Abstract
Small GTPases of the Rho family act as molecular switches, and modulation of the GTP-bound state of Rho proteins is a well-characterized means of regulating their signaling activity in vivo. In contrast, the regulation of Rho-type GTPases by posttranslational modifications is poorly understood. Here, we present evidence of the control of the Saccharomyces cerevisiae Rho-type GTPase Rho5p by phosphorylation and ubiquitination. Rho5p binds to Ste50p, and the expression of the activated RHO5(Q91H) allele in an Δste50 strain is lethal under conditions of osmotic stress. An overexpression screen identified RGD2 and MSI1 as being high-copy suppressors of the osmotic sensitivity of this lethality. Rgd2p had been identified as being a possible Rho5p GTPase-activating protein based on an in vitro assay; this result supports its function as a regulator of Rho5p activity in vivo. MSI1 was previously identified as being a suppressor of hyperactive Ras/cyclic AMP signaling, where it antagonizes Npr1p kinase activity and promotes ubiquitination. Here, we show that Msi1p also acts via Npr1p to suppress activated Rho5p signaling. Rho5p is ubiquitinated, and its expression is lethal in a strain that is compromised for proteasome activity. These data identify Rho5p as being a target of Msi1p/Npr1p regulation and describe a regulatory circuit involving phosphorylation and ubiquitination.
In Saccharomyces cerevisiae, hyperosmotic stress conditions induce a series of signaling events resulting in rapid physiological adaptation. Primary among the signaling responses is the activation of the high-osmolarity glycerol (HOG) pathway (18, 41). Two nonredundant membrane-associated proteins, Sln1p and Sho1p, regulate the activation of this pathway. These proteins activate signaling via separate mitogen-activated protein kinase (MAPK) kinase kinases, Ssk2p or Ssk22p on the Sln1p branch and Ste11p on the Sho1p branch, but converge on the MAPK kinase Pbs2p. Pbs2p then activates the MAPK Hog1p, with a resultant efflux of water from the cell and increased levels of production and accumulation of intracellular glycerol. The adaptor protein Ste50p is essential for the activation of the Sho1p branch of this pathway. Ste50p binds to the MAPK kinase kinase Ste11p through their respective sterile-alpha motif (SAM) domains (20, 44, 63). Besides the N-terminal SAM domain, Ste50p also possesses a Ras-associated (RA) domain (45); RA domains have been identified in proteins throughout eukaryotic evolution, where they are involved in binding members of the Ras superfamily, including Rho family members (43). Ste50p's RA domain binds to the C terminus of the integral membrane protein Opy2p during HOG signaling, thereby tethering the Ste50p-Ste11p module to the membrane (62). In contrast, during pseudohyphal development, the Ste50p-RA domain has been proposed to bind the Rho-type GTPase Cdc42p, resulting in the localization of Ste50p and Ste11p to the membrane, where Ste11p can be activated by Cdc42p-associated Ste20p (57).
Rho family GTPases are involved in a variety of cellular processes. Rho proteins are known principally for their involvement in controlling cell morphology via actin cytoskeleton dynamics, but they have also been implicated in the control of cell polarity, membrane transport dynamics, transcription factor activity, and cellular signaling (for a review, see reference 6). The budding yeast genome codes for six members of the Rho family of GTPases: CDC42 and RHO1 to RHO5. The essential Cdc42p protein is involved in bud site assembly, polarized growth, and cytokinesis (21). Furthermore, Cdc42p is involved in activating MAPK signaling cascades via its effector Ste20p in response to osmotic stress, mating pheromones, and nutrient deprivation (27, 36, 42, 53, 66). The other yeast Rho family members are less well characterized: Rho1p is an essential protein involved in cell wall synthesis, cell polarity, and the regulation of protein kinase C signaling (23, 33); Rho2p function is partially redundant with Rho1p (34); Rho3p and Rho4p share an uncharacterized essential function and are involved in establishing cell polarity and exocytosis (1, 39); and Rho5p has been implicated in cell wall integrity signaling (52) and in mediating the oxidative stress response (54).
Like all GTPases, Rho family members cycle between “active” and “inactive” states by binding to GTP or GDP, respectively; they bind to effectors and induce a cellular response according to their nucleotide-bound state. Intrinsic GTPase activity results in the hydrolysis of GTP to GDP and the conversion of the GTPase to the inactive state. The Rho GTPase is reactivated when GDP is exchanged for GTP. This switch mechanism is tightly regulated by a set of Rho-binding proteins, the guanine nucleotide exchange factors, which catalyze the exchange of GDP for GTP and thus result in their activation (10, 24). GTPase-activating proteins (GAPs) perform the opposite function, stimulating the rate of intrinsic GTPase hydrolysis and catalyzing the conversion of the GTPase to its inactive, GDP-bound form (8, 11). A third class of regulators, guanine nucleotide dissociation inhibitors, inhibits GDP dissociation, thereby maintaining Rho GTPases in the inactive state (7, 25). These Rho-associated elements provide targets for signaling networks and allow the regulation of Rho GTPase activity.
Phosphorylation and ubiquitin-mediated proteolysis are well-characterized methods of regulating signaling activity. These posttranslational modifications can modulate signaling activity by determining the relative activity, localization, stability, and interactions of the target substrate. Phosphorylation constitutes a major source of cellular regulation; the protein kinase complement of the human genome is estimated to contain over 500 kinases, and approximately 30% of cellular proteins are phosphorylated (4, 35). The reversible phosphorylation and dephosphorylation of a protein allow for sophisticated regulatory control involving one or many inputs and finely tuned outputs (for a review, see reference 4). Conversely, ubiquitin-mediated proteolysis constitutes an irreversible step in many signaling pathways and is thus a major regulatory mechanism (for a review, see reference 14). The regulated destruction of components of signal transduction pathways is observed, for instance, in the NF-κB pathway (2). Often, phosphorylation and ubiquitination are coordinated. Many proteins regulated by ubiquitin-mediated proteolysis contain characteristic PEST sequences. For instance, the cyclin-dependent kinase phosphorylation of residues within G1 cyclin PEST sequences specifies the protein's recognition and ubiquitination, with subsequent processing by the proteasome (29, 60, 61, 65). This coordination of signaling output via phosphorylation with the targeted destruction of signaling components illustrates a way in which posttranslational modifications result in sophisticated modulations of cellular physiology.
Despite their prevalence in signaling networks, there have been few reports of posttranslational modifications of small Rho-type GTPases. In human natural killer cells, RhoA is phosphorylated by protein kinase A, resulting in the translocation of membrane-associated RhoA toward the cytosol (28). Phosphorylated RhoA is thereby separated from its effectors, and its signaling is terminated independently of GTP/GDP cycling. In fibroblasts, RhoA associates with the E3 ubiquitin ligase Smurf1 in lamellipodia and filopodia and is targeted for destruction (58). This prevents RhoA signaling during dynamic membrane movements at the leading edge of migrating cells. An atypical Rho-type GTPase, RhoBTB2, is ubiquitinated by a Cul3-based ubiquitin ligase complex, and a missense mutation that is not recognized by the ubiquitin-proteasome system has been identified in a lung cancer cell line (59). In yeast, no experimental evidence for the phosphorylation or ubiquitination of Rho-type GTPases exists.
In this study, the yeast Rho-type GTPase Rho5p is shown to interact with Ste50p to regulate the response to osmotic stress. We have identified several means of regulating its activity. First, Rgd2p's purported role as a Rho GAP for Rho5p is confirmed in vivo. Second, Rho5p is regulated by a module involving Msi1p, phosphorylation by Npr1p, and ubiquitin-mediated proteolysis. We have identified the phosphorylation and ubiquitination of Rho5p and have linked these posttranslational modifications to its regulation in vivo.
MATERIALS AND METHODS
Materials.
Restriction endonucleases and DNA-modifying enzymes were obtained from New England Biolabs and GE Healthcare. An enhanced chemiluminescence assay system, protease inhibitor tablets, and reduced glutathione were obtained from Roche. Nitrocellulose membranes were purchased from Bio-Rad. Anti-His polyclonal antibody, anti-ubiquitin mouse antibody, and horseradish peroxidase-conjugated secondary antibodies were purchased from Santa Cruz Biotechnology; anti-hemagglutinin (HA) polyclonal antibody was purchased from Open Biosystems; and the antibody against glutathione S-transferase (GST) was described previously (63). Glutathione-Sepharose 4B beads and PreScission protease were purchased from GE Healthcare. Radioisotopes were purchased from GE Healthcare and Perkin-Elmer, and film for autoradiography was BioMax MS from Kodak. The QuikChange site-directed mutagenesis kit was purchased from Stratagene. Acid-washed glass beads (450 to 600 μm), protease inhibitors, sorbitol, and trypsin were purchased from Sigma.
Construction of plasmids.
The His-tagged STE50 construct was described previously (62). GST-tagged NPR1 was obtained from the GST-ORF library purchased from Invitrogen. Other GST-tagged constructs were obtained from a genomic collection kindly provided by Eric M. Phizicky (University of Rochester) (38).
The Rho5p overexpression plasmids pGAL-RHO5 and pGAL-RHO5Q91H and their parent plasmid (pGAL) were provided by Jurgen J. Heinisch (Universitat Osnabruck) (52). HA-tagged ubiquitin plasmid and its untagged parent were obtained from M. Hochstrasser (Yale University) (17).
Plasmid pGEX-RHO5 was constructed by amplifying the full sequence of Rho5 minus the C-terminal 4 amino acids of the membrane-localizing CAAX box (nucleotides 1 to 984) from vector pGAL-RHO5 (52) with the oligonucleotides 5′-GAGAGAATTCATGAGGTCTATTAAATGTGTGATAA-3′ and 5′-GAGAGTCGACTTACTTTGACTTCTTTTTCTTCTTGTC-3′, where the underlined nucleotides are EcoRI and SalI sites, respectively. The resultant PCR product was inserted into vector pGEX-6P-1 by cutting with both EcoRI and SalI, followed by ligation. The resultant plasmid, pGEX-RHO5, was confirmed by sequencing.
Yeast strains and manipulations.
Yeast media, culture conditions, and manipulations were described previously (47). Transformation of yeast with plasmid DNA was achieved with lithium acetate and standard protocols (47). The yeast deletion strain collection was purchased from the ATCC.
Viability assays of yeast cells were performed by performing 10-fold serial dilutions of mid-logarithmic-phase cultures onto selective plates. Plates were incubated for 3 days at 30°C.
High-copy library screening for suppressors.
Strain YCW1321 (Δste50::Natr) expressing RHO5(Q91H) was transformed with a genomic library constructed in vector YEp213 (3). Roughly 15,000 transformants were recovered on synthetic dextrose (SD) plates lacking uracil and leucine. These transformants were first replica plated onto rich-medium plates and then rereplicated onto selection plates containing 2% galactose as a carbon source and 1.25 M sorbitol. Clones that were able to grow on the hyperosmotic media after 4 days were considered to be positive.
Preparation of GST fusion or His-tagged proteins.
The expression of GST fusion proteins in S. cerevisiae strain BY4741 or W303 was induced by the addition of 0.4 mM CuSO4 for 3 h with CUP1 promoter-driven expression or with 4% galactose for 5 h with GAL1-driven expression. The preparation of total cell extracts and isolation of GST fusion proteins by binding to glutathione-Sepharose beads were performed as described previously (63). Eluted proteins were washed with storage buffer and concentrated with Centricon 30 filters before storage at −80°C.
The purification of His-tagged Ste50p was done as previously described (62).
Expression of pGEX-RHO5 was done with Escherichia coli strain BL21, which was induced with 0.4 mM isopropyl-β-d-thiogalactopyranoside for 3 h. Fusion proteins were obtained essentially as described previously (55), with the modification that the proteins were eluted by the cleavage of the GST tag with PreScission protease (GE Healthcare).
Protein kinase assays.
Kinase assays were performed as described previously (64). Approximately 1.0 μg of Rho5p or GST-Cdc42p substrates in solution was resuspended in kinase buffer supplemented with 1 μM ATP and 1 μl [γ-32P]ATP (4,500 Ci/mmol and 10 Ci/μl, respectively) and 0.5 μg GST-Npr1p or GST-Ste20p, where appropriate. Reaction mixtures were incubated for 30 min and then boiled for 5 min after the addition of Laemmli buffer. Samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), dried, and visualized by autoradiography.
Photomicroscopy.
Cells were grown under the conditions indicated and viewed with a microscope (Nikon Eclipse E800) equipped with Nomarski optics. Microscopic photographs were acquired with a 100× objective and a Nikon DXM1200 camera and ACT-1 version 2.10 software (Nikon).
RESULTS
Rho5p binds to the RA domain of Ste50p.
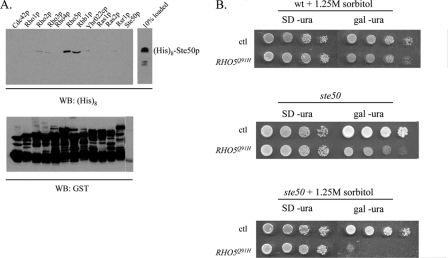
Ste50p is involved in the activation of the HOG MAPK pathway in response to hyperosmotic shock. In this pathway, Ste50p is responsible for delivering Ste11p to the plasma membrane (57, 62). We have previously shown that the C-terminal RA domain of Ste50p is required for Ste50p function in both the pheromone response and HOG pathways (62, 63). In an attempt to establish which small GTPase(s) could bind to the RA domain of Ste50p, we performed an in vitro resin-binding assay between Ste50p and yeast Ras and Rho-type small GTPases. GTPases were expressed as GST fusion proteins in yeast and were purified on glutathione-Sepharose beads according to standard procedures (64). Purified yeast GTPases were then incubated with bacterially expressed and purified His8-tagged Ste50p. The glutathione-Sepharose beads were washed extensively, and the bound proteins were resolved by SDS-PAGE, transferred onto a nitrocellulose membrane, and then revealed by Western blotting using anti-His tag antibodies and appropriate secondary antibodies, followed by chemiluminescence. As shown in Fig. 1A, Ste50p selectively binds to Rho5p and Rhb1p and weakly to Rho2p and Rho3p. Given the role of Rho GTPases in Ste50p-associated processes, we focused on the interaction between Ste50p and Rho5p.
FIG. 1.
Rho5 and Ste50 interact physically and genetically. (A) GST fusion constructs of yeast Ras superfamily GTPases were incubated with bacterially expressed His8-tagged Ste50p and were then purified with glutathione-Sepharose. Bound proteins were separated by SDS-PAGE, and the fraction of Ste50p associated with the membrane-bound GTPases was detected with antibodies to His8. (B) Yeast cells transformed with hyperactive RHO5(Q91H) were tested for their ability to grow under hyperosmotic conditions (1.25 M sorbitol) in the presence (wild type [wt]) and absence (Δste50) of endogenous Ste50p. SD −ura, SD uracil dropout medium; Gal −ura, selective medium with galactose as the carbon source; ctl, control; WB, Western blot.
Expression of RHO5(Q91H) in a Δste50 strain results in a growth defect and is osmotically lethal.
To determine the biological relevance of the physical interaction between Ste50p and Rho5p, we asked whether there is a role for Rho5p in resistance to hyperosmotic stress. The RHO5(Q91H) allele exchanges the glutamine at position 91 in loop 4 for histidine, creating an activated protein with reduced intrinsic GTPase activity (52). This mutant protein was overexpressed from the GAL10 promoter in an ste50 deletion strain (YCW1321). This strain was tested on hyperosmotic medium, which induced the HOG pathway, and on isosmotic medium, which permits normal vegetative growth. On osmotically neutral medium, the transformants exhibited only a mild growth defect compared to the wild type (Fig. 1B, middle). However, when grown on hyperosmotic medium (1.25 M sorbitol) under inducing conditions, the Δste50 RHO5(Q91H) strain died (Fig. 1B, bottom). The expression of RHO5(Q91H) in a wild-type strain generated no detectable defects under either isosmotic (data not shown) or hyperosmotic (Fig. 1B, top) conditions. Thus, it appears that Rho5p plays a role in the cellular response to hyperosmotic stress in a Ste50p-dependent manner.
Isolation of high-copy suppressors of RHO5(Q91H) Δste50 osmotic sensitivity.
To define the basis of the osmotic sensitivity of the Δste50 RHO5(Q91H) strain, we conducted a suppressor screen using the inability to grow under hyperosmotic conditions as a condition for selection. The strain was transformed with a yeast genomic library constructed in the multicopy vector YEp213 and plated onto osmotically neutral SD plates lacking uracil and leucine and then replica plated onto medium selecting for osmotic resistance. Roughly 15,000 colonies were screened.
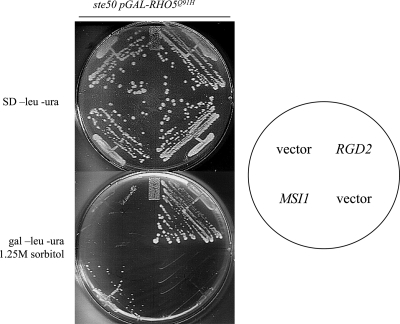
This approach identified 37 colonies that suppressed the osmotic sensitivity of the Δste50 RHO5(Q91H) strain. Plasmids were isolated from these colonies and retested, and confirmed positives were sequenced. We isolated seven genes: RGD2 (four times), MSI1 (multicopy suppressor of Δira1) (four times), PGM1 (one time), PGM2 (two times), ATX1 (one time), GAL80 (four times), and URA3 (two inserts) (20 times). ATX1 encodes a metallochaperone involved in iron absorption (31). GAL80 encodes a transcriptional regulator involved in the inhibition of expression of galactose response genes (30). It presumably suppresses the osmotic sensitivity of the strain by downregulating the expression of RHO5(Q91H) from the GAL10 promoter. The expression of URA3 on the YEp213 library plasmid presumably allows rescue via the loss of the URA3-marked RHO5(Q91H) plasmid. Both isoforms of phosphoglucomutase (PGM1 and PGM2) act as suppressors. Interestingly, phosphoglucomutase and MSI1 were both previously identified as being suppressors of hyperactive Ras pathway mutants (19, 50), suggesting a connection between Ras signaling and Rho5p activity. Based on their potential involvement in the direct regulation of Rho5p activity, RGD2 and MSI1 were chosen for further examination.
Rgd2p acts as a high-copy suppressor of activated Rho5p.
RGD2 was identified as a suppressor of the synthetic osmotic sensitivity of the Δste50 RHO5(Q91H) strain in four isolates (Fig. 2). Rgd2p was predicted to be a Rho GAP based on the identification of sequences with similarity to Rho GAP domains (49). That same study confirmed that Rgd2p stimulated the GTPase activity of Rho5p and Cdc42p in an in vitro assay. The RHO5(Q91H) allele contains a mutation that is analogous to the mutation of glutamine to histidine at position 61 in human p21 Ras, which reduces intrinsic GTP hydrolysis (26, 52). The overexpression of its cognate GAP is predicted to downregulate the activity of the activated RHO5(Q91H) allele in vivo (9). Rgd2p could suppress the synthetic osmotic sensitivity of this strain by stimulating the GTPase activity of RHO5(Q91H), and thus, in vivo, Rgd2p is likely functioning as a GAP for Rho5p.
FIG. 2.
A high-copy library screen identified RGD2 and MSI1 as being suppressors of the osmotic sensitivity of Δste50 pGAL-RHO5Q91H. SD −leucine −ura, synthetic dextrose leucine and uracil dropout medium; gal −leu −ura, leucine and uracil dropout medium with galactose as the carbon source.
MSI1 suppression of the RHO5(Q91H) Δste50 growth defect may involve Npr1p.
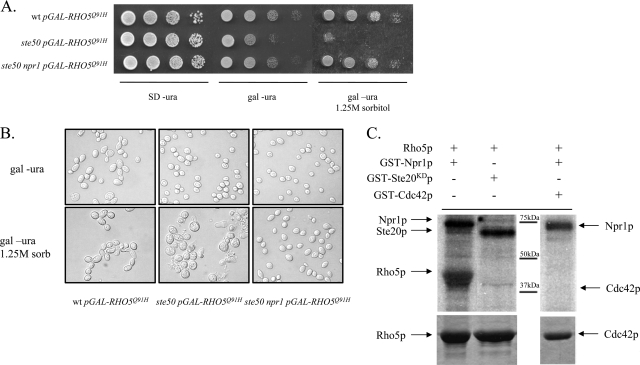
MSI1 was identified as being a suppressor in four isolates (Fig. 2). MSI1 was previously identified as being a suppressor of hyperactive Ras signaling, potentially through its ability to sequester and inhibit the kinase Npr1p (22, 50) because the deletion of NPR1 was shown to mimic the effect of MSI1 overexpression (22). To determine if MSI1 suppression of Δste50 RHO5(Q91H) osmotic sensitivity occurs via the same mechanism, we generated an Δste50 Δnpr1 strain, which we then transformed with the RHO5(Q91H) plasmid. As expected, the deletion of NPR1 rescued the growth defect of the Δste50 RHO5(Q91H) strain (Fig. 3A). Microscopic analysis of cells supports the plate phenotypes (Fig. 3B). Under isosmotic conditions, the expression of RHO5(Q91H) in a variety of strain backgrounds results in mild morphological defects. A wild-type strain overexpressing RHO5(Q91H) exhibits mild morphological defects including mildly elongated buds and occasional multibudded cells, whereas Δste50 and Δste50 Δnpr1 cells exhibit wild-type morphologies despite the slow growth exhibited by the Δste50 RHO5(Q91H) strain (Fig. 3A). Osmotic stress, however, results in more severe phenotypes. The wild-type strain expressing RHO5(Q91H) exhibits severely elongated cells. The Δste50 RHO5(Q91H) strain exhibits a variety of aberrant cell morphologies including large, round, misshapen cells and widespread cell lysis. However, combining the Δnpr1 and Δste50 mutations rescues the aberrant morphology and cell lysis defects observed when RHO5(Q91H) is expressed. Thus, the suppression of growth defects by MSI1 overexpression is mimicked by the deletion of the gene that codes for its antagonist kinase, NPR1, suggesting that, as with the suppression of hyperactive Ras signaling, this regulatory module is also involved in controlling Rho5p signaling.
FIG. 3.
MSI1 suppression of Δste50 RHO5(Q91H) osmotic sensitivity may involve Npr1p phosphorylation. (A) Serial dilutions of relevant genotypes under osmotically neutral noninducing (SD plates lacking uracil [SD −ura]), neutral inducing (uracil dropout medium with galactose as the carbon source [gal −ura]), and hyperosmotic inducing (uracil dropout medium with galactose as the carbon source plus 1.25 M sorbitol) conditions. wt, wild type. (B) Morphology of yeast cells with relevant genotypes (as indicated at the bottom) under neutral (uracil dropout medium with galactose as the carbon source) and hyperosmotic (uracil dropout medium with galactose as the carbon source plus 1.25 M sorbitol) conditions. (C) In vitro phosphorylation of Rho5p by Npr1p. Relevant proteins were resuspended in the combinations indicated in a buffered solution containing relevant divalent cofactors and [γ-32P]ATP, followed by 30 min of incubation at 30°C. Radiolabeled proteins were separated by SDS-PAGE and subsequently visualized by autoradiography (top). Loading controls of Rho5p and Cdc42p are indicated by Coomassie staining (bottom).
Npr1p is a protein kinase whose activity influences the stability of its substrates by promoting or antagonizing ubiquitin-mediated proteolysis (51, 56). We therefore investigated if Rho5p can act as a substrate for Npr1p phosphorylation by using an in vitro kinase assay. As can be seen in Fig. 3C, Rho5p is effectively labeled in vitro by GST-Npr1p (lane 1) but not by an unrelated kinase, Ste20p (lane 2). Specificity toward the phosphorylation of Rho5p is demonstrated by the inability of GST-Npr1p to phosphorylate the closely related Rho GTPase Cdc42p (Fig. 3C, lane 3). Autophosphorylation of both Npr1p and Ste20p was also observed. This demonstrates that in vitro, GST-Rho5p is a substrate of Npr1p.
Rho5p is ubiquitinated, and its overexpression is lethal in a strain with a compromised proteasome.
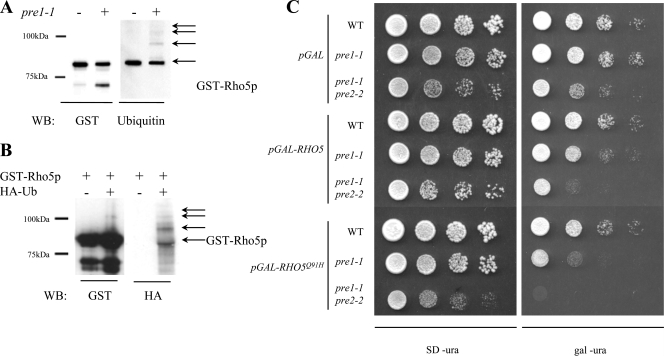
Since Npr1p kinase activity has been shown to antagonize ubiquitin-mediated proteolysis, it is possible that the Msi1p/Npr1p regulatory module is moderating Rho5p signaling via this mechanism. We therefore examined whether Rho5p is a substrate for ubiquitination. Strain WCG4-11 carries the pre1-1 mutation and results in a temperature-sensitive allele of PRE1, which codes for a core component of the 26S proteasome (13). This mutation is lethal in strains grown at 37°C but viable at the semipermissive temperature of 30°C, where it accumulates ubiquitin-protein conjugates normally targeted for proteasomal proteolysis. To determine if Rho5p is ubiquitinated, we expressed GST-Rho5p in congenic wild-type (WCG4a) and proteasome-impaired (WCG4-11a) strains. GST-Rho5p was purified and examined by Western blotting with either anti-GST or anti-ubiquitin probes. When probed with anti-GST antibodies (Fig. 4A), a strong band corresponding to GST-Rho5p was observed in both the wild-type and proteasome-deficient strains. The band observed at a low molecular weight is in greater relative abundance in the pre1-1 strain than in the wild type. This may represent a Rho5p breakdown product that is efficiently processed in wild-type cells but accumulates to higher concentrations in a strain with partially deficient proteasome function.
FIG. 4.
Rho5p is ubiquitinated. (A) GST-Rho5p purified from congenic wild-type (WT) and proteasome-impaired (pre1-1) strains was assayed for ubiquitination by probing with the anti-ubiquitin antibody. The levels of GST-Rho5p were monitored by the anti-GST antibody. (B) GST-Rho5p was expressed singly or in combination with HA-tagged ubiquitin and was subsequently purified and separated by SDS-PAGE. The presence of HA-ubiquitin was assayed by an anti-ubiquitin antibody. (C) Impaired proteasome function impairs growth of Rho5-expressing strains. Vectors containing wild-type RHO5 (pGAL-RHO5) and activated rho5 (pGAL-RHO5Q91H) were transformed into strains compromised for proteasome function due to one (pre1-1) or two (pre1-1 pre2-2) mutations to core components of the 26S proteasome. A congenic wild-type strain and empty vector (pGAL) are included as controls.
We then probed the blot with anti-ubiquitin antibodies (Fig. 4A). Interestingly, the main GST-Rho5p bands tested positive for ubiquitination, suggesting that the majority of GST-Rho5p is ubiquitinated. The estimated molecular weight indicates monoubiquitination. Monoubiquitination has been identified as being a common method of posttranslational modification involved in diverse cellular processes such as membrane trafficking and histone function (15). There is no evidence of the faint smear beneath the primary band, as seen when probed with anti-GST antibodies, suggesting that the phosphorylated form of Rho5p may correspond with monoubiquitination. Furthermore, there is no evidence of the low-molecular-weight GST-Rho5p breakdown product as seen with anti-GST probes, which implies that the processing of GST-Rho5p involves the processing of the ubiquitin-conjugated region of the molecule. In the pre1-1 strain, we observed a series of higher-molecular-weight species suggestive of a polyubiquitination “ladder.” This was not observed in the wild-type strain, suggesting that the pre1-1 strain grown at a semipermissive temperature is accumulating the polyubiquitin-GST-Rho5p conjugates, as predicted. Thus, it seems that GST-Rho5p is mono- and polyubiquitinated, although the polyubiquitination was detectable only in the proteasome-deficient strain.
To confirm this, we coexpressed GST-Rho5p with an HA-ubiquitin fusion protein in W303 cells. HA-tagged ubiquitin is efficiently utilized by the ubiquitination machinery and has been shown to recapitulate ubiquitination patterns of wild-type ubiquitin, although the HA-ubiquitin-protein conjugates are inefficiently processed by the proteasome (5). In this case, we purified GST-Rho5p and tested for the presence of HA-ubiquitin by probing Western blots with anti-HA probes. As can be seen in Fig. 4B, GST-Rho5p reacts strongly with anti-HA antibodies when purified from cells coexpressing HA-tagged ubiquitin. Again, we see the presence of a characteristic polyubiquitin ladder as well as the presence of monoubiquitinated protein. We also observed lower-molecular-weight species when blotted for HA-ubiquitin, which we ascribe to processing by the proteasome. This would explain why these species are not observed in the proteasome-compromised pre1-1 strain shown in Fig. 4A. Thus, based on these observations, we conclude that GST-Rho5p is monoubiquitinated and exhibits polyubiquitination in strains inhibited for proteasome activity.
In order to assay the importance of Rho5p ubiquitination in vivo, we overexpressed RHO5 alleles in strains compromised for proteasome activity. Strains WCG4, WCG4-11a, and WCG4-11/22a are congenic, with the latter two containing mutations (pre1-1 or pre1-1 pre2-2, respectively) in components of the 20S subunit of the 26S proteasome (12, 13). These strains are inviable at the restrictive temperature of 37°C but are viable when grown at lower temperatures (30°C). We reasoned that the inability to regulate Rho5p activity by ubiquitin-mediated proteolysis may result in an observable growth defect. We thus expressed wild-type RHO5 and the activated RHO5(Q91H) in the appropriate strains and incubated them at 30°C for 3 days. Proteasome-inhibited strains overexpressing wild-type RHO5 were noticeably impaired for growth compared to the wild-type strain and compared to a plasmid control (Fig. 4C). The expression of the activated RHO5(Q91H) allele severely impaired growth in the pre1-1 strain and was lethal in the more compromised pre1-1 pre2-2 strain (Fig. 4C). The expression of the activated allele in the congenic wild-type strains had no noticeable defect, which suggests that an active proteasome is required to fully insulate the cells from the effects of the overexpression of RHO5 alleles. Furthermore, the increased severity of the phenotype associated with the activated allele of RHO5 suggests that this is not a general overexpression defect but is associated with increased Rho5p activity. Thus, we conclude that Rho5p is subject to polyubiquitination and subsequent 26S proteasome-mediated proteolysis, and this is an important element of its regulation.
DISCUSSION
Our investigation of the function of Saccharomyces cerevisiae Rho family member Rho5p has shown that it plays a role in the osmotic shock response, and its activity is regulated by phosphorylation and ubiquitination. Rho5p binds the RA domain of Ste50p, and an activated allele of RHO5 exhibits a synthetic osmotic growth defect with an Δste50 mutation. A screen for suppressors of this defect revealed two distinct means of regulating Rho5p activity: the overexpression of its purported Rho GAP, Rgd2p, and a module involving phosphorylation and ubiquitin-mediated proteolysis controlled by the nuclear factor Msi1p. We have described a yeast Rho-type GTPase regulated by phosphorylation and ubiquitination, suggesting the possibility that other Rho-type GTPases may be similarly regulated.
Rho5p binds to Ste50p, and a potential role for Rho5p in the regulation of the osmotic stress response is suggested by the synthetic osmotic sensitivity of an Δste50 RHO5(Q91H) mutant. We have previously shown that the association between the RA domain of Ste50p and the C terminus of Opy2p results in the membrane localization of both Ste50p and its SAM domain-associated partner Ste11p under conditions of osmotic stress (62). The binding of Rho5p to Ste50p suggests another possible interaction and another role for Ste50p in coordinating the osmotic stress response. One explanation is that Rho5p may act as a direct negative regulator of HOG pathway signaling, and the combination of increased Rho5p-dependent inhibition with reduced pathway activation due to the Δste50 mutation results in a synthetic osmotic lethality. A second explanation is that the synthetic effects are indirect due to the interactions of distinct and separate pathways. Other yeast signaling pathways have exhibited synthetic interactions with HOG pathway mutants. For example, increased signaling by cyclic AMP/protein kinase A pathway results in increased osmotic sensitivity (16, 40). Also, it has been suggested that the cell integrity and HOG pathways cooperate in the same process (18). If Rho5p is involved in moderating signaling through a separate pathway that exhibits synthetic growth defects with osmotic signaling, the combination of increased Rho5p signaling and decreased HOG signaling may result in a synthetic osmotic lethality.
A screen for suppressors of this synthetic osmotic defect revealed different means of regulating Rho5p signaling activity. One suppressor, Rgd2p, was previously suggested to be a possible Rho5p Rho GAP based on in vitro studies (49). Here, we confirm its ability to downregulate Rho5p signaling in vivo. It is likely that Rgd2p is not the only Rho GAP for Rho5p; if it was, an Δrgd2 mutation should mimic the activated RHO5(Q91H) allele, yet an Δste50 Δrgd2 strain does not display the synthetic osmotic sensitivity defect characteristic of the Δste50 RHO5(Q91H) strain (data not shown). Thus, other Rho GAPs that can stimulate Rho5p GTPase hydrolysis may exist.
The suppressor screen also identified MSI1, thus suggesting a regulatory mechanism involving Npr1p phosphorylation and ubiquitin-mediated proteolysis. Npr1p is a kinase that stabilizes several membrane proteins by moderating ubiquitination and subsequent proteasomal degradation (51, 56). Phosphorylation by Npr1p affects the ubiquitination and subsequent processing by the proteasome, thereby stabilizing some substrates such as Gap1p (32, 56) while destabilizing others such as Tat2p (51). In the suppression of Ras signaling, Npr1p phosphorylation was found to act antagonistically to the ubiquitin-proteasome system (22). We found that Npr1p seems to be similarly involved in the suppression of Rho5p signaling, as an Δnpr1 mutation suppresses the hyperosmotic sensitivity of the Δste50 RHO5(Q91H) strain. Rho5p is an efficient in vitro substrate for Npr1p phosphorylation; while in vitro assays do not necessarily confirm a physiological role for phosphorylation in vivo, this result suggests that Rho5p can serve as a substrate for Npr1p. These results suggest that, as in the case of Ras signaling suppression, Msi1p, via Npr1p, suppresses the Δste50 RHO5(Q91H) synthetic osmotic sensitivity via phosphorylation and subsequent ubiquitin-mediated proteolysis of Rho5(Q91H)p.
In agreement with the model of Msi1p/Npr1p suppression of Rho5p signaling via phosphorylation and subsequent ubiquitin-mediated proteolysis, we found that Rho5p is ubiquitinated in vivo. A strain compromised for proteasome activity accumulated a characteristic high-molecular-weight ladder of GST-Rho5p species, which were detected with antibodies against ubiquitin. Furthermore, GST-Rho5p purified from a strain coexpressing HA-ubiquitin accumulated high-molecular-weight species, which were detected with antibodies against the HA tag. These results support the model that Rho5p is ubiquitinated and may be regulated by ubiquitin-mediated proteolysis. We hypothesized that the overexpression of Rho5p in a strain compromised for proteasome function may generate a growth defect and that this defect may be increased upon the expression of the activated allele in the same strain. Indeed, the expression of wild-type Rho5p in pre1-1 or pre1-1 pre2-2 temperature-sensitive strains caused mild growth defects at a semipermissive temperature. The expression of the activated allele in these backgrounds resulted in more pronounced growth defects. In support of the model of ubiquitin-mediated proteolytic regulation of Rho5p signaling, efficient proteolysis is required to mitigate the effects of Rho5p overexpression. It remains to be determined under what conditions Rho5p ubiquitination occurs whether this modification is constitutive or cell cycle regulated or occurs in response to conditions of stress.
Rho5p phosphorylation and ubiquitination may be coordinated. This suggestion is supported by the presence of a canonical PEST sequence in Rho5p. PEST sequences are characteristic of many short-lived proteins targeted for proteasome-mediated turnover (46); Rho5p is the only yeast Rho-type GTPase to possess a PEST sequence (49). These regions are often sites of regulated phosphorylation; this provides a signal for ubiquitination, thus allowing signaling networks to efficiently regulate the stability of candidate proteins. This is classically observed in the case of G1 cyclins, whose PEST sequence phosphorylation by their cognate cyclin-dependent kinases results in their targeted destruction (29, 60, 61, 65). This regulatory element is observed in numerous other systems in yeast, including membrane proteins (37, 48).
While the function of Rho5p in the osmotic stress response remains elusive, several lines of evidence suggest that it may play a role in mediating signaling by the Ras/cyclic AMP pathway. Three of the suppressors of Δste50 RHO5(Q91H) signaling (MSI1, PGM1, and PGM2) were previously identified as being suppressors of hyperactive Ras pathway mutants (19, 50). Furthermore, cells with a hyperactive Ras pathway are more sensitive to osmotic stress (16, 40). The target of the Msi1p/Npr1p-mediated suppression of Ras signaling was not identified in previous work (22, 50); Rho5p thus represents a potential target for the Msi1p/Npr1p regulation of Ras signaling as the first yeast Rho-type GTPase to be regulated by phosphorylation and ubiquitination.
Acknowledgments
We gratefully thank W. Heinemeyer for strains, J. J. Heinisch and M. Hochstrasser for plasmids, E. Phizicky for the GST-tagged yeast genomic library, and T. Chow for the YEp213 yeast genomic library.
R.B.A. was the recipient of an NSERC graduate student fellowship. This work was supported by the NRC and grants from the NCIC and CIHR to M.W. and D.Y.T.
This work is NRC publication 49561.
Footnotes
Published ahead of print on 11 July 2008.
REFERENCES
- 1.Adamo, J. E., G. Rossi, and P. Brennwald. 1999. The Rho GTPase Rho3 has a direct role in exocytosis that is distinct from its role in actin polarity. Mol. Biol. Cell 104121-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen, Z. J. 2005. Ubiquitin signalling in the NF-kappaB pathway. Nat. Cell Biol. 7758-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chow, T. Y., E. L. Perkins, and M. A. Resnick. 1992. Yeast RNC1 encodes a chimeric protein, RhoNUC, with a human rho motif and deoxyribonuclease activity. Nucleic Acids Res. 205215-5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen, P. 2000. The regulation of protein function by multisite phosphorylation—a 25 year update. Trends Biochem. Sci. 25596-601. [DOI] [PubMed] [Google Scholar]
- 5.Ellison, M. J., and M. Hochstrasser. 1991. Epitope-tagged ubiquitin. A new probe for analyzing ubiquitin function. J. Biol. Chem. 26621150-21157. [PubMed] [Google Scholar]
- 6.Etienne-Manneville, S., and A. Hall. 2002. Rho GTPases in cell biology. Nature 420629-635. [DOI] [PubMed] [Google Scholar]
- 7.Fukumoto, Y., K. Kaibuchi, Y. Hori, H. Fujioka, S. Araki, T. Ueda, A. Kikuchi, and Y. Takai. 1990. Molecular cloning and characterization of a novel type of regulatory protein (GDI) for the rho proteins, ras p21-like small GTP-binding proteins. Oncogene 51321-1328. [PubMed] [Google Scholar]
- 8.Garrett, M. D., G. N. Major, N. Totty, and A. Hall. 1991. Purification and N-terminal sequence of the p21rho GTPase-activating protein, rho GAP. Biochem. J. 276833-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gideon, P., J. John, M. Frech, A. Lautwein, R. Clark, J. E. Scheffler, and A. Wittinghofer. 1992. Mutational and kinetic analyses of the GTPase-activating protein (GAP)-p21 interaction: the C-terminal domain of GAP is not sufficient for full activity. Mol. Cell. Biol. 122050-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hart, M. J., A. Eva, T. Evans, S. A. Aaronson, and R. A. Cerione. 1991. Catalysis of guanine nucleotide exchange on the CDC42Hs protein by the dbl oncogene product. Nature 354311-314. [DOI] [PubMed] [Google Scholar]
- 11.Hart, M. J., K. Shinjo, A. Hall, T. Evans, and R. A. Cerione. 1991. Identification of the human platelet GTPase activating protein for the CDC42Hs protein. J. Biol. Chem. 26620840-20848. [PubMed] [Google Scholar]
- 12.Heinemeyer, W., A. Gruhler, V. Mohrle, Y. Mahe, and D. H. Wolf. 1993. PRE2, highly homologous to the human major histocompatibility complex-linked RING10 gene, codes for a yeast proteasome subunit necessary for chrymotryptic activity and degradation of ubiquitinated proteins. J. Biol. Chem. 2685115-5120. [PubMed] [Google Scholar]
- 13.Heinemeyer, W., J. A. Kleinschmidt, J. Saidowsky, C. Escher, and D. H. Wolf. 1991. Proteinase yscE, the yeast proteasome/multicatalytic-multifunctional proteinase: mutants unravel its function in stress induced proteolysis and uncover its necessity for cell survival. EMBO J. 10555-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67425-479. [DOI] [PubMed] [Google Scholar]
- 15.Hicke, L. 2001. Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell Biol. 2195-201. [DOI] [PubMed] [Google Scholar]
- 16.Hirata, D., S. Harada, H. Namba, and T. Miyakawa. 1995. Adaptation to high-salt stress in Saccharomyces cerevisiae is regulated by Ca2+/calmodulin-dependent phosphoprotein phosphatase (calcineurin) and cAMP-dependent protein kinase. Mol. Gen. Genet. 249257-264. [DOI] [PubMed] [Google Scholar]
- 17.Hochstrasser, M., M. J. Ellison, V. Chau, and A. Varshavsky. 1991. The short-lived MAT alpha 2 transcriptional regulator is ubiquitinated in vivo. Proc. Natl. Acad. Sci. USA 884606-4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hohmann, S. 2002. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 66300-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howard, S. C., S. J. Deminoff, and P. K. Herman. 2006. Increased phosphoglucomutase activity suppresses the galactose growth defect associated with elevated levels of Ras signaling in S. cerevisiae. Curr. Genet. 491-6. [DOI] [PubMed] [Google Scholar]
- 20.Jansen, G., F. Buhring, C. P. Hollenberg, and M. R. Rad. 2001. Mutations in the SAM domain of STE50 differentially influence the MAPK-mediated pathways for mating, filamentous growth and osmotolerance in Saccharomyces cerevisiae. Mol. Genet. Genomics 265102-117. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, D. I., and J. R. Pringle. 1990. Molecular characterization of CDC42, a Saccharomyces cerevisiae gene involved in the development of cell polarity. J. Cell Biol. 111143-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnston, S. D., S. Enomoto, L. Schneper, M. C. McClellan, F. Twu, N. D. Montgomery, S. A. Haney, J. R. Broach, and J. Berman. 2001. CAC3(MSI1) suppression of RAS2(G19V) is independent of chromatin assembly factor I and mediated by NPR1. Mol. Cell. Biol. 211784-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamada, Y., H. Qadota, C. P. Python, Y. Anraku, Y. Ohya, and D. E. Levin. 1996. Activation of yeast protein kinase C by Rho1 GTPase. J. Biol. Chem. 2719193-9196. [DOI] [PubMed] [Google Scholar]
- 24.Kjoller, L., and A. Hall. 1999. Signaling to Rho GTPases. Exp. Cell Res. 253166-179. [DOI] [PubMed] [Google Scholar]
- 25.Koch, G., K. Tanaka, T. Masuda, W. Yamochi, H. Nonaka, and Y. Takai. 1997. Association of the Rho family small GTP-binding proteins with Rho GDP dissociation inhibitor (Rho GDI) in Saccharomyces cerevisiae. Oncogene 15417-422. [DOI] [PubMed] [Google Scholar]
- 26.Krengel, U., I. Schlichting, A. Scherer, R. Schumann, M. Frech, J. John, W. Kabsch, E. F. Pai, and A. Wittinghofer. 1990. Three-dimensional structures of H-ras p21 mutants: molecular basis for their inability to function as signal switch molecules. Cell 62539-548. [DOI] [PubMed] [Google Scholar]
- 27.Lamson, R. E., M. J. Winters, and P. M. Pryciak. 2002. Cdc42 regulation of kinase activity and signaling by the yeast p21-activated kinase Ste20. Mol. Cell. Biol. 222939-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lang, P., F. Gesbert, M. Delespine-Carmagnat, R. Stancou, M. Pouchelet, and J. Bertoglio. 1996. Protein kinase A phosphorylation of RhoA mediates the morphological and functional effects of cyclic AMP in cytotoxic lymphocytes. EMBO J. 15510-519. [PMC free article] [PubMed] [Google Scholar]
- 29.Lanker, S., M. H. Valdivieso, and C. Wittenberg. 1996. Rapid degradation of the G1 cyclin Cln2 induced by CDK-dependent phosphorylation. Science 2711597-1601. [DOI] [PubMed] [Google Scholar]
- 30.Leuther, K. K., and S. A. Johnston. 1992. Nondissociation of GAL4 and GAL80 in vivo after galactose induction. Science 2561333-1335. [DOI] [PubMed] [Google Scholar]
- 31.Lin, S. J., and V. C. Culotta. 1995. The ATX1 gene of Saccharomyces cerevisiae encodes a small metal homeostasis factor that protects cells against reactive oxygen toxicity. Proc. Natl. Acad. Sci. USA 923784-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lorenz, M. C., and J. Heitman. 1998. The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J. 171236-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madaule, P., R. Axel, and A. M. Myers. 1987. Characterization of two members of the rho gene family from the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 84779-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manning, B. D., R. Padmanabha, and M. Snyder. 1997. The Rho-GEF Rom2p localizes to sites of polarized cell growth and participates in cytoskeletal functions in Saccharomyces cerevisiae. Mol. Biol. Cell 81829-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manning, G., D. B. Whyte, R. Martinez, T. Hunter, and S. Sudarsanam. 2002. The protein kinase complement of the human genome. Science 2981912-1934. [DOI] [PubMed] [Google Scholar]
- 36.Manser, E., T. Leung, H. Salihuddin, Z. S. Zhao, and L. Lim. 1994. A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature 36740-46. [DOI] [PubMed] [Google Scholar]
- 37.Marchal, C., R. Haguenauer-Tsapis, and D. Urban-Grimal. 1998. A PEST-like sequence mediates phosphorylation and efficient ubiquitination of yeast uracil permease. Mol. Cell. Biol. 18314-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martzen, M. R., S. M. McCraith, S. L. Spinelli, F. M. Torres, S. Fields, E. J. Grayhack, and E. M. Phizicky. 1999. A biochemical genomics approach for identifying genes by the activity of their products. Science 2861153-1155. [DOI] [PubMed] [Google Scholar]
- 39.Matsui, Y., and E. A. Toh. 1992. Yeast RHO3 and RHO4 ras superfamily genes are necessary for bud growth, and their defect is suppressed by a high dose of bud formation genes CDC42 and BEM1. Mol. Cell. Biol. 125690-5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Norbeck, J., and A. Blomberg. 2000. The level of cAMP-dependent protein kinase A activity strongly affects osmotolerance and osmo-instigated gene expression changes in Saccharomyces cerevisiae. Yeast 16121-137. [DOI] [PubMed] [Google Scholar]
- 41.O'Rourke, S. M., I. Herskowitz, and E. K. O'Shea. 2002. Yeast go the whole HOG for the hyperosmotic response. Trends Genet. 18405-412. [DOI] [PubMed] [Google Scholar]
- 42.Peter, M., A. M. Neiman, H. O. Park, M. van Lohuizen, and I. Herskowitz. 1996. Functional analysis of the interaction between the small GTP binding protein Cdc42 and the Ste20 protein kinase in yeast. EMBO J. 157046-7059. [PMC free article] [PubMed] [Google Scholar]
- 43.Ponting, C. P., and D. R. Benjamin. 1996. A novel family of Ras-binding domains. Trends Biochem. Sci. 21422-425. [DOI] [PubMed] [Google Scholar]
- 44.Posas, F., E. A. Witten, and H. Saito. 1998. Requirement of STE50 for osmostress-induced activation of the STE11 mitogen-activated protein kinase kinase kinase in the high-osmolarity glycerol response pathway. Mol. Cell. Biol. 185788-5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramezani-Rad, M. 2003. The role of adaptor protein Ste50-dependent regulation of the MAPKKK Ste11 in multiple signalling pathways of yeast. Curr. Genet. 43161-170. [DOI] [PubMed] [Google Scholar]
- 46.Rogers, S., R. Wells, and M. Rechsteiner. 1986. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science 234364-368. [DOI] [PubMed] [Google Scholar]
- 47.Rose, M. D., F. M. Winston, P. Hieter, F. Sherman, G. R. Fink, and J. Hicks. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 48.Roth, A. F., and N. G. Davis. 2000. Ubiquitination of the PEST-like endocytosis signal of the yeast a-factor receptor. J. Biol. Chem. 2758143-8153. [DOI] [PubMed] [Google Scholar]
- 49.Roumanie, O., C. Weinachter, I. Larrieu, M. Crouzet, and F. Doignon. 2001. Functional characterization of the Bag7, Lrg1 and Rgd2 RhoGAP proteins from Saccharomyces cerevisiae. FEBS Lett. 506149-156. [DOI] [PubMed] [Google Scholar]
- 50.Ruggieri, R., K. Tanaka, M. Nakafuku, Y. Kaziro, A. Toh-e, and K. Matsumoto. 1989. MSI1, a negative regulator of the RAS-cAMP pathway in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 868778-8782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmidt, A., T. Beck, A. Koller, J. Kunz, and M. N. Hall. 1998. The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J. 176924-6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmitz, H. P., S. Huppert, A. Lorberg, and J. J. Heinisch. 2002. Rho5p downregulates the yeast cell integrity pathway. J. Cell Sci. 1153139-3148. [DOI] [PubMed] [Google Scholar]
- 53.Simon, M. N., C. De Virgilio, B. Souza, J. R. Pringle, A. Abo, and S. I. Reed. 1995. Role for the Rho-family GTPase Cdc42 in yeast mating-pheromone signal pathway. Nature 376702-705. [DOI] [PubMed] [Google Scholar]
- 54.Singh, K., P. J. Kang, and H. O. Park. 2008. The Rho5 GTPase is necessary for oxidant-induced cell death in budding yeast. Proc. Natl. Acad. Sci. USA 1051522-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith, D. B., and K. S. Johnson. 1988. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 6731-40. [DOI] [PubMed] [Google Scholar]
- 56.Stanbrough, M., and B. Magasanik. 1995. Transcriptional and posttranslational regulation of the general amino acid permease of Saccharomyces cerevisiae. J. Bacteriol. 17794-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Truckses, D. M., J. E. Bloomekatz, and J. Thorner. 2006. The RA domain of Ste50 adaptor protein is required for delivery of Ste11 to the plasma membrane in the filamentous growth signaling pathway of the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 26912-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, H. R., Y. Zhang, B. Ozdamar, A. A. Ogunjimi, E. Alexandrova, G. H. Thomsen, and J. L. Wrana. 2003. Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science 3021775-1779. [DOI] [PubMed] [Google Scholar]
- 59.Wilkins, A., Q. Ping, and C. L. Carpenter. 2004. RhoBTB2 is a substrate of the mammalian Cul3 ubiquitin ligase complex. Genes Dev. 18856-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Willems, A. R., S. Lanker, E. E. Patton, K. L. Craig, T. F. Nason, N. Mathias, R. Kobayashi, C. Wittenberg, and M. Tyers. 1996. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell 86453-463. [DOI] [PubMed] [Google Scholar]
- 61.Won, K. A., and S. I. Reed. 1996. Activation of cyclin E/CDK2 is coupled to site-specific autophosphorylation and ubiquitin-dependent degradation of cyclin E. EMBO J. 154182-4193. [PMC free article] [PubMed] [Google Scholar]
- 62.Wu, C., G. Jansen, J. Zhang, D. Y. Thomas, and M. Whiteway. 2006. Adaptor protein Ste50p links the Ste11p MEKK to the HOG pathway through plasma membrane association. Genes Dev. 20734-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu, C., E. Leberer, D. Y. Thomas, and M. Whiteway. 1999. Functional characterization of the interaction of Ste50p with Ste11p MAPKKK in Saccharomyces cerevisiae. Mol. Biol. Cell 102425-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu, C., M. Whiteway, D. Y. Thomas, and E. Leberer. 1995. Molecular characterization of Ste20p, a potential mitogen-activated protein or extracellular signal-regulated kinase kinase (MEK) kinase kinase from Saccharomyces cerevisiae. J. Biol. Chem. 27015984-15992. [DOI] [PubMed] [Google Scholar]
- 65.Yaglom, J., M. H. Linskens, S. Sadis, D. M. Rubin, B. Futcher, and D. Finley. 1995. p34Cdc28-mediated control of Cln3 cyclin degradation. Mol. Cell. Biol. 15731-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao, Z. S., T. Leung, E. Manser, and L. Lim. 1995. Pheromone signalling in Saccharomyces cerevisiae requires the small GTP-binding protein Cdc42p and its activator CDC24. Mol. Cell. Biol. 155246-5257. [DOI] [PMC free article] [PubMed] [Google Scholar]