Figure 1.
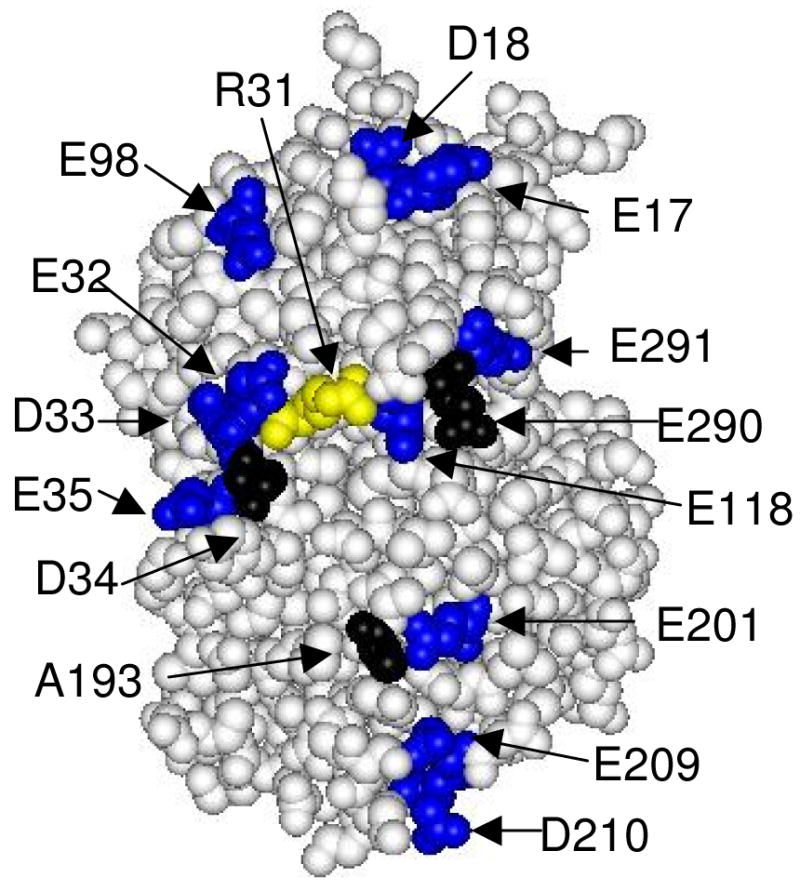
Space-filling model of the front face of CcP showing the location of residues that define the cytochrome c-binding site (black) and the location of residues mutated in this study (blue and yellow). Asp-34, Ala-193, and Glu-290 (black) define the yeast iso-1 cytochrome c binding site. The aspartate and glutamate residues on the front face of CcP, excluding Asp-34 and Glu-290, are shown in blue. The thirteen aspartate and glutamate residues shown in blue were individually mutated to lysine residues in this study. Arg-31 is shown in yellow and was mutated to a glutamate residue. The one-letter abbreviations and sequence numbers are used to identify the amino acid residues. Data from reference (9); Protein Data Bank (PDB) ID: 2PCC.
