Abstract
A pheromone-mediated signaling pathway that couples seven-transmembrane-domain (7-TMD) receptors to a mitogen-activated protein kinase module controls Candida albicans mating. 7-TMD receptors are typically connected to heterotrimeric G proteins whose activation regulates downstream effectors. Two Gα subunits in C. albicans have been identified previously, both of which have been implicated in aspects of pheromone response. Cag1p was found to complement the mating pathway function of the pheromone receptor-coupled Gα subunit in Saccharomyces cerevisiae, and Gpa2p was shown to have a role in the regulation of cyclic AMP signaling in C. albicans and to repress pheromone-mediated arrest. Here, we show that the disruption of CAG1 prevented mating, inactivated pheromone-mediated arrest and morphological changes, and blocked pheromone-mediated gene expression changes in opaque cells of C. albicans and that the overproduction of CAG1 suppressed the hyperactive cell cycle arrest exhibited by sst2 mutant cells. Because the disruption of the STE4 homolog constituting the only C. albicans gene for a heterotrimeric Gβ subunit also blocked mating and pheromone response, it appears that in this fungal pathogen the Gα and Gβ subunits do not act antagonistically but, instead, are both required for the transmission of the mating signal.
Many fungi have well-defined mating systems. Currently, the most thoroughly studied is that of the baker's or brewer's yeast Saccharomyces cerevisiae (2, 12). In this yeast, a signaling pathway has been elucidated that contains cell type-specific receptors of the seven-transmembrane-domain class that are activated by cell type-specific pheromones (7, 46). The pheromone-bound receptor in turn activates a heterotrimeric G protein (17, 43, 61). In contrast to many of the related G-protein-linked receptor signaling pathways identified in mammalian systems that use the activated α subunit to transfer the signal to the next step in the signaling cascade, the yeast pathway uses the βγ subunit as the positive activator of downstream functions (61). The role of the free βγ subunit is to bind the Ste5p scaffold protein (63) and the Ste20p p21-activated kinase (35) and trigger localization to the plasma membrane (50), as well as to direct polarized growth by binding the Far1p scaffold (8). The association of the Ste5p scaffold with the membrane (20, 21) ultimately turns on a mitogen-activated protein (MAP) kinase cascade, and the targets of the MAP kinase include critical elements in the mating response (14, 22, 33, 58).
In the yeast pathway, the Gα subunit serves a role primarily in downregulating the signaling pathway. In its GDP-bound state, it associates with and inactivates the signaling βγ subunit; the absence of Gpa1p leads to constitutive signaling and cell cycle arrest (17, 43) and, thus, to haploid-specific lethality (6). This genetic behavior is consistent with the predicted biochemical G-protein cycle; in the off state, the subunits are associated and inactive, while when activated, α and βγ have a relaxed linkage and free βγ modulates the MAP kinase pathway involved in mating. The overexpression of Gpa1p dampens down the signal (17), and the overproduction of Ste4p increases the signal (13, 48, 62). Thus, the α and βγ subunits play primarily physiologically opposite roles in this signaling process. There is also evidence that the active GTP-bound subunit may act to downregulate signaling directly (41), while other roles proposed for the Gα subunit are to interact with an RNA-binding protein, Scp160 (23), and to regulate intracellular protein trafficking (54).
Subsequently, several other fungal signaling pathways that contain heterotrimeric G-protein modules have been identified. Genome sequences from a variety of fungi suggest that fungal cells typically have two or three α subunits and usually a single βγ pair. An excess of α subunits is also found in higher eukaryotes such as mammals, in which there are multiple α subunits and fewer β or γ subunits (5). In mammals, the α subunits are believed to associate with various combinations of β and γ subunits, leading to extensive combinatorial variation (51). In the fungi, however, the unique βγ element appears to associate with only one of the α subunits, leading to specific α subunits' apparently functioning in signaling pathways in the absence of a classical βγ subunit. For example, in S. cerevisiae, Gpa1p associates with the βγ subunit and functions in the mating pathway, while a second Gα subunit, Gpa2p, which appears to lack a standard βγ subunit (24), functions in the regulation of adenylyl cyclase to control cyclic AMP (cAMP) levels (34, 45). In a second well-studied ascomycete, the fission yeast Schizosaccharomyces pombe, these relationships are reversed; the unique βγ element associates with Gpa2p as part of the regulation of adenylyl cyclase (29), while the Gpa1p subunit acts to control mating but does not have an associated βγ subunit (25).
The fungal pathogen Candida albicans has recently been shown to also have a pheromone-mediated mating pathway active in opaque-form cells. Cells of the MTLα type produce an α-factor pheromone, and the loss of this product causes cell type-specific sterility (4, 38, 49), while cells of the MTLa type express an a-factor gene that is also required for mating (18). Genes with strong sequence similarity to the majority of the elements in S. cerevisiae that make up the intracellular pheromone response signaling pathway have been identified in the genome of C. albicans. Several of these genes have been tested previously and implicated in mating (11, 39). Although the identification of the opaque cell type as the mating-competent form of C. albicans (42) opened the possibility that in these early experiments the loss-of-function mutations blocked the white-opaque switch and not mating itself, the results of recent retesting of strains defective in components of the signaling cascade that had been switched to the mating-competent opaque state confirm the expected sterility of these strains (64).
C. albicans has only two Gα subunits and, similar to other fungi such as S. cerevisiae, S. pombe, and Kluyveromyces lactis, a unique β and a unique γ subunit. It has recently been established in a previous study that the Gpa2 subunit implicated in cAMP signaling is required for proper responsiveness to the mating factor, as gpa2 mutants are hypersensitive to pheromone-mediated arrest (3). The other Gα subunit, encoded by CAG1 (52), is capable of functioning in S. cerevisiae to replace Gpa1p in the pheromone response pathway. This observation led the authors of the study to propose that the apparently asexual C. albicans may have an undiscovered mating capacity; this mating ability was formally established only recently. Here we have investigated whether the CAG1 gene product in fact functions in the C. albicans pheromone response pathway and have tested the role of the unique Gβ subunit in this process as well; independent analysis of the role of the Gβ subunit has recently been provided (64).
MATERIALS AND METHODS
Strains and culture conditions.
The C. albicans strains used in this work are described in Table 1. For general growth and maintenance of the strains in the white phase, the cells were cultured at 30°C in yeast extract-peptone-dextrose medium (1% yeast extract, 2% Bacto peptone, 2% dextrose, and 2% agar for solid medium) supplemented with uridine at 50 μg/ml (YPD). The modified one-step lithium acetate method (10) was used for transformations as described previously (19). The strains were switched to the opaque phase in two rounds of selection on plates with synthetic dextrose (SD; 0.67% yeast nitrogen base, 0.15% amino acid mix with uridine at 100 μg/ml, 2% dextrose, and 1.6% agar for solid medium) containing 0.0005% (wt/vol) phloxine B dye as described previously (19). Cultures in SD medium at 24°C were used to maintain the cells in the opaque phase, and the typical oblong cell morphology phenotype of the cells in the opaque phase was confirmed by microscopy.
TABLE 1.
C. albicans strains used in this work
| Strain | Parent | Mating type | Description | Source or reference |
|---|---|---|---|---|
| Mating type homozygous strains | ||||
| 3294 | CNC43 | a/a | his1/his1 ura3/ura3 arg5,6/arg5,6 | P. T. Magee |
| 3740 | CNC43 | α/α | his1/his1 ura3/ura3 arg5,6/arg5,6 | P. T. Magee |
| 3745 | A505 | a/a | trp1/trp1 lys2/lys2 | 49 |
| 3315 | A505 | α/α | trp1/trp1 lys2/lys2 | P. T. Magee |
| CA29 | 3294 | a/a | sst2::HIS1/sst2::HIS1 ura3/ura3 arg5,6/arg5,6 | 19 |
| Strains generated by deletion and reintegration of STE4 | ||||
| CA22 | 3740 | α/α | STE4/ste4::HIS1 ura3/ura3 arg5,6/arg5,6 | This work |
| CA26 | CA22 | α/α | ste4::HIS1/ste4::URA3 arg5,6/arg5,6 | This work |
| CA32 | CA26 | α/α | ste4::HIS1/ste4::HIS1 ura3/ura3 arg5,6/arg5,6 | This work |
| CA49 | CA32 | α/α | ste4::HIS1/ste4::HIS1 RPS1/rps1::STE4-URA3 arg5,6/arg5,6 | This work |
| CA88 | 3294 | a/a | STE4/ste4::HIS1 ura3/ura3 arg5,6/arg5,6 | This work |
| CA92 | CA88 | a/a | ste4::HIS1/ste4::URA3 arg5,6/arg5,6 | This work |
| CA100 | CA92 | a/a | ste4::HIS1/ste4::HIS1 ura3/ura3 arg5,6/arg5,6 | This work |
| CA104 | CA100 | a/a | ste4::HIS1/ste4::HIS1 RPS1/rps1::STE4-URA3 arg5,6/arg5,6 | This work |
| Strains generated by deletion and reintegration of CAG1 | ||||
| CA110 | 3294 | a/a | CAG1/cag1::HIS1 ura3/ura3 arg5,6/arg5,6 | This work |
| CA111 | 3294 | a/a | CAG1/cag1::HIS1 ura3/ura3 arg5,6/arg5,6 | This work |
| CA114 | CA110 | a/a | cag1::HIS1/cag1::URA3 arg5,6/arg5,6 | This work |
| CA116 | CA111 | a/a | cag1::HIS1/cag1::URA3 arg5,6/arg5,6 | This work |
| CA119 | CA114 | a/a | cag1::HIS1/cag1::HIS1 ura3/ura3 arg5,6/arg5,6 | This work |
| CA120 | CA116 | a/a | cag1::HIS1/cag1::HIS1 ura3/ura3 arg5,6/arg5,6 | This work |
| CA121 | CA119 | a/a | cag1::HIS1/cag1::HIS1 RPS1/rps1::CAG1-URA3 arg5,6/arg5,6 | This work |
| CA122 | CA120 | a/a | cag1::HIS1/cag1::HIS1 RPS1/rps1::CAG1-URA3 arg5,6/arg5,6 | This work |
| CA125 | 3740 | α/α | CAG1/cag1::HIS1 ura3/ura3 arg5,6/arg5,6 | This work |
| CA132 | CA125 | α/α | cag1::HIS1/cag1::URA3 arg5,6/arg5,6 | This work |
| CA137 | CA132 | α/α | cag1::HIS1/cag1::HIS1 ura3/ura3 arg5,6/arg5,6 | This work |
| CA141 | CA137 | α/α | cag1::HIS1/cag1::HIS1 RPS1/rps1::CAG1-URA3 arg5,6/arg5,6 | This work |
| Overexpression strains | ||||
| CA143 | 3294 | a/a | his1/his1 RPS1/rps1::act-STE4-URA3 arg5,6/arg5,6 | This work |
| CA144 | 3294 | a/a | his1/his1 RPS1/rps1::act-CAG1-URA3 arg5,6/arg5,6 | This work |
| CA146 | CA29 | a/a | sst2::HIS1/sst2::HIS1 RPS1/rps1::act-CAG1-URA3 arg5,6/arg5,6 | This work |
| CA147 | CA100 | a/a | ste4::HIS1/ste4::HIS1 RPS1/rps1::act-CAG1-URA3 arg5,6/arg5,6 | This work |
| CA149 | CA120 | a/a | cag1::HIS1/cag1::HIS1 RPS1/rps1::act-STE4-URA3 arg5,6/arg5,6 | This work |
Disruption of STE4 and CAG1.
The C. albicans sequence (assembly 19) from the Candida Genome Database (http://www.candidagenome.org/) was used as the reference for the genomic sequence. The two alleles of the STE4 gene (ORF19.799) and those of the CAG1 gene (ORF19.4015) were deleted from the MTLα strain 3740 and the MTLa strain 3294. All the disruptions were done with the two-step PCR method by the replacement of the first allele with HIS1 and of the second allele with URA3 as described previously (18). The oligonucleotides used in this work for the disruption of the genes and for the confirmation of the deletions are described in Table 2. Oligonucleotides 5′ste4 and 3′ste4 were used to prepare the cassettes for the deletion of the STE4 gene. The strains produced by deleting one allele of the STE4 gene in the MTLα strain 3740 and in the MTLa strain 3294 were named CA22 and CA88, respectively. The correct insertion of the HIS1 cassette at the STE4 locus was confirmed by PCR analysis of genomic DNA from strains CA22 and CA88 with oligonucleotides ste4-F-ex plus H2 and ste4-R-ex plus H1, and no PCR amplification from the wild-type parent strains was observed. Oligonucleotides ste4-F-ex and ste4-R-ex flank, and are external relative to, the recombination sites of the PCR cassettes. Oligonucleotides H1 and H2 are internal relative to the HIS1 gene of the PCR cassette. The second allele of the STE4 gene was deleted from strains CA22 and CA88 to generate the ste4 null strains CA26 (MTLα) and CA92 (MTLa). The correct insertion of the URA3 cassette at the STE4 locus was confirmed by PCR with oligonucleotides ste4-F-ex plus U2 and ste4-R-ex plus U1, and no PCR amplification from the wild-type strains or from the parent strains CA22 and CA88 was observed. Oligonucleotides U1 and U2 are internal relative to the URA3 gene of the cassette. The complete deletion of the STE4 gene from strains CA26 and CA92 was further confirmed by the absence of a PCR amplification product when oligonucleotides STE4-F-in plus STE4-R-in, ste4-F-ex plus STE4-R-in, and STE4-F-in plus ste4-R-ex were used. Oligonucleotides STE4-F-in and STE4-R-in are internal relative to the STE4 gene. The CAG1 gene was deleted using a similar strategy. Oligonucleotides 5′cag1 and 3′cag1 were used to prepare the PCR cassettes. The strains produced by deleting one allele from the parent strain 3294 were named CA110 and CA111, and the strain generated by deleting one allele from the parent strain 3740 was named CA125. The correct insertion of the HIS1 cassette at the CAG1 locus was confirmed by PCR with oligonucleotides cag1-F-ex plus H2 and cag1-R-ex plus H1. Strains CA110, CA111, and CA125 were transformed with the URA3 cassette for the deletion of the second allele to generate, respectively, the cag1 null strains CA114, CA116, and CA132. The correct insertion site of the URA3 cassette was confirmed by PCR with oligonucleotides cag1-F-ex plus U2 and cag1-R-ex plus U1. The complete deletion of the CAG1 gene was also confirmed by the absence of PCR amplification with the oligonucleotides CAG1-F-in plus CAG1-R-in, cag1-F-ex plus CAG1-R-in, and CAG1-F-in plus cag1-R-ex. Oligonucleotides cag1-F-ex and cag1-R-ex are external relative to the disruption site, and oligonucleotides CAG1-F-in and CAG1-R-in are internal relative to the CAG1 gene. Illustrations of the deletions are available at http://candida.bri.nrc.ca/chipdata/danield/cag1-deletion.pdf and http://candida.bri.nrc.ca/chipdata/danield/ste4-deletion.pdf.
TABLE 2.
Oligonucleotides used in this work
| Name | Descriptiona | Sequenceb (5′ to 3′) |
|---|---|---|
| 5′ste4 | 5′ end of PCR cassette for STE4 deletion (100 nt) | TTTGAAAAAAAAAAGAAAAACTTTAACATCCACTAAGTAGTACCCTGTGAGTTCATTCGTGGGATCTGTTTTACAAAAATtatagggcgaattggagctc |
| 3′ste4 | 3′ end of PCR cassette for STE4 deletion (100 nt) | ACATATTAAGGTGTGTGGTTAATGTACTCTTGTGCTTGAGTTTTTTTTTTCTCTACCCTCAGTCTCGCTCTTTTTACTTCgacggtatcgataagcttga |
| ste4-F-ex | STE4 forward external primer | TTACCAAGACTCAATTGTTCCCG |
| ste4-R-ex | STE4 reverse external primer | TATCTACTGTTAAGTAAACTATAACG |
| STE4-F-in | STE4 forward internal primer | ACTATACAACACCTTGCGAGGA |
| STE4-R-in | STE4 reverse internal primer | CAGTTGCCAAAGCTACACCATC |
| ste4-SalI | 5′ end of STE4 for reintegration | CTGATGGTcgACTCGAACATGTATTGTTGTTA |
| ste4-HindIII | 3′ end of STE4 for reintegration | GAATATTAAAAATGATGTAGATAACGG |
| 5′cag1 | 5′ end of PCR cassette for CAG1 deletion (100 nt) | AAAGATATTTTGGGTTTTTTTCTTAATGTACATTAAAATCTGTCTTTTAGTTTACCTTTTTTTAATACCAGTATTCAATCtatagggcgaattggagctc |
| 3′cag1 | 3′ end of PCR cassette for CAG1 deletion (100 nt) | TGACAAATAGATATAAACACAAAAAATTTAAACTGAACATTAATTGTAAAGTAAAAAAAGATATCGCCTACTTCTTGCAAgacggtatcgataagcttga |
| cag1-F-ex | CAG1 forward external primer | TTAACTTTGTATTGAGAGTAGACC |
| cag1-R-ex | CAG1 reverse external primer | AGATATTGTTATTTCTGGAACCGG |
| CAG1-F-in | CAG1 forward internal primer | ATTGAACAAAGTTTACAATTGCGTC |
| CAG1-R-in | CAG1 reverse internal primer | TCATTAGTATCGTCTGGTTTGCC |
| cag1-KpnI | 5′ end of CAG1 for reintegration | ACAATTTGgtACCAAATTCAATAATACAAGAC |
| H1 | HIS1 forward primer | TTTAGTCAATCATTTACCAGACCG |
| H2 | HIS1 reverse primer | TCTATGGCCTTTAACCCAGCTG |
| U1 | URA3 forward primer | TTGAAGGATTAAAACAGGGAGC |
| U2 | URA3 reverse primer | ATACCTTTTACCTTCAATATCTGG |
| a1-f | MTLa-a1 forward primer | TTGAAGCGTGAGAGGCAGGAG |
| a1-r | MTLa-a1 reverse primer | GTTTGGGTTCCTTCTTTCTCATTC |
| alpha-f | MTLα-α1 forward primer | TTCGAGTACATTCTGGTCGCG |
| alpha-r | MTLα-α1 reverse primer | TGTAAACATCCTCAATTGTACCCGA |
| rps1-F-ex | RPS1 forward external primer | AGTGTGTGTGTTCCAAGTCCC |
| rps1-R-ex | RPS1 reverse external primer | AATAGAGAGAAACTATATTATACAC |
| RPS1-R-in | RPS1 reverse internal primer | TTTCTGGTGAATGGGTCAACGAC |
| 561-HindIII | 5′ end of STE4 (for pl390) | AGTTCATTCGTGGGAaagcTTTTACAAAAATATGTCCG |
| 562-SalI | 3′ end of STE4 (for pl390) | CTTGAGTTTTTTTTTTgTCgACCCTCAGTCTCGCTC |
| 563-MluI | 5′ end of CAG1 (for pl391) | ACCTTTTTTTAATACgcGTATTCAATCATGGG |
| 564-NheI | 3′ end of CAG1 (for pl391) | GTAAAGTAAAAAAAGcTAgCGCCTACTTCTTGC |
| 565 | Actin promoter | TTTTCTAATTTTCACTCCTGG |
The terms forward and reverse refer to the orientation of the oligonucleotide relative to the orf coding sequence. The terms external and internal indicate the position relative to the gene disruption site.
Lowercase letters indicate a heterologous sequence in the oligonucleotide: for 5′ste4, 3′ste4, 5′cag1, and 3′cag1, they correspond to the vector sequence of the plasmid containing the HIS1 or the URA3 gene of the PCR cassette used for the disruption, and for the other oligonucleotides, they correspond to point mutations to create restriction sites (underlined) to facilitate cloning.
Reintegration.
A copy of the wild-type gene was reintegrated at the RPS1 locus (RP10) (44) in the Δste4 and Δcag1 strains for complementation experiments. The recipient strains CA26, CA92, CA114, CA116, and CA132 were treated with 5-fluoroorotic acid to recycle the URA3 marker. The resulting uridine-negative strains were named, respectively, CA32, CA100, CA119, CA120, and CA137 (Table 1). For the STE4 gene, a 2.23-kb DNA fragment from genomic DNA was amplified by PCR with Expand high-fidelity polymerase (Roche) using oligonucleotides ste4-SalI and ste4-HindIII. Oligonucleotide ste4-SalI contains an exogenous SalI restriction site, absent from the wild-type sequence, near its 5′ tail, and ste4-HindIII is located approximately 80 nucleotides (nt) from a unique HindIII site in the 3′-end noncoding sequence of the STE4 gene. The PCR fragment was digested with SalI and HindIII enzymes, the resulting 2.15-kb fragment was ligated with vector CIp10 (44) cut with SalI and HindIII, and E. coli strain DH5α was transformed with the construct. The integrity of the clone with respect to the STE4 wild-type sequence was confirmed by DNA sequencing. The selected clone for the wild-type STE4 gene was named plasmid pl383 and was digested with the enzyme StuI for the transformation of the strains CA32 and CA100. The new STE4+ strains were named CA49 and CA104, respectively. A similar strategy and protocol were used for the reintegration of the CAG1 gene. A 1.84-kb fragment was amplified with oligonucleotides cag1-KpnI and cag-R-ex. Oligonucleotide cag1-KpnI was designed with an exogenous KpnI restriction site, absent in the CAG1 gene sequence, and oligonucleotide cag1-R-ex is positioned approximately 160 nt after a unique EcoRV restriction site in the 3′-end noncoding sequence of the CAG1 gene. This PCR fragment was digested with KpnI and EcoRV enzymes, and the 1.67-kb fragment was ligated to the CIp10 vector cleaved with the same two enzymes. The integrity of the clone was also confirmed by DNA sequencing, and the selected clone with the CAG1 gene wild-type sequence was named plasmid pl388. This plasmid was digested with StuI enzyme, and Δcag1 strains CA119, CA120, and CA137 were transformed with the construct. These new CAG1+ strains were named CA121, CA122, and CA141, respectively. The integration of pl383 and pl388 at the correct site in the RPS1 locus was confirmed by PCR (data not shown).
Overexpression.
The STE4 and CAG1 genes were amplified by PCR from genomic DNA of the strain 3294 and cloned into the vector pACT1 (59) under the control of the actin ACT1 gene promoter for ectopic expression experiments. Oligonucleotides 561-HindIII and 562-SalI were used to clone the STE4 gene at the HindIII and SalI sites of the vector to create plasmid pl390. Oligonucleotides 563-MluI and 564-NheI (Table 2) were used to clone the CAG1 gene at the MluI and NheI sites of the vector to create plasmid pl391. The clones were resequenced and selected on the basis of the correct DNA sequences. Plasmids pl390 and pl391 were digested with StuI for transformation and integration at the RPS1 locus. Proper integration of the plasmids in the transformants was confirmed by PCR with oligonucleotides rps1-R-ex, RPS1-R-in, and 565 (data not shown). Strains 3294 and CA120 (Δcag1) were transformed with plasmid pl390 to create, respectively, strains CA143 and CA149. Strains 3294, CA29 (Δsst2), and CA100 (Δste4) were transformed with plasmid pl391 to create, respectively, strains CA144, CA146, and CA147 (Table 1). The strains were switched to the opaque phase for the experiments.
Microscopy.
The MTLa strains were studied by Nomarski microscopy for the formation of unconstricted projections (shmoos) in response to the α-factor pheromone. Liquid cultures from opaque colonies were grown in SD medium for 24 h at 24°C. At time zero, cells from the starter cultures were diluted to an optical density at 600 nm (OD600) of 2.0, and the α-factor pheromone 13-amino-acid synthetic peptide (49) was added in a single dose to the culture at a final concentration of 1 μg/ml. A sample from the cultures was analyzed by microscopy at time zero and after 2, 4, and 6 h of induction. For the responsive strains, such as 3294, shmoos start to be visible after 2 h of induction and are more developed after 4 or 6 h of induction.
Transcription profiling.
Transcription profiling was carried out with DNA microarrays that were obtained from the Biotechnology Research Institute microarray laboratory (http://www.bri.nrc.gc.ca/services/microarray/scanning_e.html). Cells in the opaque phase were cultured in SD medium at 24°C and were harvested at an OD600 of 0.8. For induction with α-factor, the synthetic peptide (49) was added to a final concentration of 1 μg/ml to the culture at an OD600 of 0.6 and cells were harvested 2 h later at an OD600 of about 0.8. Total RNA was extracted using the hot-phenol and glass-bead method (32) and enriched with poly(A)+ mRNA to prepare the cDNA samples as described previously (19). Standard methods were used for DNA microarray hybridization, as described previously (19, 47). Data from three biological samples, or from two biological samples in the case of the reintegrant strains CA104 and CA121 and also the strains CA143 and CA144, used for ectopic overexpression experiments, each with dye swaps, were compiled and analyzed with the GeneSpring software (Agilent Technologies). The transcription data from opaque-phase-specific genes were also used to confirm the integrity of the opaque strains. The complete data set for the 52 microarrays is accessible at http://candida.bri.nrc.ca/chipdata/danield/MicroarrayDataSet.xls either as individual microarrays or as groups organized by conditions (biological replicates).
RESULTS
The CAG1 gene of C. albicans (ORF19.4015), encoding a heterotrimeric G-protein α subunit, was identified many years ago through sequence similarity to the S. cerevisiae GPA1/SCG1 gene (52). This gene is able to functionally complement the loss of the yeast Gα subunit and is regulated properly in S. cerevisiae, suggesting that both protein function and gene regulation are conserved between the two fungi (52). The subsequent identification of a mating type locus (27) and a functional mating process (28, 40) in C. albicans raised the question of whether in fact the CAG1 locus encodes a component of the mating pathway of the pathogen. We disrupted both alleles of CAG1 in strains 3294 and 3740, the first allele with HIS1 and the second with URA3, to create a/a strain CA114 and α/α strain CA132. We then identified opaque derivatives of these cag1 strains on phloxine B plates and tested them for mating capacity.
In baker's yeast, the loss of Gpa1p leads to permanent cell cycle arrest and, thus, the GPA1 gene encodes a haploid-specific essential function (17, 43). However, the loss of Cag1p function did not cause the permanent arrest of mating-competent C. albicans cells; opaque derivatives of the a/a and α/α strains with deletions of the CAG1 gene, identified by phloxine B staining, were readily detected. Intriguingly, when the opaque cag1 mutant strain was tested in a cross-patch mating assay, no prototrophic products derived from mating were detected, suggesting that the cag1 strain was totally sterile (Fig. 1). The results of other phenotypic assays were consistent with the cag1 mutant's being sterile; the a/a cag1 strain CA114 did not generate mating projections in response to α-factor (Fig. 2), and there was no pheromone-induced transcriptional response as measured by microarray analysis (Fig. 3).
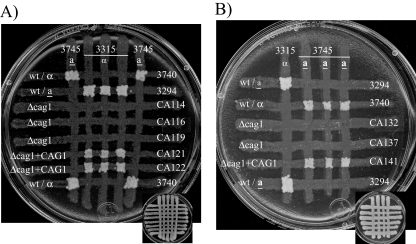
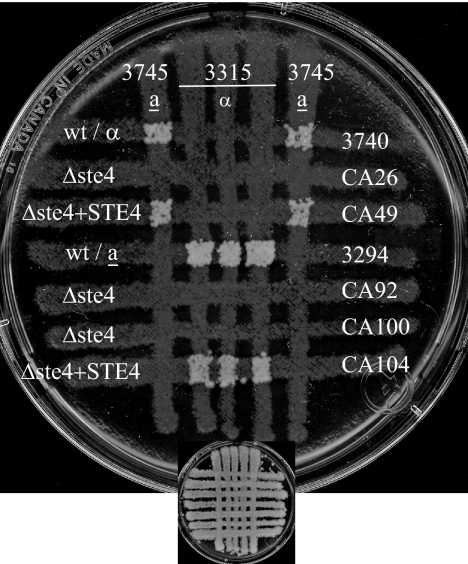
FIG. 1.
Strains with CAG1 deleted are sterile. The mating was assayed by auxotrophic marker complementation with strains of opposite mating types. Cells from opaque colonies were grown at 24°C on a YPD plate and crossed for 24 h before transfer by replica plating onto a plate lacking five amino acids for the selection of the mating products shown here after 5 days of incubation (the YPD template is shown on a reduced scale in the lower part of each panel). (A) Mating assay for MTLa Δcag1 strains and strains with CAG1 reintegrated (Δcag1+CAG1). wt, wild type. (B) Mating assay for MTLα Δcag1 strains and strains with CAG1 reintegrated. No colonies of the Δcag1 strains of both mating types (CA114, CA116, CA119, CA132, and CA137) were detectable, while the strains with CAG1 reintegrated (CA121, CA122, and CA141) reverted the sterile phenotype. Opaque cells from the Δcag1 strains used in the experiment were morphologically similar to the cells of the wild-type parent strain 3294 (data not shown).
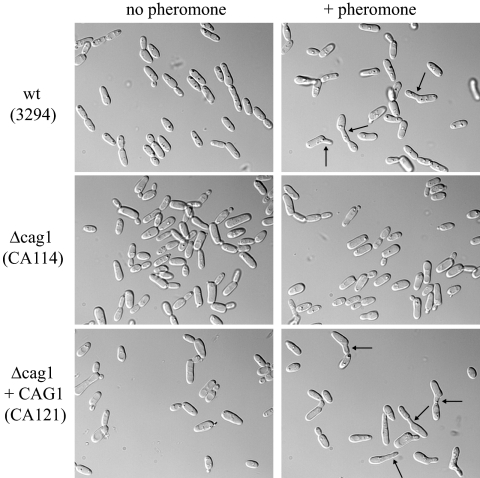
FIG. 2.
Δcag1 cells do not respond to the α-factor pheromone. Opaque cells were cultured in the presence of α-factor (+ pheromone) as described in Materials and Methods. Cells from the MTLa strain 3294 responded to the pheromone with the typical formation of unconstricted projections (shmoos). However, no morphological change was observed for the Δcag1 cells from the strain CA114. Similar results were also obtained for the other Δcag1 strain, CA116 (data not shown). Cells from the strains with CAG1 reintegrated (Δcag1+CAG1; CA121 and CA122) had a response similar to that of the wild-type (wt) parent strain 3294 (data not shown for the CA122 strain). Typical shmoos are highlighted with black arrows. The pictures were taken after 6 h of incubation with the pheromone. Similar results were also obtained after 4 h of incubation with the pheromone (data not shown).
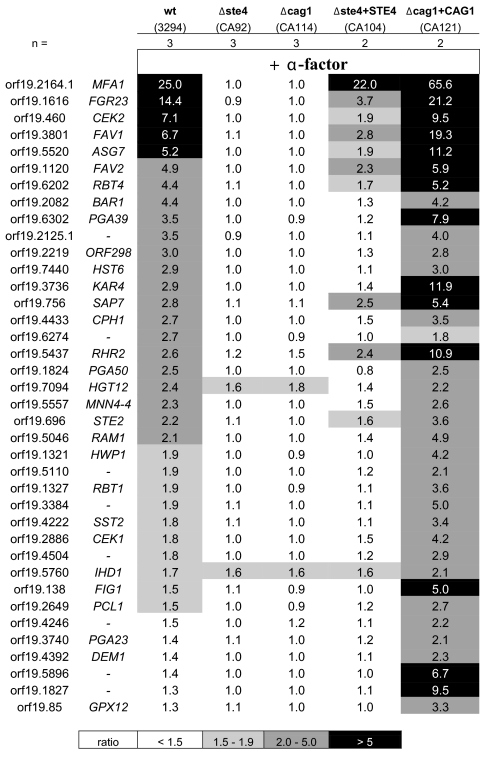
FIG. 3.
No genes in MTLa Δcag1 and Δste4 strains are induced by α-factor. DNA microarrays were used to determine the transcription profiles of the strains in the opaque phase after 2 h of incubation in the presence of the α-factor peptide. This figure presents a comparative list of the genes (no name is given for unannotated genes) with a signal ratio—relative to the signal from uninduced cells—greater than 2 (P < 0.05) under at least one condition. The Δste4 CA92 and the Δcag1 CA114 strains showed no induction of the genes induced in the wild-type (wt) 3294 strain. The induction of these genes by the pheromone was at least partially restored in the strains with reintegrated genes, CA104 and CA121. The number of biological replicates in this experiment is indicated by n. Δste4+STE4, Δste4 strain with STE4 reintegrated; Δcag1+CAG1, Δcag1 strain with CAG1 reintegrated.
The sterile phenotype was a result of the loss of Cag1p function in the mutant strain. The reintroduction of a single copy of the CAG1 gene at the RP10 locus reestablished both mating competence (Fig. 1) and pheromone responsiveness (Fig. 2). Thus, CAG1 functions as a positive component in the pheromone response pathway of C. albicans. Positive functioning of the α subunit of the heterotrimeric G protein contrasts with the situation in S. cerevisiae but is similar to the situation in S. pombe, in which the loss of the G-protein α subunit Gpa1 results in the loss of mating competence (25). However, in S. pombe, the Gpa1 subunit does not work antagonistically with respect to a Gβγ subunit, as the unique βγ element of this yeast is coupled to the Gpa2p α subunit and apparently functions in the glucose-sensing pathway (25). Thus, if the C. albicans signaling pathway followed the S. pombe paradigm, the α subunit would be essential for mating but the βγ element would not function in the mating process. Therefore, we also identified genes encoding homologs of both Gβ and Gγ proteins in C. albicans and disrupted both copies of the Gβ gene ORF19.799 (STE4). Opaque versions were identified on phloxine plates to assess the role of this protein in the mating process. Like the cag1/cag1 null strain, the ste4/ste4 strain was unresponsive to the pheromone in terms of gene induction (Fig. 3), failed to form mating projections (Fig. 4), and was unable to mate (Fig. 5). The reintroduction of a functional STE4 gene complemented all these functions, allowing mating (Fig. 5), projection formation (Fig. 4), and gene induction (Fig. 3); these data confirm and extend the recent independent characterization of this STE4 homolog (64). Thus, the function of the heterotrimeric G protein in C. albicans mating is distinct from that in either the S. cerevisiae or the S. pombe paradigm; in C. albicans, both the α and β subunits are required for mating.
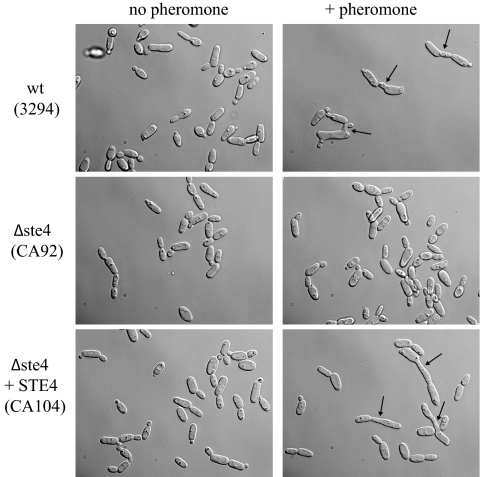
FIG. 4.
Δste4 cells do not respond to the α-factor pheromone. Opaque cells were cultured in the presence of α-factor (+ pheromone) as described in Materials and Methods. No projections (shmoos) from the MTLa Δste4 strain (CA92) were detected. Shmoo formation was restored in the strain with STE4 reintegrated (Δste4+STE4; CA104). Pictures were taken after 6 h of incubation, and black arrows highlight typical shmoos.
FIG. 5.
Δste4 strains are sterile. The mating assay was done as described for the Δcag1 strains in the legend to Fig. 1. No prototrophic colonies from the Δste4 strains of both mating types (CA26, CA92, and CA100) were detected after 5 days incubation. The reintegration of a copy of the wild-type STE4 gene (Δste4+STE4; strains CA49 and CA104) resulted in the reversion of the sterile phenotype. wt, wild type.
Although the reintegration of the CAG1 gene and the STE4 gene permitted the respective cag1 null and ste4 null strains to mate, to respond morphologically, and to undergo transcriptional activation by the pheromone, the two reintegrants were not quantitatively identical. The STE4 reintegrant showed reduced gene expression responsiveness relative to that of the wild-type strain, while the CAG1 reintegrant showed somewhat enhanced responsiveness. These subtle transcriptional differences were potentially caused by the differential expression of these elements, as the reintegrated CAG1 gene was expressed at a higher-than-wild-type level while the reintegrated STE4 gene was expressed at a lower-than-wild-type level, as reflected by the microarray data.
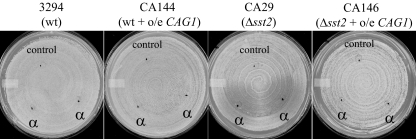
In addition to characterizing the CAG1 and STE4 null mutants, we overexpressed CAG1 and STE4 under the control of the strong ACT1 promoter (59). When introduced into otherwise wild-type cells, the ACT1 promoter permitted 15-fold overexpression of STE4 and 7-fold overexpression of CAG1, as measured from the microarray data. These enhanced expression levels did not result in any constitutive expression of pheromone-responsive genes or lead to increased shmoo formation (data not shown) or cell cycle arrest (Fig. 6) in the presence of the pheromone. As well, the overproduction of STE4 did not suppress the sterility of a cag1 mutant, while the overexpression of CAG1 was similarly ineffective in permitting an ste4 mutant to mate. However, CAG1 overexpression was able to suppress the hyperresponsiveness of an sst2 mutant to pheromone-mediated arrest; the distinct halos that were generated in response to α-factor treatment in lawn assays of the sst2 mutant were eliminated by the introduction of the ACT1-driven CAG1 construct (Fig. 6).
FIG. 6.
Halo assay of MTLa strains overexpressing CAG1. Cells in the opaque phase were spread onto an SD plate. The black dots indicate the positions where 5 μl of either the α-factor synthetic peptide (1 μg/μl; α) or the solvent, as a negative control (control), was spotted. The plates are shown after 2 days of incubation at 24°C. Halos delineate the zones of growth arrest induced by α-factor. The overexpression of the CAG1 gene (o/e CAG1) suppresses the sensitivity of the Δsst2 strain to the pheromone. wt, wild type.
DISCUSSION
C. albicans has two genes encoding homologs of Gα subunits (ORF19.4015 and ORF19.1621) and a single gene each for a Gβ homolog (ORF19.799) and a Gγ subunit (ORF19.6551.1). Gpa2p (Orf19.1621p) has been implicated previously in the regulation of cAMP signaling and in sensitivity to pheromones, perhaps indirectly through its role in signaling nutrient status (3). The expression patterns of CAG1 (ORF19.4015), STE4 (ORF19.799), and STE18 (ORF19.6551.1) are consistent with these elements' working directly in the pheromone response pathway (55), because their transcription is limited to cells that are homozygous at the MTL locus and thus do not express the a1-α2 repressor (42). In contrast, the expression of GPA2 is not influenced by the mating competence of the cell.
Recently, the STE4 product has been shown to be required for pheromone-mediated mating in C. albicans (64). Intriguingly, we have found that CAG1 is also completely essential for pheromone response and mating. Thus it appears that, like S. cerevisiae, C. albicans uses a heterotrimeric G protein to control the pheromone response pathway that is necessary for mating. However, because in C. albicans the loss of either Gα or Gβ causes total sterility, it is clear that the function of the G protein in the pathogen is not identical to that in S. cerevisiae. While many of the components of the MAP kinase cascade that is the target of the yeast mating-coupled G protein are found in C. albicans, a close homolog of the gene for the critical Ste5p scaffold is not evident in the C. albicans genome. This is significant because in yeast a key link between the G protein and the activation of the MAP kinase cascade results from the association of the free βγ subunit and Ste5p (50, 63). In C. albicans, the Far1p protein, which has limited structural similarity to the Ste5p scaffold, has been found to be necessary for all aspects of pheromone response, including the activation of gene expression in response to the mating factor (16). This finding contrasts with the yeast paradigm, in which Far1p is required only for cell cycle arrest and morphological changes in response to pheromone treatment and is not involved in transcriptional activation due to mating factor stimulation (9). Therefore, the linkages between the G protein and the MAP kinase cascade in C. albicans and S. cerevisiae must be different. It is possible that the yeast Ste5p and Far1p represent copies of a single gene that diverged dramatically after the whole-genome duplication and that Far1p of C. albicans represents the ancestral gene product. However, given the complexities of activities and associations for both Far1p and Ste5p, it is difficult to imagine a single protein fulfilling all the associated functions.
In other fungi, there are also functions for heterotrimeric G proteins in mating processes, but these functions have been found previously to have additional complexity relative to the roles identified in the budding and fission yeasts (37). For example, in Cryptococcus neoformans, there are three genes encoding Gα subunits, with a single gene for Gβ and two for Gγ. The loss of the unique Gβ subunit Gpb1p (60) or one of the Gγ subunits (26, 36) blocks mating, suggesting that the βγ subunit acts as a positive regulator of the process. Among the Gα subunits, Gpa1 is implicated in cAMP signaling (1), while Gpa2p and Gpa3p are involved in the response to pheromones. However, this involvement is multifaceted, as the loss of either subunit does not block mating but the loss of both subunits creates a bilateral mating defect and leads to the constitutive expression of mating-factor-induced genes (26, 36). Analysis of putative hyperactive alleles suggests that Gpa2p functions positively in response to pheromones and that Gpa3p acts negatively (26); this assessment is supported by the observation that the removal of Gpa3p allows the constitutive activation of the pheromone response genes. Therefore, the C. neoformans situation is different from that of C. albicans, as although both Gα and Gβ elements in C. neoformans can function positively in the mating process, it is only the loss of the βγ subunit that creates complete sterility.
K. lactis, which is evolutionarily intermediate between C. albicans and S. cerevisiae in the ascomycete lineage, also needs both Gα and Gβ for efficient mating. This fungus has two α subunits and a single copy each of β and γ subunit-encoding genes. The Gpa1p subunit is implicated in mating (53), while Gpa2p is involved in cAMP regulation (15). In this organism, which like S. cerevisiae has an Ste5p homolog required for pheromone response (30), the absence of Gα dramatically reduces, but does not eliminate, mating while the Gβ mutant is totally sterile (31). Surprisingly, however, the deletion of the putative γ subunit has been reported previously not to compromise mating (15); this distinction between the β and γ subunit deletion phenotypes is inconsistent with the generic G-protein model and will require confirmation, but it could be explained if the Gβ subunit has an independent membrane-targeting capacity.
Because the phenotypic consequences of the loss of the Gα subunit in C. albicans mating are distinct from those identified for the subunit in other fungi, it is possible that the molecular roles in the mating process are different. The loss of Gpa1p in S. cerevisiae and Gpa3p in C. neoformans leads to the constitutive activation of pheromone-induced gene expression. However, no induction of the mating pathway genes in the cag1 mutant of C. albicans is observed, and thus, Cag1p does not appear to be repressing a βγ signaling module. Also, in yeast the overproduction of the Gβ subunit leads to the constitutive activation of the pheromone signaling pathway (13, 48, 62), while in C. albicans the overproduction of either the α or the β subunit does not lead to the constitutive activation of even a subset of the pheromone-responsive genes.
In general, the loss of Gα function in the other fungi appears to result in a lack of pheromone-directed mating polarity and, thus, to cause less extreme defects than those generated by the loss of βγ signaling. In yeast, bilateral mating defects are often associated with polarity defects, and S. cerevisiae gpa1 mutants are abnormal in polarized growth since they are responding to a nonlocalized, internally generated signal (17, 43). Recent evidence suggests that a constitutively activated MAP kinase cascade does not generate proper polarity signals in yeast in the absence of Gpa1p (56). The bilateral mating defect of the gpa2 gpa3 mutant of C. neoformans may result from the failure to polarize properly, and the reduction to 5% of the wild-type level of mating in the Gα mutant of K. lactis is also consistent with a polarity defect. However, in C. albicans, the α and β subunit defects generate identical sterile phenotypes and, thus, the loss of Cag1p appears to affect more than just mating-factor-directed polarity.
Overall, heterotrimeric G proteins couple seven-transmembrane-domain receptors to a wide variety of effector pathways in eukaryotic cells, and these systems transmit an amazing diversity of signals. Our understanding of these processes depends on comprehending the way the effectors are controlled in response to the activation of the G protein. These activations follow many patterns; some signaling processes are dependent on the Gα or the Gβγ subunit, but other pathways involve both the subunits together. For example, specific isoforms of the mammalian adenylyl cyclase are activated by both the α and βγ subunits. In isoforms AC2, AC4, and AC7, the βγ subunits synergize with the Gs α subunit to stimulate adenylyl cyclase activity (57). The fungal pheromone-dependent mating systems are a diverse collection of G-protein modules directed at a common process but exhibiting significant variety at the functional level. Because of the ability to manipulate these systems at a molecular level, the fungal mating pathways provide the opportunity to dissect the logic of using specifically the α, the βγ, or various combinations of G-protein subunits to transmit the mating response signal to a downstream kinase cascade.
Acknowledgments
This work was supported in part by CIHR grant MOP-42516 and is National Research Council Canada publication number 49557.
Footnotes
Published ahead of print on 25 July 2008.
REFERENCES
- 1.Alspaugh, J. A., J. R. Perfect, and J. Heitman. 1997. Cryptococcus neoformans mating and virulence are regulated by the G-protein alpha subunit GPA1 and cAMP. Genes Dev. 113206-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bardwell, L. 2004. A walk-through of the yeast mating pheromone response pathway. Peptides 251465-1476. [DOI] [PubMed] [Google Scholar]
- 3.Bennett, R. J., and A. D. Johnson. 2006. The role of nutrient regulation and the Gpa2 protein in the mating pheromone response of C. albicans. Mol. Microbiol. 62100-119. [DOI] [PubMed] [Google Scholar]
- 4.Bennett, R. J., M. A. Uhl, M. G. Miller, and A. D. Johnson. 2003. Identification and characterization of a Candida albicans mating pheromone. Mol. Cell. Biol. 238189-8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birnbaumer, L. 2007. Expansion of signal transduction by G proteins. The second 15 years or so: from 3 to 16 alpha subunits plus βγ dimers. Biochim. Biophys. Acta 1768772-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blinder, D., S. Bouvier, and D. D. Jenness. 1989. Constitutive mutants in the yeast pheromone response: ordered function of the gene products. Cell 56479-486. [DOI] [PubMed] [Google Scholar]
- 7.Burkholder, A. C., and L. H. Hartwell. 1985. The yeast alpha-factor receptor: structural properties deduced from the sequence of the STE2 gene. Nucleic Acids Res. 138463-8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butty, A. C., P. M. Pryciak, L. S. Huang, I. Herskowitz, and M. Peter. 1998. The role of Far1p in linking the heterotrimeric G protein to polarity establishment proteins during yeast mating. Science 2821511-1516. [DOI] [PubMed] [Google Scholar]
- 9.Chang, F., and I. Herskowitz. 1990. Identification of a gene necessary for cell cycle arrest by a negative growth factor of yeast: FAR1 is an inhibitor of a G1 cyclin, CLN2. Cell 63999-1011. [DOI] [PubMed] [Google Scholar]
- 10.Chen, D. C., B. C. Yang, and T. T. Kuo. 1992. One-step transformation of yeast in stationary phase. Curr. Genet. 2183-84. [DOI] [PubMed] [Google Scholar]
- 11.Chen, J., J. Chen, S. Lane, and H. Liu. 2002. A conserved mitogen-activated protein kinase pathway is required for mating in Candida albicans. Mol. Microbiol. 461335-1344. [DOI] [PubMed] [Google Scholar]
- 12.Chen, R. E., and J. Thorner. 2007. Function and regulation in MAPK signaling pathways: lessons learned from the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 17731311-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole, G. M., D. E. Stone, and S. I. Reed. 1990. Stoichiometry of G protein subunits affects the Saccharomyces cerevisiae mating pheromone signal transduction pathway. Mol. Cell. Biol. 10510-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cook, J. G., L. Bardwell, S. J. Kron, and J. Thorner. 1996. Two novel targets of the MAP kinase Kss1 are negative regulators of invasive growth in the yeast Saccharomyces cerevisiae. Genes Dev. 102831-2848. [DOI] [PubMed] [Google Scholar]
- 15.Coria, R., L. Kawasaki, F. Torres-Quiroz, L. Ongay-Larios, E. Sanchez-Paredes, N. Velazquez-Zavala, R. Navarro-Olmos, M. Rodriguez-Gonzalez, R. Aguilar-Corachan, and G. Coello. 2006. The pheromone response pathway of Kluyveromyces lactis. FEMS Yeast Res. 6336-344. [DOI] [PubMed] [Google Scholar]
- 16.Cote, P., and M. Whiteway. 2008. The role of Candida albicans FAR1 in regulation of pheromone-mediated mating, gene expression and cell cycle arrest. Mol. Microbiol. 68392-404. [DOI] [PubMed] [Google Scholar]
- 17.Dietzel, C., and J. Kurjan. 1987. The yeast SCG1 gene: a G alpha-like protein implicated in the a- and alpha-factor response pathway. Cell 501001-1010. [DOI] [PubMed] [Google Scholar]
- 18.Dignard, D., A. L. El-Naggar, M. E. Logue, G. Butler, and M. Whiteway. 2007. Identification and characterization of MFA1, the gene encoding Candida albicans a-factor pheromone. Eukaryot. Cell 6487-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dignard, D., and M. Whiteway. 2006. SST2, a regulator of G-protein signaling for the Candida albicans mating response pathway. Eukaryot. Cell 5192-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elion, E. A. 1995. Ste5: a meeting place for MAP kinases and their associates. Trends Cell Biol. 5322-327. [DOI] [PubMed] [Google Scholar]
- 21.Elion, E. A. 2001. The Ste5p scaffold. J. Cell Sci. 1143967-3978. [DOI] [PubMed] [Google Scholar]
- 22.Elion, E. A., B. Satterberg, and J. E. Kranz. 1993. FUS3 phosphorylates multiple components of the mating signal transduction cascade: evidence for STE12 and FAR1. Mol. Biol. Cell 4495-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo, M., C. Aston, S. A. Burchett, C. Dyke, S. Fields, S. J. Rajarao, P. Uetz, Y. Wang, K. Young, and H. G. Dohlman. 2003. The yeast G protein α subunit Gpa1 transmits a signal through an RNA binding effector protein Scp160. Mol. Cell 12517-524. [DOI] [PubMed] [Google Scholar]
- 24.Harashima, T., and J. Heitman. 2002. The Gα protein Gpa2 controls yeast differentiation by interacting with kelch repeat proteins that mimic Gβ subunits. Mol. Cell 10163-173. [DOI] [PubMed] [Google Scholar]
- 25.Hoffman, C. S. 2005. Except in every detail: comparing and contrasting G-protein signaling in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Eukaryot. Cell 4495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsueh, Y. P., C. Xue, and J. Heitman. 2007. G protein signaling governing cell fate decisions involves opposing Gα subunits in Cryptococcus neoformans. Mol. Biol. Cell 183237-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hull, C. M., and A. D. Johnson. 1999. Identification of a mating type-like locus in the asexual pathogenic yeast Candida albicans. Science 2851271-1275. [DOI] [PubMed] [Google Scholar]
- 28.Hull, C. M., R. M. Raisner, and A. D. Johnson. 2000. Evidence for mating of the “asexual” yeast Candida albicans in a mammalian host. Science 289307-310. [DOI] [PubMed] [Google Scholar]
- 29.Ivey, F. D., and C. S. Hoffman. 2005. Direct activation of fission yeast adenylate cyclase by the Gpa2 Gα of the glucose signaling pathway. Proc. Natl. Acad. Sci. USA 1026108-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawasaki, L., M. Castaneda-Bueno, E. Sanchez-Paredes, N. Velazquez-Zavala, F. Torres-Quiroz, L. Ongay-Larios, and R. Coria. 2008. Protein kinases involved in mating and osmotic stress in the yeast Kluyveromyces lactis. Eukaryot. Cell 778-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawasaki, L., A. L. Savinon-Tejeda, L. Ongay-Larios, J. Ramirez, and R. Coria. 2005. The Gbeta(KlSte4p) subunit of the heterotrimeric G protein has a positive and essential role in the induction of mating in the yeast Kluyveromyces lactis. Yeast 22947-956. [DOI] [PubMed] [Google Scholar]
- 32.Kohrer, K., and H. Domdey. 1991. Preparation of high molecular weight RNA. Methods Enzymol. 194398-405. [DOI] [PubMed] [Google Scholar]
- 33.Kranz, J. E., B. Satterberg, and E. A. Elion. 1994. The MAP kinase Fus3 associates with and phosphorylates the upstream signaling component Ste5. Genes Dev. 8313-327. [DOI] [PubMed] [Google Scholar]
- 34.Kubler, E., H. U. Mosch, S. Rupp, and M. P. Lisanti. 1997. Gpa2p, a G-protein alpha-subunit, regulates growth and pseudohyphal development in Saccharomyces cerevisiae via a cAMP-dependent mechanism. J. Biol. Chem. 27220321-20323. [DOI] [PubMed] [Google Scholar]
- 35.Leeuw, T., C. Wu, J. D. Schrag, M. Whiteway, D. Y. Thomas, and E. Leberer. 1998. Interaction of a G-protein beta-subunit with a conserved sequence in Ste20/PAK family protein kinases. Nature 391191-195. [DOI] [PubMed] [Google Scholar]
- 36.Li, L., G. Shen, Z. G. Zhang, Y. L. Wang, J. K. Thompson, and P. Wang. 2007. Canonical heterotrimeric G proteins regulating mating and virulence of Cryptococcus neoformans. Mol. Biol. Cell 184201-4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li, L., S. J. Wright, S. Krystofova, G. Park, and K. A. Borkovich. 2007. Heterotrimeric G protein signaling in filamentous fungi. Annu. Rev. Microbiol. 61423-452. [DOI] [PubMed] [Google Scholar]
- 38.Lockhart, S. R., R. Zhao, K. J. Daniels, and D. R. Soll. 2003. Alpha-pheromone-induced “shmooing” and gene regulation require white-opaque switching during Candida albicans mating. Eukaryot. Cell 2847-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Magee, B. B., M. Legrand, A. M. Alarco, M. Raymond, and P. T. Magee. 2002. Many of the genes required for mating in Saccharomyces cerevisiae are also required for mating in Candida albicans. Mol. Microbiol. 461345-1351. [DOI] [PubMed] [Google Scholar]
- 40.Magee, B. B., and P. T. Magee. 2000. Induction of mating in Candida albicans by construction of MTLa and MTLα strains. Science 289310-313. [DOI] [PubMed] [Google Scholar]
- 41.Metodiev, M. V., D. Matheos, M. D. Rose, and D. E. Stone. 2002. Regulation of MAPK function by direct interaction with the mating-specific Gα in yeast. Science 2961483-1486. [DOI] [PubMed] [Google Scholar]
- 42.Miller, M. G., and A. D. Johnson. 2002. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell 110293-302. [DOI] [PubMed] [Google Scholar]
- 43.Miyajima, I., M. Nakafuku, N. Nakayama, C. Brenner, A. Miyajima, K. Kaibuchi, K. Arai, Y. Kaziro, and K. Matsumoto. 1987. GPA1, a haploid-specific essential gene, encodes a yeast homolog of mammalian G protein which may be involved in mating factor signal transduction. Cell 501011-1019. [DOI] [PubMed] [Google Scholar]
- 44.Murad, A. M., P. R. Lee, I. D. Broadbent, C. J. Barelle, and A. J. Brown. 2000. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast 16325-327. [DOI] [PubMed] [Google Scholar]
- 45.Nakafuku, M., T. Obara, K. Kaibuchi, I. Miyajima, A. Miyajima, H. Itoh, S. Nakamura, K. Arai, K. Matsumoto, and Y. Kaziro. 1988. Isolation of a second yeast Saccharomyces cerevisiae gene (GPA2) coding for guanine nucleotide-binding regulatory protein: studies on its structure and possible functions. Proc. Natl. Acad. Sci. USA 851374-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakayama, N., A. Miyajima, and K. Arai. 1985. Nucleotide sequences of STE2 and STE3, cell type-specific sterile genes from Saccharomyces cerevisiae. EMBO J. 42643-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nantel, A., D. Dignard, C. Bachewich, D. Harcus, A. Marcil, A. P. Bouin, C. W. Sensen, H. Hogues, M. van het Hoog, P. Gordon, T. Rigby, F. Benoit, D. C. Tessier, D. Y. Thomas, and M. Whiteway. 2002. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol. Biol. Cell 133452-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nomoto, S., N. Nakayama, K. Arai, and K. Matsumoto. 1990. Regulation of the yeast pheromone response pathway by G protein subunits. EMBO J. 9691-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Panwar, S. L., M. Legrand, D. Dignard, M. Whiteway, and P. T. Magee. 2003. MFα1, the gene encoding the α mating pheromone of Candida albicans. Eukaryot. Cell 21350-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pryciak, P. M., and F. A. Huntress. 1998. Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gβγ complex underlies activation of the yeast pheromone response pathway. Genes Dev. 122684-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robishaw, J. D., and C. H. Berlot. 2004. Translating G protein subunit diversity into functional specificity. Curr. Opin. Cell Biol. 16206-209. [DOI] [PubMed] [Google Scholar]
- 52.Sadhu, C., D. Hoekstra, M. J. McEachern, S. I. Reed, and J. B. Hicks. 1992. A G-protein α subunit from asexual Candida albicans functions in the mating signal transduction pathway of Saccharomyces cerevisiae and is regulated by the a1-α2 repressor. Mol. Cell. Biol. 121977-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Savinon-Tejeda, A. L., L. Ongay-Larios, J. Valdes-Rodriguez, and R. Coria. 2001. The KlGpa1 gene encodes a G-protein α subunit that is a positive control element in the mating pathway of the budding yeast Kluyveromyces lactis. J. Bacteriol. 183229-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Slessareva, J. E., S. M. Routt, B. Temple, V. A. Bankaitis, and H. G. Dohlman. 2006. Activation of the phosphatidylinositol 3-kinase Vps34 by a G protein alpha subunit at the endosome. Cell 126191-203. [DOI] [PubMed] [Google Scholar]
- 55.Srikantha, T., A. R. Borneman, K. J. Daniels, C. Pujol, W. Wu, M. R. Seringhaus, M. Gerstein, S. Yi, M. Snyder, and D. R. Soll. 2006. TOS9 regulates white-opaque switching in Candida albicans. Eukaryot. Cell 51674-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Strickfaden, S. C., and P. M. Pryciak. 2008. Distinct roles for two Gα-Gβ interfaces in cell polarity control by a yeast heterotrimeric G protein. Mol. Biol. Cell 19181-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sunahara, R. K., and R. Taussig. 2002. Isoforms of mammalian adenylyl cyclase: multiplicities of signaling. Mol. Interv. 2168-184. [DOI] [PubMed] [Google Scholar]
- 58.Tedford, K., S. Kim, D. Sa, K. Stevens, and M. Tyers. 1997. Regulation of the mating pheromone and invasive growth responses in yeast by two MAP kinase substrates. Curr. Biol. 7228-238. [DOI] [PubMed] [Google Scholar]
- 59.Tripathi, G., C. Wiltshire, S. Macaskill, H. Tournu, S. Budge, and A. J. Brown. 2002. Gcn4 co-ordinates morphogenetic and metabolic responses to amino acid starvation in Candida albicans. EMBO J. 215448-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang, P., J. R. Perfect, and J. Heitman. 2000. The G-protein beta subunit GPB1 is required for mating and haploid fruiting in Cryptococcus neoformans. Mol. Cell. Biol. 20352-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Whiteway, M., L. Hougan, D. Dignard, D. Y. Thomas, L. Bell, G. C. Saari, F. J. Grant, P. O'Hara, and V. L. MacKay. 1989. The STE4 and STE18 genes of yeast encode potential beta and gamma subunits of the mating factor receptor-coupled G protein. Cell 56467-477. [DOI] [PubMed] [Google Scholar]
- 62.Whiteway, M., L. Hougan, and D. Y. Thomas. 1990. Overexpression of the STE4 gene leads to mating response in haploid Saccharomyces cerevisiae. Mol. Cell. Biol. 10217-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Whiteway, M. S., C. Wu, T. Leeuw, K. Clark, A. Fourest-Lieuvin, D. Y. Thomas, and E. Leberer. 1995. Association of the yeast pheromone response G protein beta gamma subunits with the MAP kinase scaffold Ste5p. Science 2691572-1575. [DOI] [PubMed] [Google Scholar]
- 64.Yi, S., N. Sahni, K. J. Daniels, C. Pujol, T. Srikantha, and D. R. Soll. 2008. The same receptor, G-protein and MAP kinase pathway activate different downstream regulators in the alternative white and opaque pheromone responses of Candida albicans. Mol. Biol. Cell 19957-970. [DOI] [PMC free article] [PubMed] [Google Scholar]