Abstract
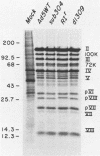
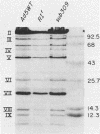
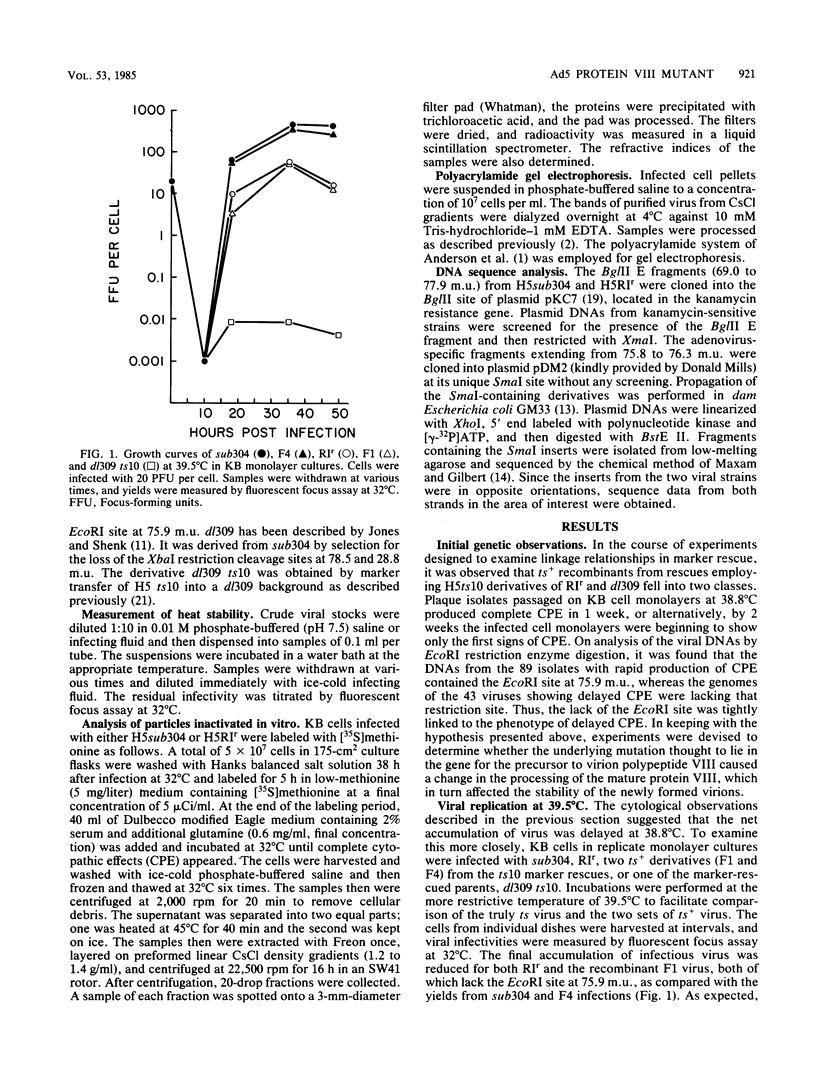
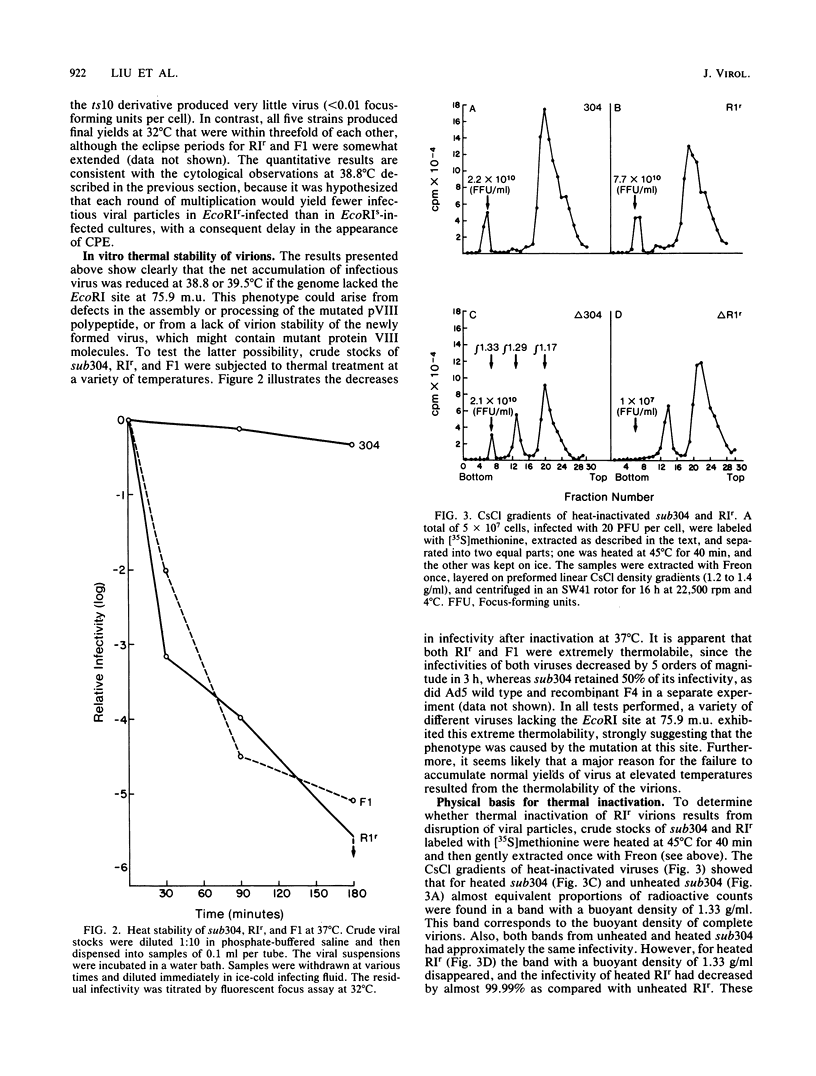
The mutant adenoviruses H5sub304 and H5RIr were isolated sequentially from adenovirus 5 wild type by selection for the loss of EcoRI restriction endonuclease sites by Jones and Shenk (Cell 13:181-188, 1978). sub304 lacks the site at 84.0 map units (m.u.), and RIr lacks both that and the site at 75.9 m.u. A set of derivatives of RIr that lack the site at 75.9 m.u. accumulated virus more slowly at 38.8 or 39.5 degrees C than those with the site present, as measured by low-multiplicity passage or single-step replication cycles, respectively. Since the EcoRI site at 75.9 m.u. is predicted to lie in the gene encoding the precursor to virion polypeptide VIII (pVIII), the failure to accumulate virus rapidly could lie either in some step in processing and assembly of virions or in an increased virion thermolability. The latter possibility was shown to be the case, as all strains mutated at the EcoRI 75.9 m.u. site were extremely thermolabile in vitro, even at 37 degrees C. CsCl equilibrium density centrifugation of heated crude stocks of RIr and sub304 demonstrated that loss of infectivity in RIr was accompanied by physical disruption of virions. Polyacrylamide gel electrophoresis of infected cell extracts or of purified virions showed that pVIII of RIr had an apparent molecular weight that was slightly greater than that of sub304, and mature RIr and sub304 virions displayed polypeptide VIIIs which appeared to be of identical molecular weights. Nucleotide sequence analysis of RIr demonstrated that it contained a 9-base-pair (bp) substitution for 6 bp found in sub304, leading to a loss of the EcoRI site and a predicted insertion of a single amino acid. Comparison of the sequence of sub304 with the published sequence of adenovirus 2 revealed two changes, a single transversion at bp 1,722 and a bp deletion at 1,749, leading to the loss of a TaqI site. The predicted reading frame change would lead to a stop codon at bp 1,885. This raises the question of whether adenovirus 2 and adenovirus 5 use the same reading fame for pVIII.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson C. W., Baum P. R., Gesteland R. F. Processing of adenovirus 2-induced proteins. J Virol. 1973 Aug;12(2):241–252. doi: 10.1128/jvi.12.2.241-252.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee-Sheung C. C., Ginsberg H. S. Characterization of a temperature-sensitive fiber mutant of type 5 adenovirus and effect of the mutation on virion assembly. J Virol. 1982 Jun;42(3):932–950. doi: 10.1128/jvi.42.3.932-950.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby W. W., Shenk T. Adenovirus type 5 virions can be assembled in vivo in the absence of detectable polypeptide IX. J Virol. 1981 Sep;39(3):977–980. doi: 10.1128/jvi.39.3.977-980.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensinger M. J., Ginsberg H. S. Selection and preliminary characterization of temperature-sensitive mutants of type 5 adenovirus. J Virol. 1972 Sep;10(3):328–339. doi: 10.1128/jvi.10.3.328-339.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everitt E., Lutter L., Philipson L. Structural proteins of adenoviruses. XII. Location and neighbor relationship among proteins of adenovirion type 2 as revealed by enzymatic iodination, immunoprecipitation and chemical cross-linking. Virology. 1975 Sep;67(1):197–208. doi: 10.1016/0042-6822(75)90417-1. [DOI] [PubMed] [Google Scholar]
- Galibert F., Hérissé J., Courtois G. Nucleotide sequence of the EcoRI-F fragment of adenovirus 2 genome. Gene. 1979 May;6(1):1–22. doi: 10.1016/0378-1119(79)90081-7. [DOI] [PubMed] [Google Scholar]
- Gingeras T. R., Sciaky D., Gelinas R. E., Bing-Dong J., Yen C. E., Kelly M. M., Bullock P. A., Parsons B. L., O'Neill K. E., Roberts R. J. Nucleotide sequences from the adenovirus-2 genome. J Biol Chem. 1982 Nov 25;257(22):13475–13491. [PubMed] [Google Scholar]
- Hérissé J., Courtois G., Galibert F. Nucleotide sequence of the EcoRI D fragment of adenovirus 2 genome. Nucleic Acids Res. 1980 May 24;8(10):2173–2192. doi: 10.1093/nar/8.10.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi M., Maizel J. V., Jr The polypeptides of adenovirus. V. Young virions, structural intermediate between top components and aged virions. Virology. 1974 Feb;57(2):409–424. doi: 10.1016/0042-6822(74)90181-0. [DOI] [PubMed] [Google Scholar]
- Jones N., Shenk T. Isolation of adenovirus type 5 host range deletion mutants defective for transformation of rat embryo cells. Cell. 1979 Jul;17(3):683–689. doi: 10.1016/0092-8674(79)90275-7. [DOI] [PubMed] [Google Scholar]
- Jones N., Shenk T. Isolation of deletion and substitution mutants of adenovirus type 5. Cell. 1978 Jan;13(1):181–188. doi: 10.1016/0092-8674(78)90148-4. [DOI] [PubMed] [Google Scholar]
- Lawrence W. C., Ginsberg H. S. Intracellular uncoating of type 5 adenovirus deoxyribonucleic acid. J Virol. 1967 Oct;1(5):851–867. doi: 10.1128/jvi.1.5.851-867.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinus M. G. Location of DNA methylation genes on the Escherichia coli K-12 genetic map. Mol Gen Genet. 1973 Dec 14;127(1):47–55. doi: 10.1007/BF00267782. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Miller J. S., Ricciardi R. P., Roberts B. E., Paterson B. M., Mathews M. B. Arrangement of messenger RNAs and protein coding sequences in the major late transcription unit of adenovirus 2. J Mol Biol. 1980 Oct 5;142(4):455–488. doi: 10.1016/0022-2836(80)90258-2. [DOI] [PubMed] [Google Scholar]
- Oberg B., Saborio J., Persson T., Everitt E., Philipson L. Identification of the in vitro translation products of adenovirus mRNA by immunoprecipitation. J Virol. 1975 Jan;15(1):199–207. doi: 10.1128/jvi.15.1.199-207.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PHILIPSON L. Adenovirus assay by the fluorescent cell-counting procedure. Virology. 1961 Nov;15:263–268. doi: 10.1016/0042-6822(61)90357-9. [DOI] [PubMed] [Google Scholar]
- Philipson L., Pettersson U., Lindberg U. Molecular biology of adenoviruses. Virol Monogr. 1975;14:1–115. doi: 10.1007/978-3-7091-8391-5_1. [DOI] [PubMed] [Google Scholar]
- Rao R. N., Rogers S. G. Plasmid pKC7: a vector containing ten restriction endonuclease sites suitable for cloning DNA segments. Gene. 1979 Sep;7(1):79–82. doi: 10.1016/0378-1119(79)90044-1. [DOI] [PubMed] [Google Scholar]
- Thiel J. F., Smith K. O. Fluorescent focus assay of viruses on cell monolayers in plastic Petri plates. Proc Soc Exp Biol Med. 1967 Jul;125(3):892–895. doi: 10.3181/00379727-125-32232. [DOI] [PubMed] [Google Scholar]
- Volkert F. C., Young C. S. The genetic analysis of recombination using adenovirus overlapping terminal DNA fragments. Virology. 1983 Feb;125(1):175–193. doi: 10.1016/0042-6822(83)90072-7. [DOI] [PubMed] [Google Scholar]
- Williams J. F. Enhancement of adenovirus plaque formation on HeLa cells by magnesium chloride. J Gen Virol. 1970 Dec;9(3):251–255. doi: 10.1099/0022-1317-9-3-251. [DOI] [PubMed] [Google Scholar]
- Young C. S. Heat-stable variant of human adenovirus type 5: characterization and use in three-factor crosses. J Virol. 1975 May;15(5):1168–1175. doi: 10.1128/jvi.15.5.1168-1175.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]