/ml). (A) Cells imaged with 3-s digital camera exposures. (B and C) Cells imaged with 4-s camera exposures.
To determine whether Cgs1p is required for stationary-phase-induced redistribution of Pka1p, the cgs1Δ pka1-GFP strain YMSM103 was cultured in YEAU medium and examined by fluorescence microscopy at late log phase and at 3-h intervals upon reaching stationary-phase density. Interestingly, we observed that the localization of Pka1-GFP in cgs1Δ cells was similar to that detected in wild-type pka1-GFP cells during the same time course, being evenly distributed between the cytoplasm and nucleus by 9 h after reaching stationary-phase density (Fig. 6C). These results suggest that the stationary-phase-induced redistribution of the S. pombe PKA catalytic subunit is not dependent on the PKA regulatory subunit.
Subcellular localization of PKA during S. pombe sexual differentiation.
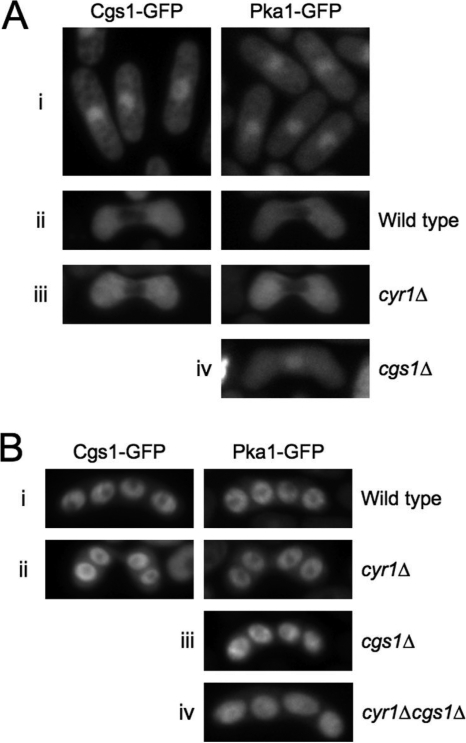
In S. pombe, sexual differentiation, namely, mating, meiosis, and sporulation, is induced by nitrogen or glucose deprivation (10). Since PKA is a repressor of sexual differentiation in S. pombe (10, 11, 24), we sought to determine whether its localization varies in response to nitrogen starvation, mating, and/or sporulation. As shown in Fig. 7Ai, nitrogen starvation alone did not result in obvious changes in either Cgs1-GFP or Pka1-GFP localization, since both proteins remained concentrated in the nucleus and diffusely present in the cytoplasm in cgs1-GFP (YMSM105) and pka1-GFP (YMSM101) cells cultured in nitrogen-deficient minimal medium. Interestingly, we observed that during at least some portion of the karyogamy phase of zygotes produced from the mating of haploid cells, both Cgs1-GFP and Pka1-GFP were detectable only in the cytoplasm (Fig. 7Aii). Not unexpectedly, in adenylate cyclase mutants (cyr1Δ) undergoing karyogamy, Cgs1-GFP and Pka1-GFP were also detectable only in the cytoplasm (Fig. 7Aiii). In contrast, in cgs1Δ zygotes undergoing karyogamy, Pka1-GFP, as in cells undergoing vegetative growth, was concentrated in the nucleus (Fig. 7Aiv).
FIG. 7.
Subcellular localization of Cgs1-GFP and Pka1-GFP during S. pombe sexual differentiation. Fluorescence photomicrographs of cgs1-GFP (left panels) and pka1-GFP (right panels) nitrogen-starved cells (Ai), zygotes (Aii to Aiv), and zygotic asci (Bi to Biii). (Ai) Cells were cultured in EMM, washed three times with EMM-nitrogen, resuspended in EMM-nitrogen at 2 × 106 cells/ml to induce nitrogen starvation, and incubated for 18 h at 28°C prior to photomicroscopy. (Aii to Aiv and B) Same conditions as for panel Ai, except the cells were resuspended at 2 × 107 cells/ml to induce mating (Aii to Aiv) and sporulation (B).
Under normal conditions, S. pombe zygotes, rather than forming diploid cells, carry out meiosis I and II and then undergo the process of spore formation to produce asci containing four haploid spores (10). Interestingly, we observed that both Cgs1-GFP and Pka1-GFP were highly enriched in postmeiotic spores (Fig. 7Bi). We determined further that cAMP is not required for the enrichment of Cgs1-GFP or Pka1-GFP in spores, since in asci produced by both cyr1Δ cgs1-GFP and cyr1Δ pka1-GFP strains, respectively, we observed that GFP fluorescence was as highly enriched in spores as in their wild-type counterparts (Fig. 7Bii). Interestingly, we also determined that Cgs1p is not required for the concentration of Pka1-GFP in postmeiotic spores, since GFP fluorescence appeared similarly enriched in spores produced by both cgs1Δ pka1-GFP spores (Fig. 7Biii) and cgs1Δ cyr1Δ pka1-GFP spores (Fig. 7Biv) as in spores of asci produced by wild-type pka1-GFP cells.
DISCUSSION
Although PKA-dependent cellular processes and signaling pathways have been studied in S. pombe for more than two decades, the present study represents what is to our knowledge the first detailed analysis of the subcellular localization of PKA regulatory and catalytic subunits in this model eukaryote. As discussed below, our results demonstrate that the subcellular distribution of PKA subunits is regulated in response to several different physiological conditions in which the protein kinase has been demonstrated to have significant and, in some cases, essential functions. Importantly, our findings also reveal that there are striking differences in the regulation of PKA localization in S. pombe and its evolutionarily distant cousin, the budding yeast, S. cerevisiae.
The regulation and function of S. pombe PKA has perhaps been most studied with respect to its role as a mediator of glucose sensing. In S. pombe, PKA activity is required for glucose-induced repression of the fructose-1,6-bisphosphatase encoding gene, fbp1 (reviewed in reference 18). Studies on the regulation of fbp1 expression suggest that S. pombe cells growing in glucose-rich medium are in a state of high PKA activity, whereas cells grown under glucose-limited conditions have comparatively lower PKA activity. In the present study, we found that in log-phase cultures of S. pombe cells grown in medium containing glucose as a primary carbon source, the PKA regulatory and catalytic subunits, Cgs1p and Pka1p, respectively, are both concentrated in the nucleus and more diffusely present in the cytoplasm. In contrast, in glucose-limited medium (glucose-derepressed condition), both PKA subunits are evenly distributed between the cytoplasm and the nucleus. Because cAMP is not required for the growth or proliferation of S. pombe cells under normal culturing conditions, we were able to unambiguously assess its role in regulating PKA localization. We found that in S. pombe cells bearing a null mutation in the adenylate cyclase gene, cyr1, the PKA regulatory and catalytic subunits are both highly enriched in the cytoplasm and largely or completely excluded from the nucleus in both glucose-repressed and glucose-derepressed conditions. We determined further that in cells lacking both adenylate cyclase and Cgs1p, the localization of Pka1p was indistinguishable from that detected in wild-type S. pombe cells, being concentrated in the nucleus and more diffusely present in the cytoplasm. Taken together, our results indicate that the catalytically inactive PKA holoenzyme complex resides in the cytoplasm of S. pombe cells during log-phase growth in both glucose-repressed and glucose-derepressed conditions. Furthermore, our findings demonstrate that Cgs1p plays a key role in the process required for nuclear exclusion of Pka1p in glucose-repressed conditions and, further, that cAMP-induced nuclear localization Pka1p occurs as a consequence of its dissociation from Cgs1p. Since our results also demonstrate that cAMP induces nuclear localization of Cgs1p during glucose-repressed growth, it will be of great interest to determine whether the same or distinct factors are involved in regulating the nuclear import of S. pombe PKA regulatory and catalytic subunits. In this regard, our finding that in glucose-repressed conditions there is little or no difference in the subcellular distribution of Pka1p in wild-type, cgs1Δ, and cyr1Δ cgs1Δ cells suggests that there are mechanisms regulating the nuclear-cytoplasmic distribution of Pka1p that operate independently of cAMP and Cgs1p even under normal, glucose-repressed growth conditions. At the same time, we found it interesting that Cgs1p is translocated to the nucleus under conditions of active PKA (e.g., in the presence of cAMP). A possible explanation for this characteristic of Cgs1p localization is that it facilitates rapid reassociation of nuclear-localized PKA catalytic and regulatory subunits as cAMP levels fall, thus providing a mechanism for tight regulation of PKA activity in the nucleus. The implication of this and related findings presented here is that at least one PKA substrate in S. pombe is a nuclear-localized protein.
Additional experiments revealed that S. pombe PKA regulatory and catalytic subunits are subcellularly redistributed in response to high osmolarity. We found that nuclear-cytoplasmic redistribution of both Cgs1p and Pka1p was triggered in response to growth of S. pombe cells in media containing 1.2 M KCl. In contrast, culturing of cells in medium containing 2 M sorbitol triggered nuclear-cytoplasmic redistribution of Cgs1p but had no discernible effect on the subcellular distribution of Pka1p. One possible explanation for these findings is that the S. pombe cAMP-PKA pathway responds differently to inorganic and organic osmolyte-induced hyperosmolarity. Alternatively, cAMP-PKA-dependent responses to high concentrations of KCl may involve adaptations not only to high osmolarity but also to other, distinct physiological perturbations, such as high ionic strength or general salinity. It is perhaps worth noting that S. pombe cyr1Δ and pka1Δ mutants undergo complete, albeit reversible growth arrest in response to culturing in medium containing high concentrations of KCl (34, 42; Y. Matsuo, unpublished results). However, these mutants are not inhibited for growth in media containing 2 M sorbitol (34; Y. Matsuo, unpublished results), despite the fact that pka1 gene expression is upregulated in response to sorbitol-induced hypertonic shock (7). Our finding that Pka1p is excluded from the nucleus in cells lacking adenylate cyclase in both normal and hyperosmotic conditions suggests that its function in hyperosmotic stress response involves critical interaction with at least one other nuclear-localized factor.
Recent studies by other investigators have demonstrated that PKA is required for normal cellular response(s) to stationary-phase growth (28, 34). We found that the S. pombe PKA regulatory and catalytic subunits each undergo nuclear-cytoplasmic redistribution in response to stationary-phase growth. Nuclear-cytoplasmic redistribution of Cgs1p was first detectable in late log phase, while redistribution of Pka1p was not detected until several hours after cultures reached stationary-phase density. Importantly, we found that stationary-phase-induced nuclear-cytoplasmic redistribution of Pka1p is not dependent on Cgs1p, thus demonstrating the existence of a regulatory subunit independent mechanism for PKA catalytic subunit nuclear export, exclusion, or both. It should be noted that the role(s) of cAMP-PKA during the stationary-phase growth of S. pombe cells has not been defined and may vary depending on the nutrient status of the culture, since in one study it was shown that adenylate cyclase mutants have reduced viability relative to wild-type cells (34), while in another, which used different culturing conditions, pka1 mutants were shown to have an increased rate of survival during stationary-phase growth (28).
Another well-characterized function of S. pombe PKA is that of a repressor of sexual differentiation (10). During non-nutrient-limited vegetative growth, Pka1p inhibits sexual differentiation via transcriptional repression of genes encoding components of the sexual response system (10). Starvation by either glucose or nitrogen deprivation triggers downregulation of adenylate cyclase, which results in derepression of genes controlling sexual response. The role of PKA as a nutrient-dependent inhibitor of sexual differentiation is illustrated by fact that S. pombe adenylate cyclase and pka1 mutants mate precociously in nutrient-rich media, while cgs1 mutants mate poorly in nitrogen- or glucose-deprived conditions that induce a high frequency of mating in cultures of wild-type S. pombe cells (11, 24, 43). We found that nitrogen starvation of S. pombe cells did not induce discernible nuclear-cytoplasmic redistribution of PKA regulatory or catalytic subunits. However, in zygotes produced by the mating of haploid cells, we observed that during at least some part of the karyogamy phase, both subunits were excluded from the nucleus. Nuclear-cytoplasmic redistribution of Pka1p was not detected in cgs1Δ cells undergoing karyogamy, suggesting that its exclusion or export from the nucleus may be required for efficient mating of S. pombe cells.
Interestingly, we found that the PKA catalytic and regulatory subunits were both highly concentrated in postmeiotic spores. Importantly, the process leading to the enrichment of Cgs1p and Pka1p in postmeiotic spores was not dependent on cAMP and, further, the enrichment of Pka1p in spores was found to not be dependent on Cgs1p. Our finding that Pka1p is enriched in postmeiotic spores is consistent with previous studies demonstrating its requirement for efficient spore germination in S. pombe (16).
We also found it interesting that S. pombe cyr1Δ cgs1-GFP cells required incubation with a much higher concentration of cAMP for a longer duration of time to induce nuclear localization of Cgs1-GFP than was required to induce the nuclear localization of Pka1-GFP in cyr1Δ pka1-GFP cells. This finding raises the possibility that there may be physiological states that trigger cAMP-dependent nuclear import of the PKA catalytic subunit but not its regulatory subunit and others that induce nuclear import of both subunits, either as an intact holoenzyme complex or as individual components. The underlying implication is that there is at least one additional factor regulating PKA localization in S. pombe and that its function or expression may be regulated by cAMP. Genetic screens to identify PKA localization defective mutants would likely lead to the identification of additional mechanisms responsible for differentially regulating S. pombe PKA regulatory and catalytic subunit localization in response to different physiological cues.
Unlike S. pombe, PKA is essential for cell viability in the budding yeast S. cerevisiae (40). The results of the present study reveal that there are also significant differences in the regulation of PKA localization in these evolutionarily distant yeasts. As noted above, our findings suggest that the catalytically inactive S. pombe PKA holoenzyme complex resides primarily in the cytoplasm during log-phase growth in glucose-repressed and glucose-derepressed conditions. In contrast, Griffioen and coworkers have demonstrated that under analogous conditions, the catalytically inactive PKA holoenzyme is concentrated in the nucleus in S. cerevisiae (14). Our results show that under normal growth conditions, cAMP induces nuclear localization of the S. pombe PKA catalytic subunit, in large part, as a consequence of its release from the regulatory subunit. In S. cerevisiae, cAMP induces nuclear export or exclusion the PKA catalytic subunit, whereas the PKA regulatory subunit remains in the nucleus (14). Thus, in S. pombe, the PKA regulatory subunit functions, in part, to keep the holoenzyme complex out of the nucleus, whereas in budding yeast, the opposite appears to be true. Despite these apparently fundamental differences the regulation of PKA holoenzyme localization in the two yeasts, there may also be similarities in regulation of PKA localization, at least in glucose-derepressed conditions, since in both yeasts growth in glucose-limited media induces nuclear-cytoplasmic redistribution of PKA regulatory and catalytic subunits (14; the present study). PKA localization has also been investigated in the dimorphic fungus, Candida albicans. In this organism, unlike S. pombe but similar to S. cerevisiae, the PKA regulatory subunit appears to tether the catalytic subunit to the nucleus (6).
Our BLAST searches of S. pombe and S. cerevisiae nucleic acid and protein sequence databases indicate that neither yeast possesses genes encoding proteins bearing significant homology to the AKAP and AKIP proteins that have been shown to contribute to regulation of PKA localization in mammalian cells. Thus, further elucidation of mechanisms in both yeasts underlying the regulation of PKA subunit localization in response to physiological stresses may provide novel insights into previously unrecognized PKA regulatory mechanisms in higher organisms.
Supplementary Material
Acknowledgments
We thank Eric Chang and Charles Hoffman for S. pombe strains and plasmid reagents.
This study was funded by National Institutes of Health grant R01GM068685 (to S.M.).
Footnotes
Published ahead of print on 11 July 2008.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Alfa, C., P. Fantes, J. Hyams, M. McLeod, and E. Warbrick. 1993. Experiments with fission yeast: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 2.Bahler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie III, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14943-951. [DOI] [PubMed] [Google Scholar]
- 3.Beene, D. L., and J. D. Scott. 2007. A-kinase anchoring proteins take shape. Curr. Opin. Cell Biol. 19192-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolte, M., P. Dieckhoff, C. Krause, G. H. Braus, and S. Irniger. 2003. Synergistic inhibition of APC/C by glucose and activated Ras proteins can be mediated by each of the Tpk1-3 proteins in Saccharomyces cerevisiae. Microbiology 1491205-1216. [DOI] [PubMed] [Google Scholar]
- 5.Budovskaya, Y. V., J. S. Stephan, F. Reggiori, D. J. Klionsky, and P. K. Herman. 2004. The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J. Biol. Chem. 27920663-20671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cassola, A., M. Parrot, S. Silberstein, B. B. Magee, S. Passeron, L. Giasson, and M. L. Cantore. 2004. Candida albicans lacking the gene encoding the regulatory subunit of protein kinase A displays a defect in hyphal formation and an altered localization of the catalytic subunit. Eukaryot. Cell 3190-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, D., W. M. Toone, J. Mata, R. Lyne, G. Burns, K. Kivinen, A. Brazma, N. Jones, and J. Bahler. 2003. Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell 14214-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Correa-Bordes, J., and P. Nurse. 1995. p25rum1 orders S phase and mitosis by acting as an inhibitor of the p34cdc2 mitotic kinase. Cell 831001-1009. [DOI] [PubMed] [Google Scholar]
- 9.Dalton, G. D., and W. L. Dewey. 2006. Protein kinase inhibitor peptide (PKI): a family of endogenous neuropeptides that modulate neuronal cAMP-dependent protein kinase function. Neuropeptides 4023-34. [DOI] [PubMed] [Google Scholar]
- 10.Davey, J. 1998. Fusion of a fission yeast. Yeast 141529-1566. [DOI] [PubMed] [Google Scholar]
- 11.DeVoti, J., G. Seydoux, D. Beach, and M. McLeod. 1991. Interaction between ran1+ protein kinase and cAMP-dependent protein kinase as negative regulators of fission yeast meiosis. EMBO J. 103759-3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gancedo, J. M. 2001. Control of pseudohyphae formation in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25107-123. [DOI] [PubMed] [Google Scholar]
- 13.Gesellchen, F., O. Bertinetti, and F. W. Herberg. 2006. Analysis of posttranslational modifications exemplified using protein kinase A. Biochim. Biophys. Acta 17641788-1800. [DOI] [PubMed] [Google Scholar]
- 14.Griffioen, G., P. Anghileri, E. Imre, M. D. Baroni, and H. Ruis. 2000. Nutritional control of nucleocytoplasmic localization of cAMP-dependent protein kinase catalytic and regulatory subunits in Saccharomyces cerevisiae. J. Biol. Chem. 2751449-1456. [DOI] [PubMed] [Google Scholar]
- 15.Griffioen, G., P. Branduardi, A. Ballarini, P. Anghileri, J. Norbeck, M. D. Baroni, and H. Ruis. 2001. Nucleocytoplasmic distribution of budding yeast protein kinase A regulatory subunit Bcy1 requires Zds1 and is regulated by Yak1-dependent phosphorylation of its targeting domain. Mol. Cell. Biol. 21511-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatanaka, M., and C. Shimoda. 2001. The cyclic AMP/PKA signal pathway is required for initiation of spore germination in Schizosaccharomyces pombe. Yeast 18207-217. [DOI] [PubMed] [Google Scholar]
- 17.Heim, R., A. B. Cubitt, and R. Y. Tsien. 1995. Improved green fluorescence. Nature 373663-664. [DOI] [PubMed] [Google Scholar]
- 18.Hoffman, C. S. 2005. Glucose sensing via the protein kinase A pathway in Schizosaccharomyces pombe. Biochem. Soc. Trans. 33257-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffman, C. S., and F. Winston. 1991. Glucose repression of transcription of the Schizosaccharomyces pombe fbp1 gene occurs by a cAMP signaling pathway. Genes Dev. 5561-571. [DOI] [PubMed] [Google Scholar]
- 20.Hohmann, S. 2002. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 66300-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawamukai, M., K. Ferguson, M. Wigler, and D. Young. 1991. Genetic and biochemical analysis of the adenylyl cyclase of Schizosaccharomyces pombe. Cell Regul. 2155-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kitching, R., H. Li, M. J. Wong, S. Kanaganayakam, H. Kahn, and A. Seth. 2003. Characterization of a novel human breast cancer associated gene (BCA3) encoding an alternatively spliced proline-rich protein. Biochim. Biophys. Acta 1625116-121. [DOI] [PubMed] [Google Scholar]
- 23.Krawchuk, M. D., and W. P. Wahls. 1999. High-efficiency gene targeting in Schizosaccharomyces pombe using a modular, PCR-based approach with long tracts of flanking homology. Yeast 151419-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maeda, T., Y. Watanabe, H. Kunitomo, and M. Yamamoto. 1994. Cloning of the pka1 gene encoding the catalytic subunit of the cAMP-dependent protein kinase in Schizosaccharomyces pombe. J. Biol. Chem. 2699632-9637. [PubMed] [Google Scholar]
- 25.Matsumoto, K., I. Uno, Y. Oshima, and T. Ishikawa. 1982. Isolation and characterization of yeast mutants deficient in adenylate cyclase and cAMP-dependent protein kinase. Proc. Natl. Acad. Sci. USA 792355-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuo, Y., E. Fisher, J. Patton-Vogt, and S. Marcus. 2007. Functional characterization of the fission yeast phosphatidylserine synthase gene, pps1, reveals novel cellular functions for phosphatidylserine. Eukaryot. Cell 62092-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 28.Roux, A. E., A. Quissac, P. Chartrand, G. Ferbeyre, and L. A. Rokeach. 2006. Regulation of chronological aging in Schizosaccharomyces pombe by the protein kinases Pka1 and Sck2. Aging Cell 5345-357. [DOI] [PubMed] [Google Scholar]
- 29.Santangelo, G. M. 2006. Glucose signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70253-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sastri, M., D. M. Barraclough, P. T. Carmichael, and S. S. Taylor. 2005. A-kinase-interacting protein localizes protein kinase A in the nucleus. Proc. Natl. Acad. Sci. USA 102349-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneper, L., A. Krauss, R. Miyamoto, S. Fang, and J. R. Broach. 2004. The Ras/protein kinase A pathway acts in parallel with the Mob2/Cbk1 pathway to effect cell cycle progression and proper bud site selection. Eukaryot. Cell 3108-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shabb, J. B. 2001. Physiological substrates of cAMP-dependent protein kinase. Chem. Rev. 1012381-2411. [DOI] [PubMed] [Google Scholar]
- 33.Smith, C. M., E. Radzio-Andzelm, Madhusudan, P. Akamine, and S. S. Taylor. 1999. The catalytic subunit of cAMP-dependent protein kinase: prototype for an extended network of communication. Prog. Biophys. Mol. Biol. 71313-341. [DOI] [PubMed] [Google Scholar]
- 34.Stiefel, J., L. Wang, D. A. Kelly, R. T. Janoo, J. Seitz, S. K. Whitehall, and C. S. Hoffman. 2004. Suppressors of an adenylate cyclase deletion in the fission yeast Schizosaccharomyces pombe. Eukaryot. Cell 3610-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tallada, V. A., R. R. Daga, C. Palomeque, A. Garzon, and J. Jimenez. 2002. Genome-wide search of Schizosaccharomyces pombe genes causing overexpression-mediated cell cycle defects. Yeast 191139-1151. [DOI] [PubMed] [Google Scholar]
- 36.Tang, Y., and M. McLeod. 2004. In vivo activation of protein kinase A in Schizosaccharomyces pombe requires threonine phosphorylation at its activation loop and is dependent on PDK1. Genetics 1681843-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taylor, S. S., C. Kim, C. Y. Cheng, S. H. Brown, J. Wu, and N. Kannan. 2008. Signaling through cAMP and cAMP-dependent protein kinase: diverse strategies for drug design. Biochim. Biophys. Acta 178416-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor, S. S., C. Kim, D. Vigil, N. M. Haste, J. Yang, J. Wu, and G. S. Anand. 2005. Dynamics of signaling by PKA. Biochim. Biophys. Acta 175425-37. [DOI] [PubMed] [Google Scholar]
- 39.Toda, T., S. Cameron, P. Sass, M. Zoller, J. D. Scott, B. McMullen, M. Hurwitz, E. G. Krebs, and M. Wigler. 1987. Cloning and characterization of BCY1, a locus encoding a regulatory subunit of the cyclic AMP-dependent protein kinase in Saccharomyces cerevisiae. Mol. Cell. Biol. 71371-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toda, T., S. Cameron, P. Sass, M. Zoller, and M. Wigler. 1987. Three different genes in Saccharomyces cerevisiae encode the catalytic subunits of the cAMP-dependent protein kinase. Cell 50277-287. [DOI] [PubMed] [Google Scholar]
- 41.Yanagida, M., Y. M. Yamashita, H. Tatebe, K. Ishii, K. Kumada, and Y. Nakaseko. 1999. Control of metaphase-anaphase progression by proteolysis: cyclosome function regulated by the protein kinase A pathway, ubiquitination, and localization. Philos. Trans. R. Soc. Lond. B Biol. Sci. 3541559-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang, P., H. Du, C. S. Hoffman, and S. Marcus. 2003. The phospholipase B homolog Plb1 is a mediator of osmotic stress response and of nutrient-dependent repression of sexual differentiation in the fission yeast Schizosaccharomyces pombe. Mol. Genet. Genomics 269116-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Young, D., M. Riggs, J. Field, A. Vojtek, D. Broek, and M. Wigler. 1989. The adenylyl cyclase gene from Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 867989-7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.